Abstract
Phyto-thermotherapy is a treatment consisting in immersing oneself in baths of self-heating alpine grass, to benefit of the heat and rich aromatic components released by the process. The aim of this study was to characterize the bacterial and fungal diversity of three phyto-thermal baths (PTB) performed in three different months, and to compare the data with the profile of the volatile organic compounds (VOCs) of the process. All the data collected showed that PTBs were structured in two stages: the first three days were characterised by an exponential rise of the temperature, a fast bacterial development, higher microbial diversity and higher concentrations of plant aliphatic hydrocarbons. The second stage was characterised by a stable high temperature, shrinkage of the microbial diversity with a predominance of few bacterial and fungi species and higher concentrations of volatiles of microbial origin. Erwinia was the dominant microbial species during the first stage and probably responsible of the self-heating process. In conclusion, PTBs has shown both similarities with common self-heating processes and important peculiarities such as the absence of pathogenic bacteria and the dominance of plant terpenoids with health characteristics among the VOCs confirming the evidence of beneficial effects in particular in the first three days.
Subject terms: Biochemistry, Ecology, Microbiology, Molecular biology
Introduction
Phyto-thermotherapy1, 2 also known as Phyto-balneotherapy3 consists of immersing people in phyto-thermal baths (PTB) containing freshly-cut mountain grass in hot fermentation for no more than 30 min. PTB is an ancient practice performed since the nineteenth century in mountain areas of Tyrol (Austria), Alto Adige and Trentino (Italy). The PTBs are usually located in Spa and loaded with mountain herbs cut at vegetative maturity. An historical area for PTB grass collection is the meadow of the Viote in Monte Bondone (1540 m a.s.l., Trentino, Italy). The grass is harvested during the summer season, cut and bailed in early mornings (to avoid sunlight drying of the dew), and brought to the spa located in Garniga Terme (Trentino, Italy). A 50 cm layer of grass is placed in baths and stays two or three days before naturally reaching the temperature of 55–60 °C, following spontaneous processes of self-heating. Among the most significant species constituting the grass pile, there are Gentiana (pumilum, vernum, germanica, kochiana, lutea and verna), Arnica montana, Hypericum maculatum, Thymus serpyllum, and Pulsatilla alpine4. By applying studied and tested techniques, it is possible to benefit from the baths also during the winter season: the collected grass is packaged and quickly brought to temperatures of − 20 °C; in this way, the thermophilic process is stopped without destroying the grass organoleptic properties.
Although PTBs are empirically known as natural treatments for rheumatic diseases2, 5, knowledge on the involved microbiota and investigations about the thermo-chemical process occurring in the PTB are totally missing and will help to improve the onset and the progress of the process.
This study had been essentially focused to microbiological aspects of PTB by means of both culture dependent and independent methods (partial 16S rRNA and ITS genes MiSeq Illumina sequencing). The main aim was to establish the microbial community structure at different stages of the PT process (i) identifying the bacterial and fungi community members; (ii) assessing the biodiversity of bacterial and fungal microbiota throughout the process; and finally (iii) coupling this data with the analysis of the volatiles organic compounds (VOCs) released during the process.
Results
Temperature and pH
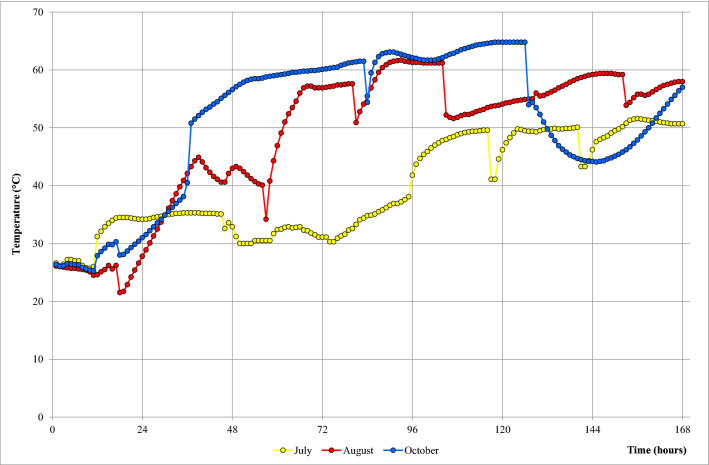
Figure 1 shows the temperature trend during the July, August and October PTBs. In the first 35 h, all the PTBs showed similar temperatures rising from 26 to 35 °C – 39 °C (Fig. 1). In the following hours, the temperature showed different trends according to the month. In July, the temperature was stable for the first four days (96 h) and then rose, reaching the max. temperature of 51 °C. In August, the temperature was stable for the first 55 h and then rose reaching the max. temperature of 61 °C. Finally, in October, after the first 35 h, the temperature rose immediately reaching 50 °C, and then was stable reaching the max. of 64.8 °C.
Figure 1.
Temperature dynamic of herbs pile bath during PTB. Temperature was recorded for seven days from day 0 to d7 each hour (24 h = d1; 48 h = d2; 72 h = d3; 96 h = d4; 120 h = d5; 144 h = d6; and 168 h = d7) at 20 cm of depth in the middle of the pool bath. In yellow is the temperature trend for July, in red August and in blue October batch.
The pH (Table 1), in the first two days, was in a range between 6.2 and 6.5 with the exception of October when pH sowed a mean value of 7.4. After d5, the pH increased significantly and stabilized in a range between 7.4 and 7.8.
Table 1.
Microbiological counts (Log CFU g−1) and pH in the herbs samples in different days and month of the PTB.
| pH | Aerobic bacteria | Mesoph. LAB | Thermoph. LAB | Entero-bacteria | Yeasts | Moulds | |
|---|---|---|---|---|---|---|---|
| July | |||||||
| day 0 | 6.2 ± 0.43a | 6.7 ± 0.18a | n. d.a | n. d.a | 3.8 ± 0.59a | n. d.a | n. d.a |
| day 2 | 6.2 ± 0.32a | 7.1 ± 0.51a | 5.3 ± 0.20b | 2.9 ± 0.53b | 4.6 ± 0.57a | n. d.a | n. d.a |
| day 3 | 6.6 ± 0.21ab | 8.4 ± 0.29b | 5.9 ± 0.60b | 4.6 ± 0.52c | 5.7 ± 1.13b | 5.9 ± 1.36c | 5.7 ± 0.89c |
| day 5 | 7.6 ± 0.26b | 8.7 ± 0.47b | 5.5 ± 0.74b | 5.1 ± 1.04c | 5.6 ± 0.67b | 2.5 ± 1.44b | 5.4 ± 0.59c |
| day 7 | 7.7 ± 0.22b | 8.7 ± 0.31b | 5.9 ± 0.80b | 5.2 ± 0.87c | 6.2 ± 0.52b | n. d.a | 3.4 ± 1.20b |
| August | |||||||
| Day 0 | 6.4 ± 0.31a | 7.7 ± 0.46a | 1.2 ± 0.80a | n. d.a | 4.3 ± 0.70a | 6.0 ± 0.64c | 5.2 ± 0.27b |
| Day 2 | 6.5 ± 0.20a | 8.1 ± 0.60 ab | 6.2 ± 0.57b | 1.6 ± 0.64a | 5.5 ± 0.99b | 5.4 ± 1.31c | 5.1 ± 0.47b |
| Day 3 | 7.0 ± 0.27ab | 8.4 ± 0.55 ab | 6.1 ± 0.65b | 4.0 ± 0.64b | 6.5 ± 0.23b | 3.8 ± 0.96b | 4.3 ± 1.09b |
| Day 5 | 7.4 ± 0.29ab | 8.2 ± 0.65 ab | 5.8 ± 1.04b | 4.3 ± 0.84b | 5.8 ± 0.67b | n. d.a | 4.5 ± 0.87b |
| Day 7 | 7.6 ± 0.15b | 8.8 ± 0.42b | 5.9 ± 0.87b | 5.7 ± 0.94c | 5.7 ± 0.71b | n. d.a | 3.2 ± 0.84a |
| October | |||||||
| Day 0 | 7.4 ± 0.32 | 9.1 ± 0.46 b | 7.4 ± 0.80bc | 3.8 ± 1.55a | 5.0 ± 0.70a | 7.8 ± 0.64 | 6.7 ± 0.27 ab |
| Day 2 | 7.4 ± 0.74 | 8.9 ± 0.60 ab | 7.2 ± 0.57bc | 4.5 ± 0.64a | 5.1 ± 0.99a | 7.7 ± 1.31 | 7.0 ± 0.47 ab |
| Day 3 | 7.0 ± 1.10 | 9.4 ± 0.55 b | 7.6 ± 0.65c | 5.6 ± 0.64b | 5.9 ± 0.23ab | 7.8 ± 0.96 | 7.7 ± 1.09 b |
| Day 5 | 7.7 ± 0.33 | 8.1 ± 0.65 a | 6.4 ± 1.04b | 6.2 ± 0.84b | 5.8 ± 0.67ab | 8.8 ± 1.48 | 6.6 ± 0.87 a |
| Day 7 | 7.8 ± 0.24 | 7.6 ± 0.42 a | 4.9 ± 0.87a | 5.1 ± 0.94ab | 6.7 ± 0.71b | 8.1 ± 0.96 | 6.4 ± 0.84 a |
Results are showed as mean values ± standard deviations of three replicates for each point of sampling (5 and 40 cm) for a total of six values. For a given column and month, microbial count or pH values with a, b and c superscripts are significantly different (p < 0.05).
n. d.: not detected or present in traces lower than 1.0 LOG cfu g−1.
Microbial counts in herbs during PT process
The counts of viable total aerobic, mesophilic and thermophilic anaerobic bacteria, enterobacteria, yeasts and moulds in July, August and October at day 0, d2, d3, d5 and d7 are shown in Table 1. The plate counts showed no significant difference for depth of sampling (p value > 0.05; data not shown). The aerobic bacteria were always high at day 0 (6.7, 7.7 and 9.1 Log CFU g−1 in July, August and October respectively). In July and August, they significantly rose until d7, reaching similar amounts (8.7 and 8.8 Log CFU g−1 respectively). The mesophilic and thermophilic lactic acid bacteria (LAB) were very low in July and August at day 0 when they were present in traces or not detected (Table 1). Mesophilic LAB rose from d2 to d7 with a similar trend to aerobic bacteria reaching 5.9 Log CFU g−1. By contrast, in October they showed different trend: as aerobic bacteria, mesophilic LAB counts were very high at day 0 and stable without significant differences until d3 and then significantly decreased until d7 reaching 4.9 Log CFU g−1. Counts of thermophilic LAB showed similar trends in all the three months: they rose until d7 reaching similar amount in July, August and October (5.2, 5.7 and 5.1 Log CFU g−1 respectively). Enterobacteria counts were lower in herbs at day 0 (3.8, 4.3 and 5.0 Log CFU g−1 in July, August and October respectively) and then significantly rose until d7 reaching 6.2, 5.7 and 6.7 Log CFU g−1 in July, August and October respectively. In July, yeasts and moulds in the first two days were not detected or present in traces, then reached their highest value at d3 and significantly decreased until d7. By contrast, their counts were very high in both August and October; in particular, moulds counts trends were similar: they significantly decreased until d7 to 3.2 and 6.4 Log CFU g−1 in August and October respectively.
Characteristics of the sequencing data
The DNA extracted from the 90 PTB samples had been all successfully amplified. After merging and quality trimming the raw data, 2,980,511 reads for bacteria and 1,227,092 reads for fungi remained for subsequent analysis (Table S1). After alignment, the remaining Operational Taxonomy Units (OTU) had been clustered at a 3% of distance.
Bacteria and fungi: alpha diversity
The number of OTUs and the Shannon diversity index were determined using QIIME2 at 97% similarity levels (Table 2), in order to analyse the bacterial and fungi community richness in samples obtained during the PT process. Regarding the sample position (5 and 40 cm depth), there was no significant difference in both observed OTUs number and Shannon diversity index for bacteria and fungi communities (p value > 0.05). It is worth noting that the degree of bacterial diversity was significantly higher in July and August than in October samples, by contrast, the degree of fungal diversity was significantly higher in July and October than in August samples (Shannon diversity index in Table 2).
Table 2.
Observed OTUs (Obs. OTUs) and Shannon diversity index (Shannon div. ind.) in the herbs at different depth, month and of day sampling of the PTB.
| Bacteria | Fungi | |||
|---|---|---|---|---|
| Obs. OTUs | Shannon div. ind. | Obs. OTUs | Shannon div. ind. | |
| Depth of sampling | ||||
| 5 cm | 340 ± 126 | 6.9 ± 1.19 | 82 ± 48 | 3.7 ± 1.75 |
| 40 cm | 259 ± 87 | 6.5 ± 1.15 | 70 ± 56 | 3.3 ± 2.02 |
| Month of sampling | ||||
| July | 381 ± 136a | 7.4 ± 0.76a | 85 ± 56 | 3.6 ± 1.00ab |
| August | 369 ± 289a | 7.2 ± 0.77a | 59 ± 39 | 2.6 ± 2.09a |
| October | 153 ± 111b | 5.5 ± 1.17b | 85 ± 38 | 4.2 ± 1.21b |
| Day of sampling | ||||
| Day 0 | 318 ± 166a | 7.0 ± 1.14a | 133 ± 16a | 5.3 ± 0.35a |
| Day 2 | 451 ± 268a | 7.1 ± 1.56a | 106 ± 38a | 4.6 ± 1.17a |
| Day 3 | 411 ± 209a | 7.1 ± 0.98a | 77 ± 49b | 3.3 ± 1.87b |
| Day 5 | 177 ± 96b | 6.2 ± 1.13b | 41 ± 35bc | 2.6 ± 1.71b |
| Day 7 | 138 ± 28b | 6.0 ± 0.62b | 22 ± 19c | 1.6 ± 1.11c |
Results are showed as mean values ± standard deviations.
For a given column and month, observed OTUs and Shannon diversity index values with a, b and c superscripts are significantly different (p < 0.05).
The variation in number of OTUs and Shannon diversity index over time indicates highest microbial diversity at day 0 and d2, and a lower microbial diversity at d5 and d7.
Bacteria and fungi: beta diversity
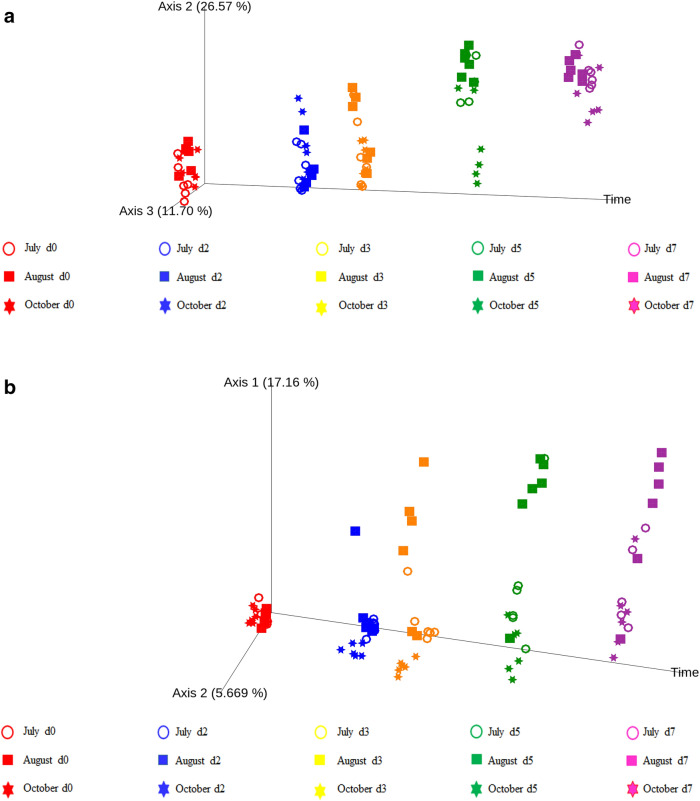
The PCoA of UniFrac and Jaccard metric indicated clear clustering of both bacterial and fungi communities according to the different PTB days (Fig. 2a,b).
Figure 2.
Principal coordinate analysis of Weighted UniFrac distances for bacterial community (a) and Bray–Curtis distances for fungi community (b) in PTB. The Time custom axis has been used to show the PCoA changes in the days. For interpretation of the symbols and colors the reader is referred to the legend.
Bacterial and fungi communities were more phylogenetically dissimilar between successional days (day 0, d2, d3, d5 and d7) or months (July, August and October) than between position of sampling (5 and 40 cm). The bacterial Weighted UniFrac PCoA (Fig. 2a, total variation explained: 52.09%) and the fungal Jaccards PCoA (Fig. 2b, total variation explained: 32.83%) revealed a clearer picture of the similarities across different days. The PCoA plots emphasized the similarities of the bacterial and fungi communities in the PTB at d2 and d3 when compared with d5 and d7.
These results were supported by the PERMANOVA statistical analysis (Table 3). The differences between position of sampling (5 and 40 cm) were not significant both for bacteria and fungi communities as the p values were 0.19 for bacteria and 0.27 for fungi community respectively. The differences among months of sampling were significant comparing July and August with October for both bacteria and fungi communities as well as the differences among microbial communities through time. The pairwise comparison (Table 3) clearly showed two significantly different stages during the PTB after day 0: the first stage including d2 and d3 (1st stage), and the second stage including d5 and d7 (2nd stage). The time effect on microbial composition (pseudo-F value) was showing a growing trend with the proceeding of the sampling days (Table 3). The bacterial pseudo-F values were smaller and smaller with progression of the days; by contrast, fungi pseudo-F values were higher. This means that the bacteria became more similar and the fungi more different in the samples from the three piles studied as the process progressed.
Table 3.
Permanova analysis (999 permutations) results for bacterial and fungi communities based on weighted unifrac and Jaccard distances respectively.
| Bacteria | Fungi | |||
|---|---|---|---|---|
| Pseudo-F | p value | Pseudo-F | p value | |
| Main effects | ||||
| Depth of sampling | 2.822 | 0.29 | 1.612 | 0.23 |
| Month of sampling | 4.447 | 0.001** | 3.600 | 0.001** |
| Day of sampling | 10.553 | 0.001** | 3.348 | 0.001** |
| Pairwise comparisons | ||||
| July versus August | 2.177 | 0.071 | 2.495 | 0.002** |
| July versus October | 4.815 | 0.003** | 4.485 | 0.001** |
| August versus October | 6.253 | 0.003** | 3.834 | 0.001** |
| Pairwise comparisons | ||||
| Day 0 versus day 2 | 11.033 | 0.001** | 1.362 | 0.015* |
| Day 0 versus day 3 | 13.459 | 0.001** | 2.633 | 0.001** |
| Day 0 versus day 5 | 17.984 | 0.001** | 5.538 | 0.001** |
| Day 0 versus day 7 | 18.172 | 0.001** | 7.545 | 0.001** |
| Day 2 versus day 3 | 0.808 | 0.495 | 1.349 | 0.096 |
| Day 2 versus day 5 | 7.957 | 0.001** | 3.630 | 0.001** |
| Day 2 versus day 7 | 15.168 | 0.001** | 5.556 | 0.001** |
| Day 3 versus day 5 | 5.392 | 0.002** | 1.825 | 0.025* |
| Day 3 versus day 7 | 12.266 | 0.001** | 3.187 | 0.001** |
| Day 5 versus day 7 | 3.421 | 0.12 | 1.097 | 0.232 |
Significance levels: *p < 0.05; **p < 0.01.
Bacterial community structure
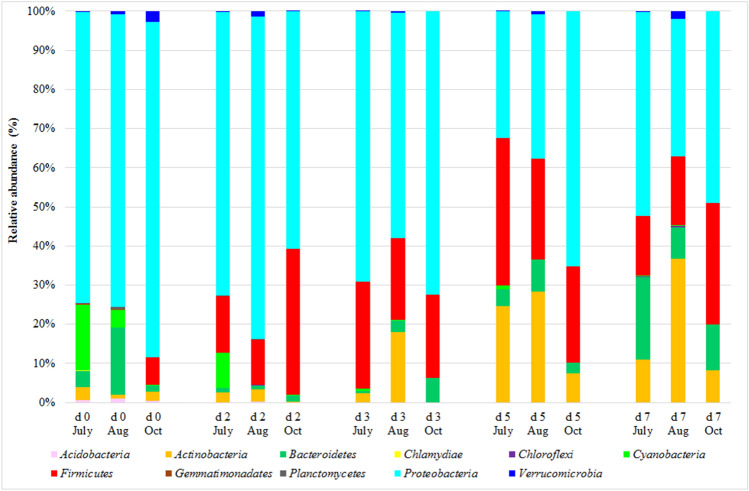
Eleven prokaryotic phyla were found in the 90 samples accounting for the total bacterial community. The predominant phylum across all bacterial communities was the Proteobacteria, accounting for 33%–83% of the OTUs in each time and month of the PTB (Fig. 3). The trend of Proteobacteria was similar in all the months with different amount of presence: at day 0 and d2 Proteobacteria abundance was between 61% and 85%; at d3, showed a slight decrease (between 58% and 72%) and reached the lower values at d5 and d7 (between 33% and 65%). Twenty-six bacterial phylotypes were found dominant across all samples, accounting for the 90% of the total bacterial community. Of these 26 phylotypes (in Fig. 4), 10 belonged to Proteobacteria. Erwinia was the most abundant genus in this phylum and reached the higher presence at d2 and d3. Other frequently sequenced genera included Acinetobacter, Pseudomonas, mainly found at day 0, and unclassified genera belonging to Xanthomonadaceae family.
Figure 3.
Phylum composition (in mean relative abundance) of herbs samples as revealed by high-throughput sequencing analysis. The samples were collected in triplicate from the same pool, for five days (day 0, d2, d3, d5, and d7) at 5 and 40 cm of depth, in July, August (Aug) and October (Oct). For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
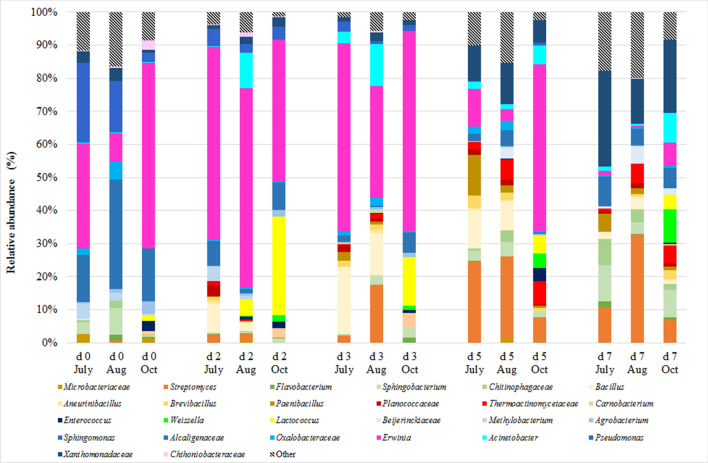
Figure 4.
Bacterial taxa groups (genus level or above) composition, in mean relative abundance, of herb samples as revealed by high-throughput sequencing analysis. The samples have been collected in triplicate from the same pool, for five days (d0, d2, d3, d5, and d7) at 5 and 40 cm of depth, in July, August (Aug) and October (Oct). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Of the Alphaproteobacteria, the genera Agrobacterium, Methylobacterium, Sphingomonas and unclassified genera of the family Beijerinckiaceae were the most abundant and dominant at day 0. All these phylotypes showed a lowering trend in time decreasing from day 0 to d7 with the only exceptions of the Beijerinckiaceae that were higher at d7 than at day 0.
Of the Betaproteobacteria, unclassified genera belonging to Alcaligenaceae and Oxalobacteraceae were the most abundant families. Their relative abundances were constant along the time and showed higher differences among the months than the sampling days (average values of relative abundance were 2.4%, 4.5% and 0.5% in July, August and October respectively).
Firmicutes accounted for 12%–38% of the OTUs from d2 to d7 of the PT process. At day 0, Firmicutes were present only in October’s samples at 7%, then increased at d2 and remained stable until the end of the process (Fig. 3). Overall, there were 10 Firmicutes in the 26 most abundant phylotypes found during the PTBs (Fig. 4). Most of these were thermophilic bacteria such as Bacillus, Thermoactinomycetaceae, Brevibacillus and Paenibacillus6. These themophilic phylotypes had never been detected at day 0, appeared at d2 and reached their higher values at d5 when their presence accounted for 29.9%, 19.4% and 8.5% in July, August and October respectively.
The Bacteroidetes constituted another dominant phylum detected in all the samples (Fig. 3). Bacteroidetes abundance at d0 was in the range of 1.4%–17.0%, decreased at d2, d3 and d5 to a range of 0.7%–8.1% and reached the highest values at d7 (between 8.1% and 21.2%). The most abundant genera belonging to this phylum were Flavobacterium and Sphingobacterium.
The abundance of Actinobacteria was 1.0%–3.3% at day 0 and increased until the end of the process reaching a maximum of 7.5–36.6% relative abundances respectively (Fig. 3). The most abundant phylotypes in the Actinobacteria phylum were Microbacteriaceae and Streptomyces (Fig. 4); in particular, Streptomyces was one of the genera dominating bacterial community biodiversity at d5 and d7.
Cyanobacteria were found at relative abundances higher than 1% only in July and August samples at day 0 and d2, with a maximum at day 0 (16.9% and 4.5% relative abundances in June and July samples respectively). After d2, Cyanobacteria decreased until the end of the process (Fig. 3). The relative abundance of Verrucomicrobia was 0.35%–2.8% at d0 and remained constant during the whole PTB process (Fig. 3). Chtoniobacteraceae was the most abundant bacterial family of the Verrucomicrobia phylum.
Further bacterial phyla had always been found at very low relative abundances (never higher than 1.0%, Fig. 3).
Fungal community structure
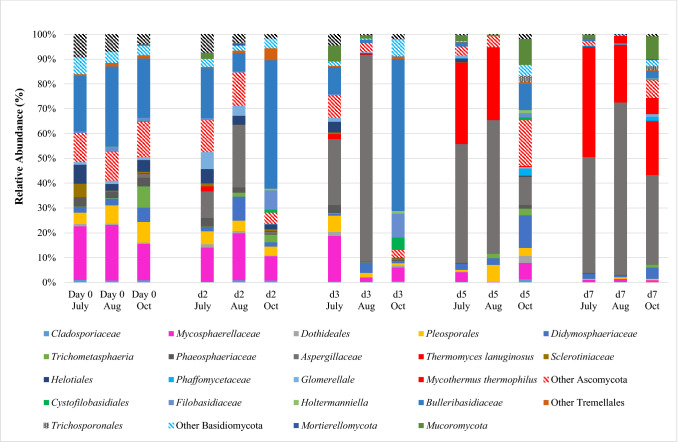
Before the PTB started, the fungal community in the herbs (day 0 in Fig. 5), was dominated by Mycosphaerellaceaes (Mycosphaerella, Ramularia and Zymoseptoria genera), representing the 21.5%, 21.4% and 15.0% of the total in July, August and October respectively, and Bulleribasidiaceaes (Vishniacozyma, and Dioszegia genera) representing the 22.3%, 32.4% and 23.4% of the total in July, August and October respectively. Other fungal taxonomic groups, mainly belonging to the Ascomycota phylum, were detected in lower relative abundance (lower than 10%). After two days (d2), the Aspergillaceae family was emerging, mainly constituted by the Aspergillus genus with traces of Penicillium in 13 out of the 90 samples. Aspergillaceae dramatically increased from day 0 (always less than 1%) to d2 in July and August trials (10.6% and 24.9% respectively), and after d3 they became the most dominant fungi (26.6% and 83.2% respectively). By October, Bullerobasidiaceae was always the dominant fungal family at d2 and d3. After five days (d5 in Fig. 5), Bulleribasidiaceaes (lower than 10.6%) and Mycosphaerellaceae (lower than 6.5%) relative abundances decreased sharply. Aspergillaceae kept their rising trend, remaining the dominant fungal family in July and August trials (47.9% and 56.6% respectively), but they represented only the 11.4% of the fungal relative abundance in October. Other thermophilic species emerged in July and August: the Trichocomaceae mainly constituted by Thermomyces lanuginosus whose relative abundance was never higher than 2.1% in the first three days and then suddenly increased to 32.8% and 23.2% in July and August respectively.
Figure 5.
Fungi taxa groups (genus level or above) composition, in mean relative abundance, of herb samples as revealed by high-throughput sequencing analysis. The samples were collected in triplicate from the same pool, for five days (d0, d2, d3, d5, and d7) at 5 and 40 cm of depth, in July, August (Aug) and October (Oct). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
After seven days (d7 in Fig. 5), the fungal community was totally dominated by Aspergillaceae (46.6%, 69.2% and 35.9% in July, August and October respectively) and Thermomyces lanuginosus (44.2%, 23.3% and 21.6% in July, August and October respectively). None of the OTUs was predominant throughout all samples.
Volatiles organic compounds (VOCs) released during the PTBs
After raw GCxGC–MS data deconvolution and pre-processing, the three dataset (July, August and October) consisted of 722, 1105 and 815 volatiles respectively. As already reported by Narduzzi et al.7, the majority of the VOCs are not in common among the months. The identified volatiles through all the three months’ datasets were matched using their InchiKey, and produced a table consisting of 295 VOCs present in all the months (Table S2). As shown in the top part of the Fig. 6, there is a cluster of 34 compounds that are the most representative of all PTB samples because contributing to the 85% of the total VOCs mass emissions. In the heatmap Fig. 6, the samples from the same month clustered together. Moreover, within each month, the samples split in two different clusters according to the stages already identified in the microbial analysis. The first cluster is composed by the samples of d1, d2 and d3 (1st stage) and the second cluster by the samples of d4, d5, and d6 (2nd stage). Looking at the differences in the days within each batch, the d1, d2 and d3 samples were richer in aliphatic hydrocarbons (heneicosane, hexadecane, tetradecane and 3-methyltridecane), alpha-terpineol, and estragole. By contrast, the d4, d5 and d6 samples were richer in 1-methylnaphthalene, nonanal, 2-nonanone, 3-octanol, m-xylene, 2,6-dimethylheptadecane and 2-ethyl-hexanol.
Figure 6.
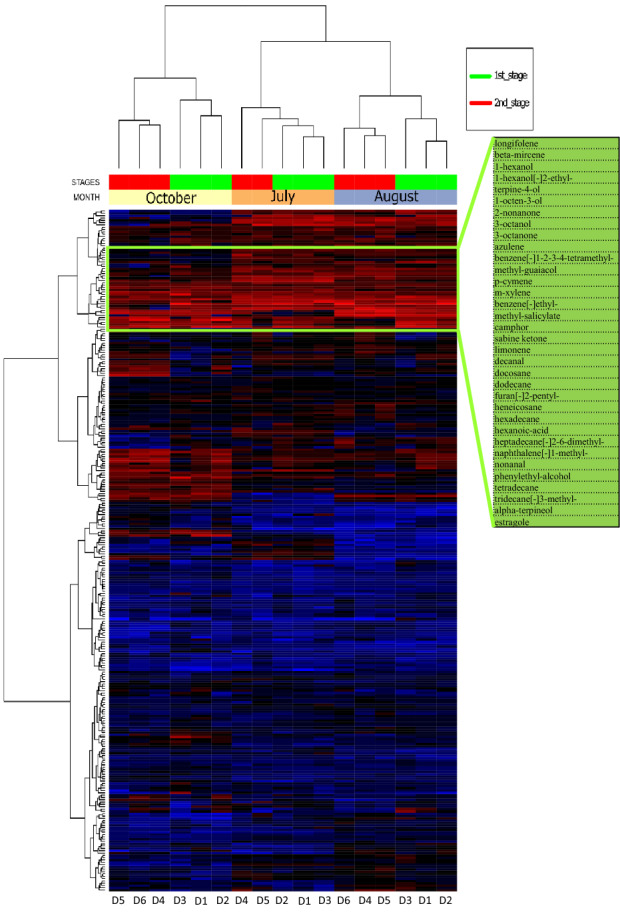
Heatmap and hierarchical clustering based on the normalized quantities of the identified VOCs, for PTB herbs in the six days (d1, d2, d3, d4, d5, and d6) and three months (July, August and October) of sampling. The highest content is in red and the lowest in blue. The values have been UV scaled and clustered according the Ward algorithm. The list of the 34 compounds highlighted in the upper side represents the most abundant (core) VOCs found. In July, d6 is missing as the sample has not been collected.
Discussion
The PTB is documented as a natural healthy treatment for human rheumatic diseases2, 5, but this is the first study investigating the microbiota and VOCs occurring in this process. The only similar processes are the self-heating ones occurring during composting8, 9 or silage fermentation10. In order to establish the different stages of PTB, the temperature, pH, microbial composition, microbial abundance, and VOCs data had been recorded for seven days in three different months: July, August, and October.
The PTB temperature was always increasing from ambient temperature (25 °C) until more than 50 °C with great variation among the different months, in particular the temperature rise over 50 °C, was very fast in August and October (never longer than 2.5 days). This trend is very similar to other self-heating processes where the initial mesophilic phase (until 42 °C) may last for only a few hours or a couple of days8. The different temperature trends among the three months suggested heterogeneity of the PTB that could reflect differences in herbs composition, compaction and aeration of the pile as well as in the active microorganism populations. The day-by-day addition of the bulking herbs determined an aeration and regeneration effect within the PTB pile, increasing substrate and oxygen availability and likely determining a boosting effect on the microbial growth and the heating process11.
In general, the pH remained in a neutral range in all the PTBs. In July and August, after d3, it increased significantly and then stabilized in a range between 7.4 and 7.8 that is suitable for most microorganisms’ life8, 9. No pH drop was recorded, contrary to what occurring in further self-heating processes where microorganisms rapidly break down soluble and easily degradable carbon sources, resulting in a pH drop from 6.5 to 6.0 due to organic acids accumulation8. This is in agreement with bacteria and fungi counts that were two orders of magnitude lower than in other self-heating processes8 and with the volatile acids data that where not among the core of the recorded VOCs. Only in October, the pH showed a (non-significant) drop, between d2 and d3. It was probably caused by the generation of organic acids from Aspergillaceae species11 that showed high relative abundance (Fig. 5) and counts (Table 1, moulds counts) already after three sampling days in October.
Regarding microbial counts, bacteria and fungi showed lower counts (at least two orders of magnitudes) than microbial population of other self-heating processes8, with the exception of October when microbial counts were at higher amount. In general, aerobic bacteria dominated the PTB microbiota (around three orders of magnitude higher than other microbial groups). Therefore, aerobic bacteria were the main actors leading the PTB self-heating, probably by means of an aerobic exothermic step already known as a bio-oxidative process typical of the first phase of the composting process12. According to our results, the microbial evolution throughout the PTB was a very dynamic process, and the temperature did not result as deleterious for bacteria as reported for other self-heating process12. Microorganisms’ kept growing albeit temperature reached values higher than 50 °C, showing that most of them were thermo-tolerant.
Fungi counts showed a different trend when compared with bacteria. In July and August, the yeasts and moulds showed always very low counts following a significant decreasing trend at d5 and d7, similar to the one reported by Woolford10 in silage fermentation. This decrease is probably due to the difficult adaptation to the high temperatures. Therefore, temperature could be a selective factor for fungal community in PTBs, similarly to the results of Zhang et al.13, that reported high temperature as the driving factor affecting the fungal population in waste pile composting.
Alpha and beta diversities were analysed to study the bacterial and fungi community richness in samples obtained during the seven days of the PTB process. The first 3 days showed the highest bacterial and fungal alpha-diversity. After d5, the bacterial and fungal diversity significantly decreased which could be due to a high competitive selection, occurring in environments with a high nutrient complexity containing similar species competing for the same resources14. The depletion of organic substrates could lead to a gradual disappear of less competitive taxa and to a microbial community convergence towards stable composition of limited diversity14. Both the OTUs number and Shannon diversity index of fungi showed a significant constant lowering trend indicating that fungal diversity decreased with temperature rise, confirming that the high temperature had a certain degree of selectivity against fungi.
All these results suggested the existence of two-stages in the PTB process after day 0. The 1st stage was characterised by temperature boosting until d3, higher microbial diversity and a predominance of the bacterial development among the microbial community, and the 2nd stage, from d5 to d7, characterised by almost constant high temperature, a minor bacterial and fungi diversity and higher pH than 1st stage.
The presence of a two-stage process has been clearly confirmed also by the beta-diversity. The microbial communities in the first three days showed significant differences when compared to the last two days of the PT process. No significant difference had been detected at sampling positions 5 and 40 cm suggesting a certain homogeneity in the pool of the PTB that makes the samples representative of the overall complete batch whatever the deep of sampling. Although the succession of microbial communities occurred with some differences in July, August and October, the overall microbial community was similar according to the PTB stage in all the months of sampling. This suggests that there is a similar bacterial and fungal phylogenetic turnover pattern during the PTB processes, alongside a comparable progression. The significant differences among the microbial communities of July, August and October could be associated to differences in herb composition, compaction and aeration of the pile that leads to differences in microbial composition and temperature trend.
Analysing the structure of the bacterial community developing during the PTB process, the predominant bacteria were belonging to Proteobacteria phylum and, in particular, Erwinia was the most abundant genus in the 1st stage. This genus is known to include phyto-pathogenic bacteria and it was never found in previous study about self-heating microbial processes8–10, confirming the peculiarity of PTBs. Acinetobacter, Pseudomonas and Xanthomonadaceae were also bacterial phylotypes belonging to Proteobacteria phylum whose sequences were abundant in the PTBs samples. Acinetobacter and Pseudomonas are generally recognised as mesophilic genera and probably this explain their higher abundance in the 1st stage and the drop following the temperature increase, as reported in others self-heating processes8, 9. Bacteria belonging to Xanthomonadaceae family can live both in thermophilic and extremely thermophilic environments9, and probably for this reason, they were dominant in the 2nd stage of the PT process when temperatures were always over 50 °C. The Firmicutes relative abundance, mainly represented by Bacillales (Bacillus and Thermoactynomycetaceae), increased after two days and then remained a constant presence in the 2nd stage, similarly to Partanen et al.15 that showed Bacillales prevalence during the thermophilic phase of self-heating composting processes. Bacteroidetes constituted another phylum detected in all the samples. The most abundant genera belonging to Bacteroidetes were Flavobacterium and Sphingobacterium, present along the overall PTB process without differences between 1st and 2nd stages. Flavobacterium and Sphingobacterium are known as active bacteria during the composting of lignocellulosic materials16, 17. The most abundant phylotypes belonging to Actinobacteria phylum were Microbacteriaceae and Streptomyces. In particular Streptomyces abundance was higher in the 2nd stage in agreement with previous studies that found this species predominant during the thermophilic stage of self-heating organic compounds processes16, 18 as they can tolerate high temperatures and are able to degrade cellulose and lignin8, 15, 19.
Analysing the structure of the fungal community developed during the PTB process, the dominant groups, before the temperature rises, were Mycosphaerellaceaes and Bulleribasidiaceaes families. Mycosphaerellaceae is one of the largest family of Ascomycota and comprises many important crop pathogens20. Bulleribasidiaceae belong to Basidiomiycota phylum and is constituted by well cold-adapted genera recovered in cold terrestrial areas like Himalayas, Andes, and European high mountains21, 22; their cold adaptation could explain why they were found in very high amount at day 0 in the fresh-cut grass from Viote area where the average year temperature is about 5.5 °C23. After two days, a new fungal family emerged when the temperatures rises: the Aspergillaceae, a family able to resist high temperatures24 and known to be one of the dominant fungi families in self-heating processes8. After three days, other thermophilic species emerged to become dominant in the 2nd stage like Thermomyces lanuginosus, whose presence in thermophilic stage of self-heating process was already documented8. Aspergillaceae and Thermomyces lanuginosus predominance in the 2nd stage was probably due to their thermophilic attitude and their potential in producing thermostable enzymes25, 26.
The majority of the VOCs released during PTB process were not in common from among the months, confirming the heterogeneity of the herbs constituting the PTBs. The different concentrations and the presence of unique VOCs in the three months of analysis might be due, not only to different herbs and microbial populations in the collected samples, but also to the competitive binding of the volatiles on the used sampling tubes, limiting the repeatability of VOCs quantification. The VOCs analysis revealed the presence of 34 compounds constituting the core of the most abundant compounds in the PTB process. Most of them were monoterpenes and sesquiterpenes such as longifolene, myrcene, camphor, limonene, terpinen-4-ol and cymene. They were constantly present in all the samples and are known to be the major chemical compounds of plant essential oils27, 28. They have been reported to possess a wide range of bacterial inhibitory potential in particular against pathogens like Escherichia and Staphylococcus genera27 and terpinen-4-ol in particular can be used in the therapy of skin infections29. The antibacterial activity of these compounds may justify the lower counts found both for bacteria and fungi when compared with generic self-heating processes8, and the absence in the PTBs of pathogenic bacterial genera, such as Escherichia, Staphylococcus, Serratia, Enterobacter and Klebsiella, that are very common in self-heating composting process8.
Considering the 1st stage of the PTB process, there was a higher concentration of aliphatic hydrocarbons (heneicosane, hexadecane, tetradecane and 3-methyltridecane), alpha-terpineol, and estragole than at the 2nd stage. By contrast, the 2nd stage was richer in 1-methylnaphthalene, nonanal, 2-nonanone, 3-octanol, m-xylene and 2,6-dimethylheptadecane. The compounds more representative of the 1st stage of PTB process were of plant origin, because common component of the volatile oils of the herbs and flowers constituting the PTB pile and in some cases have been shown to have significant health effects. Aliphatic hydrocarbons are constituents of Arnica montana buds28 and Hypericum maculatum flowers30. Estragole is a natural anise odor component of plant essential oils and has in vivo anti-inflammatory action31. Natural alpha-terpineol is a lilac odor compound that can be produced by fungal (Penicillum sp.) or bacterial (Pseudomonas sp.) biotransformation of limonene32 and has antioxidant and anti-proliferative potential33. Regarding the 2nd stage of PTB process, with the exception of nonanal that is one of the main constituents of Arnica montana buds28, the most representative compounds were of microbial origin and had been recorded among Aspergillum volatile metabolites such as 1-methylnaphthalene, 3-octanol, xylene and 2,6-dimethylheptadecane34, and Bacillus volatile metabolites such as 2-nonanone35.
In conclusion, this is the first study about the PTB process, which is a traditional treatment for rheumatic diseases widespread in some Alpine areas, but, due to its peculiarity and limited area of diffusion, has never been previously studied. The PTB process has some similarities with compost and silage self-heating processes such as: (i) the rapid rise in temperature over 50 °C, (ii) the presence of two stages, one more mesophilic and one more thermophilic, (iii) the rise in pH from 6.5 to about 7.5, (iv) the development of some microbial species in the 2nd stage, such as Streptomyces, Xanthomonadaceae, Aspergillaceae and Thermomyces. More relevant are the differences that confirm the originality of the PTB process when compared with compost and silage self-heating processes such as: (i) the lower counts of both bacteria and fungi, (ii) the absence of pathogenic bacteria, (iii) the dominance of Erwinia sp. in the 1st stage which could be the main actor of self-heating in PTB process, (iv) the dominance of plant terpenoids among the VOCs, some of them with health characteristics27, 29, (v) a 1st mesophilic stage characterized by plant VOCs with some beneficial effects27, 29, 31, and (vi) a 2nd thermophilic stage characterized by the Aspergillaceae presence and the production of VOCs of microbial origin.
Nevertheless, it is worth noting that there are some fungi, such as Aspergillaceae, that could be pathogenic to humans in particular within the 2nd stage of the PTB process and could be a real problem for operators and customers36.
Therefore, additional studies should be conducted to evaluate the possibility of extending the 1st stage in order to lower the development of Aspergillaceae and to promote the extended release of plant VOCs with beneficial characteristics. The extension of the 1st stage could be achieved setting up a system of better temperature and aeration control, also increasing the amount of bulk herb added each day (for instance from 10% to 20%). The pH and temperature record could be a direct measure to check the transition from the first to the second stage because both values are easy to detect and increase significantly between the two stages.
Based on the results of this investigation, the authors strongly recommend taking action to reduce the workers’ exposure to Aspergillaceae and harmful fungi that could be raised during total removal of the herbs at the end of the process.
Methods
The phyto-thermal bath (PTB)
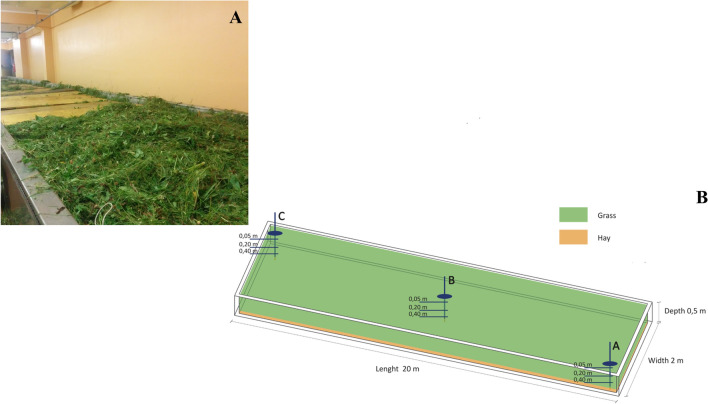
In order to study the real PTB process, traditional PTB has been performed in the original facility bath at Garniga Terme Spa, following the original protocol. The grass was harvested on July and August 2016, in Viote, (Trento, Italy, N 46°01′16″; E 11°02′11″), elevation 1540 m above sea level, sandy loam soil type, with average year precipitation of 576 mm/m2, and average year temperature of 5.5 °C24. The grass had been mowed and directly baled to be transported to the thermal facility of Garniga (Trento, Italy, S 23°38′57″; W 46°37′19″) where the PTB process was carried out following the protocol described by Narduzzi et al.7. The facility has four 20 m3 stainless steel open baths (Fig. 7) and is designed to host about 1 ton of grass for each cycle in each bath. The herbs had been layered in the baths to create a 45 cm tall pile, and covered with wooden bars. Adequate aeration of the pile had been kept by daily adding a 10% of bulk herbs, turning every day the herbs through a dedicated machine. After seven days, the PTB process within the baths had been considered finished and the material removed.
Figure 7.
(A) Picture of the grass stockpile at the beginning of the PTB with a (B) schematic representation of the grass bath.
Sample collection
About 50 g samples had been collected throughout the PTB process from the pool in three different months: July, August and October. After completion of the PTB pile, samples were collected at the arrival of the herbs at the facility (day 0) and then at d2, d3, d5 and d7 of each trial. To obtain a representative sample of the cell, for each sampling day, the pile was sampled in three different points, along the cell diagonal: two edges (A and C Fig. 7B) and in the middle (B in Fig. 7B), at 5 and 40 cm depths. Each sample was stored into two sterile 50 mL Falcon tube. One tube was immediately frozen in liquid nitrogen and stored at − 80 °C until DNA extraction. The second tube was kept at 4 °C and used for the immediate pH and microbial counts analyses.
Temperatures and pH determination
Temperature had been recorded for seven days from day 0 to d7, each hour at 20 cm in the middle of the pool bath (0.20 m point B in Fig. 7B). The temperature of the room and of the PTB pile has been determined using a Testo 175 T2 (Testo limited, Alton, UK). The pH was determined from suspensions of fresh grass samples in 0.9% sodium chloride using a pH electrode (Crison Instruments, Barcelona, Spain).
Microbial analyses
All the herbs samples were subjected to microbial analysis and prepared by adding 180 g of sterile (autoclaved for 15 min at 121 °C) peptone water to 20 g of sample. The sample was run twice for 60 s at the setting “normal speed” in a laboratory blender (Stomacher Seward 3500; Worthing, West Sussex, UK). Since this was the first work on PTBs’ microbial counts, it was not clear which kind of viable microbial groups we could find nor their respective abundance. In order to acquire more information, several microbial groups (mesophilic and psychrotrophic aerobic bacteria, mesophilic and thermophilic lactic acid bacteria, Enterobacteria, Acetobacteria, yeasts and moulds) were determined on ten subsamples as a preliminary experiment. Ten decimal dilution series were made and all plated, in order to determine the microbial groups present and the order of magnitude. Enumeration of the present microbial groups (total aerobic bacteria, mesophilic and thermophilic lactic acid bacteria, Enterobacteria, yeasts and moulds) were made from the last four decimal dilutions where the microbial group had shown to be present according the results from the preliminary experiment. Total bacteria had been cultured aerobically on Plate Count Agar (PCA) at 30 °C for 24 h. Lactic acid bacteria (LAB) were cultivated under anaerobic conditions on MRS agar plates for 72 h at 30 °C for mesophilic and 45 °C for thermophilic bacteria. Enterobacteria had been cultured on violet red bile glucose agar (VRBGA) following the overlay method at 37 °C for 24 h. Fungi had been cultured on WL agar plates, and aerobically incubated at 25 °C. Yeasts and moulds had been counted after two and five days respectively. All media had been purchased by Oxoid (Oxoid Ltd, Cambridge, UK).
DNA extraction
All the samples collected, had been subjected to DNA extraction, performed with MoBio DNA Power Soil kit (Mo Bio Laboratories Inc., Qiagen Venlo, Netherlands) following vendor’s instructions. All DNA samples were purified by PowerClean DNA Clean-up Kit (Mo Bio Laboratories Inc.) and quantified by Nanodrop3300 Fluorospectrometer (Thermo Scientific, Waltham, USA) using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen Life Technology, Thermo Scientific).
Miseq library preparation and Illumina sequencing
Amplicon library preparation, quality and quantification of pooled libraries were performed at the Sequencing Platform, Fondazione Edmund Mach (FEM, San Michele a/Adige, Italy). Briefly, for each sample, a 464-nucleotide sequence of the V3-V4 region37, 38, of the 16S rRNA gene (Escherichia coli positions 341 to 805) and ITS1F (5′-GTTTCCGTAGGTGAACCTGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) specific for the ITS1-5.8S fungi region39 were amplified for bacteria and fungi respectively. Unique barcodes were attached before the forward primers to facilitate the pooling and subsequent differentiation of samples. To prevent preferential sequencing of the smaller amplicons, the amplicons were cleaned using the Agencourt AMPure kit (Beckman coulter, Brea, USA) according to the manufacturer’s instructions; subsequently, DNA concentrations of the amplicons were determined using the Quant-iT PicoGreen dsDNA kit (Invitrogen Life Technology) following the manufacturer’s instructions. In order to ensure the absence of primer dimers and to assay the purity, the generated amplicon libraries quality was evaluated by a Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) using the High Sensitivity DNA Kit (Agilent). Following the quantitation, cleaned amplicons had been mixed and combined in equimolar ratios. Pair-end sequencing using the Illumina MiSeq system (Illumina, San Diego, USA) had been carried out at CIBIO (Center of Integrative Biology) – University of Trento (Trento, Italy).
Illumina data analysis and sequences identification by QIIME2
Raw paired-end FASTQ files were demultiplexed using idemp (https://github.com/yhwu/idemp/blob/master/idemp.cpp) and imported into Quantitative Insights Into Microbial Ecology (Qiime2, version 2018.2). Sequences were quality filtered, trimmed, de-noised, and merged using DADA240. Chimeric sequences had been identified and removed via the consensus method in DADA2. Representative bacterial sequences had been aligned with MAFFT and used for phylogenetic reconstruction in FastTree using plugins alignment and phylogeny41, 42. Alpha and beta diversity metrics had been calculated using the core-diversity plugin within QIIME2 and visualised by emperor43. For bacteria, beta diversity metric had been calculated using the beta-diversity plugin within QIIME2 and the—metric command was set on weighted_normalized_unifrac. For bacteria, taxonomic and compositional analyses were carried on by using plugins feature-classifier (https://github.com/qiime2/q2-feature-classifier). A pre-trained Naive Bayes classifier based on the Greengenes 13_8 99% Operational Taxonomic Units (OTUs) database which had been previously trimmed to the V4 region of 16S rDNA, bound by the 341F/805R primer pair, was applied to paired-end sequence reads to generate taxonomy tables. For fungi, sequences were classified to the species-level using a 97 or 99% threshold using UNITE dynamic classifier version 8.0 released for QIIME2 (UNITE QIIME release for Fungi. Version 18.11.2018. UNITE Community. https://doi.org/10.15156/BIO/786334). The data generated by MiSeq Illumina sequencing were deposited in the NCBI Sequence Read Archive (SRA) and are available under Ac. Number PRJNA588404 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA588404/https://www.ncbi.nlm.nih.gov/bioproject/PRJNA588404/).
Volatiles determination
The chemical volatiles released during the cycles were measured following the method by Narduzzi et al.7. Briefly, two Tenax cartridges (147, Radiello, Supelco), pre-conditioned under nitrogen flow for 2 h at 300 °C, were inserted in their dedicated air filters and placed on the top of the wooden bars of the grass baths for 24 h, to passively collect the volatiles released by the fermentation process. After 24 h, each cartridge was sealed in its glass tube and stored in the dark at 4 °C until analysis. In July, the d6 sample had been lost.
The cartridges were desorbed using a Thermal Desorber (Unity 2, Markes) and desorbed gases were injected in a comprehensive GCxGC–TOF–MS (LECO Corporation, St. Joseph, MI, USA). The chromatograms were acquired and de-convoluted using the ChromaTOF software (Version 4.22; LECO) pre-processed using the package GCxGC Leco.analyser7 and normalized using the VSN algorithm44 through the web service NOREVA45.
Statistical analysis
A normality test (Shapiro–Wilk W) was performed, as well as a nonparametric tests (Kruskal–Wallis) analyzing the day of collection as independent variables and the microbial plate counts as dependent variables. All the tests on plate counts were performed using the STATISTICA data analysis software system, version 9.1 (StatSoft, Inc. 2010 ww.statsoft.com).
Differences in diversity indices (OTUs number and Shannon diversity index) of different samples were tested by Kruskal–Wallis test by a plug-in implemented in QIIME2. The overall structural changes of bacterial and fungal communities were visualised by principal coordinates analysis (PCoA) based on Weighted Unifrac for bacterial and Jaccard Distance Matrix for fungi community. In order to aid the ordination of the samples across time, the day of sampling has been considered as a custom-axis in both PCoAs by means of a plug-in implemented in QIIME2. The statistical significance of communities among all samples was assessed via the non-parametric PERMANOVA (permutational multivariate analysis of variance) by means of plug-in implemented in QIIME2.
The heat map of the volatiles compounds dataset was built using Euclidean score distance and Ward clustering algorithm in R environment.
Supplementary information
Acknowledgements
The authors would like to thank Garniga Terme S.p.A. for providing the facilities, the operators and the precious information to replicate the original and traditional process of the phytothermal baths.
Author contributions
E.F. participated in the design of the experimental plane, performed pH measurements, Illumina data analysis and sequences identification by QIIME2 as well as extensive manuscript writing. L.N. performed samples collection, temperature determination and volatile analysis. S.C. performed volatile instrumental determination. A.P. performed plate counts analysis and DNA extraction. K.T. participated in the design of the experimental plane. A.B. participated in the conception of the project. F.M. participated in the conception of the project and in the design of the experimental plane. All authors partecipated in the extensive revision of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74586-9.
References
- 1.Petraglia A, Bellisai B, Manica P, Fioravanti A. Phytobalneotherapy (“Hay Baths”): between tradition and modern medicine. Presse Therm. Clim. 2009;146:283–290. [Google Scholar]
- 2.Tenti S, Manica P, Galeazzi M, Fioravanti A. Phytothermotherapy in fibromyalgia and osteoarthritis: Between tradition and modern medicine. Eur. J. Integr. Med. 2013;5(3):248–253. doi: 10.1016/j.eujim.2012.12.002. [DOI] [Google Scholar]
- 3.Miori R, Paolazzi G, Albertazzi R. Phytothermotherapy with fermenting alpine grass in knee osteoarthritis: mid-long term results. Reumatismo. 2008;60:282–9. doi: 10.4081/reumatismo.2008.282. [DOI] [PubMed] [Google Scholar]
- 4.Talamucci P, Piemontese S, Coser P. La gestione dell'erba per i “Bagni di fieno”. Risultati di un triennio di studi. Rep. CEALP. 1996;6(2):17–62. [Google Scholar]
- 5.Fioravanti A, Bellisai B, Iacoponi F, Manica P, Galeazzi M. Phytothermotherapy in osteoarthritis: a randomized controller clinical trial. J. Altern. Complement. Med. 2011;17:407–412. doi: 10.1089/acm.2010.0294. [DOI] [PubMed] [Google Scholar]
- 6.Wrighton KC, et al. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2:1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- 7.Narduzzi L, et al. Applying novel approaches for GC × GC-TOF-MS data cleaning and trends clustering in VOCs time-series analysis: following the volatiles fate in grass baths through passive diffusion sampling. J. Chromatogr. B. 2018;1096:56–65. doi: 10.1016/j.jchromb.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Ryckeboer J, et al. A survey of bacteria and fungi occurring during composting and self-heating processes. Ann. Microbiol. 2003;53:349–410. [Google Scholar]
- 9.Insam H, de Bertoldi M, et al. Microbiology of the composting process. In: Díaz LF, et al., editors. Waste Management Series No 8. Compost Science and Technology. Amsterdam: Elsevier; 2007. pp. 25–48. [Google Scholar]
- 10.Woolford, M. K. Bacterial developments their implications for silage production and aerobic stability In Biotechnology in the Feed Industry, Proceedings of the 14”’ Annual Symposium (Nottingham University Press, 1998).
- 11.Gerardi, M. H. The microbiology of anaerobic digesters. In Fermentation 43–50 (Wiley, New Jersey, 2003).
- 12.Ferreira JA, Mahboubi A, Lennartsson PR, Taherzadeh MJ. Waste biorefineries using filamentous ascomycetes fungi: present status and future prospects. Bioresour. Technol. 2016;215:334–345. doi: 10.1016/j.biortech.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, et al. Effects of physico-chemical parameters on the bacterial and fungal communities during agricultural waste composting. Bioresour. Technol. 2011;102:2950–2956. doi: 10.1016/j.biortech.2010.11.089. [DOI] [PubMed] [Google Scholar]
- 14.Ghoul M, Mitri S. The ecology and evolution of microbial competition. Trends Microbiol. 2016;24:833–845. doi: 10.1016/j.tim.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Partanen P, Hultman J, Paulin L, Auvinen P, Romantschuk M. Bacterial diversity at different stages of the composting process. BMC Microbiol. 2010 doi: 10.1186/1471-2180-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-González JA, et al. Dynamics of bacterial microbiota during lignocellulosic waste composting: studies upon its structure, functionality and biodiversity. Bioresour. Technol. 2015;175:406–416. doi: 10.1016/j.biortech.2014.10.123. [DOI] [PubMed] [Google Scholar]
- 17.Karadag D, et al. Profiling of bacterial community in a full-scale aerobic composting plant. Int. Biodeter. Biodegr. 2013;7:85–90. doi: 10.1016/j.ibiod.2012.10.011. [DOI] [Google Scholar]
- 18.Jurado M, et al. Exploiting composting biodiversity: study of the persistent and biotechnologically relevant microorganisms from lignocellulose-based composting. Bioresour. Technol. 2014;162:283–293. doi: 10.1016/j.biortech.2014.03.145. [DOI] [PubMed] [Google Scholar]
- 19.Taylor CR, et al. Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 2012;9:521–530. doi: 10.1111/j.1365-2672.2012.05352.x. [DOI] [PubMed] [Google Scholar]
- 20.Braun U, Nakashima C, Crous PW. Cercosporoid fungi (Mycosphaerellaceae) 1 species on other fungi Pteridophyta and Gymnospermae. IMA Fungus. 2013;4(2):265–345. doi: 10.5598/imafungus.2013.04.02.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buzzini P, Turchetti B, Yurkov A. Extremophilic yeasts: the toughest yeasts around? Yeast. 2018;35:487–497. doi: 10.1002/yea.3314. [DOI] [PubMed] [Google Scholar]
- 22.Sannino C, Tasselli G, Filippucci S, Turchetti B, Buzzini P, et al. Yeasts in nonpolar cold habitats. In: Buzzini P, et al., editors. Yeasts in Natural Ecosystems: Diversity. Berlin: Springer; 2017. pp. 366–396. [Google Scholar]
- 23.https://www.climatrentino.it/binary/pat_climaticamente/osservatorio_trentino_clima/Libro_CLIMA_.1355921619.pdf.
- 24.Miao Y, et al. Genome-wide transcriptomic analysis of a superior biomass-degrading strain of A-fumigatus revealed active lignocellulose-degrading genes. BMC Genom. 2015;16(4599):1–20. doi: 10.1186/s12864-015-1658-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S, Madlala AM, Prior BA. Thermomyces lanuginosus: properties of strains and their hemicellulases. FEMS Microbiol. Rev. 2003;27(1):3–16. doi: 10.1016/S0168-6445(03)00018-4. [DOI] [PubMed] [Google Scholar]
- 26.McHunu NP, et al. Xylanase Superproducer: genome sequence of a compost-loving thermophilic fungus thermomyces lanuginosus strain SSBP. Genome Announc. 2013 doi: 10.1128/genomeA00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tariq S, et al. A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug resistant microbial pathogens. Microb Pathog. 2019 doi: 10.1016/j.micpath.2019.103580. [DOI] [PubMed] [Google Scholar]
- 28.Kowalski R, Sugier D, Sugier P, Kołodziej B. Evaluation of the chemical composition of essential oils with respect to the maturity of flower heads of Arnica montana L. and Arnica chamissonis Less. cultivated for industry. Ind. Crops Prod. 2015;76:857–865. doi: 10.1016/j.indcrop.2015.07.029. [DOI] [Google Scholar]
- 29.Loughlin R, Gilmore BF, McCarron PA, Tunney MM. Comparison of the cidal activity of tea tree oil and terpinen-4-ol against clinical bacterial skin isolates and human fibroblast cells. Lett. Appl. Microbiol. 2008;46:428–433. doi: 10.1111/j.1472-765X.2008.02334.x. [DOI] [PubMed] [Google Scholar]
- 30.Stojanovic G, Palic R, Tarr CH, Reddy CM, Marinkovic O. n-alkanes and fatty acids of Hypericum perforatum, Hypericum maculatum and Hypericum olympicum. Biochem. Syst. Ecol. 2003;31:223–226. doi: 10.1016/S0305-1978(02)00076-5. [DOI] [Google Scholar]
- 31.Bicas JL, Fontanille P, Pastore GM, Larroche C. A bioprocess for the production of high concentrations of R-(+)-a-terpineol from R-(+)-limonene. Process Biochem. 2010;45:481–486. doi: 10.1016/j.procbio.2009.11.007. [DOI] [Google Scholar]
- 32.Bicas JL, Neri-Numa IA, Ruiz AL, De Carvalho JE, Pastore GM. Evaluation of the antioxidant and antiproliferative potential of bioflavors. Food Chem Toxicol. 2011;49:610–615. doi: 10.1016/j.fct.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Bezerra-Rodrigues L, et al. Anti-inflammatory and antiedematogenic activity of the Ocimum basilicum essential oil and its main compound estragole: in vivo mouse models. Chem. Biol. Interact. 2016;257:14–25. doi: 10.1016/j.cbi.2016.07.026. [DOI] [PubMed] [Google Scholar]
- 34.Siddiquee S, Al Azad S, Abu Bakar F, Naher L, Vijay Kumar S. Separation and identification of hydrocarbons and other volatile compounds from cultures of Aspergillus niger by GC–MS using two different capillary columns and solvents. J. Saudi Chem. Soc. 2015;19:243–256. doi: 10.1016/j.jscs.2012.02.007. [DOI] [Google Scholar]
- 35.Fincheira P, Parra L, Mutis A, Parada M, Quiroz A. Volatiles emitted by Bacillus sp. BCT9 act as growth modulating agents on Lactuca sativa seedlings. Microbiol. Res. 2017;203:47–56. doi: 10.1016/j.micres.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Kanj A, Abdallah N, Soubani AO. The spectrum of pulmonary aspergillosis. Resp. Med. 2018;141:121–131. doi: 10.1016/j.rmed.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 37.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardes M, Bruns TD. ITS primers with enhanced specificity for higher fungi and basidiomycetes: application to identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 40.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- 45.Li B, et al. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.