ABSTRACT
Seipin, an evolutionary conserved protein, plays pivotal roles during lipid droplet (LD) biogenesis and is associated with various human diseases with unclear mechanisms. Here, we analyzed Caenorhabditis elegans mutants deleted of the sole SEIPIN gene, seip-1. Homozygous seip-1 mutants displayed penetrant embryonic lethality, which is caused by the disruption of the lipid-rich permeability barrier, the innermost layer of the C. elegans embryonic eggshell. In C. elegans oocytes and embryos, SEIP-1 is associated with LDs and is crucial for controlling LD size and lipid homeostasis. The seip-1 deletion mutants reduced the ratio of polyunsaturated fatty acids (PUFAs) in their embryonic fatty acid pool. Interestingly, dietary supplementation of selected n-6 PUFAs rescued the embryonic lethality and defective permeability barrier. Accordingly, we propose that SEIP-1 may maternally regulate LD biogenesis and lipid homeostasis to orchestrate the formation of the permeability barrier for eggshell synthesis during embryogenesis. A lipodystrophy allele of seip-1 resulted in embryonic lethality as well and could be rescued by PUFA supplementation. These experiments support a great potential for using C. elegans to model SEIPIN-associated human diseases.
KEY WORDS: Seipin, Eggshell, Permeability barrier, PUFAs, Fatty acid, Lipid droplet
Summary: Mutagenic analyses reveal that seipin plays a crucial role in permeability barrier synthesis during embryogenesis in Caenorhabditis elegans, and loss of seipin results in osmotically sensitive and dysfunctional embryos.
INTRODUCTION
Lipid droplets (LDs) are ubiquitous cellular organelles for neutral lipid storage. Formation of this organelle requires an evolutionarily conserved protein, seipin, the deficiency of which causes aberrant LD morphology (Cao et al., 2019; Cartwright and Goodman, 2012; Fei et al., 2011, 2008; Grippa et al., 2015; Szymanski et al., 2007; Wang et al., 2016a; Wolinski et al., 2011). Seipin functions as a transmembrane protein residing within subdomains of the endoplasmic reticulum (ER) in which LDs emerge. Structural studies indicate that seipin forms a ring-shaped oligomer with its lumenal domain folded into a hydrophobic pocket, presumably mediating lipid binding, and that seipin anchors LDs to the ER bilayer through the cytosolic and lumenal helices (Sui et al., 2018; Yan et al., 2018). In addition to anchoring LDs to the ER, seipin might mediate lipid synthesis and sorting to promote LD biogenesis and assembly. In human, SEIPIN (also known as BSCL2) is highly expressed in various tissues including brain, testis and adipose (Magré et al., 2001). Intriguingly, failure in SEIPIN functions have been identified in patients diagnosed with Beradinelli-Seip congenital generalized lipodystrophy type 2 (BSCL2), motor neuropathy, Silver syndrome and defects in reproduction resulting in infertility (Chen et al., 2009; El Zowalaty et al., 2015; Fei et al., 2011; Ito et al., 2008; Jiang et al., 2014; Magré et al., 2001; Walther et al., 2017; Windpassinger et al., 2004; Zowalaty and Ye, 2017). How the cellular and molecular mechanisms of the SEIPIN gene are associated with these genetic disorders remain largely unresolved.
Caenorhabditis elegans stores fats, mostly in the form of triacylglycerol (TAG), within LDs found in the intestinal and hypodermal cells (Mak, 2012). Several LD-localized proteins have been characterized in C. elegans, such as TAG synthesis enzyme DGAT-2, fatty acid CoA synthetase ACS-22 and a major LD protein PLIN-1 that contributes to LD biogenesis and lipolysis (Golden et al., 2009; Li et al., 2016; Shi et al., 2013; Vrablik et al., 2015; Xu et al., 2012). A seipin ortholog was also identified in C. elegans. A deletion allele of seip-1 reduced the size of a subset of LDs in intestinal cells, whereas overexpression had the opposite effect (Cao et al., 2019). SEIP-1 is enriched in ER subdomains, called peri-LDs, and this localization is dependent on polyunsaturated fatty acids (PUFAs) (Cao et al., 2019). Together, these findings have placed C. elegans as an attractive model for addressing fundamental issues about the metabolism and functions of LDs at the organismal level. However, this organism has yet to be employed as a model for human diseases associated with lipids or LD disorders, such as that caused by SEIPIN mutations.
Lipids and lipid metabolism influence diverse growth and developmental decisions. Failure of proper lipid delivery to the C. elegans oocyte impairs oocyte growth, fertilization and the development of early embryos (Kimble and Sharrock, 1983; Sturmey et al., 2009). In addition to intestinal and hypodermal cells, LDs were identified in C. elegans oocytes and embryos (Shi et al., 2016). Yolk granules, structural mimics of mammalian lipoproteins, are synthesized in the C. elegans intestine and transferred to maturing oocytes during reproduction (Hall et al., 1999; Kimble and Sharrock, 1983). The yolk vitellogenin protein VIT-2 might contribute to LD formation and regulation during reproduction (Vrablik et al., 2015; Wang et al., 2016b; Zhang et al., 2012). Thus, together with yolk granules, LDs also likely regulate lipid metabolism in oocytes and embryos. However, the exact contribution of these two lipid-enriched structures during germ cell and embryonic development in C. elegans remains obscure.
In C. elegans, a multilayered extracellular matrix, the eggshell, begins to form when an oocyte enters the spermatheca where it is fertilized. The synthesis of the eggshell occurs in a hierarchical pattern, with each layer being sequentially added, starting with the outermost layer first. A lipid-rich layer, known as the permeability barrier, is the fifth layer of the eggshell to be synthesized and is responsible for preventing osmotic and mechanical stress from harming developing embryos (Johnston and Dennis, 2012; Olson et al., 2012; Stein and Golden, 2018). Embryos lacking this layer are osmotically sensitive. The inhibition of a large number of C. elegans genes, many of which are involved in lipid biosynthesis, cause the osmotic integrity defective (OID) phenotype (Benenati et al., 2009; Rappleye et al., 2003; Tagawa et al., 2001). Interestingly, depletion of the yolk receptor protein RME-2 also causes the OID phenotype, which implies that yolk lipids and/or LDs might contribute to permeability barrier formation (Gunsalus et al., 2005).
In this study, we report that homozygous seip-1 mutants produce osmotically sensitive embryos, many of which fail to hatch. Taking advantage of high resolution confocal and transmission electron microscopy (TEM), we document that seip-1 mutants accumulate abnormally sized LDs in oocytes and embryos, and exhibit severe defects in the formation of the permeability barrier of the eggshell. Lipid analysis data revealed that lipidomic perturbations, consistent with fatty acid imbalance, was specific to the seip-1 embryos. Interestingly, supplementing the diet of seip-1 mutants with selected PUFAs rescued the OID phenotype and the associated embryonic lethality through restoring permeability barrier synthesis. These findings support the idea that seipin is a crucial factor for maintaining fatty acid homeostasis, which is necessary for permeability barrier synthesis during C. elegans embryogenesis. We further document that a lipodystrophy allele of the seip-1 mutant, like seip-1 null mutants, displayed OID and embryonic lethality phenotypes, which supports the notion that C. elegans is a feasible system not only to investigate mechanisms of SEIP-1 in modulating LDs but also to model seipin-associated human diseases.
RESULTS
C. elegans SEIP-1 complements Saccharomyces cerevisiae sei1Δ mutants
A sole SEIPIN ortholog, seip-1 (R01B10.6), was identified in the C. elegans genome. SEIP-1 shares ∼20% amino acid identity with both human SEIPIN/BSCL2 and S. cerevisiae seipin (Sei1p). Depletion of the SEI1 gene (previously termed FLD1) in yeast resulted in enlarged and small clustered LDs (Fei et al., 2008; Szymanski et al., 2007). Expression of C. elegans SEIP-1 fused with a GFP tag (GFP-Seip-1p) in yeast was found to abut or surround the LDs labeled by the LD marker Erg6-mCherry (Fig. S1A-H). Importantly, GFP-Seip-1p significantly rescued the aberrant LD population in sei1Δ cells (Fig. S1E-I), suggesting expression of the heterologous C. elegans seip-1 gene functionally complements the loss of SEI1 gene in yeast.
seip-1 mutants disrupt permeability barrier formation in newly fertilized embryos
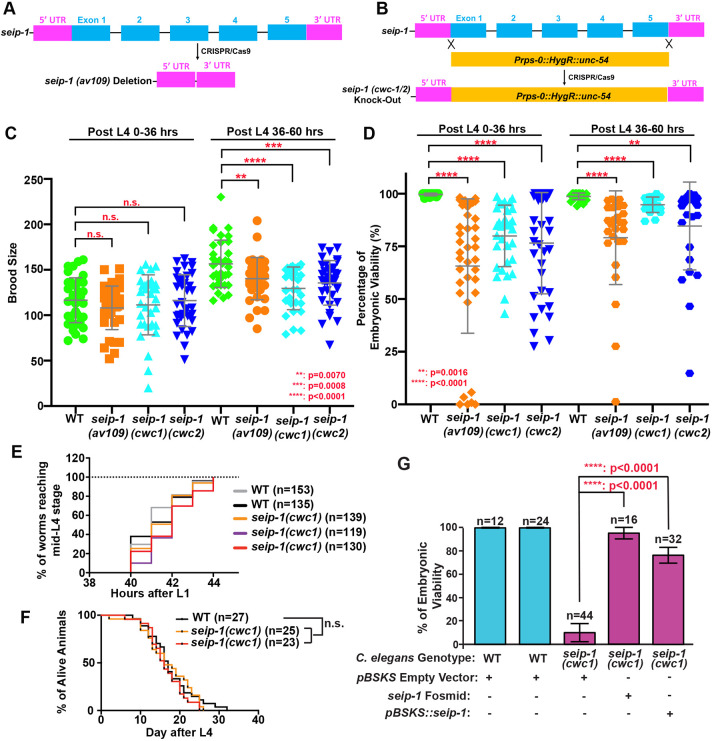
To investigate the biological functions of SEIP-1 in C. elegans, three candidate null alleles were generated by CRISPR/Cas9 genome editing; one allele was a deletion of the whole seip-1 gene [seip-1(av109)] (Fig. 1A; Fig. S2A) and the other two alleles [seip-1(cwc1) and seip-1(cwc2)] were generated by replacing the seip-1 gene with an expression cassette of a hygromycin B resistance gene (HygR) (Radman et al., 2013) (Fig. 1B; Fig. S2A). A small seip-1 deletion allele, seip-1(tm4221), was also characterized in this study (Fig. S2B). The number of F1 progeny (brood size) was significantly reduced in all tested homozygous deletion mutants compared with wild type (Fig. 1C). In addition, the embryonic viability of F1 progeny was dramatically decreased in all tested seip-1 mutants compared with wild type (Fig. 1D). On average, 10%-35% of the tested seip-1 mutant embryos failed to hatch. Surprisingly, we did not observe any obvious defects during later developmental processes, such as the timing of larval development (Fig. 1E) and longevity (Fig. 1F) of the hatched seip-1 mutants. As the expression of seip-1 genomic DNA plasmids in the seip-1(cwc1) mutants significantly rescued the embryonic lethality of F1 progeny, our results demonstrate that seip-1 has an essential function during early embryogenesis (Fig. 1G).
Fig. 1.
Deletion of the seip-1 gene causes hatching defects. (A) Diagram of deleted regions in the seip-1(av109) mutant or (B) replacement of the seip-1 gene with a hygromycin-resistant gene (HygR) cassette in seip-1(cwc1) and seip-1(cwc2) mutants. (C) Brood size was compared between wild-type (WT) and seip-1 mutants over two time periods post L4. (D) The percentage of viable embryos was reduced in the seip-1 deletion animals. (E) The developmental time required for synchronized L1 larvae to reach L4 and (F) the longevity of viable animals in the population were similar to wild-type animals. (G) Expression of the seip-1 containing fosmid and plasmid in the seip-1(cwc1) mutant significantly rescues the embryonic lethality. Data are mean±s.d. Statistical significance was determined using an unpaired two-tailed Student's t-test. n.s., not significant.
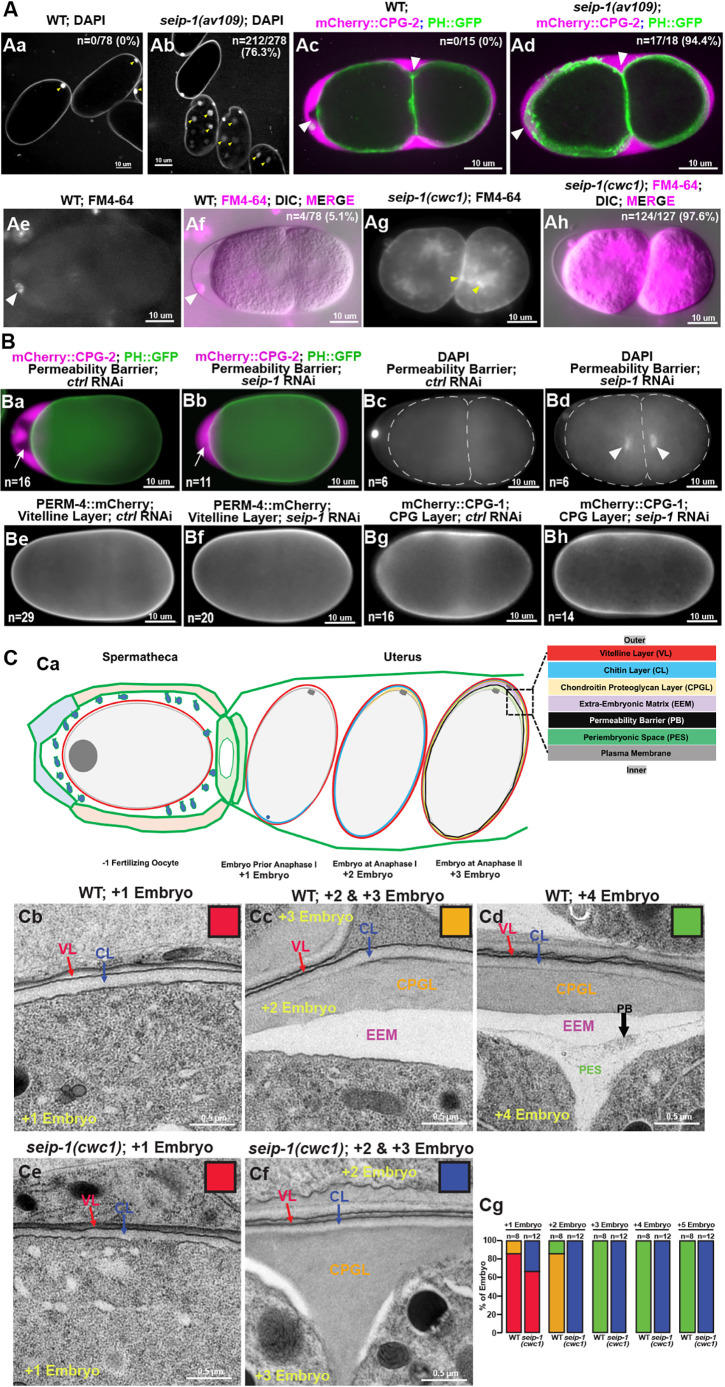
Many of the seip-1 mutant embryos displayed a shrunken and misshapen morphology (Fig. S2C), which is suggestive of eggshell formation defects. As small molecules, such as DAPI and the lipophilic dye FM4-64, are able to penetrate into fertilized embryos with defective eggshells, seip-1 mutant embryos were incubated with DAPI and FM4-64 to test the integrity of their eggshells. Zygotic chromatin, labeled by DAPI, and cytoplasmic membranes, labeled by FM4-64, were frequently observed in the postmeiotic embryos of the seip-1 mutants (Fig. 2Ab,Ag,Ah). In contrast, only the first polar body was stained by DAPI and no membranes were stained by FM4-64 in wild-type embryos (Fig. 2Aa,Ae,Af). Additionally, mCherry::CPG-2 was used to visualize the permeability barrier in this study. The lipid-rich permeability barrier, which defines the innermost layer of the eggshell, prevents diffusion of mCherry::CPG-2 beyond the space between the outer layers of the eggshell and the permeability barrier (Olson et al., 2012). In wild type, the white arrowheads highlight the extra-embryonic space (marked by mCherry::CPG-2; Fig. 2Ac) excluded by the permeability barrier. By contrast, mCherry::CPG-2 fills the entire space between the outer eggshell and the embryonic surface in seip-1 null mutants due to the disruption of the permeability barrier (Fig. 2Ad). The depletion of SEIP-1 by RNAi and a small deletion seip-1(tm4221) mutant displayed identical eggshell permeability defects (Fig. 2Ba-Bd; Fig. S2Bb,Bc). Collectively, these data suggest that loss of SEIP-1 causes severe defects in eggshell formation, which may account for the embryonic lethality observed in these mutants.
Fig. 2.
Deletion of the seip-1 gene causes severe eggshell formation defects. (A) DAPI staining of zygotic chromatin (yellow arrows, Ab) was apparent in seip-1 embryos, whereas only the first polar body (yellow arrows, Aa) stained in wild-type (WT) embryos (n=number of embryos with zygotic chromatin stained with DAPI/number of embryos imaged). mCherry::CPG-2 was used to visualize the integrity of the permeability barrier. In wild type, the inner layer of the eggshell prevents diffusion of mCherry::CPG-2 beyond the space between the outer layers of the eggshell and the embryo surface (white arrowheads, Ac). However, in seip-1(av109) embryos, mCherry::CPG-2 penetrates inside the entire space between the eggshell and the embryo surface (white arrowheads, Ad) (n=number of embryos with mCherry::CPG-2 penetration/number of embryos imaged). Only the plasma membrane surrounding the first polar body (white arrowheads) was stained by FM4-64 (magenta) in wild-type embryos (Ae,Af), whereas staining was observed at the plasma membrane and cytosol (yellow arrowheads) in seip-1(cwc1) mutant embryos (Ag,Ah) (n=number of embryos with FM4-64 penetration/number of embryos imaged). (B) Depletion of SEIP-1 by RNAi resulted in defective permeability barrier formation (Ba,Bb), allowing mCherry::CPG-2 (white arrows in Ba and Bb) to fill the space between the outer layers of the eggshell and the embryo surface (n=number of embryos with mCherry::CPG-2 penetration), and DAPI (white arrowheads, Bd) to penetrate inside the embryos to stain zygotic chromatin (n=number of embryos with DAPI penetration). The eggshell proper appears mainly intact when SEIP-1 was depleted by RNAi, as markers for the vitelline layer (PERM-4::mCherry, Be,Bf) and the CPG layer (mCherry::CPG-1, Bg,Bh) are properly localized compared with embryos treated by control RNAi (n=the number of embryos examined). (C) Diagram of the sequential steps during eggshell assembly (Ca). At anaphase of meiosis II, the permeability barrier is formed between the outer eggshell and the plasma membrane. TEM micrographs of high-pressure frozen embryos demonstrate that the vitelline layer (VL), chitin layer (CL) and the chondroitin proteoglycan layer (CPGL) are easily observed in both wild-type and seip-1(cwc1) embryos that have completed meiosis I (Cb,Cc,Ce). A clear edge of the permeability barrier (PB, black arrow) was visualized in every wild-type embryo that completed meiosis II (Cd). EEM defines the space between the chondroitin proteoglycan layer and the permeability barrier, whereas the periembryonic space (PES) bridges the permeability barrier and the embryo proper. However, no permeability barrier was found at any stage of the seip-1(cwc1) embryo (Ce,Cf). The white patches in Cf show the severely compromised EEM. Quantification of the embryos with the forming eggshell in both WT and seip-1(cwc1) embryos is presented in Cg. The embryos containing different components of the eggshell were categorized into four groups, which are represented by the colored squares inserted at the top right of each panel (Cb-Cf). Red, embryos containing the vitelline layer and chitin layer; blue, embryos with the vitelline layer, chitin layer and chondroitin proteoglycan layer; orange, embryos with four layers, including the vitelline layer, chitin layer, chondroitin proteoglycan layer and EEM; green, embryos with all five eggshell layers (vitelline layer, chitin layer, chondroitin proteoglycan layer, EEM and permeability barrier). n=the number of embryos examined.
We next performed thin-sectioned electron microscopy (EM) to directly examine the ultrastructure of forming eggshells in newly fertilized embryos, which reside in the uteri of gravid hermaphrodites. Eggshell formation occurs in a hierarchical pattern, with each layer being sequentially added (Olson et al., 2012). The whole process starts from the unfertilized oocyte (which is at prophase of meiosis I) and ends at anaphase II of the meiotic embryo (Fig. 2Ca). The examined embryos were labeled as +1 to +5, which represents the positioning relative to the spermatheca (+1; adjacent to the spermatheca) and to the vulva (+5) (Fig. 2C). The EM results indicated that the youngest newly fertilized +1 embryo had an outermost vitelline layer (synthesized pre-fertilization) and a chitin layer (formed immediately after fertilization but before anaphase I of meiosis), whereas the +2 embryo exhibited an additional forming chondroitin proteoglycan (CPG) layer (fully formed after anaphase of meiosis I), in both wild-type and seip-1(cwc1) mutants (Fig. 2Cb-Cf). The permeability barrier in wild type was initially synthesized during anaphase of meiosis II and properly formed in the older embryos (Fig. 2Cd). Indeed, all of the examined mitotic embryos (+3 to ∼+5 embryos) contained a properly formed permeability barrier (Fig. 2Cd,Cg) in wild-type embryos. However, we could not identify the permeability barrier structure in any seip-1(cwc1/2) embryos by EM, including in the mitotic embryos (Fig. 2Ce-Cg). The protein-rich and fluid-filled structure termed extra-embryonic matrix (EEM) sat between the CPG layer and the permeability barrier in wild-type embryos (Fig. 2Cc,Cd). By contrast, we observed a severely compromised and reduced EEM in seip-1 mutant embryos (Fig. 2Cf). Thus, only the first three outer layers (vitelline layer, chitin layer and CPG layer) of the eggshell were properly synthesized in seip-1 mutant embryos (Fig. 2Ce-Cg). This is consistent with the observation that the vitelline layer marker, PERM-4::mCherry (Fig. 2Be) (González et al., 2018), and CPG layer marker, mCherry::CPG-1 (Fig. 2Bg), were properly localized when seip-1 was depleted by RNAi (Fig. 2Bf,Bh). Overall, we conclude that SEIP-1 is essential for the proper formation of the permeability barrier layer of the eggshell.
seip-1 mutants have abnormal LDs
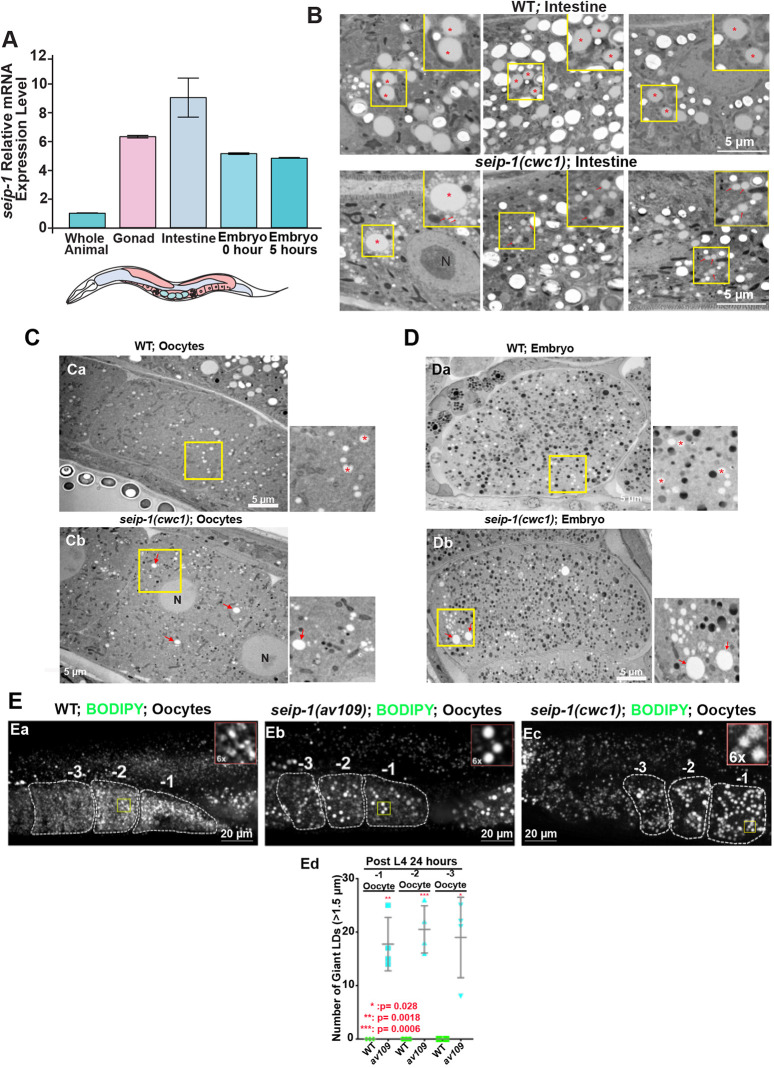
The permeability barrier is a lipid-rich layer, in which glycolipids might play an important role in its formation (Olson et al., 2012; Stein and Golden, 2018; Watts et al., 2018). However, the exact lipid composition and mechanisms to build this structure are still unknown. Given that seipin provides structural support and regulates lipid flux into LDs, we investigated whether SEIP-1 might be required for LD functions in C. elegans. We first employed qPCR to examine the expression of seip-1 mRNA in various tissues dissected from C. elegans. High expression of seip-1 mRNA was detected in the gonad, intestine and embryos of wild-type animals (Fig. 3A). Most cells are capable of shaping their LDs into a similar size, probably to maintain homeostasis of this organelle to cope with cellular lipid metabolism. Consistent with a previous study in C. elegans (Cao et al., 2019), thin-section EM results indicated that the seip-1 null animals accumulated abnormally sized LDs in intestinal cells, in which LDs are abundant (Fig. 3B). Remarkably, EM examination showed that aberrant LDs, either abnormally large or small ones, exist in both seip-1(cwc1) oocytes and embryos (Fig. 3C,D). When LDs were marked by BODIPY493/503, all three seip-1 deletion alleles showed enlarged LDs in the oocytes and embryos (Fig. 3Eb,Ec). More than ten enlarged LDs (>1.5 μm) were readily detectable in the −1 to −3 oocytes in each of these mutants (Fig. 3Ed). In contrast, no such enlarged LDs were observed in oocytes in wild type (Fig. 3Ea). Thus, these observations are consistent with previous studies in mammalian and yeast cells that demonstrate that seip-1 dysfunction results in aberrant LDs and establish that SEIP-1 plays a conserved role in LD assembly in the oocytes and embryos of C. elegans.
Fig. 3.
SEIP-1 depletion alters LD size in the oocytes and embryos. (A) qRT-PCR was performed using RNAs isolated from whole animals, dissected tissues or embryos immediately prepared (0 h) from gravid adults or embryos allowed to develop for 5 h after dissection from gravid adults. seip-1 is highly expressed in the C. elegans germline, embryos and intestine. (B) TEM micrographs of high-pressure frozen animals displayed normal LDs in wild-type and abnormal LDs in seip-1(cwc1) intestinal cells. Red asterisks and arrows mark the LDs. A 1.6× magnification of the regions marked with a yellow square are also shown in upper right insets in each panel. (C,D) TEM micrographs of high-pressure frozen animals show normal LDs (red asterisks) in wild-type oocytes (C) and embryos (D), as well as enlarged (red arrows) and tiny LDs in the seip-1(cwc1) oocytes and embryos. A 2.5× magnification of the regions marked with a yellow square are also shown to the right of each panel. (E) BODIPY-stained LDs in the −1 to −3 oocytes of wild-type (Ea) and seip-1 (Eb,Ec) animals. A 6× magnification of the yellow squares are shown on the top right corner of each panel. Quantification of the enlarged LDs (diameter >1.5 μm) inside the −1 to −3 oocytes of both wild-type (wt; green) and seip-1 mutants (blue) is presented in Ed. Data are mean±s.d. Statistical significance was determined using an unpaired two-tailed Student's t-test. N, nucleus.
SEIP-1::mScarlet expresses in multiple C. elegans tissues and associates with LDs
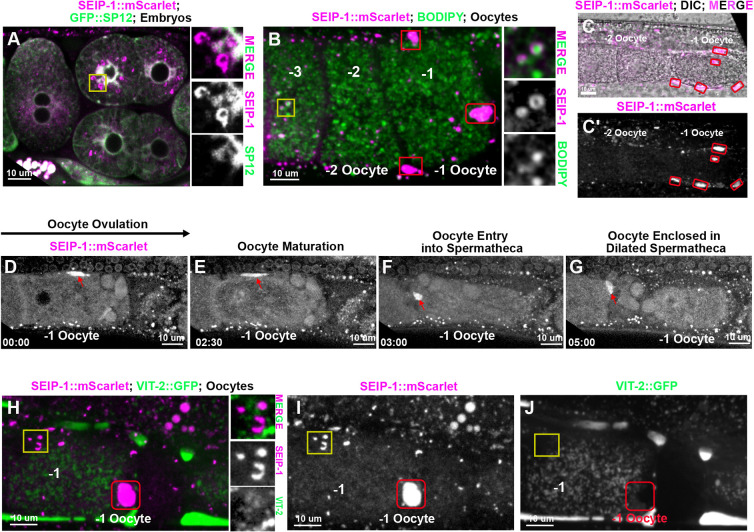
To determine the subcellular localization of SEIP-1 in C. elegans, we directly knocked-in a monomeric red fluorescent reporter gene, mScarlet, into the seip-1 endogenous locus using CRISPR/Cas9. N-terminal mScarlet insertions (mScarlet::SEIP-1) caused embryonic lethality, probably as a consequence of disrupted SEIP-1 functions. Animals with C-terminally tagged SEIP-1 (SEIP-1::mScarlet) did not reveal embryonic lethality, indicative of SEIP-1 functionality. The protein was widely expressed from embryonic stages through adulthood (Fig. 4; Fig. S3). In embryos, SEIP-1::mScarlet colocalized with the ER marker GFP::SP12 (Poteryaev et al., 2005) in the regions surrounding embryonic nuclear envelopes (Fig. 4A). In maturing oocytes, SEIP-1::mScarlet was adjacent to or present on ring structures surrounding a subset of BODIPY-stained LDs (Fig. 4B). Similar colocalization patterns were found in the embryos and different tissues of larvae and adult animals (Fig. S3), supporting the conserved role of seipin in promoting LD biogenesis and assembly in the ER subdomain.
Fig. 4.
SEIP-1 localization in C. elegans oocytes and embryos. (A) SEIP-1::mScarlet observed in embryos. Right inserts represent the magnified area of the yellow square, showing that SEIP-1::mScarlet is adjacent to the ER labeled by GFP::SP12. (B) In the oocytes, SEIP-1::mScarlet is either adjacent to or surrounds the LDs stained by BODIPY. Right inserts represent the magnified area of the yellow square, showing that SEIP-1::mScarlet localizes to the surface of the BODIPY stained LDs. −1, −2 and −3 indicate oocyte from the most proximal to the distal to the spermathecal. (B-C′) SEIP-1::mScarlet localizes to the pseudocoelomic space (red squares in panel B; red circles in panels C,C′) surrounding the ovulating oocytes (-1 oocyte). (D-G) Representative images of SEIP-1::mScarlet localization during ovulation and fertilization. A strong SEIP-1::mScarlet signal was observed in the fifth sheath cell (red arrows) that surrounds the −1 oocyte (D) until the oocyte is ovulated and enters the spermatheca (E-G). (H-J) A vitellogenin reporter (VIT-2::GFP) transgene was used to monitor yolk lipoprotein localization, showing no colocalization with SEIP-1::mScarlet either in the LDs (yellow squares) or the pseudocoelomic space (red squares). Amplified images of the yellow square are shown to the right of H.
Intriguingly, a strong SEIP-1::mScarlet signal was observed in the fifth gonadal sheath cell that surrounds the maturing oocytes before ovulation into the spermatheca (Fig. 4D-G; Movie 1). In C. elegans, the pseudocoelomic lipoprotein pool was identified as yolk, which is secreted from the intestine and travels through the gonadal basal lamina and sheath pores, and is then endocytosed by the developing oocytes (Paupard et al., 2001). When the yolk marker VIT-2::GFP was co-expressed in the animals containing SEIP-1::mScarlet, we did not detect VIT-2::GFP on the SEIP-1::mScarlet-labeled droplets in the intestine and oocytes (Fig. 4H-J). In addition, as seen by EM, the number and size of the electron-dense yolk particles in oocytes or embryos of seip-1(cwc1) mutants was similar to that in wild type (Fig. 3C,D). Thus, SEIP-1::mScarlet likely regulates only LDs, not yolk delivery to maturing oocytes, and thus the permeability barrier defect of seip-1 mutants might not be caused by disrupted yolk functions. Collectively, the strong SEIP-1::mScarlet signals seen in the oocytes and sperm (Fig. S4Aa,Ab,Cb,Cd) supports roles for SEIP-1 in C. elegans reproduction.
The fatty acid synthesis pathway is disrupted in seip-1 mutants
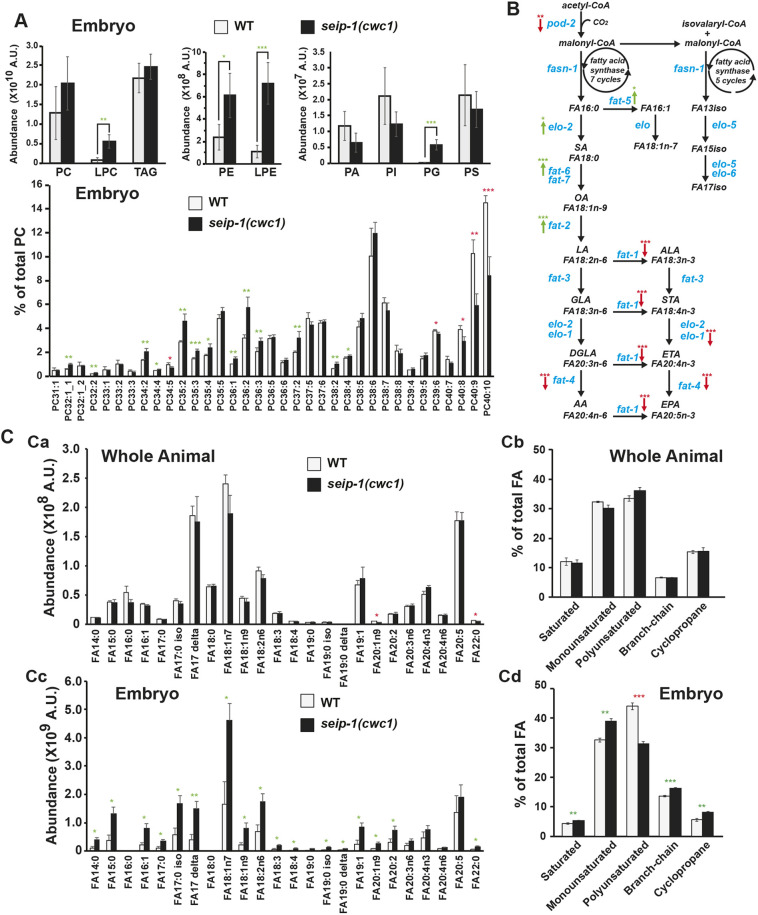
To explore whether SEIP-1 regulates lipid homeostasis and how SEIP-1 affects reproduction, we took advantage of lipidomic approaches to decipher the potential variations of the lipid profile in seip-1 mutants. Liquid chromatography-mass spectrometry (LC-MS) assays identified only subtle variations of lipidomic change in whole seip-1(cwc1) animals (Fig. S5B), in contrast to a profound lipidomic perturbation observed in isolated embryos of seip-1 mutants compared with wild type (Fig. 5A; Fig. S5A). In seip-1 mutant embryos, phosphatidylethanolamine, phosphatidylglycerol, and lysophospholipids showed marked increases in overall level, whereas others, such as phosphatidylcholine, phosphatidylinositol, and phosphatidylserine and TAG, were minimally affected (Fig. 5A; Fig. S5). Interestingly, most lipids, like phosphatidylcholine present in Fig. 5A, displayed a similar pattern in which the level of lipid species with more fatty acid double bonds were reduced, whereas those with fewer fatty acid double bonds were increased in seip-1 mutant embryos (Fig. 5A; Fig. S5). Thus, the lipidomic analyses revealed widespread variations of the lipidome in seip-1 mutant embryos, suggesting the importance of seip-1 in maintaining lipid homeostasis during embryogenesis.
Fig. 5.
Lipidomic perturbations and fatty acid imbalances were observed in the seip-1 mutant embryos. (A) Lipidomic analysis of the early embryos indicated that phospholipid levels were broadly affected in seip-1(cwc1) mutant embryos compared with wild-type embryos. Detailed phosphatidylcholine (PC) species in the lipidome of wild-type and the seip-1(cwc1) mutant were compared based on their ratio normalized to the total PC level. Green and red asterisks indicate those values that are significantly increased or reduced, respectively, in the seip-1(cwc1) mutant compared with wild type. (B) The de novo fatty acid synthesis pathway of C. elegans. Enzymes (blue fonts) that catalyze each of the biochemical steps (black fonts) are indicated. Expression of the de novo fatty acid synthetic genes were either upregulated (elo-2, fat-2, fat-5 and fat-6, green arrows) or downregulated (pod-2, elo-1, fat-1, fat-4, red arrows) in seip-1(cwc1) mutant embryos (see also Fig. S6). Green and red arrows indicate that gene expression is significantly increased or reduced in seip-1(cwc1) mutant embryos compared with wild type. (C) The absolute abundance of different fatty acid (FA) species (Ca) or the fatty acid species in subgroups (Cb) were not affected in seip-1(cwc1) whole animals measured by GC/MS. However, the absolute abundance of most fatty acid species was increased in seip-1(cwc1) embryos (Cc). Fatty acids were divided into subgroups and compared based on the ratio normalized to the total fatty acid level. The percentages of MUFAs, branch-chain and cyclopropane were increased in the isolated seip-1 mutant embryos (Cd). In contrast, the percentage of PUFAs was significantly decreased in seip-1 mutant embryos. Green and red asterisks indicate those values that are significantly increased or reduced, respectively, in the seip-1(cwc1) mutant compared with wild type. Data are mean±s.d. Statistical significance was determined using an unpaired two-tailed Student's t-test. *P<0.05; **P<0.01; ***P<0.001. A.U., arbitrary units.
It has been suggested that de novo lipid synthesis machinery is crucial for early embryonic development, whereas later larval development use fatty acid precursors from their bacterial diet (Watts et al., 2018). The assembly of long chain fatty acids requires key enzymatic reactions (Fig. 5B). Given that the lipidomic perturbation observed in seip-1 mutant embryos might be caused by fatty acid imbalance, we next performed gas chromatography-mass spectrometry (GC-MS) to analyze fatty acid contents. Consistent with the LC-MS results, only a subtle variation of fatty acid was identified in whole seip-1(cwc1) animals compared with wild-type animals (Fig. 5Ca,Cb). By contrast, most fatty acids, except for PUFAs, showed significant increases in their absolute level in seip-1 mutant embryos compared with wild-type embryos (Fig. 5Cc). The C. elegans fatty acids were further organized into saturated, monounsaturated, PUFA, branch-chain and cyclopropane. When normalized to the total fatty acid level, the seip-1 mutant embryos displayed a substantial reduction in the proportion of PUFAs when the proportion of other types of fatty acids were slightly increased (Fig. 5Cd).
As a wide range of PUFAs are synthesized in C. elegans through the de novo fatty acid pathway (Watts and Ristow, 2017), a genome-wide microarray, followed by qRT-PCR, was carried out to analyze maternal fatty acid gene expression pattern during early embryogenesis (Fig. 5B, Fig. S6). The qRT-PCR results indicated that the expression of PUFA synthesis genes, such as elo-1, fat-1 and fat-4 were significantly reduced in the seip-1 mutant embryos, supporting the reduced PUFA ratio (Fig. S6C). Other genes, such as elo-2, fat-2 and fat-6, were upregulated in seip-1 deletion mutant embryos, which might be explained as a compensatory effect. Interestingly, the genes acox-1.1, acox-3, daf-22 and dhs-28, which play roles in fatty acid β-oxidation, were significantly upregulated in seip-1 mutants (Fig. S6C), supporting the perturbation of fatty acid homeostasis in the seip-1 mutant embryos.
The C. elegans permeability barrier was identified as a lipid-rich layer, which primarily consists of fatty acid derivatives (Olson et al., 2012), and this study links LDs to permeability barrier formation. Fatty acids are esterified to form TAG and sterol esters (SE), which are the primary contents of LDs. The depletion of genes implicated in TAG and SE synthesis, such as the DGAT2-related acyltransferase dgtr-1, which might convert diacylglycerol (DAG) to TAG, and the cytochrome P450s cyp-31a2/a3 and the putative hydroxysteroid dehydrogenase/isomerase perm-1, which might modify sterol esters, have been shown to give rise to the OID phenotype and impaired permeability barrier formation (Olson et al., 2012). In seip-1 mutant embryos, transcriptional expression of cyp-31A2/3, perm-1 and dgtr-1 were tested by qRT-PCR and were found to be significantly downregulated compared with wild-type embryos, suggesting that dysfunction of seip-1 might disturb the synthesis of lipids traditionally stored within LDs (Fig. S6D). Together, the reduced gene expression observed in the seip-1 mutant embryos likely resulted in the deficit of lipid content crucial to permeability barrier formation. Given that loss of SEIP-1 disrupted LD morphology, fatty acid balance and de novo fatty acid synthesis, we conclude that SEIP-1 plays an essential role in maintaining lipid homeostasis during C. elegans germ cell and embryonic development. Though the changes that affect the permeability barrier are clearly maternal, some of the changes observed could reflect maternal and/or zygotic changes in lipid biosynthesis that affect later events of embryonic development.
Dietary supplementation of PUFAs rescued the permeability barrier defects
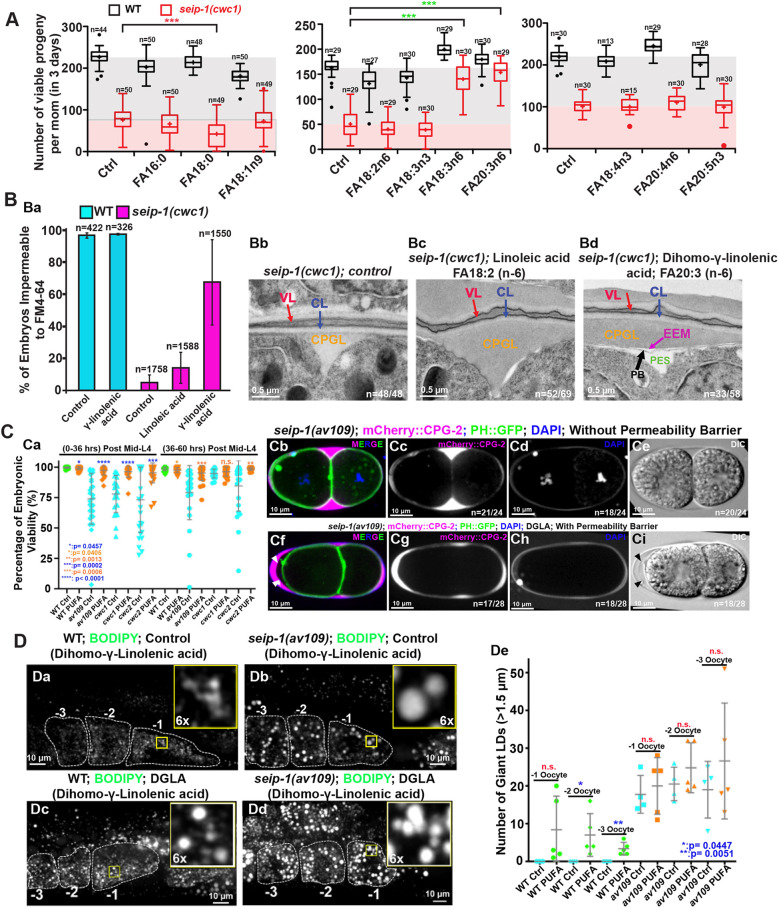
From the lipidomic analysis, we hypothesized that fatty acid imbalance could, in part, be responsible for the defective formation of the permeability barrier in seip-1 mutants. To test this hypothesis, different fatty acids were added into the diet of the seip-1 deletion mutants. Interestingly, feeding animals a standard nematode growth media (NGM) diet containing 300 µM n-6 PUFA, namely gamma-linolenic acid (C18:3n-6, GLA) and dihomo-gamma-linolenic acid (C20:3n-6, DGLA), significantly improved progeny numbers in the seip-1(cwc1) mutant, whereas feeding animals other types of PUFAs or monounsaturated fatty acids (MUFAs) did not have the same effect or worsened the progeny number in the seip-1(cwc1) mutant (Fig. 6A). Although fatty acid supplementation in wild-type animals also influenced progeny numbers, the effects were relatively minor. Remarkably, dietary supplementation of DGLA, but not its precursor linoleic acid (C18:2n-6, LA), resulted in embryos impermeable to the fluorescent dye FM4-64 in the population (Fig. 6Ba). Consistently, EM data showed that the defective permeability barrier was significantly rescued in the seip-1(cwc1) mutant after dietary supplementation with DGLA, but not with LA or mock (Fig. 6Bb-Bd).
Fig. 6.
Dietary supplementation of PUFAs rescued the permeability barrier defects but not the abnormal LDs in seip-1 mutants. (A) The total number of progeny of wild-type (WT) and seip-1 animals after feeding with NGM supplemented with 300 µM of the indicated fatty acids for 3 days were counted and compared. n=the number of L4 larvae used for the experiment. ***P<0.001. The boxes extend from the 25th to 75th percentiles. The whiskers extend from minimum to maximum. (B) Dietary supplementation of DGLA (FA18:3n6), but not LA (FA18:2n6), reduced the population of penetrant seip-1 embryos stained with FM4-64 (Ba). TEM micrographs of high-pressure frozen embryos showing the permeability barrier (black arrow) formed after seip-1 mutant animals were fed with DGLA, but not formed when fed with LA (Bb-Bd). n=number of embryos showing representative eggshell layers/number of embryos imaged. (C) The percentage of viable embryos was significantly rescued in the DGLA-fed seip-1 deletion animals (wild type, green; mutant, blue; DGLA-fed, orange) (Ca). The penetration of DAPI, the expression pattern of CPG2::mCherry, and DIC images, demonstrated that the permeability barrier was defective in seip-1 mutants embryos (Cb-Ci), which can be restored after the diet of seip-1 mutants animals was supplemented with DGLA (Cf-Ci). The GFP::PH transgene was used to indicate the plasma membranes. n=number of embryos with abnormal DAPI penetration and abnormal CPG2::mCherry pattern/number of embryos imaged. The permeability barrier can be easily seen in panel Cf (white arrowheads) and panel Ci (black arrowheads). (D) Enlarged LDs were frequently observed in the −1 to −3 oocytes of both wild-type (Dc) and seip-1(av109) (Dd) animals fed with DGLA. Right top insets represent 6× amplified images of the inlaid yellow squares (Da-Dd). Quantification of enlarged LDs (diameter >1.5 μm) in −1 to −3 oocytes of both PUFA-fed wild-type and seip-1(av109) animals is shown in De. Data are mean±s.d. Statistical significance was determined using an unpaired two-tailed Student's t-test. n.s., not significant.
When all three seip-1 null animals were tested with the modified Youngren's only bactopeptone (MYOB) diet, DGLA supplementation greatly improved the hatch ratio to more than 95% (Fig. 6C), albeit there was a reduction in brood sizes (not shown). Additionally, a significantly higher percentage of seip-1(av109) mutant embryos formed proper permeability barriers, readily detectable with differential interference contrast (DIC) microscopy (Fig. 6Ce), to prevent the penetration of DAPI, FM4-64 and mCherry::CPG-2 into the cytosol and peri-embryonic space after dietary supplementation of DGLA (Fig. 6Cf-Ci). By contrast, no obvious permeability barriers were observed in the seip-1(av109) mutants without DGLA supplementation (Fig. 6Cb,Cd). As the number of enlarged LDs was slightly increased in both wild-type and seip-1(av109) oocytes after supplementation with DGLA (Fig. 6Dc,Dd), DGLA taken up by the animals might be either metabolized or channeled to neutral lipid production in germ cells. The result also implies that the permeability barrier defect, the primary phenotype of seip-1 deficiency, was not correlated with LD size.
Collectively, our data indicate that fatty acid imbalance compromises permeability layer formation in the seip-1 mutants, and thus led to embryonic lethality. This lethality can be reversed when the diet is supplemented with selected n-6 PUFAs. Loss of SEIP-1 might disrupt the availability of PUFAs by either disrupting their synthesis or by interfering with their ability to directly become incorporated into the permeability barrier. The embryonic lethality of the seip-1 mutants is likely due to the malformation of a robust permeability barrier, leading to osmotic stress during embryogenesis.
Modeling human BSCL2 genetic diseases in C. elegans
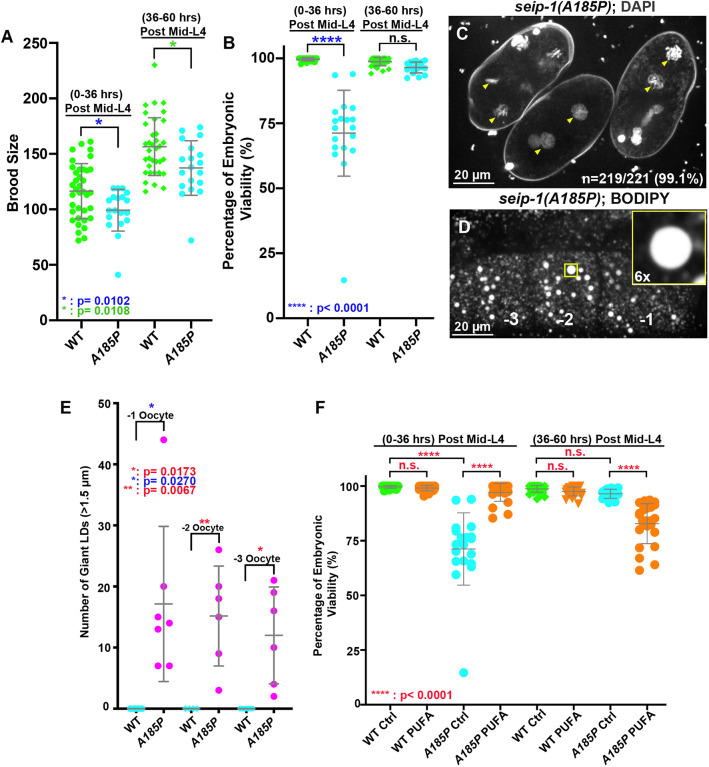
This study has provided strong evidence that seip-1 is essential for C. elegans embryogenesis, eggshell formation and fatty acid synthesis and metabolism. Given the striking phenotypes associated with seip-1 deletions, we further tested whether it is plausible to model human SEIPIN-associated diseases with SEIPIN missense mutations using C. elegans embryos. Of the many missense mutations associated with disease, we chose to study a conserved alanine (A212P) in the human SEIPIN gene. Patients bearing this variant have been diagnosed with the autosomal recessive BSCL2 syndrome (Magré et al., 2001), characterized by a total lack of adipose tissue in the body and a very muscular appearance. Fat storage thus takes place in other organs in the body, such as the liver, which can lead to hepatomegaly and liver failure (Garg, 2004; Simha and Garg, 2003). Previous studies have shown that this A212P missense mutation is a loss-of-function mutation in SEIPIN (Salo et al., 2016; Szymanski et al., 2007). Bioinformatics analysis indicated that alanine 185 (A185) in C. elegans SEIP-1 is the homologous alanine residue to human SEIPIN A212. Using CRISPR/Cas9, we generated this missense mutation in C. elegans and named the allele seip-1(av160[A185P]).
Homozygous seip-1(A185P) animals displayed embryonic lethality (Fig. 7A,B), defective permeability barrier formation (Fig. 7C) and enlarged LDs (Fig. 7D,E), similar to those observed in our seip-1 null mutants. Dietary supplementation with DGLA significantly rescued the lethality of seip-1(A185P) embryos laid by mothers that were fed DGLA on day 1 (0 h to 36 h post L4) (Fig. 7F). However, day 2 and 3 (36 h to 60 h post L4) DGLA-fed seip-1(A185P) animals revealed an increase in embryonic lethality compared with wild type (Fig. 7F), suggesting that the patient-specific allele is sensitive to excessive DGLA. Overall, these observations strongly support the idea that C. elegans is an appropriate model system to study seipin-associated diseases. Using patient-specific alleles, we will be able to perform genetic suppressor screens in the future to help identify other potential genetic interactors that, when mutated, restore viability to seip-1 embryos.
Fig. 7.
A seipin patient-specific allele disrupts eggshell formation and displays enlarged LDs in C. elegans. (A) A conserved patient-specific allele seip-1(A185P) was generated and resulted in reduced brood sizes. (B) seip-1(A185P) causes embryonic lethality in the day 1 (post L4 0-36 h) adult animals. However, embryonic viability of seip-1(A185P) is not affected in the day 2 and 3 (post L4 36-60 h) animals. (C) DAPI staining of zygotic chromatin (yellow arrowheads) in seip-1(A185P) mutant embryos is indicative of a defective permeability barrier. (D) seip-1(A185P) exhibits enlarged LDs in the −1 to −3 oocytes marked by BODIPY. The panel inset shows a 6× enlarged view of a single enlarged LD indicated by the yellow box in the main panel. (E) Quantification of enlarged LDs (diameter >1.5 μm) in −1 to −3 oocytes of seip-1(A185P) animals. (F) The percentage of viable embryos was significantly rescued in the day 1 (post L4 0-36 h) DGLA (PUFA)-fed seip-1(A185P) animals compared with the control. In contrast, the percentage of viable embryos was reduced in the day 2 and 3 animals (post L4 36-60 h) fed with PUFAs. Data are mean±s.d. Statistical significance was determined using an unpaired two-tailed Student's t-test. n.s., not significant.
DISCUSSION
Seipin has been perceived as a mysterious protein in LD biology. This ER-integral membrane protein plays a key role at LD-forming subdomains in the ER during LD biogenesis and assembly, yet its exact molecular mechanisms remain largely elusive. Intriguingly, dysfunction of SEIPIN affects both adipocyte development and lipid storage in non-adipose tissues, which causes various human diseases, such as lipodystrophy, neurological seipinopathies and reproductive infertility (Chen et al., 2009; Jiang et al., 2014; Magré et al., 2001; Windpassinger et al., 2004). However, how SEIPIN dysfunction affects lipid biosynthesis, metabolism and utilization in building the lipid-rich organelles that lead to the disease progression remains largely unknown.
In this study, we present a comprehensive in vivo analysis of C. elegans seipin, seip-1. We provide strong evidence to support the idea that C. elegans seip-1 has an evolutionarily conserved function in the formation of LDs. As in other seipin-deficiency systems (human, mice and yeast), loss of SEIP-1 was sufficient to impair lipid biosynthetic gene expression, lipid balance and LD morphology (Figs 3,5; Figs S5,S6). Furthermore, we suggest that the functional role of SEIP-1 is to maintain the fatty acid homeostasis in C. elegans embryos, which is crucial for the proper formation of the permeability barrier of the eggshell that forms early in embryogenesis (Figs 2,6). To our knowledge, this is the first report of seipin in both the gametes and embryos of C. elegans, and might represent an organismal model to study seipin function in non-adipose tissues, particularly in germ cells and reproductive tissues.
The C. elegans permeability barrier, built during anaphase of meiosis II, prevents the entry of potential toxic molecules and protects the embryos from osmotic damage. Depletion of POD-2, FASN-1 and other fatty acid synthetic enzymes in the germline impaired permeability barrier formation, suggesting that germline-derived fatty acids are likely components of the permeability barrier (Rappleye et al., 2003; Tagawa et al., 2001). Consistent with previous studies in mice and humans showing that seipin dysfunction caused defects in fatty acid biosynthesis and metabolism (Chen et al., 2009; Jiang et al., 2014; Payne et al., 2008), our findings revealed that C. elegans seip-1 mutants disrupted fatty acid homeostasis in embryos, resulting in lipidomic perturbation (Fig. 5, Fig. S5). Thus, SEIP-1 shares similar functions in maintaining fatty acid homeostasis with SEIPIN in humans.
Previous findings have also revealed the involvement of CYP-31A2/A3, DGTR-1 and PERM-1 for permeability barrier synthesis and suggest that the fatty acids used for permeability barrier synthesis might be used in the synthesis of lipid derivatives commonly stored within LDs (Olson et al., 2012). Intriguingly, the transcriptional analysis indicated that perm-1 and dgtr-1 expression was reduced in the seip-1 mutant embryos (Fig. S6D), which might result in the deficit of lipid content crucial to permeability barrier formation. One possibility is that fatty acids, such as GLA or DGLA, may either be modified to the relevant lipid derivative found in the permeability barrier, or delivered alone to the extracellular matrix for building the permeability barrier. If this hypothesis is true, LDs might have a role in permeability barrier synthesis. Combined with the high expression of seip-1 in the sheath cells surrounding the ovulating oocyte (Fig. 4C,C′), it will be interesting to determine whether SEIP-1 delivers LDs with C20 long chain fatty acid derivatives to the ovulating oocyte to prepare for permeability barrier formation. The direct involvement of LDs for de novo membrane synthesis has at least been established in diploid yeast during prospore membrane formation during anaphase of meiosis II, leading to sporulation (Hsu et al., 2017).
Fatty acids are building blocks of membranes and storage lipids. Fatty acids were known to affect cell-type-specific gene expression that impacts metabolic pathways, including lipid metabolism (Nakamura et al., 2004; Georgiadi and Kersten, 2012; Pégorie et al., 2004). Fatty acids might also be indirectly involved in regulating gene expression through their effects on specific enzyme-mediated pathways, membrane compositions or signaling molecules. Although we cannot completely rule out the contribution of other fatty acids, our findings seem to favor a role for PUFAs in permeability barrier synthesis. We provide evidence that the seip-1 mutants reduced the proportion of PUFAs in the total fatty acid pool and that dietary supplementation with GLA or DGLA significantly rescued the embryonic lethality, and restored the defective permeability barrier (Figs 5,6). Although the precise role of SEIP-1 in regulating fatty acid metabolism has not been clear, it is key to understanding the de novo fatty acid synthesis pathway during germ cell development and embryogenesis. Equally important, as SEIP-1 is a crucial factor for LDs, is to determine whether or not LDs might contribute to cell- or non-cell-autonomous regulation of fatty acid metabolism.
Clinical reports indicate that either loss-of-function or gain-of-function mutations in the human SEIPIN gene cause a variety of disorders. However, the molecular mechanisms underlying all SEIPIN-associated disorders are still far from being understood. Disorders involving SEIPIN have been identified in different tissues, including both adipose and non-adipose tissues. Recessive homozygous SEIPIN loss-of-function mutations in human cause BSCL2, a recessive disease. Affected individuals display a severe reduction in adipose tissue. Additionally, in these individuals, SEIPIN mutations also caused spermatozoid morphology defects and teratozoospermia. However, these phenotypes have not been well studied in individuals with SEIPIN mutations.
In this study, our data support for the first time that it is plausible to model BSCL2 or other SEIPIN-associated diseases in C. elegans. The C. elegans seip-1 gene strongly expresses in oocyte and sperm, and mutants have severe reproductive defects, including smaller brood sizes and defects in eggshell formation (Fig. 1). These phenotypes allowed us to investigate the role of seip-1 in eggshell formation (Fig. 2), a crucial process in embryogenesis and for viability. However, we did not observe any obvious defects in sperm morphology (Fig. S4D), spermatogenesis (Fig. S4E) or sperm fertility (Fig. S4F) in seip-1 mutant hermaphrodites. Our observation that a BSCL2 patient-specific allele in the C. elegans seip-1 gene displayed defects similar to our deletion alleles, suggests that this allele is loss-of-function (Fig. 7). Importantly, the embryonic lethality and defective permeability barrier in seip-1 mutants could be significantly rescued by dietary supplementation with the n-6 PUFAs GLA and DGLA (Fig. 6), suggesting that C. elegans is a powerful model system to screen potential genetic and pharmaceutical suppressors for SEIPIN1 diseases.
In summary, C. elegans seipin is required for reproduction. We chose to focus on the most striking phenotypes, the formation of the permeability barrier and fatty acid homeostasis, in which defects in each result in smaller brood sizes and embryonic lethality. The discovery of dietary rescue provides an avenue to study the molecular mechanisms of seipin in vivo. Further studies will focus on understanding the molecular mechanisms of seipin in regulating lipid biosynthesis during the reproductive process. The chemical suppressor screen and/or genetic screens to identify extragenic suppressor mutations that restore viability to seip-1 mutant embryos might help provide a direction for future clinical therapy.
MATERIALS AND METHODS
C. elegans and S. cerevisae strains used in this study
C. elegans strains were maintained with standard protocols. Strain information is listed in Table S1. AG547 and AG548 were created by crossing AG444 (seip-1::mScarlet) males with hermaphrodites containing ocfIs2[pie-1p:GFP::SP12::pie-1 3′UTR+unc-119(+)] and pwIs23 [vit-2::GFP], respectively. We screened the F3 adults for the presence of the SEIP-1::mScarlet with GFP::SP12 and VIT-2::GFP transgenes, respectively, by microscopy. AG549 and AG550 were created by crossing OD344 males with hermaphrodites containing seip-1(av109) and seip-1(tm4221), respectively. F3 adults expressing mCherry::CPG-2 and GFP::PH transgenes were genotyped by PCR for the seip-1(av109) and seip-1(tm4221) alleles. The S. cerevisae strain CWY3115 (sei1Δ::HIS ERG6-mCherry::KAN BY4742) was generated by a PCR-based integration method (Longtine et al., 1998). The yeast expression plasmids pRS416-PGPD-GFP or pRS416-PGPD-GFP::seip-1 were transformed into the yeast cells. The transformed cells were grown in synthetic complete dextrose (SCD) media (0.67% yeast nitrogen base, amino acids and 2% glucose) to log phase (OD600=1.0) at 30°C. A further 24 h growth period allowed the cells to reach the stationary phase for visualization by microscopy.
RNAi treatment
The RNAi feeding constructs were obtained from the Ahringer and Vidal libraries (Fraser et al., 2000; Rual et al., 2004). RNAi bacteria were grown until log phase was reached and spread on MYOB plates containing 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 25 μg/ml carbenicillin, and incubated overnight at 22°C. To silence the target genes, mid-L4 hermaphrodites were picked onto plates with the IPTG-induced bacteria. Animals were grown on RNAi plates at 20°C.
CRISPR design
We used the Bristol N2 strain as the wild type for CRISPR/Cas9 editing. The gene-specific 20 nucleotide sequences for crRNA synthesis were selected with the help of a guide RNA design checker from Integrated DNA Technologies (www.idtdna.com) and were ordered as 20 nmol or 4 nmol products from Dharmacon (www.dharmacon.horizondiscovery.com), along with tracRNA. Repair template design followed the standard protocols (Paix et al., 2015; Vicencio et al., 2019). Young gravid animals (∼30) were injected with the prepared CRISPR/Cas9 injection mix as described in the literature (Paix et al., 2015). seip-1(av109) was generated by CRISPR/Cas9 mixes that contained two guide RNAs at flanking regions of the seip-1 coding region. Heterozygous seip-1 deletion animals were first screened by PCR and then homozygosed in subsequent generations. mScarlet insertions at the seip-1 C-terminus were performed by nested CRISPR (Vicencio et al., 2019). Homozygous nest-1 edited animals were confirmed by PCR and restriction enzyme digestion, and selected for the secondary CRISPR/Cas9 editing. Full-length mScarlet insertion animals were screened by PCR and visualized by fluorescence microscopy. All homozygous animals edited by CRISPR/Cas9 were confirmed by Sanger sequencing (Eurofins). The detailed sequence information of the repair template and guide RNAs are listed in Table S2. Two SEIP-1 (R01B10.6) deletion strains were custom made by Knudra Transgenics (currently InVivo Biosystems) using three proprietary vectors, the locus-targeting guide RNA pNU1244, the locus-targeting guide RNA PNU1245, and the donor homology plasmid pNU1246. Along with standard fluorescence markers and Cas9 plasmid (pNU792), the DNA mix was injected into gonads of the N2 strain, and the injected animals were screened for resistance to 0.25 mg/ml hygromycin followed by a heat shock counter screen to remove animals bearing extrachromosomal arrays. Two verified transgenic lines received from Knudra were backcrossed with N2 four times to obtain the seip-1(cwc1) and seip-1(cwc2) strains that were confirmed by PCR. The three deletion strains did appear to have different levels of embryonic viability that we attribute to the protocols and the different wild-type genetic backgrounds in which they were generated, and by the media and incubators in which they were grown.
Brood size determinations and embryonic viability assays
Single mid-L4 hermaphrodites were picked onto 35 mm MYOB plates seeded with 10 μl of OP50 bacteria and allowed to lay eggs for 36 h (plate one contained the brood from 0 h to 36 h post mid-L4). The same hermaphrodite was moved to a new 35 mm MYOB plate to lay eggs for another 24 h and was removed from the plate (this plate contains the brood from 36 h to 60 h post mid-L4). After removing the mothers, only fertilized embryos and larvae were counted 24 h later to determine the brood size. Brood sizes were determined at 36 h and 60 h. The percentage of embryonic viability=(the number of hatched larva / the total number of hatched and unhatched animals)×100%. The embryonic lethality of cwc1 and cwc2 alleles was much higher (∼75-80%) than that of the av109 deletion allele. It is possible that the difference in lethality observed was partly because of the culturing conditions used between two laboratories. The cwc1 and cwc2 alleles were grown on standard NGM plates, whereas the av109 allele was grown on MYOB plates. They differ in pH and in some salts. In addition, we occasionally observed adaptive rescue of lethality (or the acquisition of spontaneous suppressors) after maintaining homozygous strains over a period of a few months. Thus, the deletion strains were revived from frozen stocks every 3 months. Such spontaneous suppressors have been observed for unrelated temperature-sensitive embryonic lethal mutations (K. O'Connell, personal communication).
Mating assay with the fem-1 mutant
Ten mid-L4 BA17 fem-1(hc17ts) females raised from embryos at the non-permissive temperature of 25°C were mated with ∼30 N2, seip-1(av109) or seip-1(cwc1) adult males for 24 h at 25°C. Single mated females and three to five males were then transferred to a fresh 35 mm growth plate and allowed to mate for another 24 h at 25°C before all adults were removed from the plates. As a control, ten unmated BA17 hermaphrodites grown at 25°C were kept on ten individual plates. The brood sizes and embryonic viability were determined 24 h after the removal of all adults.
BODIPY 493/503 staining
BODIPY 493/503 (Invitrogen, D3922) was dissolved in 100% DMSO to 1 mg/ml. BODIPY stock was diluted by M9 to 6.7 μg/ml BODIPY (final concentration of DMSO was 0.8%) as the working stock. Hermaphrodites were washed in M9 three times and incubated in 6.7 μg/ml BODIPY for 20 min, and washed again in M9 at least three times. All washes and incubations were performed in a concavity slide (Thermo Fisher Scientific, S99369). The stained hermaphrodites were anesthetized with 0.1% tricaine and 0.01% tetramisole in M9 buffer for 15 min to 30 min. The anesthetized animals were then transferred to 5% agarose pads for imaging. Image acquisition was conducted using a Nikon 60×1.2 NA water objective with 0.5 μm z-step size.
DAPI staining of embryos
Hermaphrodites were washed in M9 three times and dissected with 23 G×3/4″ needles. Different stage embryos were transferred to a hanging drop chamber filled with blastomere culture medium (BCM) (Edgar and Goldstein, 2012). The BCM was made fresh with 5 μg/ml DAPI. The hanging drop chamber was sealed by molten Vaseline before imaging. Image acquisition was conducted using a Nikon 60×1.2 NA water objective with 1 μm z-step size.
DAPI staining of whole animals
To quantify sperm numbers, animals were washed twice in 1 ml PBS containing 0.01% Tween 20 (PBST) and then fixed in 1 ml methanol at −20°C for 5 min, followed by washing with 1 ml PBST. Animals were then stained with 1 ml PBST and 2 μg/ml DAPI in the dark for 5 min. After another wash with 1 ml PBST, animal pellets were resuspended in 75% glycerol on agarose pads for viewing with a microscope. Images were taken using a Zeiss LSM 880 microscope with a 40× water objective (NA1.2) with ∼20 z-steps, focused around the spermetheca area.
FM4-64 staining of embryos
To check eggshell integrity, embryos were dissected from both wild-type and seip-1(cwc1) animals in 5 μl egg buffer [4 mM HEPES (pH 7.4), 94 mM NaCl, 32 mM KCl, 2.7 mM CaCl2 and 2.7 mM MgCl2] containing 6.6 μM FM4-64 (Thermo Fisher Scientific, T3166). Samples were mounted in a chamber circled by vacuum grease on a 2% agar pad (15×15 mm) for imaging. The images were acquired on a Zeiss Z1 fluorescence microscopy system that used an EC Plan-Neofluar 40× objective lens (NA0.75), an EC Plan-Neofluar M27 20× objective lens (NA0.5), a mCherry filter set (Ex 540-552 nm, Em 575-640 nm) and a Zeiss AxioChem 506 charge-coupled device (CCD) camera. The images were acquired and analyzed using Zeiss ZEN software.
Live imaging of ovulation
For imaging SEIP-1::mScarlet expression during ovulation, animals were immobilized on 4% agar pads with anesthetic (0.1% tricaine and 0.01% tetramisole in M9 buffer). DIC and mScarlet image acquisition were conducted using a Nikon 60×1.2 NA water objective with 1-2 μm z-step size; 10 to 15 z planes were captured. The time interval for ovulation imaging was every 30 s, and the duration of imaging was 10-15 min.
Imaging of yolk proteins
Day 1 adults (24 h post mid-L4) of AG548 were immobilized on 4% agar pads with anesthetic (0.1% tricaine and 0.01% tetramisole in M9 buffer). The acquisition of both DIC and 561 nm images was performed using our confocal imaging system (see below) with a Nikon 60×1.2 NA water objective. Ten to 20 z planes were captured with 1 μm z-step size. Images were generated using custom Fiji code using Image>Stacks>Z Project.
Imaging of spermatozoa
Day 1 adults (24 h post mid-L4) of AG444 males were dissected and mounted in BCM with 10 µg/ml Hoechst 33342 by the hanging drop method (Edgar and Goldstein, 2012). The acquisition of DIC, 408 nm, 488 nm and 561 nm were performed using our confocal imaging system (see below) with a Nikon 60×1.2 NA water objective. Ten z planes were captured with 1 μm z-step size. Images were generated using custom Fiji code using Image>Stacks>Z Project.
Imaging of LDs in yeast cells
To image yeast cells, yeast cells were cultured in SCD medium to stationary phase and were imaged using a Zeiss Z1 fluorescence microscopy system that uses a Zeiss Plan-Apochromat 100× objective lens (NA 1.4), a GFP filter set (Ex 450-490 nm, Em 500-550 nm), a mCherry filter set (Ex 540-552 nm, Em 575-640 nm) and a Zeiss AxioChem 506 CCD camera. The images were acquired and analyzed using Zeiss ZEN software.
PUFA dietary supplementation
PUFA dietary supplementation was modified from a previous report (Yang et al., 2006). LA (C18:2n-6, Sigma-Aldrich, L1376), GLA (C18:3n-6, Sigma-Aldrich, L2378) and DGLA (C20:3n-6, Cayman Chemical, 90230) were added to NGM or MYOB medium to a final concentration of 300 μM. Single mid-L4 hermaphrodites were picked onto 35 mm PUFA-supplemented MYOB and control MYOB plates seeded with 10 μl of OP50 bacteria, and allowed to lay eggs for 36 h (plate one contained the brood from 0 h to 36 h post mid-L4). The same hermaphrodite was moved to a new 35 mm MYOB plate to lay eggs for another 24 h and was removed from the plate (this plate contains the brood from 36 h to 60 h post mid-L4). Twenty-four hours after removing the mothers, only fertilized embryos and larvae were counted to determine the brood size. Brood sizes were determined at 36 h and 60 h. The percentage of embryonic viability=(the number of hatched larva/the total number of hatched and unhatched animals)×100%. For embryo imaging, single mid-L4 hermaphrodites were allowed to grow on PUFA-supplemented MYOB for 24 h. These gravid adults were then dissected in 5 μl of 0.7× egg buffer or BCM containing either 6.6 μM FM4-64 or 5 μg/ml DAPI. The embryos were mounted in a hanging drop chamber and images were acquired as described above.
Electron microscopy
Approximately 60 gravid hermaphrodites (60 h after L1) were picked (10-12 animals per grid) and mixed with fresh yeast, and were immediately subjected to cryofixation by high-pressure freezing with a high-pressure freezer EM HPM100 (Leica). An AFS2 EM (Leica) was used for the subsequent freeze substitution and low temperature embedding. Freeze substitution was performed in 2% osmium tetroxide and 0.1% uranyl acetate in acetone following the program of −90°C for 16 h, −85°C for 50 h, −50°C for 10 h, −20°C for 10 h and 0°C for 6 h. Temperature increased by a rate of 2.5°C/h for each time slope. The samples were then washed in acetone and subjected to infiltration with 5%, 10%, 25%, 50%, 75% and, finally, 100% Spurr resin. After polymerization at 60°C for 48 h, the blocks were cut into 70-90 nm ultrathin sections on an EM UC6 ultramicrotome (Leica) with an ultra 45° diamond knife (DiATOME). Sections were placed on 0.4% Formvar-coated slot grids (EMS) and imaged at 120 kV on a TEM (FEI Tecnai G2 Spirit TWIN) with a digital CCD camera (832 Orius SC1000B). The images were processed and analyzed using Photoshop CS4 software (Adobe).
Microarray
Embryos were isolated from gravid adults by hypochlorite disruption (0.5 N NaOH and 1% NaOCl) followed by M9 washes. After overnight incubations in M9 without food, synchronized L1 were then seeded on a 5.5 cm NGM plate (∼150 L1/plate). The synchronized young adults (52 h post L1 feeding) were harvested and washed by M9 buffer and pelleted. Embryos were isolated by hypochlorite disruption from young gravid adults (52 h after L1), washed with M9 and pelleted. To extract total RNA, synchronized young adults or embryos derived from ∼40,000 young adults were resuspended in 500 μl TRIzol Reagent, followed by flash freezing in liquid nitrogen. Samples were thawed at 42°C and frozen in liquid nitrogen and the cycle was repeated six to seven times. Chloroform (100 μl) was mixed with the sample and spun at 12,000 g for 15 min at 4°C. The corresponding aqueous layer was harvested and mixed with an equal volume of 70% ethanol. RNeasy Mini kits (Qiagen) were used to obtain pure RNA for microarray analysis. Total RNA (0.2 μg) was subjected to Cy3 labeling by in vitro transcription using a Low Input Quick Amp Labeling kit (Agilent). Subsequently, Cy3-labeled cRNA (1.65 μg) was incubated at 60°C for 30 min, followed by hybridization to the Agilent C. elegans V2 4×44K Microarray chip (Agilent) at 65°C for 17 h. After washing and drying, the microarray chip was scanned with the microarray scanner (Agilent) at 535 nm for Cy3. The array image was read by Feature Extraction software version 10.7.1.1, and data was subjected to further analysis by GeneSpring software (Agilent). The average value of N2 sample was used as the control; a statistics filter was set to select for seip-1/N2 gene expression at ≥1.5 fold change and P≤0.05. Genes that passed these criteria were subjected to Kyoto encyclopedia of genes and genome pathway annotation using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/home.jsp).
qPCR
C. elegans animals were first synchronized by the hypochlorite disruption method as described above. Expression levels of SEIP-1 were carried out in whole animals or tissues dissected from animals. RNA samples prepared immediately (0 h) or 5 h after embryos were dissected from animals were compared. Embryos isolated from ∼40,000 young adults (52 h after L1) were resuspended in 500 μl TRIzol Reagent followed by flash freezing in liquid nitrogen. Samples were thawed at 42°C and frozen in liquid nitrogen, and the cycle was repeated six or seven times. Chloroform (100 μl) was mixed with the sample, which was then spun at 12,000 g for 15 min at 4°C. The corresponding aqueous layer was harvested and mixed with an equal volume of 70% ethanol. RNeasy Mini kits were used to extract pure RNA. RNA (∼15 μg ) in nuclease-free water was treated by DNase I. DNaseI-treated RNA (2 μg) was used as the template to synthesize cDNAs with the SuperScript III First-Strand System (Thermo Fisher Scientific). The real-time PCR reactions were performed by the Applied Biosystems QuantStudio 12K Flex system using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). The thermal cycling conditions followed an initial denaturation step at 95°C for 10 min, 40 cycles at 95°C for 10 s, and 60°C for 1 min, followed by a continuous melt curve stage at 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. Three independent experiments were replicated for each target gene. The ΔCT value was determined by QuantStudio 12K Flex software (Thermo Fisher Scientific). Gene expression levels were calculated as 2^[−ΔΔC(T)], which was normalized to the expression of the act-1 gene. Statistical analyses were calculated based on four biological replicates and data were represented as mean±s.d. The sequences for the qPCR primers are listed in Table S3.
Lipidomic analyses
Synchronized young adults (51 h after synchronized L1) were washed off plates and ∼30,000 worms were collected as a pellet. Early embryos were also isolated from ∼80,000 synchronized gravid adults (51 h after L1). The animal debris was washed by M9 buffer at least three times. The animal or embryo pellets were washed with 20 mM NaN3 three times, followed by immediate immersion in liquid nitrogen for at least 1 h. Samples were thawed and sonicated with a 20 s pulse/40 s rest cycle on ice four times. A small portion of the lysate was quantified by Bradford assay to determine the total protein concentration. The extraction of lipid was followed by the Folch method (Folch et al., 1957). Animal or embryo lysate (∼150 μl), 2 ml ice-cold chloroform/methanol (2:1 v/v) and an internal control (fatty acid 13:0) were mixed in a conical glass tube followed by overnight incubation at 4°C. Hajra's solution (0.2 M H3PO4 and 1 M KCl) was mixed into the incubated lysate. The solution was spun down for 10 min at 10,000 g to separate aqueous and organic layers. The organic layer was isolated and dried using a Thermo SpeedVac concentrator SPD111v (Thermo Scientific). The aqueous layer was treated with chloroform to re-separate the organic phase. Dried organic lipids were dissolved in 150 μl chloroform/methanol (2:1 v/v). For LC/MS/MS, we used a linear Orbitrap Elite Ion Trap-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled online with a ACQUITY UPLC/UHPLC system (Waters). Lipid sample (4 µl) in chloroform/methanol (2:1 v/v) was separated by an ACQUITY UPLC HSS T3 column, 1.8 µm, 2.1 mm×100 mm (Waters) at a flow rate of 0.5 ml/min using a cycle of 40-99.9% solvent B over 0-10 min, holding at 99.9% B for 2 min, and re-equilibration at 40% B. Solvent A contained acetonitrile/water (40:60) with 10 mM ammonium acetate (pH 5.0) and solvent B contained isopropanol/acetonitrile (90:10) with 10 mM ammonium acetate (pH 5.0). The MS was operated with either positive or negative ion modes using the full Fourier transform-mass spectrometry scan at 100-1200 m/z, resolution 60,000. The phospholipid species was identified by LC/MS/MS, and the sphingolipid species was determined based on two previous papers (Cutler et al., 2014; Mosbech et al., 2013). Lipid abundance was quantified with Xcalibur software (Thermo Scientific). The statistical analysis was performed in Excel (Microsoft). To analyze fatty acids with GC-MS, 50 μl of lipid samples in chloroform/methanol (2:1 v/v) were subjected to derivatization with BF3/methanol to prepare for the fatty acid methyl esters (FAMEs) based on a method described previously (Morrison and Smith, 1964). BF3 (∼14%) in methanol was mixed with lipid samples and heated up at 95°C for 10 min. The samples were cooled down at room temperature and mixed with benzene to heat up to 95°C for 10 min. Treated samples were cooled down to room temperature and treated with double distilled H2O and petroleum ether for phase separation. The organic phase was recovered and concentrated with the Thermo SpeedVac concentrator SPD111v (Thermo Scientific). Samples were dissolved in 100 μl chloroform/methanol (2:1 v/v) and loaded for GC/MS using an Agilent 7890A GC system equipped with 5975C inert MSD. FAME sample (1 µl) was injected at the split ratio of 2:1. FAMEs were separated with an Agilent DB-5MS column (30 m×0.25 mm×0.25 μm) at a flow rate of 1.1 ml/min. The inlet temperature was 250°C. The column temperature was initially held at 80°C for 1 min, followed by an increase to 128°C at the rate of 8°C/min, to 188°C at the rate of 10°C/min, to 222°C at the rate of 2°C/min, to 228°C at the rate of 3°C/min, and finally to 278°C at the rate of 5°C/min. The data were acquired at the scan mode (27-540 m/z) at 70 eV. Target search and data quantification were performed using Agilent ChemStation data analysis software and the NIST Mass Spectral search library.
Supplementary Material
Acknowledgements
We thank the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40OD010440), for providing strains for this study. We thank Dr Shohei Mitani for sharing the FX14734 [seip-1(tm4221)/hT1V] strain. We thank Dr Orna Cohen-Fix for generously sharing the GFP::SP-12 strain. We are grateful to the members of the Golden laboratory: Dr Peter Kropp, Dr Tao Cai, Rosie Bauer and Isabella Zafra, for productive discussions and preparing reagents. We thank Dr Rey-Huei Chen [Institute of Molecular Biology (IMB), Academia Sinica] for sharing lab resources and improving the manuscript. We are grateful to Tzu-Han Hsu (IMB, Academia Sinica) for advice on EM; Ji-Ying Huang and Mei-Jane Fang at the Cell Biology Core Lab [Institute of Plant and Microbial Biology (IPMB), Academia Sinica] for advice on microscopy and qPCR analysis; Shu-Jen Chou at the Genomic Technology Core Lab (IPMB, Academia Sinica) for help with microarray; and Dr Wen-Dar Lin at the IPMB Bioinformatic Core Lab for consulting on genomic analysis. We especially thank Dr William Prinz and Dr Alexandre Toulmay for critical inputs on the project and feedback on the manuscript. We thank all members of the National Institutes of Health and Baltimore Worm Clubs for providing feedback on and suggestions for our investigations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: X.B., S.K.O., A.G., C.-W.W.; Methodology: X.B., L.-J.H., S.-W.C., B.N., B.W., A.G., C.-W.W.; Software: X.B., L.-J.H., S.-W.C.; Validation: X.B., L.-J.H., S.-W.C., C.-W.W.; Formal analysis: X.B., L.-J.H., S.-W.C.; Investigation: X.B., L.-J.H., S.-W.C., B.N., B.W., C.-W.W.; Resources: J.W., A.G., C.-W.W.; Data curation: X.B., L.-J.H., S.-W.C.; Writing - original draft: X.B., C.-W.W.; Writing - review & editing: X.B., A.G., C.-W.W.; Visualization: X.B., C.-W.W.; Supervision: S.K.O., A.G., C.-W.W.; Project administration: S.K.O., A.G., C.-W.W.; Funding acquisition: S.K.O., A.G., C.-W.W.
Funding
This work was supported, in part, by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (X.B., B.N. and A.G.); the Academia Sinica Intramural Funds and Career Development Award (CDA-104-L11 to C.-W.W.); a National Science Foundation CAREER Award (1846563 to S.K.O.); and a Research Corporation for Scientific Advancement Cottrell College Award (22670 to S.K.O.). Deposited in PMC for release after 12 months.
Data availability
Raw microarray raw data have been deposited in GEO under accession number GSE157800.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.192997.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.192997.reviewer-comments.pdf
References
- Benenati G., Penkov S., Müller-Reichert T., Entchev E. V. and Kurzchalia T. V. (2009). Two cytochrome P450s in Caenorhabditis elegans are essential for the organization of eggshell, correct execution of meiosis and the polarization of embryo. Mech. Dev. 126, 382-393. 10.1016/j.mod.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Cao Z., Hao Y., Fung C. W., Lee Y. Y., Wang P. F., Li X. S., Xie K., Lam W. J., Qiu Y. F., Tang B. Z. et al. (2019). Dietary fatty acids promote lipid droplet diversity through seipin enrichment in an ER subdomain. Nat. Commun. 10. 10.1038/s41467-019-10835-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B. R. and Goodman J. M. (2012). Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man Seipin: from human disease to molecular mechanism. J. Lipid Res. 53, 1042-1055. 10.1194/jlr.R023754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yechoor V. K., Chang B. H., Li M. V., March K. L. and Chan L. (2009). The human lipodystrophy gene product Berardinelli-Seip congenital lipodystrophy 2/seipin plays a key role in adipocyte differentiation. Endocrinology 150, 4552-4561. 10.1210/en.2009-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler R. G., Thompson K. W., Camandola S., Mack K. T. and Mattson M. P. (2014). Sphingolipid metabolism regulates development and lifespan in Caenorhabditis elegans. Mech. Ageing Dev. 143-144, 9-18. 10.1016/j.mad.2014.11.002 [DOI]
- Edgar L. G. and Goldstein B. (2012). Culture and manipulation of embryonic cells. Method Cell Biol. 107, 153-175. 10.1016/B978-0-12-394620-1.00005-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zowalaty A. E., Baumann C., Li R., Chen W., De La Fuente R. and Ye X. (2015). Seipin deficiency increases chromocenter fragmentation and disrupts acrosome formation leading to male infertility. Cell Death Dis. 6, e1817 10.1038/cddis.2015.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W. H., Shui G. H., Gaeta B., Du X. M., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G. and Yang H. Y. (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473-482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W. H., Li H., Shui G. H., Kapterian T. S., Bielby C., Du X. M., Brown A. J., Li P., Wenk M. R., Liu P. S. et al. (2011). Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J. Lipid Res. 52, 2136-2147. 10.1194/jlr.M017566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M. and Stanley Sloane, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497-509. [PubMed]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M. and Ahringer J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325-330. 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- Garg A. (2004). Acquired and inherited lipodystrophies. N. Engl. J. Med. 350, 1220-1234. 10.1056/NEJMra025261 [DOI] [PubMed] [Google Scholar]
- Georgiadi A. and Kersten S. (2012). Mechanisms of gene regulation by fatty acids. Adv. Nutr. 3, 127-134. 10.3945/an.111.001602 [DOI]
- Golden A., Liu J. and Cohen-Fix O. (2009). Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122, 1970-1978. 10.1242/jcs.044743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González D. P., Lamb H. V., Partida D., Wilson Z. T., Harrison M.-C., Prieto J. A., Moresco J. J., Diedrich J. K., Yates J. R. III and Olson S. K. (2018). CBD-1 organizes two independent complexes required for eggshell vitelline layer formation and egg activation in C. elegans. Dev. Biol. 442, 288-300. 10.1016/j.ydbio.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A., Buxó L., Mora G., Funaya C., Idrissi F.-Z., Mancuso F., Gomez R., Muntanyá J., Sabidó E. and Carvalho P. (2015). The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829-844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus K. C., Ge H., Schetter A. J., Goldberg D. S., Han J.-D. J., Hao T., Berriz G. F., Bertin N., Huang J., Chuang L.-S. et al. (2005). Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature 436, 861-865. 10.1038/nature03876 [DOI] [PubMed] [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., Rose K. L., Hobert O. and Greenstein D. (1999). Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: Relations between the germ line and soma. Dev. Biol. 212, 101-123. 10.1006/dbio.1999.9356 [DOI] [PubMed] [Google Scholar]
- Hsu T.-H., Chen R.-H., Cheng Y.-H. and Wang C.-W. (2017). Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 28, 440-451. 10.1091/mbc.e16-06-0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D., Fujisawa T., Iida H. and Suzuki N. (2008). Characterization of seipin/BSCL2, a protein associated with spastic paraplegia 17. Neurobiol. Dis. 31, 266-277. 10.1016/j.nbd.2008.05.004 [DOI] [PubMed] [Google Scholar]
- Jiang M., Gao M., Wu C., He H., Guo X., Zhou Z., Yang H., Xiao X., Liu G. and Sha J. (2014). Lack of testicular seipin causes teratozoospermia syndrome in men. Proc. Natl. Acad. Sci. USA 111, 7054-7059. 10.1073/pnas.1324025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston W. L. and Dennis J. W. (2012). The eggshell in the C. elegans oocyte-to-embryo transition. Genesis 50, 333-349. 10.1002/dvg.20823 [DOI] [PubMed] [Google Scholar]
- Kimble J. and Sharrock W. J. (1983). Tissue-specific synthesis of yolk proteins in Caenorhabditis-elegans. Dev. Biol. 96, 189-196. 10.1016/0012-1606(83)90322-6 [DOI] [PubMed] [Google Scholar]
- Li S. W., Xu S. B., Ma Y. L., Wu S., Feng Y., Cui Q. P., Chen L. F., Zhou S., Kong Y. Y., Zhang X. Y. et al. (2016). A genetic screen for mutants with supersized lipid droplets in Caenorhabditis elegans. G3 6, 2407-2419. 10.1534/g3.116.030866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A. III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P. and Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T., Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Lathrop M. et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28, 365-370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Mak H. Y. (2012). Lipid droplets as fat storage organelles in Caenorhabditis elegans: thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J. Lipid Res. 53, 28-33. 10.1194/jlr.R021006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison W. R. and Smith L. M. (1964). Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J. Lipid Res. 5, 600-608. [PubMed]
- Mosbech M. B., Kruse R., Harvald E. B., Olsen A. S. B., Gallego S. F., Hannibal-Bach H. K., Ejsing C. S. and Faergeman N. J. (2013). Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PloS One8, e70087. 10.1371/journal.pone.0070087 [DOI] [PMC free article] [PubMed]
- Nakamura M. T., Li Cheon Y., Y. and Nara T. Y. (2004). Mechanisms of regulation of gene expression by fatty acids. Lipids39, 1077-1083. 10.1007/s11745-004-1333-0 [DOI]
- Olson S. K., Greenan G., Desai A., Müller-Reichert T. and Oegema K. (2012). Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J. Cell Biol. 198, 731-748. 10.1083/jcb.201206008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D. and Seydoux G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 Ribonucleoprotein complexes. Genetics 201, 47 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupard M.-C., Miller A., Grant B., Hirsh D. and Hall D. H. (2001). Immuno-EM localization of GFP-tagged yolk proteins in C-elegans using microwave fixation. J. Histochem. Cytochem. 49, 949-956. 10.1177/002215540104900803 [DOI] [PubMed] [Google Scholar]
- Payne V. A., Grimsey N., Tuthill A., Virtue S., Gray S. L., Nora E. D., Semple R. K., O'Rahilly S. and Rochford J. J. (2008). The human lipodystrophy gene BSCL2/Seipin may be essential for normal adipocyte differentiation. Diabetes 57, 2055-2060. 10.2337/db08-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegorier J. P., Le May C. and Girard J. (2004). Control of gene expression by fatty acids. J. Nutr. 134, 2444S-2449S. 10.1093/jn/134.9.2444S [DOI]
- Poteryaev D., Squirrell J. M., Campbell J. M., White J. G. and Spang A. (2005). Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell 16, 2139-2153. 10.1091/mbc.e04-08-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman I., Greiss S. and Chin J. W. (2013). Efficient and rapid C. elegans transgenesis by bombardment and hygromycin B selection. PLoS ONE 8, e76019 10.1371/journal.pone.0076019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C. A., Tagawa A., Le Bot N., Ahringer J. and Aroian R. V. (2003). Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev. Biol. 3, 8 10.1186/1471-213X-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A. S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S. H., Hill D. E., van den Heuvel S. et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162-2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo V. T., Belevich I., Li S. Q., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E. et al. (2016). Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35, 2699-2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Li J., Zou X. J., Greggain J., Rødkær S. V., Faergeman N. J., Liang B. and Watts J. L. (2013). Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J. Lipid Res. 54, 2504-2514. 10.1194/jlr.M039669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Tarazona P., Brock T. J., Browse J., Feussner I. and Watts J. L. (2016). A Caenorhabditis elegans model for ether lipid biosynthesis and function. J. Lipid Res. 57, 265-275. 10.1194/jlr.M064808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simha V. and Garg A. (2003). Phenotypic heterogeneity in body fat distribution in patients with congenital generalized lipodystrophy caused by mutations in the AGPAT2 or seipin genes. J. Clin. Endocrinol. Metab. 88, 5433-5437. 10.1210/jc.2003-030835 [DOI] [PubMed] [Google Scholar]
- Stein K. K. and Golden A. (2018). The C. elegans eggshell. WormBook 2018, 1-36. 10.1895/wormbook.1.179.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmey R. G., Reis A., Leese H. J. and McEvoy T. G. (2009). Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reproduction in domestic animals=Zuchthygiene 44 Suppl. 3, 50-58. 10.1111/j.1439-0531.2009.01402.x [DOI] [PubMed] [Google Scholar]
- Sui X. W., Arlt H., Brock K. P., Lai Z. W., DiMaio F., Marks D. S., Liao M. F., Farese R. V. and Walther T. C. (2018). Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217, 4080-4091. 10.1083/jcb.201809067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W.-P., Agarwal A. K., Garg A., Anderson R. G. W. and Goodman J. M. (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U.S.A. 104, 20890-20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa A., Rappleye C. A. and Aroian R. V. (2001). Pod-2, along with pod-1, defines a new class of genes required for polarity in the early Caenorhabditis elegans embryo. Dev. Biol. 233, 412-424. 10.1006/dbio.2001.0234 [DOI] [PubMed] [Google Scholar]
- Vicencio J., Martínez-Fernández C., Serrat X. and Cerón J. (2019). Efficient generation of endogenous fluorescent reporters by nested CRISPR in Caenorhabditis elegans. Genetics 211, 1143-1154. 10.1534/genetics.119.301965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrablik T. L., Petyuk V. A., Larson E. M., Smith R. D. and Watts J. L. (2015). Lipidomic and proteomic analysis of Caenorhabditis elegans lipid droplets and identification of ACS-4 as a lipid droplet-associated protein. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1851, 1337-1345. 10.1016/j.bbalip.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., Chung J. and Farese R. V. Jr. (2017). Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33, 491-510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. J., Becuwe M., Housden B. E., Chitraju C., Porras A. J., Graham M. M., Liu X. R. N., Thiam A. R., Savage D. B., Agarwal A. K. et al. (2016a). Seipin is required for converting nascent to mature lipid droplets. elife5, e16582, 1-28. 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed]
- Wang H. Z., Jiang X., Wu J. Y., Zhang L. Q., Huang J. F., Zhang Y. R., Zou X. J. and Liang B. (2016b). Iron overload coordinately promotes ferritin expression and fat accumulation in Caenorhabditis elegans. Genetics 203, 241 10.1534/genetics.116.186742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L. and Ristow M. (2017). Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 207, 413-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. S., Morton D. G., Kemphues K. J. and Watts J. L. (2018). The biotin-ligating protein BPL-1 is critical for lipid biosynthesis and polarization of the Caenorhabditis elegans embryo. J. Biol. Chem. 293, 610-622. 10.1074/jbc.M117.798553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windpassinger C., Auer-Grumbach M., Irobi J., Patel H., Petek E., Hörl G., Malli R., Reed J. A., Dierick I., Verpoorten N. et al. (2004). Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat. Genet. 36, 271-276. 10.1038/ng1313 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Kolb D., Hermann S., Koning R. I. and Kohlwein S. D. (2011). A role for seipin in lipid droplet dynamics and inheritance in yeast. J. Cell Sci. 124, 3894-3904. 10.1242/jcs.091454 [DOI] [PubMed] [Google Scholar]
- Xu N. Y., Zhang S. B. O., Cole R. A., McKinney S. A., Guo F. L., Haas J. T., Bobba S., Farese R. V. and Mak H. Y. (2012). The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198, 895-911. 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R. H., Qian H. W., Lukmantara I., Gao M. M., Du X. M., Yan N. E. and Yang H. Y. (2018). Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
- Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z.-Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S. et al. (2006). An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700-704. 10.1038/nature04942 [DOI] [PubMed] [Google Scholar]
- Zhang P., Na H. M., Liu Z. L., Zhang S. Y., Xue P., Chen Y., Pu J., Peng G., Huang X., Yang F. Q. et al. (2012). Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol. Cell. Proteomics 11, 317-328. 10.1074/mcp.M111.016345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowalaty A. E. E. and Ye X. (2017). Seipin deficiency leads to defective parturition in mice. Biol. Reprod. 97, 378-386. 10.1093/biolre/iox088 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.