Abstract
Background
Although simplified clinicopathological features and serum tumour markers (STMs) were reported to be associated with the status of mismatch repair (MMR) in colorectal cancer (CRC) patients, their predictive value alone or in combination for MMR status remains unknown.
Methods
A retrospective analysis of 3274 participants with MMR testing and STMs measurements from two institutions was conducted. The prediction model was developed in the primary cohort that consisted of 1964 participants. Best subset regression was applied to select the most useful predictors from the primary dataset. The performance of the nomogram was evaluated with respect to its calibration, discrimination, and clinical usefulness. External validation was performed in an independent validation cohort of 1310 consecutive CRC patients.
Findings
Among the ten simplified clinicopathological features, seven variables were selected as the best subset of risk factors to develop pathology-based model, including age, tumour diameters, histology, tumour location, perineural invasion, the number of sampled lymph nodes (LNs) and positive LNs. The model showed good calibration and discrimination, with an AUC of 0.756 (95% CI, 0.722 to 0.789) in the primary cohort and 0.754 (95% CI, 0.715 to 0.793) in the validation cohort. After the addition of CEA and CA 72-4, the performance of pathology-based model was significantly improved in in both the primary cohort (AUC: 0.805 (0.774-0.835) vs. 0.756 (0.722-0.789), P < 0.001) and validation cohort (AUC: 0.796 (0.758-0.835) vs. 0.754 (0.715-0.793), P < 0.001). The results of decision curve analysis revealed that using our models to predict the status of MMR would add more benefit than either the detect-all-patients scheme or the detect-none scheme.
Interpretation
The models based on simplified clinicopathological features alone or in combination with STMs can be conveniently used to facilitate the postoperative individualized prediction of MMR status in CRC patients.
Keywords: Nomogram, Mismatch repair proteins, Clinicopathological features, Serum tumour markers
Research in context.
Evidence before this study
Though several prediction models based on pathological features have been developed, the complex pathological characteristics included in previous models required detailed pathological diagnosis, which not only caused a huge burden on the pathologists, but also lacked of instructive value for clinicians.
Added value of this study
To our knowledge, our study is the first to investigate the predictive value of STMs and incorporate them into the prediction model for MMR status. In addition, we extremely simplify previously complex pathological features and make the MMR risk prediction model available to both clinicians and pathologists.
Implication of all the available evidence
The nomograms based on simplified clinicopathological features alone or in combination with STMs can be conveniently used to facilitate the postoperative individualized prediction of MMR status in CRC patients.
Alt-text: Unlabelled box
1. Introduction
Colorectal cancer (CRC) is the third most common type of cancer and the second leading cause of cancer-related deaths worldwide [1]. Though complete surgical resection is the primary treatment for patients with locoregional CRC, it has been estimated that approximately half of patients with localized CRC will develop metastases [2]. Adjuvant chemotherapy and immune checkpoint inhibitors has been recommended to improve prognosis of patients with high-risk stage II and stage III/IV CRC [3, 4]. However, accumulating evidence demonstrated that individual treatment response of CRC patients is significantly associated with its molecular characteristics [5], [6], [7].
Microsatellite instability (MSI) is the abnormal shortening or lengthening of DNA by 1-6 repeating base pair units, which is caused by the inactivation of the DNA mismatch repair (MMR) system [8]. An increasing body of evidence suggested that CRC patients with MSI are not only highly correlated with a better prognosis [9], higher incidence of Lynch syndrome [10] and a high response to immune checkpoint blockade [11], but less likely to benefit from 5-fluorouracil-based chemotherapy [12]. As a result, genetic testing for MMR or MSI in all CRC patients is recommended by the National Comprehensive Cancer Network (NCCN) guidelines to optimize the treatment and management of CRC [13, 14]. However, the recommendation is not feasible in clinical practice, especially in developing countries. It is reported that only approximately 5%-15% CRC patients presented with deficient MMR (dMMR) and have high levels of microsatellite instability [15, 16], which means 85%-95% CRC patients could not benefit directly from genetic testing. Therefore, there is an urgent need to develop a model for the prediction of MMR status in CRC patients based on phenotypic characteristics.
Previous studies have demonstrated that MMR status of CRC is significantly correlated with their clinicopathological features [17, 18] and serum tumour biomarkers (STMs) [19, 20]. Though several prediction models based on pathological features have been developed, the complex pathological characteristics included in previous models required detailed pathological diagnosis [[21], [22], [23]], which not only caused a huge burden on the pathologists, but also lacked of instructive value for clinicians. STMs are both important prognostic factors and the indicators of therapeutic effect and recurrence in patients with CRC [19, 24], whereas their predictive value for MMR status has not been assessed in previous studies. The aim of our study was to develop MMR prediction models based on simplified clinicopathological features alone or in combination with STMs.
2. Methods
2.1. Study design and patient cohort
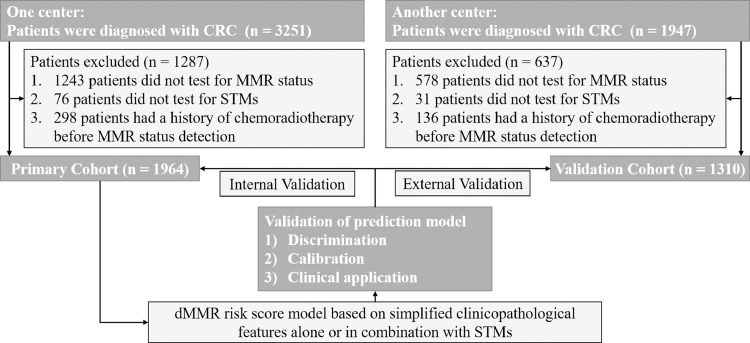
This study retrospectively reviewed 3251 who were diagnosed with CRC and underwent curative surgical resection at Union Hospital, Tongji Medical College, Huazhong University of Science and Technology between May 2017 and December 2019. Patients with the following conditions were excluded from the study: i) no results about MMR status; ii) no results about STMs; iii) the history of chemoradiotherapy before MMR status detection. A total of 1964 patients met these criteria and were included in the primary cohort (1169 males and 795 females; mean age, 57.1 ± 11.7 years, range from 17 to 92 years). From March 2018 to December 2019, an independent validation cohort of 1310 patients (785 males and 525 females; mean age, 57.3 ± 11.8 years, range from 16 to 89 years) was screened from 1947 patients using the same criteria at China-Japan Union Hospital of Jilin University. The flow diagram of developing and validating the prediction model was demonstrated in Fig. 1. This study protocol was approved by the ethics committee of Tongji Medical College, Huazhong University of Science and Technology (No. 2018-S377). All patients signed an informed consent regarding their understanding of the procedure and its potential complications as well as their approval of participation in the research.
Fig. 1.
The flow diagram of developing and validating the prediction model.
Baseline clinicopathologic data was obtained from medical records, including age, gender, tumour diameters, tumour location, pathological type, histology, T-stage, the number of sampled lymph nodes (LNs), the number of positive LNs, and perineural invasion (PNI). Proximal colon cancers were defined as those occurred in the cecum, ascending colon, and transverse colon; distal colon cancers as those in the descending and sigmoid colon; and rectal cancers as those in the rectosigmoid junction and rectum [17]. The depth of invasion and lymph node metastasis were classified according to the 8th AJCC tumour-node-metastasis classification. Perineural invasion was defined as tumour cells found within the perineural space or the infiltration of cancer cells into the endoneurium [25]. Laboratory analysis of STMs was done via routine blood tests within one week before surgery. The normal upper limits of STMs were as follows: carcinoembryonic antigen (CEA), 5 µg/L; carbohydrate antigen (CA) 19-9, 37 U/ml; CA 72-4, 6.9 U/ml. Tumour marker values above these thresholds were considered positive, otherwise, the sample was deemed as negative. The assessment of MMR status was performed through immunohistochemical (IHC) staining as described previously [8]. MMR status was determined by four markers, including MSH2, MSH6, MLH1, PMS2. Tumours displaying loss of expression of one or more MMR proteins was considered to be dMMR, whereas tumours with intact MMR proteins were classified as proficient MMR (pMMR).
2.2. Predictor selection and the development of prediction model based on clinicopathological features alone or in combination with STMs
Best subset regression was applied to select the most useful predictive factors from the primary dataset, using Akaike's information criterion (AIC) and the likelihood ratio test as the stopping rule [26]. The AIC value for the final model was minimized with the fewest number of variables. A predictive score calculated for each patient via a linear combination of selected features that were weighted by their respective coefficients [27]. To provide clinicians with a quantitative tool to predict individual probability of MMR status, we built the nomogram on the basis of selected variables.
2.3. The validation of prediction model
The nomogram's accuracy was required to be validated by 1000 times bootstrapping and cross-validation measures internally and externally. The fitting degree was evaluated by the area under the receiver operating characteristic (ROC) curve (AUC) and calibration plots. Calibration curves were plotted to assess the calibration of the nomogram, which consisted of two lines: one was a 45- degree reference line, and the other line represented the actual line [28]. The interval between the two lines reflected the accuracy of the nomogram. Hosmer-Lemeshow test was used to evaluate the calibration of prediction model and a significant test statistic implies that the model does not calibrate perfectly. In addition, decision curve analysis (DCA) was conducted to determine the clinical usefulness of the nomogram by quantifying the net benefits at different threshold probabilities [29, 30].
2.4. Statistical analysis
Statistical analysis was conducted with STATA 15.0 (StataCorp, Texas, USA) and R version 4.0.0 (R Foundation for Statistical Computing; http://www.r-project.org/). The R packages used in our study were demonstrated in the Supplementary Table 1. Data are presented as number and percentages for categorical variables, and continuous data were expressed as mean ± standard deviation, unless otherwise specified. The continuous variables were transformed into binary variables by applying inflexion points of ROC curves as the cut-offs. Patient characteristics were compared using t tests for continuous variables and x2 or Fisher exact tests for categorical variables. All statistical tests were two-sided, with statistical significance set at 0.05.
2.5. Role of funding source
This study was funded by the Fundamental Research Funds for the Central Universities of China (2020kfyXGYJ079). The funder Kailin Cai is responsible for designing the study and reviewing the manuscript.
3. Results
3.1. Patient clinical characteristics
The clinical characteristics of CRC patients in the primary and validation cohort were given in Table 1. Although the detection rate of MMR status in the primary cohort was significantly lower than that in the validation cohort (60.4% vs. 67.3%, P = 0.023), there was no significant difference between both cohorts in the incidence of dMMR (10.9% vs. 11.0%, P = 0.937). In both cohorts, dMMR was found to be significantly associated with age, tumour diameters, tumour location, the number of sampled LNs, the number of positive LNs, PNI, CEA and CA 72-4. Though pathological type (P = 0.010) and T-stage (P = 0.003) were found to be significantly correlated with MMR status in the validation cohort, no significant differences were observed in the primary cohort.
Table 1.
The clinical characteristics of CRC patients in the primary and validation cohort.
| Primary Cohort |
Validation Cohort |
|||||
|---|---|---|---|---|---|---|
| characteristics | pMMR (n = 1749) | dMMR (n = 215) | P value | pMMR (n = 1166) | dMMR (n = 144) | P value |
| Age (years) | <0.001 | <0.001 | ||||
| < 53 | 573 (32.8) | 98 (45.6) | 362 (31.1) | 67 (46.5) | ||
| >= 53 | 1176 (67.2) | 117 (54.4) | 804 (68.9) | 77 (53.5) | ||
| Gender | 0.464 | 0.189 | ||||
| Male | 1046 (59.8) | 123 (57.2) | 706 (60.6) | 79 (54.9) | ||
| Female | 703 (40.2) | 92 (42.8) | 460 (39.4) | 65 (45.1) | ||
| Primary location | <0.001 | <0.001 | ||||
| Proximal colon | 343 (19.6) | 95 (44.2) | 221 (19.0) | 60 (41.7) | ||
| Distal colon | 512 (29.3) | 83 (38.6) | 365 (31.3) | 53 (36.8) | ||
| Rectum | 894 (51.1) | 37 (17.2) | 580 (49.7) | 31 (21.5) | ||
| Tumor diameters (cm) | <0.001 | <0.001 | ||||
| < 4.6 | 1266 (72.4) | 121 (56.3) | 807 (69.2) | 44.4) | ||
| >= 4.6 | 483 (27.6) | 94 (43.7) | 359 (30.8) | 80 (55.6) | ||
| Pathological type | 0.145 | 0.010 | ||||
| non-adenocarcinoma | 274 (15.7) | 42 (19.5) | 184 (15.8) | 35 (24.3) | ||
| adenocarcinoma | 1475 (84.3) | 173 (80.5) | 982 (84.2) | 109 (75.7) | ||
| Histology | 0.324 | 0.621 | ||||
| poor | 83 (4.8) | 7 (3.3) | 38 (3.3) | 6 (4.2) | ||
| Well/moderate | 1666 (95.3) | 208 (96.7) | 1128 (96.7) | 138 (95.8) | ||
| T-stage | 0.096 | 0.003 | ||||
| I/II | 365 (20.9) | 30 (13.9) | 295 (19.8) | 19 (10.9) | ||
| III/IV | 1384 (79.1) | 185 (86.1) | 1194 (80.2) | 156 (89.1) | ||
| No. of sampled LNs (n) | <0.001 | <0.001 | ||||
| <23 | 1381 (78.9) | 133 (61.9) | 928 (79.6) | 95 (66.0) | ||
| >=23 | 368 (21.1) | 82 (38.1) | 238 (20.4) | 49 (34.0) | ||
| No. of Positive LNs (n) | 2.1 ± 3.8 | 1.2 ± 2.8 | <0.001 | 2.0 ± 3.8 | 1.2 ± 3.1 | 0.011 |
| Perineural invasion | <0.001 | <0.001 | ||||
| No | 1232 (70.4) | 177 (82.3) | 812 (69.6) | 122 (84.7) | ||
| Yes | 517 (29.6) | 38 (17.7) | 354 (30.4) | 22 (15.3) | ||
| CEA | <0.001 | <0.001 | ||||
| Negative | 944 (53.9) | 156 (72.6) | 591 (50.7) | 105 (72.9) | ||
| Positive | 805 (46.1) | 59 (27.4) | 575 (49.3) | 39 (27.1) | ||
| CA 19-9 | 0.211 | 0.966 | ||||
| Negative | 1451 (83.0) | 171 (79.5) | 941 (80.7) | 116 (80.6) | ||
| Positive | 298 (17.0) | 44 (20.5) | 225 (19.3) | 28 (19.4) | ||
| CA 72-4 | <0.001 | <0.001 | ||||
| Negative | 1471 (84.1) | 125 (58.1) | 976 (83.7) | 89 (61.8) | ||
| Positive | 278 (15.9) | 90 (41.9) | 190 (16.3) | 55 (38.2) | ||
Abbreviations: LNs, lymph nodes; Categorical variables, n (%); Continuous data, mean ± standard deviation.
3.2. The development of prediction model based on simplified clinicopathological features
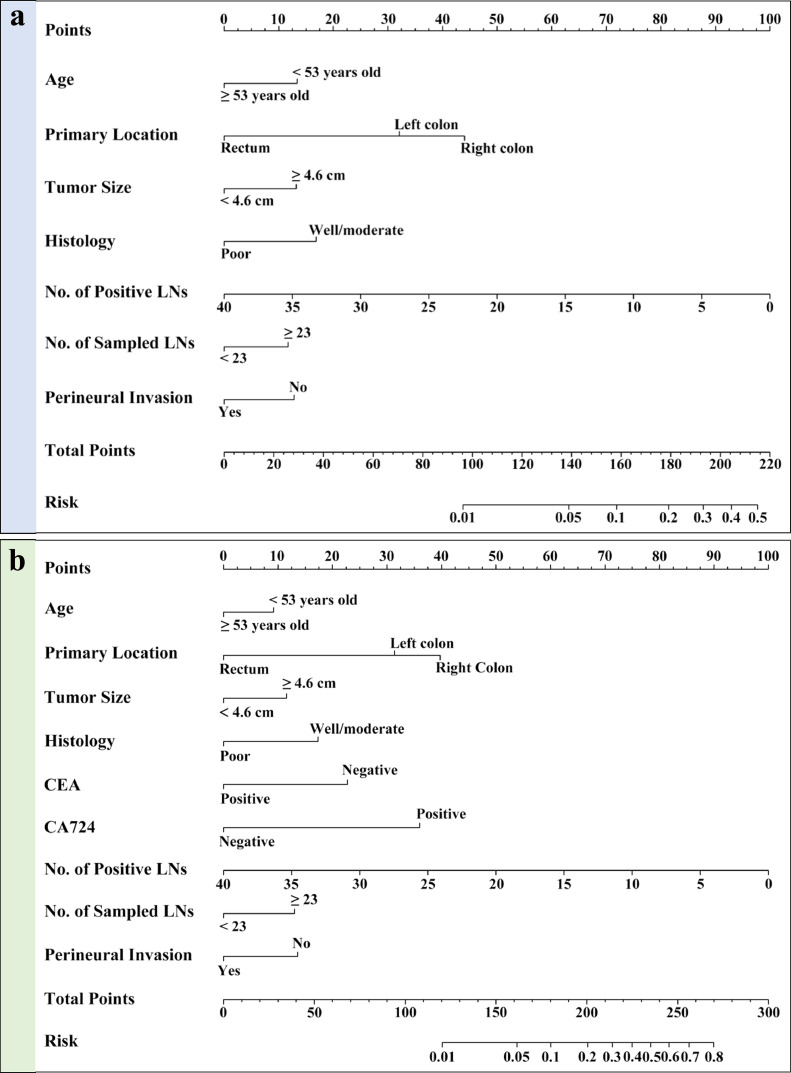
Among the ten simplified clinicopathological features, seven variables were selected as the best subset of risk factors to develop prediction model, including age, tumour diameters, histology, tumour location, the number of sampled LNs, the number of positive LNs and PNI (Table 2). Using the regression coefficients of multivariate logistic regression models to weight each feature in our models [27], we developed a risk score formula to predict MMR status: risk score = -4.281 - 0.581 (if Age < 53 years old) + 0.511 (if tumour diameters ≥ 4.6 cm) + 0.652 (if well or moderate differentiation) - (0.097 × number of positive LNs) + 0.455 (if number of sampled LNs ≥ 23) - 0.496 (if perineural invasion is negative) + (1.242, if primary location is distal colon; 1.705, if primary location is proximal colon). Predicted risk = 1/(1 + e−risk score). The model that incorporated the above predictors was developed and presented as the nomogram (Fig. 2a).
Table 2.
Risk factors for deficient MMR in Colorectal Cancer.
| Model 1 |
Model 2 |
|||||
|---|---|---|---|---|---|---|
| Intercept and variable | β | 95% OR | P value | β | 95% OR | P value |
| Intercept | -4.281 | <0.001 | -4.090 | <0.001 | ||
| Age | -0.518 | 0.595 (0.440 to 0.805) | <0.001 | -0.367 | 0.693 (0.505 to 0.951) | 0.023 |
| Tumor size | 0.511 | 1.667 (1.225 to 2.268) | 0.001 | 0.463 | 1.588 (1.148 to 2.198) | 0.005 |
| Histology | 0.652 | 1.920 (0.845 to 4.365) | 0.119 | 0.693 | 2.000 (0.864 to 4.632) | 0.106 |
| No. of positive LNs | -0.097 | 0.908 (0.855 to 0.964) | 0.002 | -0.100 | 0.905 (0.852 to 0.961) | 0.001 |
| No. of sampled LNs | 0.455 | 1.576 (1.141 to 2.177) | 0.006 | 0.521 | 1.683 (1.200 to 2.362) | 0.002 |
| Perineural invasion | -0.496 | 0.609 (0.414 to 0.895) | 0.012 | -0.543 | 0.581 (0.389 to 0.868) | 0.008 |
| Primary location | ||||||
| Rectum | reference | reference | ||||
| Distal colon | 1.242 | 3.464 (2.301 to 5.213) | <0.001 | 1.256 | 3.511 (2.310 to 5.335) | <0.001 |
| Proximal colon | 1.705 | 5.502 (3.634 to 8.329) | <0.001 | 1.591 | 4.906 (3.202 to 7.516) | <0.001 |
| CEA | NA | NA | NA | -0.910 | 0.403 (0.285 to 0.586) | <0.001 |
| CA 72-4 | NA | NA | NA | 1.440 | 4.222 (3.012 to 5.920) | <0.001 |
model 1: based on simplified clinicopathological characteristics alone.
model 2: based on simplified clinicopathological features and serum tumor biomarkers.
Abbreviations: OR, odds ratio; LNs, lymph nodes; CEA, carcinoembryonic antigen; CA, carbohydrate antigen.
Fig. 2.
The nomograms to predict the probability of dMMR in CRC patients from the training cohort. (a) The nomogram based on simplified clinicopathological features alone; (b) The nomogram based on simplified clinicopathological features and serum tumour markers.
3.3. The validation of prediction model based on simplified clinicopathological features
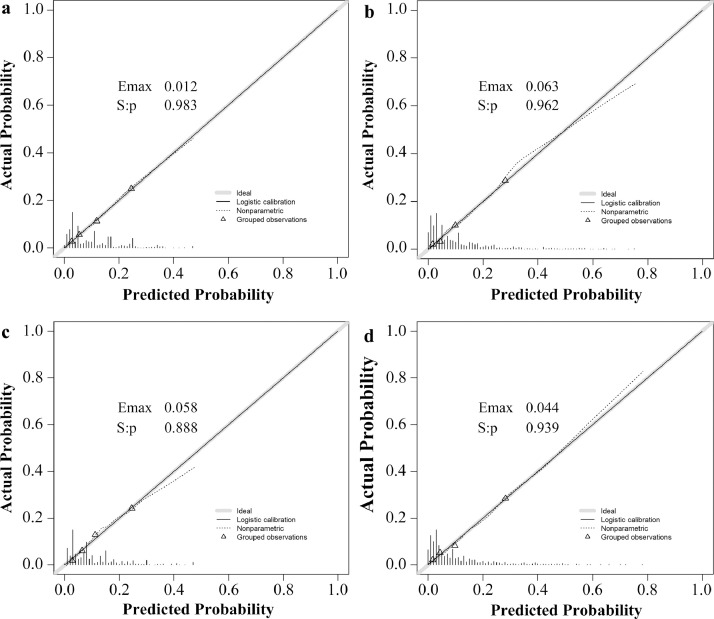
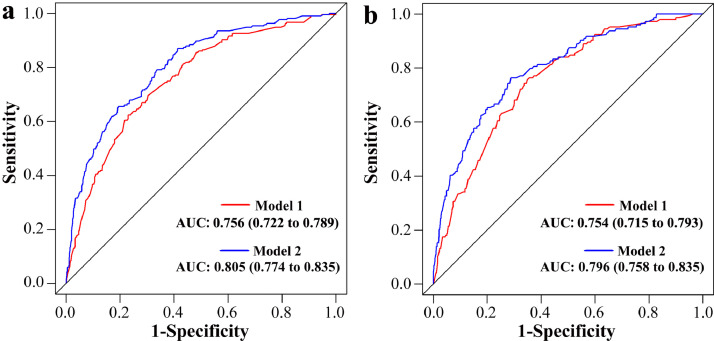
Internal validation: The calibration curve of the nomogram for the probability of dMMR showed good agreement between prediction and observation in the primary cohort (Fig. 3a). The Hosmer-Lemeshow test yielded a nonsignificant statistic (P = 0.983), which suggested that there was no departure from perfect fit. The ROC curve yielded an AUC of 0.756 (95% CI, 0.722 to 0.789) for the primary cohort (Fig. 4a).
Fig. 3.
Calibration curves of the prediction models in each cohort. Calibration curves depict the calibration of prediction models in terms of the agreement between the predicted risks of dMMR and observed outcomes of dMMR. The x-axis represents the predicted dMMR risk and the y-axis represents the actual dMMR rate. The diagonal solid line represents a perfect prediction by an ideal model. The dotted line represents the performance of our prediction models. A closer fit to the diagonal solid line represents a better prediction. (a) and (b) represents the calibration curve of pathology-based model in the primary cohort and validation cohort; (c) and (d) represents the calibration curve of the combined models in the primary cohort and validation cohort.
Fig. 4.
Receiver operating characteristic (ROC) curve of the prediction models in each cohort. The red line and blue line represent the pathology-based model and the combined model, respectively. (a) represents ROC curve of our prediction models in the primary cohort; (b) represents the ROC curve of models in the validation cohort.
Independent validation: Good calibration was also observed in the validation cohort (Fig. 3b) and the Hosmer-Lemeshow test yielded a nonsignificant statistic (P = 0.962). The discriminative ability of the nomogram in an independent cohort was 0.754 (95% CI, 0.715 to 0.793) (Fig. 4b).
3.4. Incremental predictive value of STMs to the above model
To evaluate the additional predictive value of STMs, three STMs, including CEA, CA 19-9 and CA 72-4, together with simplified clinicopathological features, were used to develop MMR prediction model. Finally, nine variables were selected as the best subset of risk factors, including age, tumour diameters, histology, tumour location, the number of sampled LNs, the number of positive LNs, PNI, CEA and CA 72-4 (Table 2). The risk score formula of the combined model was as follows: risk score = -4.090 - 0.367 (if Age < 53 years old) + 0.463 (if tumour diameters ≥ 4.6 cm) + 0.693 (if well or moderate differentiation) – (0.100 × number of positive LNs) + 0.521 (if number of sampled LNs ≥ 23) - 0.543 (if perineural invasion is negative) + (1.256, if primary location is distal colon; 1.591, if primary location is proximal colon) - 0.910 (if CEA is negative) + 1.440 (if CA 72-4 is positive). Predicted risk = 1/(1 + e−risk score). The model that incorporated the above predictors was developed and presented as the nomogram (Fig. 2b). The calibration curve for the probability of dMMR demonstrated good agreement between prediction and observation in the primary cohort (P = 0.888) and validation cohort (P = 0.939) (Fig. 3c and Fig. 3d). After the addition of CEA and CA 72-4, the discrimination ability of pathology-based model was significantly improved in the primary cohort (AUC: 0.805 (95% CI, 0.774 to 0.835) vs. 0.756 (95% CI, 0.722 to 0.789), P < 0.001) (Fig. 4a) and validation cohort (AUC: 0.796 (95% CI, 0.758 to 0.835) vs. 0.754 (95% CI, 0.715 to 0.793), P < 0.001) (Fig. 4b).
3.5. Clinical usefulness
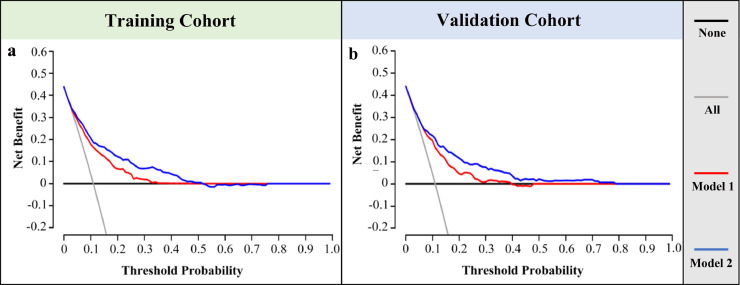
The decision curve analysis for the nomogram based on simplified clinicopathological features alone or in combination with STMs was presented in Fig. 5. Decision curve analysis is conducted to determine the clinical usefulness of the nomograms via quantifying the net benefits at different threshold probabilities [29]. The net benefit was calculated by subtracting the proportion of all patients who are false positive from the proportion who are true positive, weighting by the relative harm of forgoing detection compared with the negative consequences of an unnecessary detection [30, 31]. Here, the relative harm was calculated by (Pt/(1-Pt)). “Pt” (threshold probability) is where the expected benefit of detection is equal to the expected benefit of forgoing detection; at which time a patient will opt for IHC testing informs us of how a patient weighs the relative harms of false-positive results and false-negative results ((a-c)/(b-d) = (1-Pt)/Pt) (a, b, c and d represent the value of true positive, false positive, false negative, and true negative, respectively. (a – c) is the harm from a false-negative result; (b – d) is the harm from a false-positive result). By applying our pathology-based prediction models, higher net benefit in the number of false-positives than the strategy of conducting IHC-testing for every patient or none could be achieved when the risk thresholds range from 11% to 37%. Similarly, if the threshold probability of a patient is between 11% and 52%, using the nomogram incorporating STMs and simplified clinicopathological features would add more benefit than either the detect-all-patients scheme or the detect-none scheme. For example, at the 10% risk cutoff in the validation cohort, the net benefit was 4% in the detect-all model, 20% in the pathology-based model, and 22% in the combined model. The net benefit of the pathology-based model was equivalent to performing 20 detection per 100 men without negative results, 16 more than detect-all model. Moreover, the addition of STMs significantly added the net benefit of pathology-based model (22% vs. 20%, P = 0.002).
Fig. 5.
The decision curve of the nomograms for the prediction of MMR status. The x-axis and y-axis represent the threshold probability and the net benefit, respectively. The red line and blue line represent the pathology-based model and the combined model, respectively. The grey line and black line represent the strategy of conducting IHC-testing for every patient and none. (a) and (b) represent the decision curve of our nomograms in the primary and validation cohort.
4. Discussion
Though molecular testing for MMR status might significantly improve the treatment and management of CRC patients, the rate of MMR detection was far below expected [[32], [33], [34]]. The consensus criteria for the diagnosis of dMMR proteins are based on selecting CRC patients who fulfill the revised Bethesda guidelines (RBG), followed by MSI testing and/or IHC staining of MMR proteins [35]. Although this method is cost-effective, its age limit and low sensitivity have been shown to miss a substantial number of CRC patients with dMMR [36]. In addition, RBG recommended that all CRC patients between the ages of 50 and 59 years should test for MMR status when they have one or more of the following pathological features: Crohn's-like lymphocytic reaction, mucin/signet ring cell differentiation, medullary growth pattern, tumour-infiltrating lymphocytes [35]. However, it could not provide details on the predictive value of these pathological features, either alone or in combination. On the basis of RBG, Jenkins et al developed the MsPath model to quantify the predictive value of these pathological features in CRC patients before age 60 years [21]. Though MsPath score ≥ 1.0 had a sensitivity of 93% and a specificity of 55% for dMMR, it could not provide guidance for those patients over 60 years. Moreover, the model required detailed pathological diagnosis for complex pathological features, which caused a huge burden on the pathologists. Afterwards, a few prediction models attempted to improve MsPath model by simplifying pathological features [22, 23, 37]. Though several models achieved high sensitivity and specificity for the prediction of MMR status, other molecular targets, such as p53 [37] and BRAF [22], included in their models reduced their practical value in clinical practice. Therefore, the detailed predictive value of simplified clinicopathological features alone for MMR status remains unclear.
Using the data from 3274 participants in two institutions, we assessed the predictive value of simplified clinicopathological features in the CRC patients of all ages. The findings of our study showed that simplified clinicopathological features possessed a strong discrimination ability for MMR status. In addition, our study is the first to explore the predictive value of simplified clinicopathological features in combination with STMs for MMR status. The results demonstrated that the addition of CEA and CA 72-4 could significantly improve the discriminative ability of pathology-based model in the primary cohort (AUC: 0.805 (0.774-0.835) vs. 0.756 (0.722-0.789), P < 0.001) and validation cohort (AUC: 0.796 (0.758-0.835) vs. 0.754 (0.715-0.793), P < 0.001). The information obtained in our study may greatly help clinicians to screen the CRC patients who should conduct IHC staining or MSI testing.
Several clinicopathological features, such as age, tumour diameters, histology and tumour location, has been reported to possess a discrimination ability for MMR status in previous studies [22, 23]. Consistent with these findings, seven simplified clinicopathological features were brought into MMR risk prediction model in our study, including age, tumour diameters, histology, tumour location, number of harvested LNs, number of positive LNs and PNI. Among these variables, number of harvested LNs, number of positive LNs and PNI were the first time to be incorporated into models for the prediction of MMR status. As an independent prognostic factor of CRC, PNI has been recommended by NCCN guidelines as a routine inspection item to assess the risk of stage II CRC [38]. Several retrospective observation studies also reported that PNI was prone to occur in CRC patients with pMMR [39, 40]. In our study, more harvested LNs and less positive LNs were found to be associated with dMMR, which represent a larger extent of LNs dissection and low risk of metastasis. Therefore, our results supported that dMMR is a positive prognostic factor for CRC.
Previous studies have demonstrated that preoperative STMs are important prognostic factor that is independent of clinicopathological parameters [[41], [42], [43]]. However, the predictive value of STMs in combination with clinicopathological features for MMR status in CRC patients remains unknown. Although CEA and MMR proteins belong to different protein superfamilies, Shih-Ching et al. reported a correlation between a normal serum CEA level and dMMR in 213 CRC patients [20]. Johanna et al. also reported that elevated serum CA 72-4 level was correlated with worse survival of CRC [41]. In line with these findings, the results of our study showed that CRC patients with dMMR are prone to have normal serum CEA and elevated CA 72-4. Moreover, after the addition of CEA and CA 72-4, the discrimination ability of pathology-based model was significantly improved.
In conclusion, our study presents two nomograms based on simplified clinicopathological features alone or in combination with STMs, which can be conveniently used to facilitate the postoperative individualized prediction of MMR status in CRC patients. However, its limitations also deserve commentary. First, this was a nonrandomized retrospective analysis, and as such, there were potential biases for comparison, such as patient inclusion and sample selection biases. Second, radiomics, focusing on the relationship between imaging phenotypes and genomics [44], were not considered in this study. However, our findings will hopefully be integrated with radiomics or other markers to achieve a stronger predictive ability of MMR status in the future.
Data Sharing Statement
The dataset generated during the current study is available from the corresponding author on reasonable request.
Declaration of Interests
The authors have declared that no competing interests exist.
Acknowledgements
Thank Songqing Ke and Sufei Wang for the support of statistical analysis.
Footnotes
Yinghao Cao and Tao Peng contributed equally to this work.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103060.
Contributor Information
Kailin Cai, Email: caikailin@hust.edu.cn.
Ning Zhao, Email: zhaoning_hust_whuh@126.com.
Appendix. Supplementary materials
References
- 1.Bray F, Ferlay J, Soerjomataram I. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Goding Sauer A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Chu JN, Choi J, Ostvar S. Cost-effectiveness of immune checkpoint inhibitors for microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer. Cancer. 2019;125(2):278–289. doi: 10.1002/cncr.31795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordlinger B, Sorbye H, Glimelius B. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14(12):1208–1215. doi: 10.1016/S1470-2045(13)70447-9. [DOI] [PubMed] [Google Scholar]
- 5.Woolston A, Khan K, Spain G. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during Anti-EGFR treatment in colorectal cancer. Cancer Cell. 2019;36(1):35–50. doi: 10.1016/j.ccell.2019.05.013. e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seligmann JF, Elliott F, Richman S. Clinical and molecular characteristics and treatment outcomes of advanced right-colon, left-colon and rectal cancers: data from 1180 patients in a phase III trial of panitumumab with an extended biomarker panel. Ann Oncol. 2020;31(8):1021–1029. doi: 10.1016/j.annonc.2020.04.476. [DOI] [PubMed] [Google Scholar]
- 7.Tokunaga R, Xiu J, Johnston C. Molecular profiling of appendiceal adenocarcinoma and comparison with right-sided and left-sided colorectal cancer. Clin Cancer Res. 2019;25(10):3096–3103. doi: 10.1158/1078-0432.CCR-18-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen R, Buhard O, Cervera P. Clinical and molecular characterisation of hereditary and sporadic metastatic colorectal cancers harbouring microsatellite instability/DNA mismatch repair deficiency. Eur J Cancer. 2017;86:266–274. doi: 10.1016/j.ejca.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 9.Blaker H, Alwers E, Arnold A. The association between mutations in BRAF and colorectal cancer-specific survival depends on microsatellite status and tumour stage. Clin Gastroenterol Hepatol. 2019;17(3):455–462. doi: 10.1016/j.cgh.2018.04.015. .e456. [DOI] [PubMed] [Google Scholar]
- 10.Latham A, Srinivasan P, Kemel Y. Microsatellite instability is associated with the presence of lynch syndrome pan-cancer. J Clin Oncol. 2019;37(4):286–295. doi: 10.1200/JCO.18.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overman MJ, McDermott R, Leach JL. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klingbiel D, Saridaki Z, Roth AD. Prognosis of stage II and III colon cancer treated with adjuvant 5-fluorouracil or FOLFIRI in relation to microsatellite status: results of the PETACC-3 trial. Ann Oncol. 2015;26(1):126–132. doi: 10.1093/annonc/mdu499. [DOI] [PubMed] [Google Scholar]
- 13.Benson AB, 3rd, Venook AP, Cederquist L. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(3):370–398. doi: 10.6004/jnccn.2017.0036. [DOI] [PubMed] [Google Scholar]
- 14.Benson AB, Venook AP, Al-Hawary MM. Rectal cancer, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2018;16(7):874–901. doi: 10.6004/jnccn.2018.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawes DA, Pearson T, Sengupta S. Is MSI-H of value in predicting the development of metachronous colorectal cancer? Eur J Cancer. 2006;42(4):473–476. doi: 10.1016/j.ejca.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Oh JR, Kim DW, Lee HS. Microsatellite instability testing in Korean patients with colorectal cancer. Fam Cancer. 2012;11(3):459–466. doi: 10.1007/s10689-012-9536-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Lo CH, He X. Risk factor profiles differ for cancers of different regions of the colorectum. Gastroenterology. 2020;159(1):241–256. doi: 10.1053/j.gastro.2020.03.054. .e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Z, Sanhueza CT, Johnson B. Outcome of mismatch repair-deficient metastatic colorectal cancer: the mayo clinic experience. Oncologist. 2018;23(9):1083–1091. doi: 10.1634/theoncologist.2017-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JP, Lin JX, Ma YB. Prognostic significance of pre- and post-operative tumour markers for patients with gastric cancer. Br J Cancer. 2020;123(3):418–425. doi: 10.1038/s41416-020-0901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang SC, Lin JK, Yang SH. Relationship between genetic alterations and prognosis in sporadic colorectal cancer. Int J Cancer. 2006;118(7):1721–1727. doi: 10.1002/ijc.21563. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins MA, Hayashi S, O'Shea AM. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133(1):48–56. doi: 10.1053/j.gastro.2007.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiyoshi K, Yamaguchi T, Kakuta M. Predictive model for high-frequency microsatellite instability in colorectal cancer patients over 50 years of age. Cancer Med. 2017;6(6):1255–1263. doi: 10.1002/cam4.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyde A, Fontaine D, Stuckless S. A histology-based model for predicting microsatellite instability in colorectal cancers. Am J Surg Pathol. 2010;34(12):1820–1829. doi: 10.1097/PAS.0b013e3181f6a912. [DOI] [PubMed] [Google Scholar]
- 24.Gulhati P, Yin J, Pederson L. Threshold change in CEA as a predictor of non-progression to first-line systemic therapy in metastatic colorectal cancer patients with elevated CEA. J Natl Cancer Inst. 2020 doi: 10.1093/jnci/djaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liebig C, Ayala G, Wilks JA. Perineural invasion in cancer: a review of the literature. Cancer. 2009;115(15):3379–3391. doi: 10.1002/cncr.24396. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z. Variable selection with stepwise and best subset approaches. Ann Transl Med. 2016;4(7):136. doi: 10.21037/atm.2016.03.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Royston P, Moons KG, Altman DG. Prognosis and prognostic research: developing a prognostic model. BMJ. 2009;338:b604. doi: 10.1136/bmj.b604. [DOI] [PubMed] [Google Scholar]
- 28.Coutant C, Olivier C, Lambaudie E. Comparison of models to predict nonsentinel lymph node status in breast cancer patients with metastatic sentinel lymph nodes: a prospective multicenter study. J Clin Oncol. 2009;27(17):2800–2808. doi: 10.1200/JCO.2008.19.7418. [DOI] [PubMed] [Google Scholar]
- 29.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. JAMA. 2015;313(4):409–410. doi: 10.1001/jama.2015.37. [DOI] [PubMed] [Google Scholar]
- 30.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers AJ, Cronin AM, Elkin EB. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eriksson J, Amonkar M, Al-Jassar G. Mismatch repair/microsatellite instability testing practices among Us physicians treating patients with advanced/metastatic colorectal cancer. J Clin Med. 2019;8(4):558. doi: 10.3390/jcm8040558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noll A, P JP, Zhou M. Barriers to lynch syndrome testing and preoperative result availability in early-onset colorectal cancer: a national physician survey Study. Clin Transl Gastroenterol. 2018;9(9):185. doi: 10.1038/s41424-018-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cenin DR, Naber SK, Lansdorp-Vogelaar I. Costs and outcomes of Lynch syndrome screening in the Australian colorectal cancer population. J Gastroenterol Hepatol. 2018;33(10):1737–1744. doi: 10.1111/jgh.14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umar A, Boland CR, Terdiman JP. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bessa X, Alenda C, Paya A. Validation microsatellite path score in a population-based cohort of patients with colorectal cancer. J Clin Oncol. 2011;29(25):3374–3380. doi: 10.1200/JCO.2010.34.3947. [DOI] [PubMed] [Google Scholar]
- 37.Colomer A, Erill N, Vidal A. A novel logistic model based on clinicopathological features predicts microsatellite instability in colorectal carcinomas. Diagn Mol Pathol. 2005;14(4):213–223. doi: 10.1097/01.pas.0000177800.65959.48. [DOI] [PubMed] [Google Scholar]
- 38.Liebig C, Ayala G, Wilks J. Perineural invasion is an independent predictor of outcome in colorectal cancer. J Clin Oncol. 2009;27(31):5131–5137. doi: 10.1200/JCO.2009.22.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan E, Khaw YL, Creavin B. Tumour budding and PDC grade are stage independent predictors of clinical outcome in mismatch repair deficient colorectal cancer. Am J Surg Pathol. 2018;42(1):60–68. doi: 10.1097/PAS.0000000000000931. [DOI] [PubMed] [Google Scholar]
- 40.Williams DS, Mouradov D, Newman MR. Tumour infiltrating lymphocyte status is superior to histological grade, DNA mismatch repair and BRAF mutation for prognosis of colorectal adenocarcinomas with mucinous differentiation. Mod Pathol. 2020;33(7):1420–1432. doi: 10.1038/s41379-020-0496-1. [DOI] [PubMed] [Google Scholar]
- 41.Louhimo J, Carpelan-Holmstrom M, Alfthan H. Serum HCG beta, CA 72-4 and CEA are independent prognostic factors in colorectal cancer. Int J Cancer. 2002;101(6):545–548. doi: 10.1002/ijc.90009. [DOI] [PubMed] [Google Scholar]
- 42.Thomsen M, Skovlund E, Sorbye H. Prognostic role of carcinoembryonic antigen and carbohydrate antigen 19-9 in metastatic colorectal cancer: a BRAF-mutant subset with high CA 19-9 level and poor outcome. Br J Cancer. 2018;118(12):1609–1616. doi: 10.1038/s41416-018-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stiksma J, Grootendorst DC, van der Linden PW. CA 19-9 as a marker in addition to CEA to monitor colorectal cancer. Clin Colorectal Cancer. 2014;13(4):239–244. doi: 10.1016/j.clcc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12(8):862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.