Abstract
Background
The study objective was to determine whether unlabeled datasets can be used to further train and improve the accuracy of a deep learning system (DLS) for the detection of tuberculosis (TB) on chest radiographs (CXRs) using a two-stage semi-supervised approach.
Methods
A total of 111,622 CXRs from the National Institute of Health ChestX-ray14 database were collected. A cardiothoracic radiologist reviewed a subset of 11,000 CXRs and dichotomously labeled each for the presence or absence of potential TB findings; these interpretations were used to train a deep convolutional neural network (DCNN) to identify CXRs with possible TB (Phase I). The best performing algorithm was then used to label the remaining database consisting of 100,622 radiographs; subsequently, these newly-labeled images were used to train a second DCNN (phase II). The best-performing algorithm from phase II (TBNet) was then tested against CXRs obtained from 3 separate sites (2 from the USA, 1 from China) with clinically confirmed cases of TB. Receiver operating characteristic (ROC) curves were generated with area under the curve (AUC) calculated.
Results
The phase I algorithm trained using 11,000 expert-labelled radiographs achieved an AUC of 0.88. The phase II algorithm trained on images labeled by the phase I algorithm achieved an AUC of 0.91 testing against a TB dataset obtained from Shenzhen, China and Montgomery County, USA. The algorithm generalized well to radiographs obtained from a tertiary care hospital, achieving an AUC of 0.87; TBNet’s sensitivity, specificity, positive predictive value, and negative predictive value were 85%, 76%, 0.64, and 0.9, respectively. When TBNet was used to arbitrate discrepancies between 2 radiologists, the overall sensitivity reached 94% and negative predictive value reached 0.96, demonstrating a synergistic effect between the algorithm’s output and radiologists’ interpretations.
Conclusions
Using semi-supervised learning, we trained a deep learning algorithm that detected TB at a high accuracy and demonstrated value as a CAD tool by identifying relevant CXR findings, especially in cases that were misinterpreted by radiologists. When dataset labels are noisy or absent, the described methods can significantly reduce the required amount of curated data to build clinically-relevant deep learning models, which will play an important role in the era of precision medicine.
Keywords: Artificial intelligence (AI), deep learning system (DLS), tuberculosis (TB), chest radiography (CXR)
Introduction
Tuberculosis (TB) is an infectious disease that affects nearly one-third of the world, largely in developing countries (1-3). TB is, in fact, the ninth-leading cause of death worldwide and the leading infectious cause of death globally (4). Current TB screening programs depend heavily on the chest radiograph (CXR), which is relatively inexpensive and widely available in the United States (1-3,5). Nevertheless, consistent and comprehensive TB screening in the United States has been difficult to achieve, owing to federal gaps and state variation in TB screening policies, as well as limited resources at both levels for such screening programs (6,7). Furthermore, radiologists charged with interpreting TB screening CXRs can be overwhelmed by the high volume of studies that are a necessary consequence of widespread screening programs (8). Computer aided detection (CAD) of TB on screening CXR can prioritize review of likely positive cases and reduce the time to diagnosis, thereby facilitating more consistent TB screening initiatives worldwide.
Deep learning, an artificial intelligence (AI) technique, has shown great promise for automated medical image analysis and interpretation (8-10), and could be helpful in improving TB screening efforts. Deep learning systems (DLS) are actively studied for potential applications in various medical fields, from diabetic retinopathy screening in ophthalmology to drug discovery tools that predict molecular interactions (11-14). Specifically, Lakhani et al. recently described a promising DLS for detection of TB on CXRs, achieving AUC of 0.99 (8). This study, however, was limited by a small dataset (training set: 857 patients; testing set: 150 patients) and absence of an external test dataset that is completely separate from the training/validation dataset. Similarly, Hwang et al. developed a DLS for TB detection using a relatively homogeneous population of 10,848 Korean patients, achieving an AUC of 0.88 to 0.96 (15). However, to create a robust screening DLS that can be implemented globally, it is critical to utilize large diverse datasets and demonstrate generalizability to external populations (16).
Performance of a DLS chiefly depends on two factors: the dataset quality in terms of size and diversity, and the accuracy of labels that correspond to the images. Training a clinically-relevant DLS requires a large dataset of radiographs, in the order of tens of thousands of images, with reliable ground truth labels indicating presence or absence of the disease of interest. However, confirming data label accuracy would require significant time investment by radiologists, which is often the rate-limiting step in DLS development.
In this study, we utilized a novel two-stage semi-supervised approach to develop a TB-detecting deep convolutional neural network (DCNN) by first using a small number of radiologist-reviewed radiographs, then incorporating semi-supervised learning to analyze a larger number of unlabeled radiographs.
Methods
Datasets
This retrospective study was approved by the institutional research board. Publicly-available datasets did not contain patient-identifiers. Images obtained from our tertiary care center [Johns Hopkins Hospital (JHH), Baltimore, MD] were de-identified and compliant with the Health Insurance Portability and Accountability Act (HIPAA).
We obtained CXRs from the publicly-available NIH chest X-ray 14 database (16), comprised of 112,120 frontal CXRs from 30,805 patients. NIH CXR14 database contains labels for 14 thoracic diseases, extracted from radiology reports using Natural Language Processing. Since TB is not included as one of the 14 labelled thoracic diseases, a fellowship-trained cardiothoracic radiologist with five years of post-graduate experience (C.L.) individually reviewed 11,000 radiographs from the NIH database, determining whether each radiograph had an imaging appearance that could be present in pulmonary TB. Phase I training and validation were completed using the radiologist-labelled radiographs. A total of 498 radiographs from the entire database were excluded from analysis due to suboptimal quality of radiographs, which consisted of abdominal radiographs, radiographs with incomplete visualization of lung fields, blank radiographs, digital artifacts, inverted, and lateral CXRs.
External datasets were used for testing of DCNN performance, comprised of 662 CXRs from Shenzhen, China [336 (50.8%) with TB, 326 (49.2%) without TB], 138 CXRs from Montgomery County, USA [58 (42.0%) with TB, 80 (58.0%) without TB], and 100 CXRs from JHH [35 (35.0%) with TB, 65 (65.0%) without TB] (Table 1) (17). Radiographs from JHH were selected based on ICD code of TB, documented positive acid-fast bacillus stain on sputum results on electronic medical record, and availability of a contemporaneous CXR on our picture archiving and communication system (PACS). All images were saved in lossless Portable Network Graphics (PNG) format and resized to a 224×224 matrix.
Table 1. Datasets used for phase I deep convolutional neural network training and validation.
| Dataset | Development phase | Total number of radiographs used | Number with suspected TB (%) | Number without suspected TB (%) |
|---|---|---|---|---|
| NIH CXR | Training/validation | 11,000 | 5,381 (48.9)* | 5619 (51.1)* |
| Montgomery County, USA | Testing | 138 | 58 (42.0) | 80 (58.0) |
| Shenzhen, China | Testing | 662 | 336 (50.8) | 326 (49.2) |
| Johns Hopkins Hospital, Baltimore, MD, USA | Testing | 100 | 35 (35.0) | 65 (65.0) |
*, ground truth determined by a cardiothoracic radiologist. CXR, chest radiograph; TB, tuberculosis.
Phase I neural network training, validation, and testing
We randomly assigned 80% of 11,000 labelled NIH radiographs into the “training” dataset and 20% of the data into the “validation” dataset, ensuring no overlap in images between these datasets (Table 1). Briefly, the training phase utilizes the majority of the available data to train DCNNs to classify images into pre-defined categories by identifying image features specific to each category. The validation phase utilizes a smaller proportion of available data to test the DCNNs trained in the training phase and select the highest-performing algorithms. The final testing phase consists of assessing the diagnostic performance of the best-performing algorithm(s) on a dataset that was not utilized in either the training or validation phase.
We utilized the ResNet-50 (18) DCNN pretrained on 1.2 million color images of everyday objects from ImageNet (http://www.image-net.org/) prior to training on the CXRs. This technique is known as transfer learning and allows for modification of pretrained neural network architectures to be used for classification of different datasets not used in training of the original network (8,19,20). The solver parameters used for our DCNN training were as follows: 50 training epochs; stochastic gradient descent (SGD) with a learning rate of 0.001, momentum of 0.9, and weight decay of 1×10−5. During each training epoch, each image was augmented by a random rotation between −5 and 5 degrees, random cropping, and horizontal flipping. To identify the distinguishing image features used by the DCNN for classification, we created heat maps via Class Activation Mapping (CAM) (21).
Phase II neural network training, validation, and testing
The best-performing algorithm from phase I development was used to generate its predictions on the remaining 100,622 radiographs from the CXR14 database (Table 2). Resulting images were split 80% into training phase, and 20% into validation phase following the same methodology described above. The best-performing algorithm in Phase II, given the name TBNet, was selected by testing the DCNNs using radiographs obtained from Asia (Shenzhen, China) and USA (Montgomery County, MD and JHH at Baltimore, MD).
Table 2. Datasets used for phase II deep convolutional neural network training and validation.
| Dataset | Development phase | Total number of radiographs used | Number with suspected TB (%) | Number without suspected TB (%) |
|---|---|---|---|---|
| NIH CXR | Training/validation | 100,622 | 44,521 (44.2)+ | 56,101 (55.8)+ |
| Montgomery County, USA | Testing | 138 | 58 (42.0) | 80 (58.0) |
| Shenzhen, China | Testing | 662 | 336 (50.8) | 326 (49.2) |
| Johns Hopkins Hospital, Baltimore, MD, USA | Testing | 100 | 35 (35.0) | 65 (65.0) |
+, ground truth determined by phase I deep learning system (DLS). CXR, chest radiograph; TB, tuberculosis.
To assess TBNet’s efficacy as a CAD tool, majority vote analysis was performed. TBNet’s output was used to arbitrate any differences in radiologists’ interpretations.
Statistical analysis
Receiver operating characteristic (ROC) curves with area under the curve (AUC) were generated and statistically compared between DCNNs using the DeLong parametric method (22,23). Sensitivity, specificity, positive predictive value, and negative predictive value was calculated based on the algorithm’s performance against JHH radiographs.
Computer hardware & software specifications
All DCNN development and testing was performed using PyTorch framework (https://pytorch.org) on a 2.5 GHz Intel Haswell dual socket (12-core processors) (Intel, Santa Clara, CA) with 128 GB of RAM and 2 NVIDIA K80 GPUs (NVIDIA Corporation, Santa Clara, CA).
Radiologist interpretation
A board-certified cardiothoracic radiologist with 5 years of post-fellowship experience (C.L., Radiologist 1) and a PGY-4 diagnostic radiology resident (P.Y., Radiologist 2) interpreted the 100 CXRs obtained from JHH for potential TB.
Results
Our highest-performing DCNN from Phase I, trained using 11,000 images curated by an expert radiologist, achieved AUC of 0.88 for detection of TB in patients with clinically and pathologically-confirmed TB. After Phase II training, TBNet achieved an improved AUC of 0.91 when tested against the same dataset (P<0.05). When tested using radiographs obtained from JHH, TBNet performed with an AUC of 0.87 (Figure 1). The algorithm reached a sensitivity of 85% and a specificity of 76%, with positive predictive value of 0.64 and negative predictive value of 0.9 (Table 3). These results were comparable to those of a cardiothoracic radiologist and superior to those of a PGY-4 radiology resident. In majority vote analysis, sensitivity reached 94% and negative predictive value reached 0.96, demonstrating a synergistic effect between the algorithm’s output and radiologists’ interpretations (Table 3).
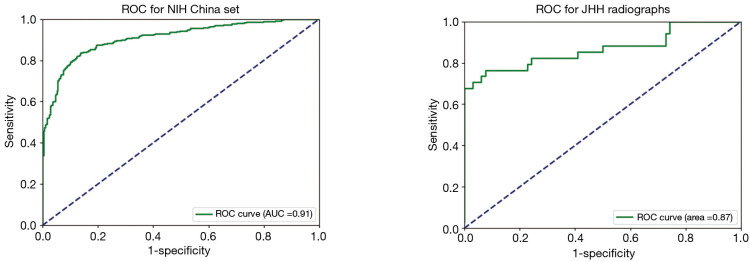
Figure 1.
Receiver operator curve of TBNet for detection of tuberculosis. Area under the ROC curve (AUC) is 0.91 for detection of tuberculosis when tested against dataset China, and 0.87 when tested against radiographs obtained from Johns Hopkins Hospital (JHH). ROC, receiver operating characteristic curve.
Table 3. Comparison of TBNet and radiologist performance on radiographs obtained from Johns Hopkins Hospital.
| Reader | Sensitivity | Specificity | Positive predictive value (PPV) | Negative predictive value (NPV) |
|---|---|---|---|---|
| TBNet | 85% | 76% | 0.64 | 0.9 |
| Radiologist 1 | 83% | 81% | 0.7 | 0.9 |
| Radiologist 2 | 77% | 75% | 0.63 | 0.86 |
| Majority vote | 94% | 85% | 0.76 | 0.96 |
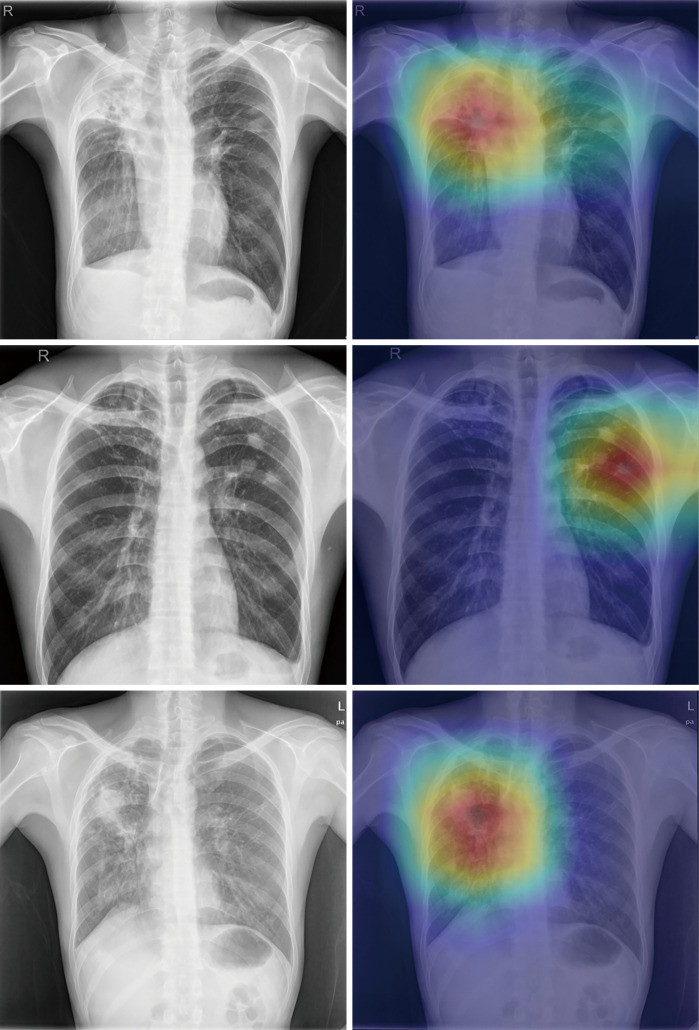
Heatmaps revealed that the DCNN appropriately emphasized the same regions of interest as the human radiologist, such as cavitary and non-cavitary parenchymal nodules/masses and hilar lymphadenopathy (Figure 2).
Figure 2.
Three chest radiographs from the test sets (left images) showing findings of pulmonary tuberculosis with their corresponding prediction outputs using class activation mapping (right images), demonstrating concordance between radiographic findings and machine-derived features.
Discussion
In this study we utilized a novel method of dataset curation and developed a reliable screening algorithm for TB detection. While 10% of the dataset required radiologist interpretation, the remaining 90% of the training dataset were analyzed using semi-supervised learning. Using this approach, we were able to develop a TB-screening algorithm that generalized well to radiographs obtained from diverse settings. None of the training cases had clinical confirmation of TB, therefore the CXR findings detected by the DCNN are non-specific and mimicked the radiologist’s impression of what could be seen in TB. This may be a viable strategy when developing a DLS to detect diseases with a low prevalence.
The preliminary algorithm developed from radiologist-generated labels subsequently created meaningful labels from the unreviewed dataset, as evidenced by the improved performance of the deep learning algorithm that was trained using DCNN-generated labels. This highlights the concept of semi-supervised learning, where the DLS is capable of further discerning features particular to TB in a previously naïve training dataset, resulting in an overall positive effect on the performance.
With increasing number of open-source algorithms, the majority of resources required for DLS development is spent on obtaining large datasets with accurate labels. PACS provides developers with no shortage of deidentified digital radiographs; however, annotating them with radiologist-level accuracy is a time-consuming and expensive task. Utilizing a 2-tiered approach can save significant amount of time and resources in deep learning algorithm development. On average, a radiologist spends 1.4 minutes interpreting a plain-film radiograph, which would translated into 2,348 hours interpreting 100,622 images that were used for phase II development (24). In comparison, we were able to generate machine-generated labels for 100,622 images within 5 minutes using our phase I algorithm.
Furthermore, phase II training yielded a more predictive algorithm with a statistically significant increase in AUC from 0.88 to 0.91 when tested against the same testing dataset (Figure 1). Our algorithm was generalizable to radiographs obtained from diverse settings including a tertiary academic medical center in the US, demonstrating a slight drop in AUC from 0.91 to 0.87 when tested against radiographs obtained from JHH.
Multiple prospective studies assessing clinical value of deep learning algorithms demonstrated significantly limited performance of algorithms compared to their original studies (25). Such findings raise concerns for overfitting in reporting outcomes for deep learning algorithms. Our algorithm has reached a clinically relevant AUC and is generalizable to radiographs obtained from very different population from Shenzhen, China and JHH (Baltimore, MD).
TBNet demonstrated added value in cases that were misinterpreted by either radiologist, demonstrating the value of this algorithm as a CAD tool. In the majority vote analysis, TBNet was able to detect 6 additional positive radiographs from the radiographs that had disparate interpretations between the radiologists, increasing the overall sensitivity to 0.94 (Table 2). The algorithm’s high sensitivity and negative predictive value can significantly reduce radiologists’ workload in screening for TB by triaging radiographs with potential findings indicative of TB.
One limitation of this study is that phase I algorithm outputs can be incorrect. However, AUC of the resulting algorithm increased from 0.88 to 0.91 after Phase II training, which validates the utility of sacrificing individual label accuracy for increased size of the training set. Another concern is the validity of publicly available datasets that were used for training, validation, and testing of our algorithms. To address this concern, we individually reviewed the entire CXR14 database and filtered 498 radiographs that were suboptimal in quality.
Our algorithm was not specific for TB, which has implications for clinical deployment. TBNet was designed to maximize sensitivity as a screening algorithm; however, in a population with a low prevalence of TB, our algorithm would likely have a low positive predictive rate. Operating threshold on the AUC curve should be adjusted for specific populations, and further prospective study is warranted to determine the ideal ROC threshold for maximizing the algorithm’s performance in target population.
Conclusions
Using semi-supervised learning, we trained a deep learning algorithm that detected TB at a high accuracy and demonstrated value as a CAD tool by identifying relevant CXR findings, especially in cases that were misinterpreted by radiologists. When dataset labels are noisy or absent, the described methods can significantly reduce the required amount of curated data to build clinically-relevant deep learning models, which will play an important role in the era of precision medicine.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by Radiological Society of North America R&E Foundation (RMS1816 to TK Kim).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was approved by the institutional research board.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Ammar Chaudhry) for the series “Role of Precision Imaging in Thoracic Disease” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2019.08.34). The series “Role of Precision Imaging in Thoracic Disease” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.Hoog AH, Meme HK, Van Deutekom H, et al. High sensitivity of chest radiograph reading by clinical officers in a tuberculosis prevalence survey. Int J Tuberc Lung Dis 2011;15:1308-14. 10.5588/ijtld.11.0004 [DOI] [PubMed] [Google Scholar]
- 2.Charles M, Pape JW. Tuberculosis and HIV: Implications in the developing world. Curr HIV/AIDS Rep 2006;3:139-44. 10.1007/BF02696658 [DOI] [PubMed] [Google Scholar]
- 3.Nachiappan AC, Rahbar K, Shi X, et al. Pulmonary Tuberculosis: Role of Radiology in Diagnosis and Management. RadioGraphics 2017;37:52-72. 10.1148/rg.2017160032 [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. Global tuberculosis report 2017. World Health Organization; 2017. [Google Scholar]
- 5.Melendez J, Sánchez CI, Philipsen RHHM, et al. An automated tuberculosis screening strategy combining X-ray-based computer-aided detection and clinical information. Sci Rep 2016;6:25265. 10.1038/srep25265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer PM, Noppert GA, Jenkins CH. Gaps in Federal and State Screening of Tuberculosis in the United States. Am J Public Health 2017;107:1750-2. 10.2105/AJPH.2017.304076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reves R, Daley CL. Screening for Latent Tuberculosis Infection. JAMA Intern Med 2016;176:1439. 10.1001/jamainternmed.2016.5464 [DOI] [PubMed] [Google Scholar]
- 8.Lakhani P, Sundaram B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology 2017;284:574-82. 10.1148/radiol.2017162326 [DOI] [PubMed] [Google Scholar]
- 9.Yi PH, Kim TK, Wei J, et al. Automated semantic labeling of pediatric musculoskeletal radiographs using deep learning. Pediatr Radiol 2019;49:1066-70. 10.1007/s00247-019-04408-2 [DOI] [PubMed] [Google Scholar]
- 10.Kim TK, Yi PH, Wei J, et al. Deep Learning Method for Automated Classification of Anteroposterior and Posteroanterior Chest Radiographs. J Digit Imaging 2019;32:925-30. 10.1007/s10278-019-00208-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gawehn E, Hiss JA, Schneider G. Deep Learning in Drug Discovery. Mol Inform 2016;35:3-14. 10.1002/minf.201501008 [DOI] [PubMed] [Google Scholar]
- 12.Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316:2402. 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 13.Ting DS, Yi PH, Hui FK. Clinical Applicability of Deep Learning System in Detecting Tuberculosis Using Chest Radiography. Radiology 2018;286:729-31. 10.1148/radiol.2017172407 [DOI] [PubMed] [Google Scholar]
- 14.Wong TY, Bressler NM. Artificial Intelligence With Deep Learning Technology Looks Into Diabetic Retinopathy Screening. JAMA 2016;316:2366. 10.1001/jama.2016.17563 [DOI] [PubMed] [Google Scholar]
- 15.Hwang S, Kim HE, Jeong J, et al. A novel approach for tuberculosis screening based on deep convolutional neural networks. SPIE Medical Imaging 2016;97852W.
- 16.Wang X, Peng Y, Lu L, et al. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017; 3462-71. [Google Scholar]
- 17.Jaeger S, Candemir S, Antani S, et al. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg 2014;4:475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. 2015;arXiv:1512.03385 [cs.CV].
- 19.Lakhani P. Deep Convolutional Neural Networks for Endotracheal Tube Position and X-ray Image Classification: Challenges and Opportunities. J Digit Imaging 2017;30:460-8. 10.1007/s10278-017-9980-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DH, MacKinnon T. Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol 2018;73:439-45. 10.1016/j.crad.2017.11.015 [DOI] [PubMed] [Google Scholar]
- 21.Zhou B, Khosla A, Lapedriza A, et al. Learning Deep Features for Discriminative Localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition 2016;2921-9. [Google Scholar]
- 22.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988;44:837-45. 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 23.Youden WJ. Index for rating diagnostic tests. Cancer 1950;3:32-5. [DOI] [PubMed] [Google Scholar]
- 24.Fleishon HB, Bhargavan M, Meghea C. Radiologists’ reading times using PACS and using films: One practice’s experience. Acad Radiol 2006;13:453-60. 10.1016/j.acra.2005.12.014 [DOI] [PubMed] [Google Scholar]
- 25.Kanagasingam Y, Xiao D, Vignarajan J, et al. Evaluation of Artificial Intelligence-Based Grading of Diabetic Retinopathy in Primary Care. JAMA Netw Open 2018;1:e182665. 10.1001/jamanetworkopen.2018.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as