Summary
The recruitment of Polycomb repressive complex 2 (PRC2) to gene promoters is critical for its function in repressing gene expression in murine embryonic stem cells (mESCs). However, previous studies have demonstrated that although the expression of early lineage-specific genes is largely repressed, the genome-wide PRC2 occupancy is unexpectedly reduced in naive mESCs. In this study, we provide evidence that fibroblast growth factor/extracellular signal-regulated kinase signaling determines the global PRC2 occupancy through regulating the expression of PRC2-recruiting factor JARID2 in naive mESCs. At the transcriptional level, the de-repression of bivalent genes is predominantly determined by the presence of cell signaling-associated transcription factors but not the status of PRC2 occupancy at gene promoters. Hence, this study not only reveals a key molecular mechanism by which cell signaling regulates the PRC2 occupancy in mESCs but also elucidates the functional roles of transcription factors and Polycomb-mediated epigenetic mechanisms in transcriptional regulation.
Subject Areas: Molecular Biology, Cell Biology, Stem Cells Research, Developmental Biology
Graphical Abstract
Highlights
-
•
FGF/ERK signaling positively regulates Jarid2 expression in mESCs
-
•
Reduced JARID2 causes global reduction of PRC2 occupancy in naive mESCs
-
•
Reduced PRC2 occupancy alone is insufficient to induce transcriptional activation
-
•
Cell signaling-associated transcription factors drive bivalent gene expression
Molecular Biology; Cell Biology; Stem Cells Research; Developmental Biology
Introduction
Polycomb repressive complexes (PRCs) are critical for maintaining mESCs in an undifferentiated state by repressing lineage-specific gene expression (Boyer et al., 2006; Lee et al., 2006; Aloia et al., 2013). Previous studies have shown that both PRC1 and PRC2 are recruited to gene promoters and mediate monoubiquitylation of histone H2A lysine 119 (H2AK119ub1) and trimethylation of histone H3 lysine 27 (H3K27me3), respectively (Cao et al., 2002; Wang et al., 2004; Di Croce and Helin, 2013), which are essential for their function in repressing gene expression (Cao et al., 2002; Blackledge et al., 2019; Tamburri et al., 2019).
Although the genome-wide binding sites of PRCs in mESCs have been well documented, the molecular mechanisms for recruiting PRCs to specific genomic loci in mammalian cells have not been fully elucidated (Laugesen et al., 2019). A variety of factors, such as JARID2 and MTF2, have been reported to mediate the PRC2 recruitment (Shen et al., 2009; Landeira et al., 2010; Pasini et al., 2010; Casanova et al., 2011; Li et al., 2017; Oksuz et al., 2018). On the other hand, histone lysine demethylase 2B (KDM2B) recruits a non-canonical PRC1.1 (ncPRC1.1) variant to unmethylated CpG islands (CGIs) in mammalian cells through its CxxC-zinc finger (CxxC-ZF) domain (Farcas et al., 2012; He et al., 2013; Wu et al., 2013). Recently, KDM2B was also reported to recruit PRC2 though the interaction of JARID2/PRC2 with KDM2B/ncPRC1.1-mediated histone H2AK119ub1 modification (Cooper et al., 2014).
The repression of differentiation gene expression is essential for mESCs to maintain their pluripotency (Smith, 2001). However, differentiation signals such as the extracellular signal-regulated kinase (ERK) signaling triggered by fibroblast growth factors (FGFs) in serum induce stochastic expression of early lineage-specific genes. After being cultured in 2i serum-free medium in which the FGF/ERK signaling is inhibited and the Wnt/β-catenin signaling is activated, mESCs turn into a naive state in which the basal expression of early lineage-specific genes is reduced (Silva et al., 2008; Ying et al., 2008). Along with the gene expression changes, naive mESCs display two major genome-wide epigenetic changes (Takahashi et al., 2018): (1) a global reduction of DNA methylation mainly due to decreased UHRF1 expression and loss of DNA methylation maintenance (Leitch et al., 2013; von Meyenn et al., 2016) and (2) both genome-wide PRC2 occupancy and its mediated histone H3K27me3 modification are drastically reduced (Marks et al., 2012; Galonska et al., 2015; Guo et al., 2016). Although the underlying molecular mechanisms for the global reduction of PRC2 occupancy are unclear, several recent studies suggest it might be due to re-location of PRC2 to new DNA de-methylated non-CGI regions (Walter et al., 2016; Kumar and Elsasser, 2019; van Mierlo et al., 2019) or reduced chromatin accessibility to PRC2 (Tee et al., 2014).
To examine the molecular mechanisms leading to the reduced PRC2 occupancy and histone H3K27me3 modification in naive mESCs, we performed RNA-sequencing (RNA-seq) analyses to compare the gene expression in wild-type mESCs cultured in serum versus 2i medium. The result showed that the expression of PRC2-recruiting factor JARID2 was significantly reduced in naive mESCs. Re-activation of FGF/ERK signaling increased the expression of Jarid2, suggesting the FGF/ERK signaling positively regulated its expression. Similarly, genetic deletion of ERK signaling molecules ERK1 and ERK2 reduced the Jarid2 expression, which could be rescued by ectopic expression of wild-type ERK2. ChIP-seq analyses showed that the global occupancy of EZH2 and histone H3K27me3 modification were largely reduced at CGIs in both naive and Erk1/Erk2 double-knockout (Erk1/Erk2-dKO) mESCs, which correlated with the global reduction of JARID2 occupancy. Importantly, ectopic expression of Jarid2 fully restored the global PRC2 occupancy and histone H3K27me3 modification at CGIs in both naive and Erk1/Erk2-dKO mESCs. At the transcriptional level, although the PRC2 occupancy and histone H3K27me3 modification were reduced at bivalent promoters, the FGF/ERK-regulated lineage-specific genes remained silenced, while the Wnt signaling-regulated genes were de-repressed in naive mESCs, suggesting the presence of transcription factors, but not the status of PRC2 occupancy, played a predominant role in determining transcriptional activation. Thus, this study not only revealed a main molecular mechanism by which the FGF/ERK signaling regulated the global PRC2 occupancy in mESCs but also elucidated a fundamental question regarding the functional roles of transcription factors and Polycomb-mediated epigenetic mechanisms in transcriptional regulation.
Results
Jarid2 Expression Is Significantly Reduced in Naïve mESCs
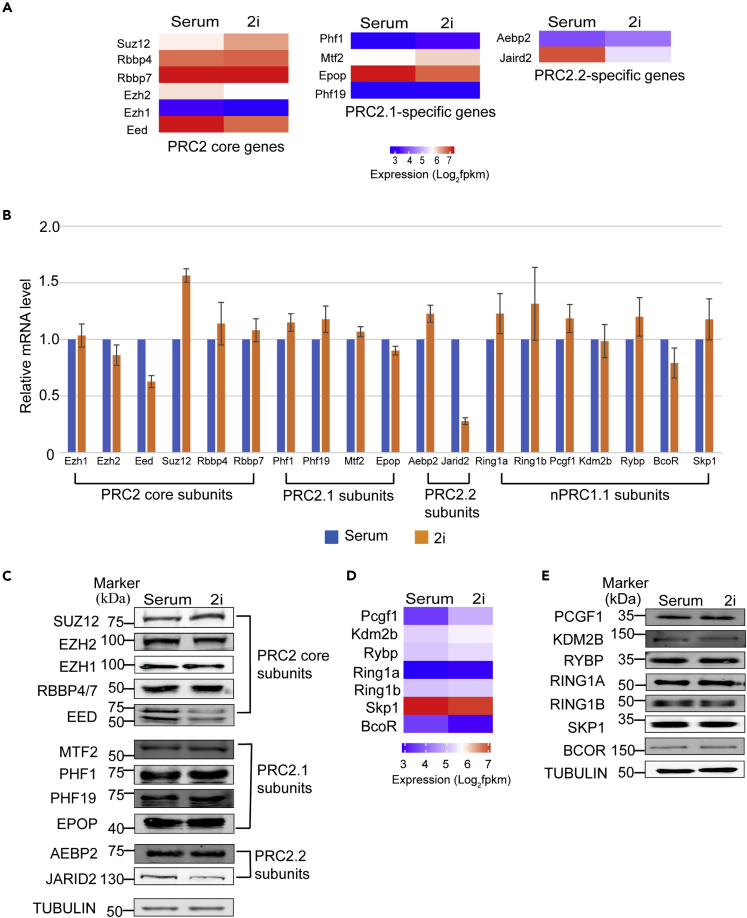
To examine whether the reduced global PRC2 occupancy in naive mESCs is caused by reduced expression of PRC2 components, we performed RNA-seq analyses to examine the differential gene expression between wild-type E14 mESCs cultured in serum-containing medium (ESC-S) and 2i medium (ESC-2i). The results identified 1,905 upregulated genes and 2,371 downregulated genes in ESC-2i, respectively (cutoff: 1.5-fold expression change and q < 0.05) (Figures S1A and S1B). Consistent with previous reports, the expression of Nanog, Klf4, and Prdm14 was found to be upregulated in ESC-2i, validating the proper 2i culture condition and the ground state of mESCs in this study (Figure S1C) (Guo et al., 2009, Munoz Descalzo et al., 2012; Yamaji et al., 2013). Among all PRC2 core and PRC2.1-/PRC2.2-specific genes, Jarid2 expression was found to be significantly reduced in mESC-2i compared to that in mESC-S (cutoff: 1.5-fold expression change and q < 0.05) (Figure 1A), which was further confirmed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and Western blot (WB) analyses at both mRNA and protein levels (Figures 1B, 1C, and S1D). Eed, to a less extent, also showed significant reduced expression in mESC-2i (Figure 1A). To corroborate our findings, we re-analyzed the expression of Jarid2 and Eed in naive mESCs from a published RNA-seq data set (Marks et al., 2012). Consistent with our findings, Jarid2 expression was largely reduced in both E14 and TNGA mESC lines cultured in 2i medium while the change of Eed expression varied in these two mESC lines (Figures S1E and S1F). Therefore, the observed reduced Eed expression in the E14 mESC line was likely to be a cell line-specific effect. In contrast to reduced Jarid2 expression, Suz12 expression was found to be increased in mESC-2i (cutoff: 1.5-fold expression change and q < 0.05). Other PRC2 genes including catalytic subunits Ezh1 and Ezh2 did not show statistically significant difference in expression between mESC-S and mESC-2i (Figures 1A–1C).
Figure 1.
Jarid2 Expression Is Significantly Reduced in naive mESC
(A) Heatmap showing the expression of PRC2 core and PRC2.1-/PRC2.2-specific genes analyzed by RNA-seq.
(B) qRT-PCR analysis showing the expression levels of PRC2 core, PRC2.1-/PRC2.2-specific genes, and ncPRC1.1 genes in mESC-S and mESC-2i. The results were normalized against levels of Gapdh, and the expression level in mESC-S was arbitrarily set to 1. The error bars represent the standard deviation (n = 3).
(C) Western blot analysis showing the protein levels of PRC2 core and PRC2.1-/PRC2.2-specific components.
(D) Heatmap showing the expression of ncPRC1.1 genes analyzed by RNA-seq.
(E) Western blot analysis showing the protein levels of ncPRC1.1 components.
In addition to JARID2 and MTF2-mediated PRC2 recruitment, ncPRC1.1 was reported to recruit PRC2 (Cooper et al., 2014). However, RNA-seq analysis did not reveal any significant expression difference of ncPRC1.1 genes in naive mESCs, which was further confirmed by qRT-PCR and WB analysis (Figures 1B, 1D, and 1E). Based on these results, we concluded that the expression of PRC2-recruiting factor JARID2 was significantly reduced in naive mESCs.
FGF/ERK Signaling Positively Regulates Jarid2 Expression in mESCs
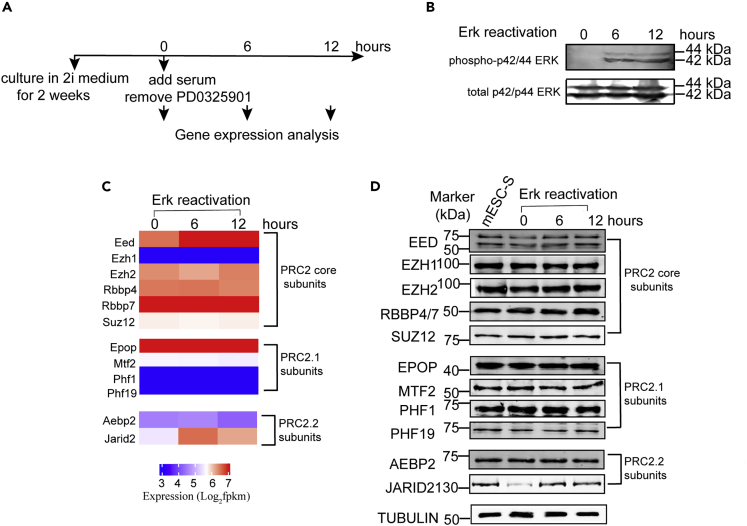
The FGF/ERK signaling is blocked by the removal of serum and supplement of mitogen-activated extracellular signal-regulated kinase (MEK) inhibitor PD0325901 in 2i medium. To examine whether the reduced expression of Jarid2 was caused by the deficiency of FGF/ERK signaling in 2i medium, we re-activated the FGF/ERK signaling by adding serum and removing the MEK inhibitor PD0325901 from 2i medium. RNA-seq analyses were performed to examine the gene expression at different time points after the culture condition switched (Figure 2A). The re-activation of FGF/ERK signaling was monitored by WB analyses on the phosphorylated MAPK p42/44 (ERK1/2). The results showed that the phosphorylated MAPK p42/44 was not detected in ESC-2i. However, the phosphorylated proteins quickly increased and reached to a constant level in 6 hr after the medium switched, confirming the FGF/ERK signaling was re-activated (Figure 2B). RNA-seq analyses identified 309 genes upregulated in response to the activation of FGF/ERK signaling (Figure S2A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the upregulated genes were enriched in MAPK signaling pathway, signaling involved in regulating stem cell pluripotency, signaling pathway in cancer, and hypoxia-inducible factor (HIF) signaling pathway (cutoff: Benjamini adjusted p < 0.05), further confirming that the FGF/ERK signaling pathway was properly re-activated in mESCs after the culture condition changed (Figure S2B and Table S1). Importantly, RNA-seq results showed that the Jarid2 expression was increased in response to the re-activation of FGF/ERK signaling (Figure 2C). The increased Jarid2 expression was further confirmed by qRT-PCR and WB analyses at both mRNA and protein levels (Figures 2D, S2C, and S2D). Other PRC2.1 and PRC2.2 subunits did not show significant difference in expression at both mRNA and protein levels in response to the re-activation of FGF/ERK signaling (Figures 2C and 2D).
Figure 2.
FGF/ERK Signaling Positively Regulates Jarid2 Expression in mESCs
(A) Schematic chart showing the experimental design for the analysis of gene expression in response to the re-activation of FGF/ERK signaling.
(B) Western blot analysis showing phosphorylated p42/44 ERKs were increased in response to the re-activation of FGF/ERK signaling.
(C) Heatmap showing the expression of PRC2 core and PRC2.1-/PRC2.2-specific genes in response to the re-activation of FGF/ERK signaling.
(D) Western blot analysis showing the protein levels of PRC2 core and PRC2.1-/PRC2.2-specific components in mESCs in response to the re-activation of FGF/ERK signaling.
To examine whether the increased Jarid2 expression was caused by cell cycle changes, we performed the cell cycle analysis during the re-activation of FGF/ERK signaling. The results showed that there were no significant changes in cell cycle within 12 hr after the FGF/ERK signaling re-activation (Figure S2E). In contrast, Jarid2 expression increased quickly and reached to a stable level in 6 hr after the ERK signaling re-activation (Figures 2C, 2D, S2C, and S2D). The results suggested the changed Jarid2 expression in mESC-2i was less likely to be caused by cell cycle difference. Based on these results, we concluded that the FGF/ERK signaling positively regulated the expression of Jarid2 and the inhibition of this signaling pathway in 2i medium reduced the Jarid2 expression at both mRNA and protein levels in naive mESCs.
Knockout of Erk1/Erk2 Reduces Jarid2 Expression in mESCs
A previous study reported the genetic depletion of MAPK signaling molecules ERK1 and ERK2 in mESCs led to reduced PRC2 occupancy and histone H3K27me3 modification at bivalent promoters (Tee et al., 2014). Since the study did not examine the Jarid2 expression in the Erk1/Erk2-depleted mESCs, we asked whether the observed reduction of PRC2 occupancy at bivalent promoters in the Erk1/Erk2-depleted mESCs was due to reduced JARID2 expression as observed in naive mESCs.
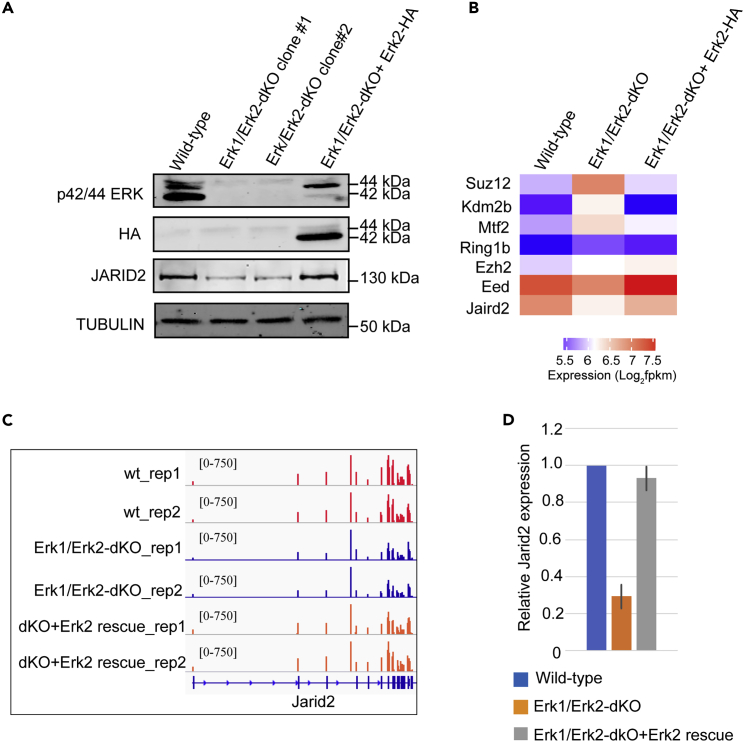
To examine this possibility, we established multiple Erk1/Erk2-dKO mESC lines by the Crispr-Cas9-mediated gene knockout approach. The depletion of ERK1/ERK2 proteins in two independent Erk1/Erk2-dKO lines was confirmed by WB analyses (Figure 3A, lane 1–3). Compared to wild-type mESCs, the Erk1/Erk2-dKO mESCs cultured in serum-containing medium supplemented with a GSK3 inhibitor CHIR99021 were able to maintain normal colony morphology (Figure S3A). RNA-seq analyses showed that compared to mESC-S, the Erk1/Erk2-dKO mESCs had similar expression of Prdm14, Nanog, Klf4, c-myc, Pou5f1, and Sox2 as naive mESCs (Figures S1C and S3B). Importantly, RNA-seq analyses showed that Jarid2 expression was significantly reduced while other key PRC1 and PRC2 genes had similar or increased expression in the Erk1/Erk2-dKO mESCs (Figures 3B and 3C). The decreased Jarid2 expression in the Erk1/Erk2-dKO cells is confirmed by qRT-PCR and WB analyses at both mRNA and protein levels (Figures 3A lane 1–3, 3D). To corroborate the findings, we rescued the ERK signaling by lentiviral vectors expressing wild-type ERK2 fused with a self-cleaved P2A peptide-linked puromycin-resistant protein. After transduction, the transduced cells were selected by puromycin and the expression of exogenous ERK2 was confirmed by WB analysis (Figure 3A lane 4). RNA-seq analysis showed the Jarid2 expression was restored in the wild-type Erk2-rescued cells (Figures 3B and 3C), which was further confirmed by qRT-PCR and WB analyses (Figures 3A lane 4, 3D).
Figure 3.
Knockout of Erk1/Erk2 Reduces Jarid2 Expression in mESCs
(A) Western blot analysis showing p42/44 ERKs, ectopically expressed wild-type ERK2-HA, and JARID2 proteins in wild-type, Erk1/Erk2-dKO, and Erk1/Erk2-dKO rescued with wild-type ERK2-HA (Erk1/Erk2-dKO + Erk2-HA) mESCs.
(B) Heatmap showing the expression of Polycomb core genes in wild-type, Erk1/Erk2-dKO, and Erk1/Erk2-dKO + Erk2-HA mESCs.
(C) IGV genome browser view of Jarid2 expression in wild-type, Erk1/Erk2-dKO, and Erk1/Erk2-dKO rescued with wild-type ERK2-HA mESCs.
(D) qRT-PCR analysis showing the Jarid2 expression in wild-type, Erk1/Erk2-dKO, and Erk1/Erk2-dKO rescued with wild-type ERK2-HA mESCs. The results were normalized against levels of Gapdh, and the expression level in wild-type mESCs was arbitrarily set to 1. The error bars represent standard deviation (n = 3).
Based on these results, we concluded that the Jarid2 expression in mESCs was positively regulated by the ERK signaling. Same as the chemical inhibition of FGF/ERK signaling in 2i medium, the genetic disruption of ERK signaling led to reduced Jarid2 expression in mESCs cultured in serum-containing medium.
The Global PRC2 Occupancy at CGIs Is Largely Reduced in Naïve mESCs
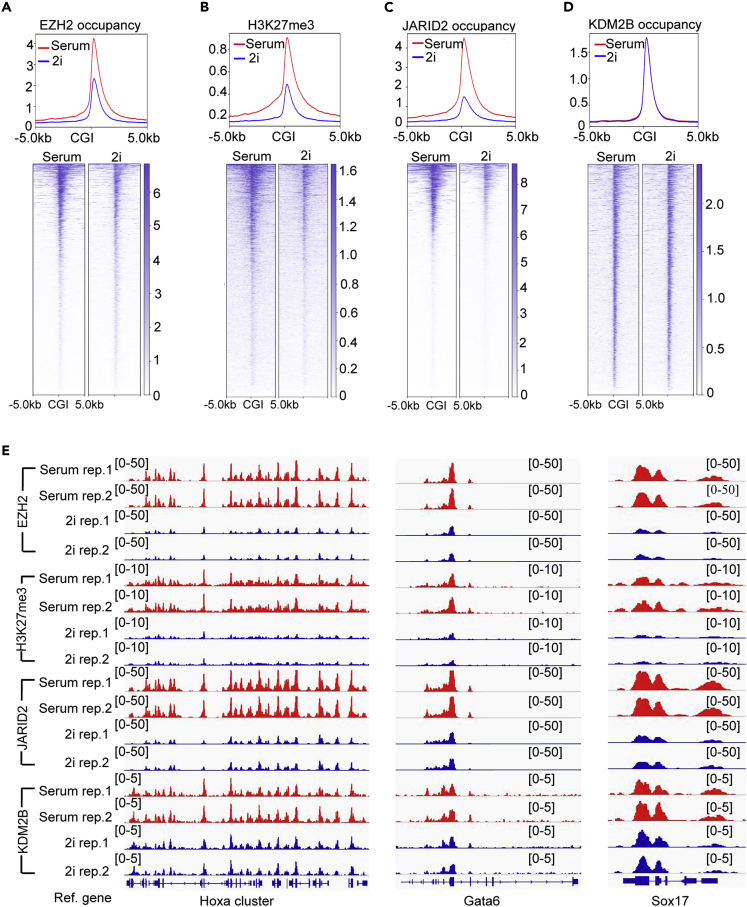
To examine the global epigenetic changes in naive mESCs, we performed ChIP-seq analyses to compare the genome-wide occupancy of PRC2 components JARID2/EZH2, ncPRC1.1 components KDM2B/RING1B, and PRC2-mediated histone H3K27me3 modification as well as Trithorax MLL1/MLL2 complexes-mediated histone H3K4me3 modification in mESC-S and mESC-2i. Since Polycomb complexes and Trithorax MLL1/MLL2 complexes were globally recruited to CGIs in mammalian cells, we measured the overall Polycomb occupancy and histone H3K27me3/H3K4me3 modifications by calculating the total normalized mapped reads within CGIs and 10-kb CGI-flanking regions. Consistent with previous reports (Marks et al., 2012; Guo et al., 2016), both genome-wide PRC2 occupancy and histone H3K27me3 modification were largely reduced at CGIs in naive mESCs, which correlated well with the global decreased JARID2 occupancy (Figures 4A–4C and 4E). However, KDM2B, one ncPRC1.1 component that binds to unmethylated CGIs through its CxxC-ZF domain, had similar genome-wide occupancy at CGIs in mESC-S and mESC-2i (Figures 4D and 4E). In contrast to the reduced histone H3K27me3 modification, the global histone H3K4me3 modification at CGIs was slightly increased in mESC-2i (Figure S4A). Also, in contrast to the reduced EZH2 occupancy at both GGIs and 10-kb CGI-flanking regions, the occupancy of PRC1 core component RING1B was slightly reduced at the CGI-flanking regions but not the CGI center (Figure S4B).
Figure 4.
The Global PRC2 Occupancy at CGIs Is Largely Reduced in naive mESCs
(A–D) Plot (upper) and heatmap (bottom) showing EZH2 occupancy, histone H3K27me3 modification, JARID2 occupancy, and KDM2B occupancy at CGIs and 10-kb CGI-flanking regions in mESC-S and mESC-2i.
(E) IGV genome browser view of EZH2, JARID2, KDM2B occupancy, and histone H3K27me3 modification at the representative Hoxa gene cluster (left panel), Gata6 (middle panel), and Sox17 (right panel) in mESC-S and mESC-2i.
To examine whether the PRC2 occupancy and H3K27me3 modification were re-distributed to non-CGI regions in naive mESCs (Marks et al., 2012; van Mierlo et al., 2019), we calculated the overall occupancy of JARID2/EZH2 and histone H3K27me3 modification over extended 50-kb CGI-flanking regions. The results did not reveal any significant increased PRC2 occupancy and histone H3K27me3 modification within 50-kb CGI-flanking regions (Figures S4C and S4E). Since JARID2 is a known PRC2-recruiting factor (Peng et al., 2009; Shen et al., 2009; Pasini et al., 2010), these results suggested that reduced Jarid2 expression was likely to be a key molecular mechanism leading to the reduced global PRC2 occupancy and histone H3K27me3 modification at CGIs in naive mESCs.
Ectopic Expression of Jarid2 Restores the Global PRC2 Occupancy in Naïve mESCs
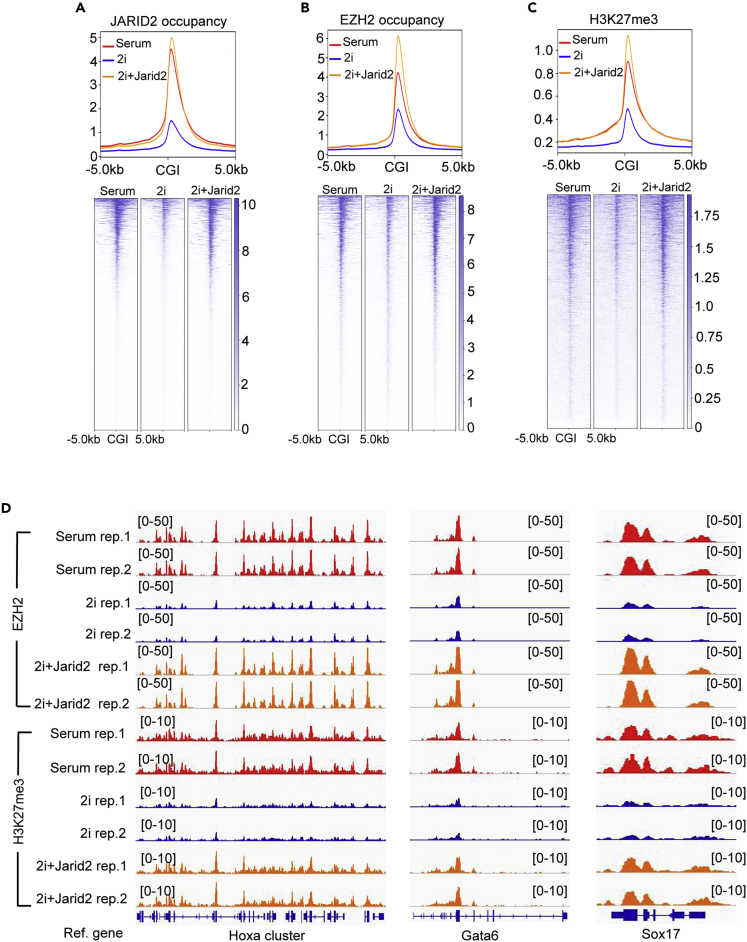
To further examine whether the reduced Jarid2 expression was the main reason causing the global reduction of PRC2 occupancy at CGIs in naive mESCs, we rescued the Jarid2 expression in naive mESCs by lentiviral viruses expressing JARID2 fused with a self-cleaved P2A peptide-linked puromycin-resistant protein. After transduction, the transduced cells were selected by puromycin in the medium. RNA-seq analyses showed that the Jarid2 expression increased to a comparable level as that in mESC-S (Figure S5A). The rescued Jarid2 expression is confirmed by qRT-PCR and WB analyses at both mRNA and protein levels (Figures S5B and S5C). The mESCs with ectopic Jarid2 expression displayed normal mESC morphology under both serum-containing and 2i medium, as well as had similar expression of pluripotent genes and early lineage-specific genes as wild-type mESCs (Figures S5D–S5F). The ChIP-seq analyses demonstrated that the ectopic Jarid2 expression fully restored both EZH2/JARID2 occupancy and histone H3K27me3 modification at CGIs in naive mESCs (Figures 5A–5D). Next, we extracted a total of 2,830 unique bivalent promoters containing both H3K27me3 and H3K4me3 modifications and analyzed the JARID2/EZH2 occupancy as well as histone H3K27me3 modification at these promoters (Figure S5G). Same as CGIs, the bivalent promoters had reduced JARID2/EZH2 occupancy and histone H3K27me3 modification in naive mESCs, which could be fully rescued by the ectopic expression of Jarid2 (Figures S5H–S5J). Based on these results, we concluded that the reduced Jarid2 expression was the main molecular mechanism leading to the reduction of global PRC2 occupancy and histone H3K27me3 modification at both CGIs and bivalent promoters in naive mESCs.
Figure 5.
Ectopic Expression of Jarid2 Restores the Global PRC2 Occupancy in naive mESCs
(A–C) Plot (upper) and heatmap (bottom) showing JARID2 occupancy, EZH2 occupancy, and histone H3K27me3 modification at CGIs and 10-kb CGI-flanking regions in mESC-S (serum), mESC-2i (2i), and mESC-2i with ectopically expressed Jarid2 (2i + Jarid2).
(D) IGV genome browser view of EZH2 and histone H3K27me3 modification at the representative Hoxa gene cluster (left panel), Gata6 (middle panel), and Sox17 (right panel) in mESC-S (serum), mESC-2i (2i), and mESC-2i with ectopically expressed Jarid2 (2i + Jarid2).
Ectopic Expression of Jarid2 Restores the Global PRC2 Occupancy in the Erk1/Erk2-dKO mESCs
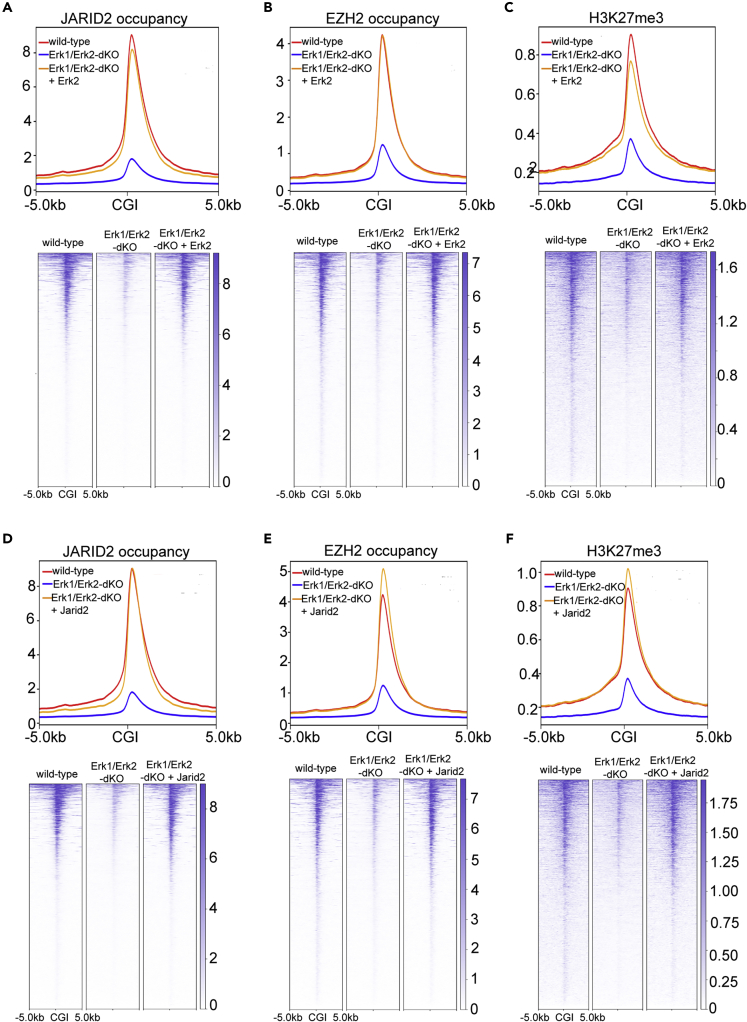
A previous study reported that genetic depletion of Erk1/Erk2 resulted in reduced PRC2 occupancy and histone H3K27me3 modification at bivalent promoters in mESCs (Tee et al., 2014). Using the established Erk1/Erk2-dKO mESCs (Figure 3), we performed ChIP-seq analyses to examine the genome-wide occupancy of JARID2/EZH2 and histone H3K27me3 modification in these cells. The results showed that the occupancy of JARID2/EZH2 and H3K27me3 modification were largely reduced not only at the bivalent promoters (Figures S6A–S6C) but more broadly at CGIs (Figures 6A–6C). Importantly, the ectopic expression of wild-type Erk2 in the Erk1/Erk2-dKO mESCs not only increased the Jarid2 expression (Figure 3) but also restored the JARID2/EZH2 occupancy and histone H3K27me3 modification at both bivalent promoters and CGIs (Figures 6A–6C and S6A–S6C).
Figure 6.
Ectopic Expression of Erk2 or Jarid2 Restores the Global PRC2 Occupancy in Erk1/Erk2-dKO mESCs
(A–C) Plot (upper) and heatmap (bottom) showing JARID2 occupancy, EZH2 occupancy, and histone H3K27me3 modification at CGIs and 10-kb CGI-flanking regions in wild-type, Erk1/Erk2-dKO, and Erk1/Erk2-dKO rescued with wild-type Erk2 (Erk1/Erk2-dKO + Erk2) mESCs.
(D–F) Plot (upper) and heatmap (bottom) showing JARID2 occupancy, EZH2 occupancy, and histone H3K27me3 modification at CGIs and 10-kb CGI-flanking regions in wild-type, Erk1/Erk2-dKO, and Erk1/Erk2-dKO with ectopically expressed Jarid2 (Erk1/Erk2-dKO + Jarid2) mESCs.
To further examine whether the reduced JARID2-mediated PRC2 recruitment led to the reduction of JARID2/EZH2 occupancy and histone H3K27me3 modification at CGIs in the Erk1/Erk2-dKO cells, we ectopically expressed Jarid2 in the Erk1/Erk2-dKO cells (Figure S6D). ChIP-seq analyses showed that the occupancy of JARID2/EZH2 and histone H3K27me3 modification at both CGIs and bivalent promoters were fully restored in the ectopically Jarid2-expressed Erk1/Erk2-dKO mESCs (Figures 6D–6F and S6E–S6G). Thus, same as in naive mESCs, the reduced genome-wide PRC2 occupancy at CGIs and bivalent promoters appeared to be mainly caused by the reduced Jarid2 expression and decreased JARID2-mediated PRC2 recruitment in the Erk1/Erk2-dKO mESCs.
Activation of bivalent genes appears to be determined by the presence of signaling-associated transcription factors but not the status of PRC2 occupancy in naive mESCs
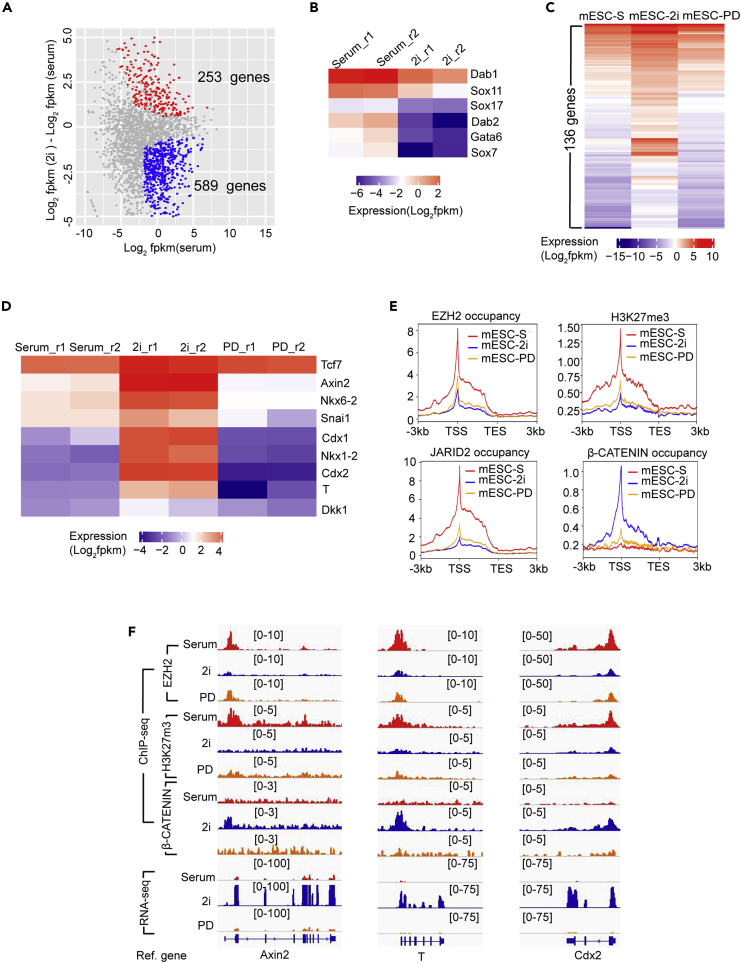
To examine whether the reduced PRC2 occupancy led to transcriptional changes of its regulated genes in naive mESCs, we compared the expression of 2,830 bivalent genes in mESC-S versus mESC-2i by RNA-seq analyses (Figure S5G). The results identified 589 downregulated genes and 253 de-repressed genes in mESC-2i (cutoff: 1.5-fold expression change and q < 0.05) (Figures 7A, S7A, and S7B). Since all bivalent gene promoters had the same reduced PRC2 occupancy and histone H3K27me3 modification in mESC-2i (Figures S5G–S5J), the distinct transcriptional status of these two gene groups suggested that the transcriptional activation was determined by factors other than the PRC2 occupancy at promoters. The gene ontology (GO) analysis showed that the downregulated genes had enriched GO terms involved in cell differentiation, organ morphogenesis, and multicellular development (cutoff: Benjamini adjusted p < 0.05), which included multiple primitive endoderm-specific genes such as Gata6, Dab1, Dab2, Sox7, and Sox17 that were known to be activated by the FGF/ERK signaling (Figures 7B and S7C and Table S2). On the other hand, the enriched GO terms of de-repressed genes were involved in multicellular development, transcriptional regulation, and canonical WNT signaling (cutoff: Benjamini adjusted p < 0.05) (Figure S7D and Table S3). Notably, multiple known Wnt/β-catenin direct target genes such as Axin2, T, Tcf7, Snai1, Cdx1, and Cdx2 were de-repressed in naive mESCs (Roose et al., 1999; Lickert et al., 2000; Yan et al., 2001; Lustig et al., 2002; ten Berge et al., 2008; Horvay et al., 2011), suggesting the activation of these genes might be induced by transcription factors associated with the activated Wnt/β-catenin signaling in 2i medium.
Figure 7.
Activation of Bivalent Genes Is Determined by the Presence of Signaling-Associated Transcription Factors but Not the Status of PRC2 Occupancy in naive mESCs
(A) Plot showing 253 upregulated and 589 downregulated bivalent genes in mESC-2i compared to mESC-S.
(B) Heatmap showing the expression of representative primitive endoderm-specific genes in mESC-S and mESC-2i.
(C) Heatmap showing 136 bivalent genes de-repressed in mESC-2i and re-silenced after inactivation of Wnt/β-catenin signaling (mESC-PD).
(D) Heatmap showing the representative Wnt/β-catenin direct target genes de-repressed in mESC-2i and re-silenced after inactivation of Wnt/β-catenin signaling (mESC-PD).
(E) Plot showing EZH2 occupancy, JARID2 occupancy, histone H3K27me3 modification, and β-CATENIN occupancy at 136 Wnt/β-catenin signaling target genes in mESC-S, mESC-2i, and mESC-PD. TSS: transcription starting sites; TES: transcription ending sites.
(F) IGV genome browser view of EZH2 occupancy, histone H3K27me3 modification, β-CATENIN occupancy, and gene expression of representative Wnt/β-catenin signaling direct target genes Axin2, T, and Cdx2 in mESC-S, mESC-2i, and mESC-PD.
To further examine whether these genes were activated by the Wnt/β-catenin signaling in naive mESCs, we inactivated the Wnt/β-catenin signaling by removing GSK3 inhibitor CHIR99021 and leaving a single MEK inhibitor PD0325901 in the medium (mESC-PD). RNA-seq analysis showed that 136 out of 253 genes that were de-repressed in mESC-2i, including all known Wnt/β-catenin direct target genes such as Axin2, T, Tcf7, Snai1, Cdx1, and Cdx2, lost the transcriptional activation and were re-silenced in mESC-PD (Figures 7C and 7D). Further ChIP-seq analysis showed that compared to mESC-S, both mESC-2i and mESC-PD had reduced EZH2/JARID2 occupancy and histone H3K27me3 modification at these 136 gene promoters, while the β-CATENIN occupancy at gene promoters increased in mESC-2i but not in mESC-S or mESC-PD, supporting the transcriptional activation of these genes in mESC-2i relied on the transcription factor binding to the promoters (Figures 7E and 7F).
Hence, all these results suggested that the reduced PRC2 occupancy at gene promoters alone was insufficient to activate transcription of bivalent genes in naive mESCs. Compared to the reduced PRC2 occupancy at gene promoters, the presence of transcription factors appeared to be necessary and play a predominant role in inducing transcriptional activation.
Discussion
In mammalian cells, PRC1 and PRC2 are recruited to unmethylated CGIs and CGI-associated bivalent gene promoters. The occupancy of Polycomb complexes and their mediated covalent histone modifications at gene promoters are critical for maintaining mESCs in an undifferentiated state by preventing stochastic transcription of lineage differentiation genes.
Previous studies have revealed that both JARID2 and MTF2 are required for the recruitment of PRC2 (Shen et al., 2009; Landeira et al., 2010; Pasini et al., 2010; Casanova et al., 2011; Li et al., 2017; Oksuz et al., 2018), while KDM2B recruits a ncPRC1.1 variant to unmethylated CGIs through its CxxC-ZF domain (Farcas et al., 2012; He et al., 2013; Wu et al., 2013). Although the exact molecular mechanisms mediating the PRC2 recruitment to its targets are not fully elucidated, it is believed that the preferential binding to unmethylated CG-rich DNA sequences by the Polycomb recruiting factors JARID2, MTF2, and KDM2B is important to determine the genome-wide Polycomb occupancy at CGIs in mammalian cells (Deaton and Bird, 2011; Laugesen et al., 2019).
Compared to mESCs cultured in serum-containing medium, naive mESCs in 2i medium have reduced basal expression of early differentiation genes (Silva et al., 2008; Ying et al., 2008). Unexpectedly, the PRC2-mediated histone H3K27me3 modification, which functions as a major epigenetic mechanism mediating gene silencing in mESCs, has been found to have a global reduction in naive mESCs (Marks et al., 2012; Guo et al., 2016). Previous studies suggested the reduced PRC2 occupancy could be caused by re-location of PRC2 from CGIs to non-CGI DNA de-methylated regions in naive mESCs, or reduced chromatin accessibility to PRC2 at bivalent promoters in the ERK signaling-deficient mESCs (Tee et al., 2014; Walter et al., 2016; Kumar and Elsasser, 2019; van Mierlo et al., 2019).
For the first time, this study provides compelling evidence to demonstrate that the genome-wide decreased PRC2 occupancy at CGIs and bivalent promoters in naive mESCs is mainly caused by the reduced JARID2-mediated PRC2 recruitment. First, the RNA-seq, qRT-PCR, and WB analyses show that the Jarid2 expression is significantly reduced in naive mESCs (Figures 1 and S1), and the reduced JARID2 expression is not due to different isoforms expressed in naive mESCs since WB analyses only detect a single long isoform protein (Figures S1D and S2D) (Al-Raawi et al., 2019). JARID2 is a known factor to recruit PRC2 to CGIs in mammalian cells (Peng et al., 2009; Shen et al., 2009; Pasini et al., 2010). Therefore, the decreased JARID2 expression provides an important molecular basis for the global reduced PRC2 occupancy in naive mESCs. Second, the FGF/ERK signaling positively regulates the Jarid2 expression since the re-activation of FGF/ERK signaling in naive mESCs upregulates the Jarid2 expression at both mRNA and protein levels (Figures 2 and S2). Third, similar to the chemical inhibition of ERK signaling in 2i medium, the genetic disruption of ERK signaling largely reduces the Jarid2 expression in the mESCs cultured in serum-containing medium, which can be rescued by the ectopic expression of wild-type Erk2, further confirming that ERK signaling positively regulates the expression of Jarid2 (Figures 3 and S3). Fourth, the ChIP-seq analyses show the global reduced EZH2 occupancy and histone H3K27me3 modification at CGIs and bivalent promoters correlate with the global reduced JARID2 occupancy, which can be fully rescued by the ectopic expression of Jarid2 in both naive and Erk1/Erk2-dKO mESCs (Figures 4, 5, 6, and S4–S6). Finally, although the PRC2 core component Suz12 showed increased expression in mESC-2i, the findings that ectopic Jarid2 expression fully restores the PRC2 occupancy in mESCs-2i suggest the reduced PRC2 occupancy in naive mESCs is mainly due to the reduced JARID2-mediated PRC2 recruitment, but unlikely to be caused by changed expression of Suz12 or other PRC2 components. Collectively, all these results strongly indicate the FGF/ERK signaling determines the global PRC2 occupancy at CGIs and bivalent promoters mainly through regulating the Jarid2 expression in mESCs.
Several recent studies report that although the H3K27me3 modification is reduced at CGIs, the overall abundance of PRC2 and H3K27me3 in naive mESCs is increased (Kumar and Elsasser, 2019; van Mierlo et al., 2019). Furthermore, the H3K27me3 is re-distributed to non-CGI euchromatin and heterochromatin regions, which correlates with the local CpG density (van Mierlo et al., 2019). Since naive mESCs have global DNA de-methylation, these results suggest the reduced PRC2 occupancy at CGIs in naive mESCs could be caused by re-location of PRC2 from CGIs to new DNA de-methylated regions (van Mierlo et al., 2019). Although our results do not reveal any re-location of EZH2/JARID2 and H3K27me3 modification to 50-kb CGI-flanking regions (Figures S4C–S4E), our current study does not preclude these conclusions due to the practical difficulty in mapping ChIP-seq reads to heterochromatin. To monitor whether there exists Polycomb re-distribution in response to the global DNA de-methylation in naive mESCs, we include KDM2B in our ChIP-seq assay since its CxxC-ZF domain has a high binding affinity to unmethylated CG-rich DNA such as CGIs, and importantly KDM2B is known to bind DNA de-methylated pericentric heterochromatin in the DNA methyltransferases triple knockout (DNMT-TKO) cells (Blackledge et al., 2010; He et al., 2013; Cooper et al., 2014). However, our study finds both mESC-S and mESC-2i have the same KDM2B occupancy at CGIs (Figure 4D). Since KDM2B has the same expression level in mESC-S and mESC-2i (Figures 1 and S1), the results suggest that KDM2B is unlikely to have a significant re-distribution or to mediate PRC2 re-location to non-CGI de-methylated regions in naive mESCs. In addition, the histone H3K4me3 modification at CGIs, which is mediated by MLL2 binding to CGIs through a similar CxxC-ZF domain as KDM2B (Long et al., 2013; Denissov et al., 2014), is increased at CGIs in naive mESCs (Figure S4), further arguing against that the CxxC-ZF domain-containing proteins are re-located from CGIs to new DNA de-methylated regions. Finally but importantly, the ectopic expression of Jarid2 in naive mESCs fully restores the global PRC2 occupancy and the histone H3K27me3 modification at CGIs (Figures 5 and S5), further supporting that the reduced PRC2 occupancy at CGIs in naive mESCs is mainly caused by the reduced JARID2-mediated PRC2 recruitment.
A previous study has reported that ERK1/ERK2 determine the PRC2 occupancy at bivalent promoters through regulating local chromatin accessibility to PRC2 (Tee et al., 2014). Consistent with this report, we find that the global PRC2 occupancy and H3K27me3 modification are reduced in the Erk1/Erk2-dKO mESCs, which is restored by ectopic expression of wild-type Erk2 (Figure 6). However, we find that same as naive mESCs, Erk1/Erk2-dKO mESCs have significant reduced Jarid2 expression, which can be rescued by ectopic expression of wild-type Erk2 (Figure 3). Importantly, the ectopic expression of Jarid2 fully restores the EZH2/JARID2 occupancy and H3K27me3 modification at both CGIs and bivalent promoters in the Erk1/Erk2-dKO mESCs (Figures 6 and S6). Since majority of bivalent promoters are associated with CGIs, our study suggests that the reduced JARID2-mediated PRC2 occupancy at CGIs is the main molecular mechanism leading to the reduction of PRC2 occupancy at bivalent promoters in the Erk1/Erk2-depleted mESCs. On the other hand, Tee et al. reported that the depletion of Jarid2 impaired the phosphorylation of ERK1/ERK2, suggesting there might exist mutual regulation of JARID2 expression and ERK signaling activation (Tee et al., 2014). It is worth to examine how the interaction of JARID2 and ERK signaling affects the global PRC2 occupancy and gene expression in mESCs in future studies.
Depletion of Polycomb induces de-repression of some bivalent genes in mESCs cultured in serum-containing medium, in which mESCs express low-level transcription factors associated with FGF/ERK signaling (He et al., 2013; Illingworth et al., 2016), suggesting Polycomb binding to promoters is essential for setting high transcriptional thresholds to prevent stochastic transcription (Laugesen et al., 2016). However, previous studies showed that majority of bivalent genes remain silenced although the PRC2 occupancy at their promoters are reduced in naive mESCs (Silva et al., 2008; Marks et al., 2012; Riising et al., 2014; Galonska et al., 2015). To solve this unexpected discrepancy, our study separates two groups of bivalent genes with distinct transcriptional states, either downregulated or de-repressed, in naive mESCs (Figures 7A, S7A, and S7B). Since all bivalent promoters have the same reduced PRC2 occupancy in mESC-2i (Figures S5G–S5J), the distinct transcriptional status of these two gene groups suggests that the transcriptional activation of bivalent genes is determined by factors other than the PRC2 occupancy at their promoters in naive mESCs. Further analyses reveal that the downregulated gene group contains multiple primitive endoderm-specific genes known to be activated by the FGF signaling (Chazaud et al., 2006; Illingworth et al., 2016; Hamilton et al., 2019), while the de-repressed gene group includes all known direct targets of Wnt/β-catenin signaling, suggesting the activation of these genes is induced by transcription factors associated with the activated Wnt/β-catenin signaling in 2i medium (Figures 7B–7D, S7C, and S7D) (Roose et al., 1999; Lickert et al., 2000; Yan et al., 2001; Lustig et al., 2002; ten Berge et al., 2008; Horvay et al., 2011). Notably, more than half de-repressed genes in naive mESCs, including all known Wnt/β-catenin direct target genes, are re-silenced after inactivation of Wnt/β-catenin signaling by removal of GSK3 inhibitor from 2i medium (Figures 7C and 7D). In addition, the activation of Wnt signaling target genes in naive mESCs correlates with the increased β-CATENIN occupancy at their promoters (Figures 7E and 7F), further supporting that the presence of transcription factors, but not the reduced PRC2 occupancy at promoters, plays a predominant role in activating bivalent genes in naive mESCs. Of note, these results are consistent with previous reports that the promoter-bound PRC2 does not actively repress transcription but rather sets high transcriptional thresholds for gene activation in cells (Riising et al., 2014; Laugesen et al., 2016).
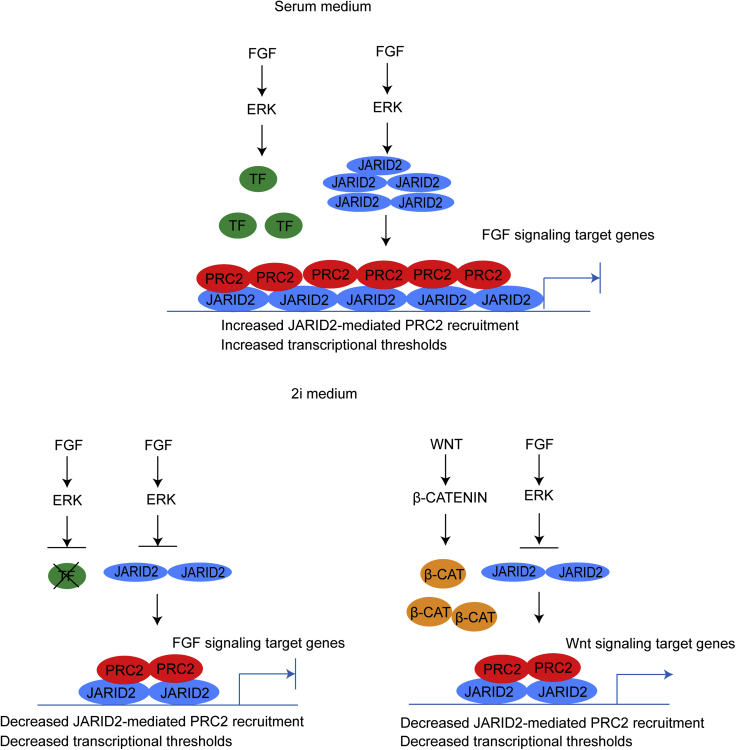
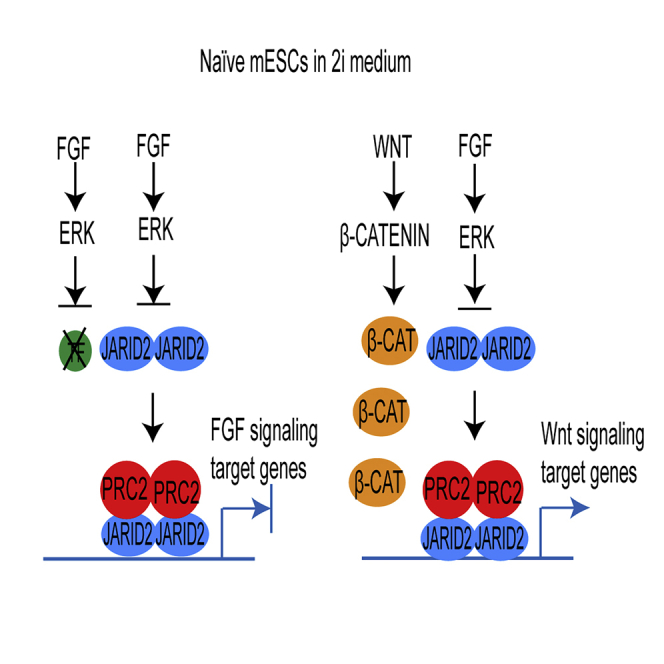
In summary, our study provides compelling evidence to reveal a key molecular mechanism by which the FGF/ERK signaling regulates the global PRC2 occupancy at CGIs in naive mESCs and to elucidate a fundamental question regarding the function of transcription factors and PRC2-mediated epigenetic mechanisms in transcriptional regulation. Based on the evidence, we propose a model to explain how cell signaling coordinates both global PRC2 recruitment and developmental gene expression in mESCs. In this model, the FGF/ERK signaling positively regulates the Jarid2 expression and promotes the JARID2-mediated PRC2 recruitment to CGIs in mESCs cultured in serum-containing medium. The PRC2 binding to promoters increases the overall thresholds for transcriptional activation, which is essential for preventing stochastic transcription of lineage differentiation genes induced by the FGF/ERK signaling in serum-containing medium. In naive mESCs, the chemical inhibition of FGF/ERK signaling reduces the Jarid2 expression and leads to a global reduction of JARID2-mediated PRC2 recruitment to CGIs and bivalent promoters, which subsequently reduces the overall transcriptional thresholds of bivalent genes. However, in the absence of FGF/ERK signaling-associated transcription factors, the reduced PRC2 occupancy at promoters alone is insufficient to activate FGF/ERK signaling target genes. In contrast, the Wnt/β-catenin signaling target genes are de-repressed by the activated Wnt/β-catenin signaling and its associated transcription factors in naive mESCs (Figure 8).
Figure 8.
Proposed Model: Cell Signaling Coordinates PRC2 Recruitment and Developmental Gene Expression in mESCs
In serum-containing medium, FGF/ERK signaling increases both Jarid2 expression and JARID2-mediated PRC2 recruitment to bivalent promoters, which increases the thresholds for transcriptional activation and prevents stochastic expression of FGF signaling target genes (upper panel). In 2i medium, the deficient FGF/ERK signaling decreases the Jarid2 expression and JARID2-mediated PRC2 recruitment to bivalent promoters, which reduces the thresholds for transcriptional activation. In the absence of FGF/ERK signaling-associated transcriptional factors, the FGF/ERK signaling target genes remain silenced. In contrast, the activated Wnt/β-catenin signaling and its transcription factor β-CATENIN (β-CAT) activate the Wnt signaling target genes (lower panel).
Limitations of the Study
Previous studies show in naive mESCs, the H3K27me3 is re-distributed to non-CGI euchromatin and heterochromatin regions, which correlates with the local CpG density, suggesting the reduced PRC2 occupancy at CGIs in naive mESCs could be caused by the re-location of PRC2 from CGIs to new DNA de-methylated regions including pericentric heterochromatin (van Mierlo et al., 2019). Due to the practical difficulty in mapping ChIP-seq reads to heterochromatin, in this study we were unable to quantify the change of PRC2 occupancy at heterochromatin and further determined the extent of PRC2 re-distribution to heterochromatin in naive mESCs. Mass spectrometry-assisted quantitative measurement of heterochromatin-bound PRC2 and H3K27me3 modification in naive mESCs should be conducted to address this question in future studies.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jin He (hejin1@msu.edu).
Materials Availability
This study did not generate any unique reagents.
Data and Code Availability
All ChIP-seq and RNA-seq reported in this paper have been deposited to NCBI Gene Expression Omnibus (GEO), GEO accession: GSE157747, GSE157748, GSE157749. Other RNA-seq samples used in the analysis are available on NCBI GEO: GSE23943.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Drs. David Arnosti and Amy Ralston for their critical reading of the manuscript. MSU genomics core facility processed the next-generation sequencing. This work was supported by the National Institutes of Health grant R01GM127431.
Author Contributions
J.H. conceived the project. M.B.A., Y.G., Y.W., and J.H. designed and performed the experiments. J.H. and G.I.M. performed the sequencing data analysis. J.H. interpreted the data and wrote the manuscript.
Declaration of Interests
Authors declare no competing interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101646.
Supplemental Information
References
- Al-Raawi D., Jones R., Wijesinghe S., Halsall J., Petric M., Roberts S., Hotchin N.A., Kanhere A. A novel form of JARID2 is required for differentiation in lineage-committed cells. EMBO J. 2019;38:e98449. doi: 10.15252/embj.201798449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L., Di Stefano B., Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- Blackledge N.P., Fursova N.A., Kelley J.R., Huseyin M.K., Feldmann A., Klose R.J. PRC1 catalytic activity is central to polycomb system function. Mol. Cell. 2019;77:857–874.e9. doi: 10.1016/j.molcel.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N.P., Zhou J.C., Tolstorukov M.Y., Farcas A.M., Park P.J., Klose R.J. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Casanova M., Preissner T., Cerase A., Poot R., Yamada D., Li X., Appanah R., Bezstarosti K., Demmers J., Koseki H., Brockdorff N. Polycomblike 2 facilitates the recruitment of PRC2 Polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development. 2011;138:1471–1482. doi: 10.1242/dev.053652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N.P., De Marco V., Elderkin S., Koseki H., Klose R. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S., Hofemeister H., Marks H., Kranz A., Ciotta G., Singh S., Anastassiadis K., Stunnenberg H.G., Stewart A.F. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Helin K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Farcas A.M., Blackledge N.P., Sudbery I., Long H.K., McGouran J.F., Rose N.R., Lee S., Sims D., Cerase A., Sheahan T.W. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galonska C., Ziller M.J., Karnik R., Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Pinello L., Han X., Lai S., Shen L., Lin T.W., Zou K., Yuan G.C., Orkin S.H. Serum-based culture conditions provoke gene expression variability in mouse embryonic stem cells as revealed by single-cell analysis. Cell Rep. 2016;14:956–965. doi: 10.1016/j.celrep.2015.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W.B., Mosesson Y., Monteiro R.S., Emdal K.B., Knudsen T.E., Francavilla C., Barkai N., Olsen J.V., Brickman J.M. Dynamic lineage priming is driven via direct enhancer regulation by ERK. Nature. 2019;575:355–360. doi: 10.1038/s41586-019-1732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Shen L., Wan M., Taranova O., Wu H., Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvay K., Casagranda F., Gany A., Hime G.R., Abud H.E. Wnt signaling regulates Snai1 expression and cellular localization in the mouse intestinal epithelial stem cell niche. Stem Cells Dev. 2011;20:737–745. doi: 10.1089/scd.2010.0188. [DOI] [PubMed] [Google Scholar]
- Illingworth R.S., Holzenspies J.J., Roske F.V., Bickmore W.A., Brickman J.M. Polycomb enables primitive endoderm lineage priming in embryonic stem cells. Elife. 2016;5:e14926. doi: 10.7554/eLife.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B., Elsasser S.J. Quantitative multiplexed ChIP reveals global alterations that shape promoter bivalency in ground state embryonic stem cells. Cell Rep. 2019;28:3274–3284.e5. doi: 10.1016/j.celrep.2019.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D., Sauer S., Poot R., Dvorkina M., Mazzarella L., Jorgensen H.F., Pereira C.F., Leleu M., Piccolo F.M., Spivakov M. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat. Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A., Hojfeldt J.W., Helin K. Role of the polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med. 2016;6:a026575. doi: 10.1101/cshperspect.a026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A., Hojfeldt J.W., Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell. 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulator's by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liefke R., Jiang J., Kurland J.V., Tian W., Deng P., Zhang W., He Q., Patel D.J., Bulyk M.L. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H., Domon C., Huls G., Wehrle C., Duluc I., Clevers H., Meyer B.I., Freund J.N., Kemler R. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 2000;127:3805–3813. doi: 10.1242/dev.127.17.3805. [DOI] [PubMed] [Google Scholar]
- Long H.K., Blackledge N.P., Klose R.J. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013;41:727–740. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., van de Wetering M., Clevers H., Schlag P.M., Birchmeier W., Behrens J. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Descalzo S., Rue P., Garcia-Ojalvo J., Martinez Arias A. Correlations between the levels of Oct4 and Nanog as a signature for naive pluripotency in mouse embryonic stem cells. Stem Cells. 2012;30:2683–2691. doi: 10.1002/stem.1230. [DOI] [PubMed] [Google Scholar]
- Oksuz O., Narendra V., Lee C.H., Descostes N., LeRoy G., Raviram R., Blumenberg L., Karch K., Rocha P.P., Garcia B.A. Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell. 2018;70:1149–1162.e5. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Cloos P.A., Walfridsson J., Olsson L., Bukowski J.P., Johansen J.V., Bak M., Tommerup N., Rappsilber J., Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Peng J.C., Valouev A., Swigut T., Zhang J., Zhao Y., Sidow A., Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising E.M., Comet I., Leblanc B., Wu X., Johansen J.V., Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Roose J., Huls G., van Beest M., Moerer P., van der Horn K., Goldschmeding R., Logtenberg T., Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Shen X., Kim W., Fujiwara Y., Simon M.D., Liu Y., Mysliwiec M.R., Yuan G.C., Lee Y., Orkin S.H. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.G. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Kobayashi S., Hiratani I. Epigenetic differences between naive and primed pluripotent stem cells. Cell Mol. Life Sci. 2018;75:1191–1203. doi: 10.1007/s00018-017-2703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburri S., Lavarone E., Fernandez-Perez D., Conway E., Zanotti M., Manganaro D., Pasini D. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell. 2019;77:840–856.e5. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee W.W., Shen S.S., Oksuz O., Narendra V., Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Koole W., Fuerer C., Fish M., Eroglu E., Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo G., Dirks R.A.M., De Clerck L., Brinkman A.B., Huth M., Kloet S.L., Saksouk N., Kroeze L.I., Willems S., Farlik M. Integrative proteomic profiling reveals PRC2-dependent epigenetic crosstalk maintains ground-state pluripotency. Cell Stem Cell. 2019;24:123–137.e8. doi: 10.1016/j.stem.2018.10.017. [DOI] [PubMed] [Google Scholar]
- von Meyenn F., Iurlaro M., Habibi E., Liu N.Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z. Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell. 2016;62:983. doi: 10.1016/j.molcel.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Teissandier A., Perez-Palacios R., Bourc'his D. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife. 2016;5 doi: 10.7554/eLife.11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wu X., Johansen J.V., Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Yamaji M., Ueda J., Hayashi K., Ohta H., Yabuta Y., Kurimoto K., Nakato R., Yamada Y., Shirahige K., Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yan D., Wiesmann M., Rohan M., Chan V., Jefferson A.B., Guo L., Sakamoto D., Caothien R.H., Fuller J.H., Reinhard C. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc. Natl. Acad. Sci. U S A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ChIP-seq and RNA-seq reported in this paper have been deposited to NCBI Gene Expression Omnibus (GEO), GEO accession: GSE157747, GSE157748, GSE157749. Other RNA-seq samples used in the analysis are available on NCBI GEO: GSE23943.