Abstract
Background
Persistent cardiac hypertrophy threatens health worldwide. Long non-coding RNAs (lncRNAs) attracted lots of attention in cardiac diseases such as cardiac hypertrophy. In this study, we aimed to study the function of KCNQ1OT1 in cardiac hypertrophy.
Methods
We first used qRT-PCR to detect the expression of KCNQ1OT1 in Ang-II-induced cardiomyocytes and mouse cardiac hypertrophy models. The function of KCNQ1OT1 was investigated by a loss-of-function test. Analysis of the luciferase reporter gene and RNA pulldown confirmed the interaction between KCNQ1OT1 and miR-2054. The target gene of miR-2054 was predicted by bioinformatics analysis and confirmed by luciferase reporter gene detection. Rescue experiments were performed to evaluate the role of miR-2054/AKT3 in the function of KCNQ1OT1.
Results
Our results suggested that KCNQ1OT1 was up-regulated in Ang-II-induced cardiomyocytes and transverse aortic constriction (TAC) mice. Knocking down of KCNQ1OT1 can reduce cell size and downregulate the expression of ANF, BNP and α-MHC in response to Ang-II. KCNQ1OT1 has been shown to compete competitively with miR-2054 and has a negative correlation with its expression. The combination of miR-2054 can reverse the effect of the KCNQ1OT1 combination in Ang-II-induced cardiomyocytes. In addition, AKT3 is a target of miR-2054 and mediates its effect on Ang-II-induced cardiomyocytes.
Conclusions
Knockdown of KCNQ1OT1 could attenuate cardiac hypertrophy through modulation of the miR-2054/AKT3 axis.
Keywords: KCNQ1OT1, cardiac hypertrophy, miR-2054, AKT3
Introduction
The increasing mass and volume of the heart due to hemodynamic changes in response to excessive load is referred to cardiac hypertrophy (1). With the increase of age, the prevalence of cardiac hypertrophy has increased significantly (2). Cardiac cell hypertrophy is the adaptive adjustment of the heart to overload, which can maintain the normal functions of heart in the early stage (3). Persistent myocardial hypertrophy is accompanied by maladaptive cardiac remodeling, which can reduce cardiac synergy and increase the risk of heart failure and sudden death (4). Chronic cardiac hypertrophy threatens health worldwide and may cause many cardiovascular diseases such as heart failure (5). Therefore, early diagnosis and effective treatment for cardiac hypertrophy are very important to reduce the hospitalization rate and improve the long-term survival rate of patients.
Long-chain non-coding RNAs (lncRNAs) are non-coding RNAs (ncRNAs) with transcripts greater than 200 nucleotides in length and have no protein-coding function (6). LncRNAs have been reported to be closely related to the development of the heart and the occurrence of various cardiovascular diseases (7). Changes in the expression of lncRNAs such as MIAT (6), ROR (8) and TINCR (9) have been observed in cardiomyocyte hypertrophy. Exploring the mechanism of action of lncRNAs in cardiomyocyte hypertrophy will help develop more effective treatments, slow or reverse the progression of cardiomyocyte hypertrophy, and improve the quality of life of patients. Several studies have shown that lncRNAs are involved in the regulation of cardiomyocyte hypertrophy (10). For example, lncRNA myocardial hypertrophy-related factor inhibits the targeted binding of miR-489 to Myd88 and induces cardiomyocyte hypertrophy (7). LncRNA KCNQ1OT1 has been demonstrated to participate in cardiovascular diseases. A recent study reported that silencing of lncRNA KCNQ1OT1 alleviated pyroptosis and fibrosis in diabetic cardiomyopathy (11). However, its role in cardiac hypertrophy remains unknown.
MicroRNAs (miRNAs) are a group of non-coding RNAs that do not have protein-coding functions (12). There are approximately 22 highly conserved small fragments of non-coding RNAs that regulate gene expression after gene transcription and play important roles in organ and tissue generation, differentiation, and cell proliferation and apoptosis (13). MiR-2054 was demonstrated to play an important role in the regulation of heart failure (14). Our preliminary bioinformatic studies showed that lncRNA KCNQ1OT1 could bind with miR-2054. However, the changes of the expression of miR-2054 in cardiac hypertrophy and the role of miR-2054 in regulating the occurrence and development of cardiac hypertrophy have not been studied. In this research, we investigated the associations of lncRNA KCNQ1OT1 and miR-2054 in cardiac hypertrophy. Our study may provide new insight into the diagnostic and treatment of cardiac hypertrophy.
Methods
Samples
Male C57BL/6 mice (SPF, n=20, 8 weeks old, 25 g) were provided by Weta River, China. Experiments were performed under a project license (NO.: SHJT-CM74839) granted by institutional ethics board of Shanghai Children’s Medical Center Affiliated to Shanghai Jiaotong University School of Medicine, in compliance with national and institutional guidelines for the care and use of animals.
All animal experiments were carried out according to the guideline of the Animal Use Committee of Shanghai Children’s Medical Center Affiliated to Shanghai Jiaotong University School of Medicine. Transverse aortic constriction (TAC) was conducted to construct a mouse model with cardiac hypertrophy. Briefly, after placing a mouse model in a mouse model with intraperitoneal ketamine (100 mg/kg) and xylazine (10 mg/kg), the thorax was dissected. After 7 days, mice with a pressure gradient of ≥60 nm were included in the research. After obtaining echocardiographic images of the aortic arch, mice with an anatomical M-mode echocardiogram were used to check the stenosis across the aorta. The sham surgery was conducted on corresponding matched mice, which underwent the same process as the above mouse model, without aortic seams.
Treatment
Cardiomyocyte was cut into small pieces and tissues were digested. After centrifugation, cells were resuspended in DMEM/F12 and placed on collagen-coated silicon wafers. Twenty-four hours after seeding, cells were cultured for an additional 24 h in serum-free medium. For cardiac hypertrophy, cardiac myocytes were incubated with 1 mmol/L angiotensin II (Ang-II, Sigma) for 48 h.
Cell transfections
KCNQ1OT1 shRNAs and miR-2054 were purchased from Santa Cruz Biotechnology, USA. For overexpression of KCNQ1OT1 and AKT3, the entire sequence of KCNQ1OT1 and AKT3 was subcloned into an adenoviral vector. MiR-2054 mimics and miR-2054 inhibitors were purchased from RiboBio (Guangzhou). All transfections were conducted using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
Quantitative RT-PCR
TRIZOL reagent (Invitrogen) was used to isolate total RNAs from neonatal mouse cardiomyocytes. Reverse transcription was conducted using the 5X All-In-One RT MasterMix (abmGood, Vancouver, Canada). The expression levels of KCNQ1OT1, miR-2054, AKT3, ANF, BNP and MHC mRNAs were determined with GAPDH or U6 as the endogenous control. Data were analyzed by the 2−ΔΔCt method. Primers used in this study are as following:
KCNQ1OT1-F: 5'- GAAGCTCCAGCCCTGACAAT-3' KCNQ1OT1-R: 5'- GGGAGCTGTTGTCCCTTACC-3' miR-2054-F: 5'- CTGTAATATAAATTTA-3' miR-2054-R: 5'- CTGTAATACATTTTTT-3' AKT3-F: 5'-TTTTCTCTATTATTTGGGCTGAGTC-3' AKT3-R: 5'-CCCCTCTTCTGAACCCAACC-3' ANF-F: 5'-TGCAGCTTCCTGTCAACACT-3' ANF-R: 5'-AGAGGCGAGGAAGTCACCAT-3' BNP-F: 5'-CTTTCCTGGGAGGTCGTTCC-3' BNP-R: 5'-CTGTAACCCGGACGTTTCCA-3' β-MHC-F: 5'-CTTTCTCCCCAGGCCGAAG-3' β-MHC-R: 5'-GAACCCGTGAAGCTCCGAA-3' GAPDH-F: 5'-AGGGCTGCTTTTAACTCTGGT-3' GAPDH-R: 5'-CCCCACTTGATTTTGGAGGGA-3' U6-F: 5'-CTCGCTTCGGCAGCACATATACT-3' U6-R: 5'-ACGCTTCACGAATTTGCGTGTC-3'
Cell surface measurements
The cardiomyocytes were fixed, infiltrated, and stained with α-actin (Abcam, Cambridge, UK) at 4 °C overnight. Cells were then incubated with Daylight 594 goat anti-mouse antibody at room temperature for 1 h. Prior to immunofluorescence capture, cells were treated with DAPI for 10 min. Immunofluorescence images were recorded. The average values were utilized for analysis. The cell surface areas were measured using Image-Pro Plus 6.0 software.
Western blotting
Cardiomyocytes were lysed, and protein concentrations were measured using a BCA kit (Beyotime, China). Proteins were processed in 10% SDS-PAGE and transferred to a membrane. The membrane was then treated with anti-ANF (1:1,000; Santa Cruz, USA), anti-BNP (1:1000; Santa Cruz), and anti-AKT3 (1:1,000, Santa Cruz). GAPDH was used as the control. ECL was utilized to quantify the blots.
Luciferase reporter gene assay
Several putative binding sites of miR-2054 containing KCNQ1OT1 wild-type or mutant were cloned into the pGL4 plasmid. The luciferase reporter plasmid and Adv-miR-2054 vector (or empty vector) were transfected into myocardial cells with lipofectamine 2000. The relative luciferase activity was measured using the Dual-luciferase Assay System (Promega, USA) after 48 h. The interaction between AKT3 3'-UTR and miR-2054 was also confirmed.
RNA pull-down assay
Human cardiomyocytes were transfected with biotin-labeled negative controls, biotin-labeled miR-2054, and biotin-labeled miR-2054-Mut for 48 h. The whole-cell extract was incubated with M-280 streptavidin magnetic beads (Invitrogen, USA) at 4 °C for 4 h. Co-precipitated RNA was then isolated using a lysis buffer containing proteinase K (Invitrogen) and 10% SDS. RNA expression was detected by qRT-PCR analysis.
Data analysis
Results were expressed as mean ± SD and analyzed using GraphPad. P<0.05 indicates statistical significance. Spearman correlation was used to analyze the correlation between genes. Student t tests and one-way analysis of variance were used to explore differences between two groups or among multiple groups, respectively.
Results
KCNQ1OT1 was up-regulated in hypertrophic cardiomyocytes
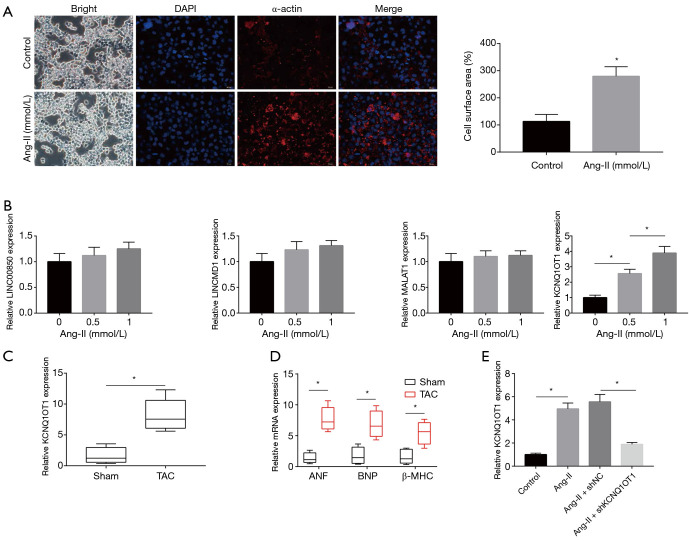
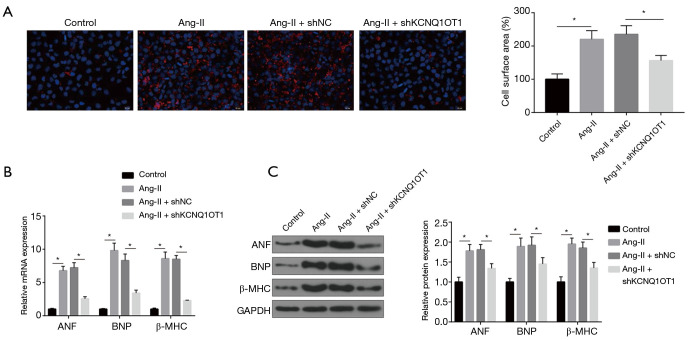
The effects of Ang-II on cardiomyocytes’ hypertrophy were investigated by quantifying cell surface areas. As shown in Figure 1A, Ang-II treatment significantly increased the cell surface area of cardiomyocytes. In other words, Ang-II treatment established a cell model of cardiac hypertrophy. In addition, only the expression levels of KCNQ1OT1 among 4 lncRNAs (LINC00850, LINCMD1, MALAT1, KCNQ1OT1) gradually increased with the increase of Ang-II concentration (Figure 1B). Compared to the sham operation group, the expression levels of KCNQ1OT1 were markedly higher in TAC mice (Figure 1C). We examined the expression levels of hypertrophy markers of ANF, BNP, and β-MHC in the TAC and the sham group. All 3 markers in the TAC group were up-regulated (Figure 1D). As shown in Figure 1E, KCNQ1OT1-specific shRNA effectively reduced Ang-II-induced up-regulation of KCNQ1OT1. Immunofluorescence images showed that knockdown of KCNQ1OT1 eliminated the effect of Ang-II on the surface areas of cardiomyocytes (Figure 2A). Furthermore, qRT-PCR and Western blotting showed that silencing of KCNQ1OT1 reduced Ang-II-induced up-regulation of shKCNQ1OT1 and pcDNA-VDR, respectively (Figure 2B,C). Herein, we demonstrated that silencing of KCNQ1OT1 inhibits the hypertrophic response of cardiomyocytes treated with Ang-II.
Figure 1.
LncRNA KCNQ1OT1 is up-regulated in hypertrophic cardiomyocytes. (A) Immunofluorescence results for the effect of Ang-II on cell surface areas. (B) Changes in the expression of LINC00850, LINCMD1, MALAT1, and KCNQ1OT1 in cardiomyocytes under Ang-II. (C) Detection of the expression of KCNQ1OT1 in TAC and sham mice. (D) The mRNA expression of hypertrophy markers by RT-PCR. (E) Transfection efficiency of shKCNQ1OT1 by RT-PCR. *, P<0.05. TAC, transverse aortic constriction.
Figure 2.
Knockdown of KCNQ1OT1 inhibits cardiomyocytes’ hypertrophy induced by Ang-II. (A) Immunofluorescence results for the effect of KCNQ1OT1 knockdown on cell surface areas. Scale bar: 200 microns. (B,C) mRNA or protein expression of ANF, BNP, and β-MHC by shKCNQ1OT1. *, P<0.05.
LncRNA KCNQ1OT1 participates in the regulation of the miR-2054/AKT3 axis
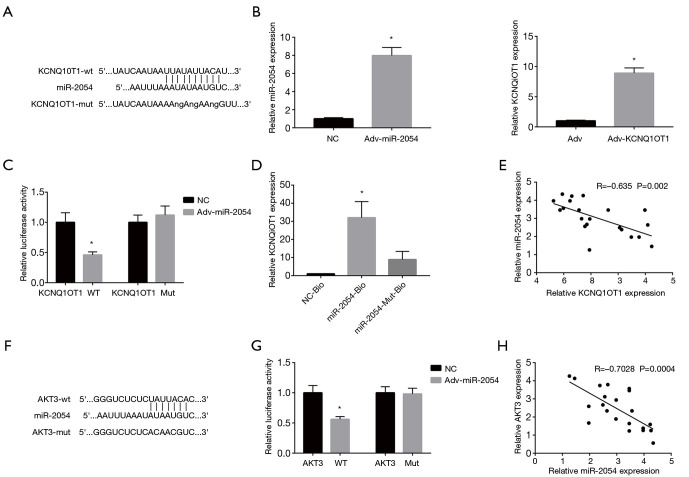
Next, we analyzed the interaction of KCNQ1OT1 and miR-2054 in cardiomyocytes. The complementary sequence between KCNQ1OT1 and miR-2054 was obtained from the online program starbase v2.0 (http://starbase.sysu.edu.cn/, Figure 3A). To determine whether KCNQ1OT1 directly regulates miR-2054, miR-2054 and KCNQ1OT1 were overexpressed in cardiomyocytes (Figure 3B). We generated a KCNQ1OT1 RNA (KCNQ1OT1-wt) and a mutant form (KCNQ1OT1-mut) of a luciferase reporter gene construct (Figure 3C). These results indicated that overexpression of miR-2054 significantly inhibited the luciferase activity of KCNQ1OT1-wt. No effect was observed in the mutant form (Figure 3C), indicating that KCNQ1OT1 directly interacts with miR-2054 at this putative binding site. In addition, RNA pull-down analysis showed that miR-2054-Bio can significantly enrich KCNQ1OT1, while miR-2054-Mut-Bio cannot enrich KCNQ1OT1. (Figure 3D). The expression of miR-2054 was inversely related to the expression of KCNQ1OT1 (Figure 3E). In addition, potential targets for miR-2054 were also predicted by Targetscan Human 7.2 (http://www.targetscan.org/, Figure 3F). The direct interaction between miR-2054 and AKT3 was confirmed by luciferase reporter detection (Figure 3G). The expression of AKT3 was also negatively correlated with the expression of miR-2054 (Figure 3H). In summary, our results indicated that lncRNA KCNQ1OT1 is involved in the regulation of the miR-2054/AKT3 axis.
Figure 3.
Directional interaction between KCNQ1OT1 and miR-2054, miR-2054 and AKT3. (A) The complementary sequence between KCNQ1OT1 and miR-2054 was obtained from the online program starbase v2.0 (http://starbase.sysu.edu.cn/). (B) Overexpression of KCNQ1OT1 and miR-2054 in cardiomyocytes transfected by recombinant adenovirus. (C) Perform a luciferase reporter gene test to detect luciferase activity at the binding site on KCNQ1OT1 when miR-2054 is overexpressed. (D) RNA pull-down test showed that lncRNA KCNQ1OT1 can be enriched by biotin-labeled miR-2054, but not by biotin-labeled miR-2054 mutant. (E) Spearman correlation analysis was used to analyze the correlation between KCNQ1OT1 level and miR-2054 level in the TAC group. (F) The complementary sequence between miR-2054 and AKT3 was obtained from Targetscan Human 7.2 (http://www.targetscan.org/). (G) Luciferase reporter assay is conducted to detect the luciferase activity of the binding site on AKT3. (H) The correlation between the levels of miR-2054 and AKT3 in the TAC group was analyzed by Spearman correlation analysis. *, P<0.05. TAC, transverse aortic constriction.
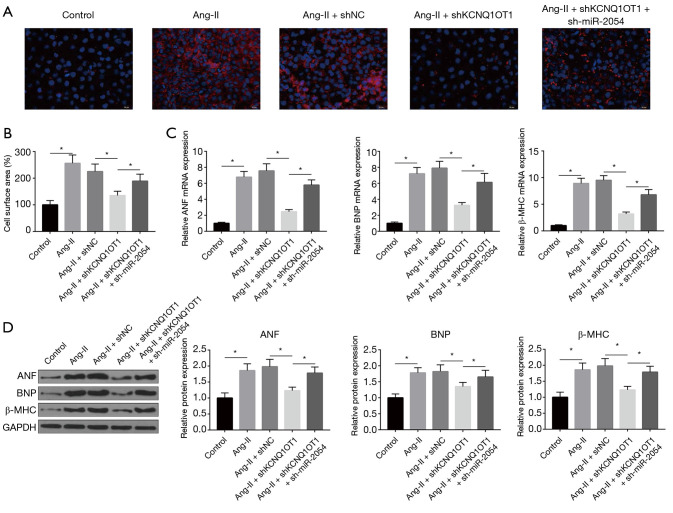
miR-2054 mediates the pro-hypertrophic effect of KCNQ1OT1 on cardiomyocytes
To further investigate whether KCNQ1OT1 plays a role in myocardial hypertrophy by regulating its target miR-2054, shKCNQ1OT1 or sh-miR-2054 was transfected into cardiomyocytes and then treated with Ang-II. As shown in Figure 4A,B, sh-miR-2054 could partially reverse the inhibitory effect of shKCNQ1OT1 on Ang-II-induced myocardial hypertrophy. In addition, sh-miR-2054 also upregulated the expression of ANF, BNP, and γ-MHC at both mRNA and protein levels (Figure 4C,D). These results indicated that KCNQ1OT1 exerted its hypertrophic effect by regulating miR-2054 in an Ang-II-induced hypertrophy model.
Figure 4.
LncRNA KCNQ1OT1 acts as CeRNA by competing with miR-2054. (A,B) Immunofluorescence for cell surface areas of Ang-II-induced cardiomyocytes under shKCNQ1OT1 and sh-miR-2054. (C) mRNA expression of ANF, BNP and β-MHC under shKCNQ1OT1 and sh-MIR-2054. (D) ANF, BNP, and β-MHC protein levels under shKCNQ1OT1 and sh-miR-2054. *, P<0.05.
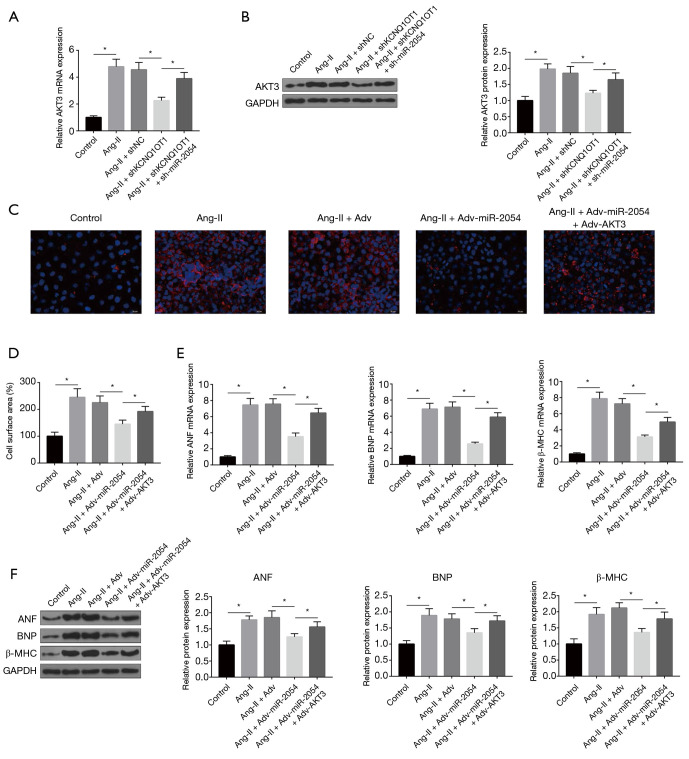
miR-2054 attenuates Ang-II-induced cardiac hypertrophy by targeting AKT3
The effects of KCNQ1OT1 and miR-2054 on the expression of AKT3 were then investigated. As shown in Figure 5A,B, the expression levels of AKT3 were significantly increased in the Ang-II-induced hypertrophy model, and shKCNQ1OT1 could down-regulate its expression. Although the effect of shKCNQ1OT1 on the expression of AKT3 has been reduced by sh-miR-2054. In order to further explore whether miR-2054 plays a role in cardiac hypertrophy by regulating its target gene AKT3, cardiomyocytes were transfected with Adv-miR-2054 alone or with Adv-AKT3 and then treated with Ang-II. As shown in Figure 5C,D,E,F, exogenous expression of miR-2054 significantly reduced Ang-II-induced hypertrophy, as revealed by smaller cell surface serum. However, after the overexpression of AKT3, the miR-2054-mediated anti-hypertrophic effects were largely eliminated in Ang-II-treated cardiomyocytes (Figure 5C,D,E,F). All these results indicated that miR-2054 exerted its anti-hypertrophic effect by regulating AKT3 in an in vitro Iso-induced hypertrophy model.
Figure 5.
miR-2054 attenuates Ang-II-induced cardiac hypertrophy by targeting AKT3. (A,B) Expression of AKT3 under shKCNQ1OT1 and sh-miR-2054. (C) and (D) Cell surface area under Adv-miR-2054 and Adv-AKT3. (E) mRNA expression of ANF, BNP and β-MHC under Adv-miR-2054 and Adv-AKT3. (F) ANF, BNP, and β-MHC protein expression were under Adv-miR-2054 and Adv-AKT3. *, P<0.05.
Discussion
Reducing myocardial adaptive hypertrophy has been considered as a therapeutic method for heart failure. One study found that lncRNA QCHRF was significantly upregulated in angiotensin II-induced hypertrophic cardiomyocytes, and the same effect was observed in mouse heart aorta contraction models and human heart failure samples. In this study, among LINC00850, LINCMD1, MALAT1, KCNQ1OT1, only the expression levels of KCNQ1OT1 increased as the increase of the concentration of Ang-II. The expression levels of KCNQ1OT1 were markedly higher in TAC mice. In addition, the hypertrophy markers of ANF, BNP, and β-MHC in the TAC group were significantly up-regulated. The immunofluorescence results confirmed that knockdown of KCNQ1OT1 inhibited the effect of Ang-II on the surface areas of cardiomyocytes. As far as we know, very limited researches were conducted regarding the genesis of cardiac hypertrophy. Our results revealed that KCNQ1OT1 inhibited the hypertrophic response of cardiomyocytes treated with Ang-II, which bridges the gap.
Previous studies have demonstrated that KCNQ1OT1 could interact with a series of miRNAs to exert its biological functions. For instance, KCNQ1OT1 was reported to promote the expression of BMP2 to regulate osteogenic differentiation by sponging miRNA-214 (15). KCNQ1OT1 can also bind with miR-211-5p (16), miR-27b-3p (17), and miR-370 (18). In our experiments, we found that overexpression of miR-2054 significantly inhibited the luciferase activity of KCNQ1OT1-wt. The expression of miR-2054 was inversely related to the expression of KCNQ1OT1. The direct interaction between miR-2054 and AKT3 was also confirmed by luciferase reporter detection. As a novel supplementary to the lncRNA-miRNA interactions, our results indicate that lncRNA KCNQ1OT1 is involved in the regulation of the miR-2054/AKT3 axis.
It was reported that the expression of miR-2054 was up-regulated in patients with congestive heart failure (19). Another study also discovered that miR-2054 might be associated with the pathological development of acute cardiovascular diseases (14). Sh-miR-2054 can partially reverse the inhibitory effect of shKCNQ1OT1 on Ang-II-induced myocardial hypertrophy. In addition, sh-miR-2054 also upregulated the expression of ANF, BNP, and γ-MHC at the mRNA and protein levels. For the first time, we found that KCNQ1OT1 exerted its hypertrophic effect by regulating miR-2054 in an Ang-II-induced hypertrophy model.
AKT3 has been found to be a critical factor in heart diseases. The deletion of Akt1 was reported to cause heart defects and abnormal cardiomyocyte proliferation (20). In consistence with the previous study, we found that AKT3 was significantly upregulated in the Ang-II-induced hypertrophy model. However, shKCNQ1OT1 could down-regulate its expression. Furthermore, the effect of shKCNQ1OT1 on the expression of AKT3 has been diminished by sh-miR-2054. Moreover, overexpression of AKT3 could eliminate the miR-2054-mediated anti-hypertrophic effects in Ang-II-treated cardiomyocytes. Interestingly, we revealed that miR-2054 attenuates Ang-II-induced cardiac hypertrophy by targeting AKT3.
Conclusions
Our experiments and analysis revealed that knockdown of KCNQ1OT1 could attenuate cardiac hypertrophy through modulation of the miR-2054/AKT3 axis.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was support from the Foundation of Shanghai Municipal Health Commission (No.20174Y0106); The National Natural Science Foundation of China (No. 81470443, No. 81770332); Shanghai Excellent Youth Diplomate Training Assistance Scheme (No.201817).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (NO.: SHJT-CM74839) granted by institutional ethics board of Shanghai Children’s Medical Center Affiliated to Shanghai Jiaotong University School of Medicine, in compliance with national and institutional guidelines for the care and use of animals.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-203
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-203). The authors have no conflicts of interest to declare.
References
- 1.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613-8. 10.1038/nm1582 [DOI] [PubMed] [Google Scholar]
- 2.Hunter JJ, Chien KR. Signaling pathways for cardiac hypertrophy and failure. N Engl J Med 1999;341:1276-83. 10.1056/NEJM199910213411706 [DOI] [PubMed] [Google Scholar]
- 3.Molkentin JD, Lu JR, Antos CL, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 1998;93:215-28. 10.1016/S0092-8674(00)81573-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan HE, Baker KM. Cardiac hypertrophy. Mechanical, neural, and endocrine dependence. Circulation 1991;83:13-25. 10.1161/01.CIR.83.1.13 [DOI] [PubMed] [Google Scholar]
- 5.Sen S, Tarazi RC, Khairallah PA, et al. Cardiac hypertrophy in spontaneously hypertensive rats. Circ Res 1974;35:775-81. 10.1161/01.RES.35.5.775 [DOI] [PubMed] [Google Scholar]
- 6.Zhu XH, Yuan YX, Rao SL, et al. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur Rev Med Pharmacol Sci 2016;20:3653-60. [PubMed] [Google Scholar]
- 7.Wang K, Liu F, Zhou LY, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 2014;114:1377-88. 10.1161/CIRCRESAHA.114.302476 [DOI] [PubMed] [Google Scholar]
- 8.Jiang F, Zhou X, Huang J. Long Non-Coding RNA-ROR Mediates the Reprogramming in Cardiac Hypertrophy. PLoS One 2016;11:e0152767. 10.1371/journal.pone.0152767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao M, Chen G, Lv F, et al. LncRNA TINCR attenuates cardiac hypertrophy by epigenetically silencing CaMKII. Oncotarget 2017;8:47565-73. 10.18632/oncotarget.17735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Wang J, Sun L, et al. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol 2018;818:508-17. 10.1016/j.ejphar.2017.11.031 [DOI] [PubMed] [Google Scholar]
- 11.Yang F, Qin Y, Lv J, et al. Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis 2018;9:1000. 10.1038/s41419-018-1029-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ucar A, Gupta SK, Fiedler J, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078. 10.1038/ncomms2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong DL, Chen C, Huo R, et al. Reciprocal repression between microRNA-133 and calcineurin regulates cardiac hypertrophy: a novel mechanism for progressive cardiac hypertrophy. Hypertension 2010;55:946-52. 10.1161/HYPERTENSIONAHA.109.139519 [DOI] [PubMed] [Google Scholar]
- 14.Ali SS, Kala C, Abid M, et al. Pathological microRNAs in acute cardiovascular diseases and microRNA therapeutics. J Acute Disease 2016;5:9-15. 10.1016/j.joad.2015.08.001 [DOI] [Google Scholar]
- 15.Wang CG, Liao Z, Xiao H, et al. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp Mol Pathol 2019;107:77-84. 10.1016/j.yexmp.2019.01.012 [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis 2018;9:742. 10.1038/s41419-018-0793-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Z, Yang P, Qiu X, et al. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J Cell Physiol 2019;234:11304-14. 10.1002/jcp.27788 [DOI] [PubMed] [Google Scholar]
- 18.Gong W, Zheng J, Liu X, et al. Knockdown of Long Non-Coding RNA KCNQ1OT1 Restrained Glioma Cells' Malignancy by Activating miR-370/CCNE2 Axis. Front Cell Neurosci 2017;11:84. 10.3389/fncel.2017.00084 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Sarli B, Topsakal R, Kaya EG, et al. Tenascin-C as predictor of left ventricular remodeling and mortality in patients with dilated cardiomyopathy. J Investig Med 2013;61:728-32. 10.2310/JIM.0b013e3182880c11 [DOI] [PubMed] [Google Scholar]
- 20.Chang Z, Zhang Q, Feng Q, et al. Deletion of Akt1 causes heart defects and abnormal cardiomyocyte proliferation. Dev Biol 2010;347:384-91. 10.1016/j.ydbio.2010.08.033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as