Abstract
The pathophysiology of polycystic ovary syndrome (PCOS) is characterized by granulosa cell (GC) dysfunction. m6A modification affects GC function in patients with premature ovarian insufficiency (POI), but the role of m6A modification in PCOS is unknown. The purpose of the prospective comparative study was to analyse the m6A profile of the luteinized GCs from normovulatory women and non‐obese PCOS patients following controlled ovarian hyperstimulation. RNA m6A methylation levels were measured by m6A quantification assay in the luteinized GCs of the controls and PCOS patients. Then, m6A profiles were analysed by methylated RNA immunoprecipitation sequencing (MeRIP‐seq). We reported that the m6A level was increased in the luteinized GCs of PCOS patients. Comparative analysis revealed differences between the m6A profiles from the luteinized GC of the controls and PCOS patients. We identified FOXO3 mRNA with reduced m6A modification in the luteinized GCs of PCOS patients. Selectively knocking down m6A methyltransferases or demethylases altered expression of FOXO3 in the luteinized GCs from the controls, but did not in PCOS patients. These suggested an absence of m6A‐mediated transcription of FOXO3 in the luteinized GCs of PCOS patients. Furthermore, we demonstrated that the involvement of m6A in the stability of the FOXO3 mRNA that is regulated via a putative methylation site in the 3’‐UTR only in the luteinized GCs of the controls. In summary, our findings showed that altered m6A modification was involved in up‐regulated expression of FOXO3 mRNA in the luteinized GCs from non‐obese PCOS patients following controlled ovarian hyperstimulation.
Keywords: FOXO3, luteinized granulosa cells, mRNA decay, N6‐methyladenosine, polycystic ovary syndrome
1. INTRODUCTION
Polycystic ovary syndrome (PCOS) is a common endocrine disorder and a common cause of female infertility in women of reproductive age. 1 One of the main characteristics of the syndrome is granulosa cell (GC) dysfunction, which largely contributes to hyperandrogenism, abnormal follicle development and anti‐Mullerian hormone excess. 2 Aberrant gene expression profile was found in GCs of PCOS patients. 3 Multiple differential expressed genes were high related to the pathogenesis of PCOS. However, the regulation mechanisms of these genes were largely unknown.
Forkhead Box O3 (FOXO3) plays important roles in diverse cellular processes including apoptosis, metabolism, cell proliferation and cell survival. 4 FOXO3 is regulated at several mechanistic levels, such as transcriptional activity, cellular localization, mRNA expression and protein stability. Oxidative stress induces FOXO activation and nuclear translocation by c‐Jun N‐terminal kinase (JNK) or mammalian Ste20‐like kinase 1 (MST1) activation despite phosphorylation by protein kinase B (Akt). 5 , 6 After energy deprivation, the increased AMP/ATP ratio leads to AMP‐activated protein kinase (AMPK) activation. 7 AMPK activates FOXO3 activity by phosphorylation at six different residues. 8 Insulin‐like growth factor‐1 (IGF‐I)/insulin and phosphoinositide 3‐kinase (PI3K)/Akt signalling pathway inactivate FOXOs by phosphorylation resulting in FOXOs nuclear exclusion. 9 Zhao et al reported that elevated wnt family member 5A (WNT5a) activates PI3K/Akt signalling in GCs of PCOS patients. 10 By contrast, Rice and colleagues showed metabolic insulin resistance in GCs of PCOS patients, suggesting that PI3K/Akt signalling is impaired. 11 While many studies have been focused on the regulation of FOXO3 activity by post‐translational modifications, the regulation of FOXO3 expression is largely unknown. FOXO3 is one of the differential expressed genes which have higher expression in PCOS. 3 Its overexpression in GCs is associated with higher apoptosis in PCOS. 12 To date, the factors that up‐regulate and activate FOXO3 in GCs of PCOS patients are still unclear.
m6A is the most prevalent modification of mRNA in higher eukaryotes. 13 The modifications are reversible and dynamically regulated by the m6A modulators. m6A modulators consist of the ‘writers’, the ‘erasers’ and the ‘readers’. Briefly, m6A modifications are installed by the ‘writers’ (Methyltransferase like 3 (METTL3) and Methyltransferase like 14 (METTL14)), removed by the ‘erasers’ (Fat mass and obesity‐associated protein (FTO) and alkB homolog 5(ALKBH5)), and recognized by the ‘readers’ (YTH domain‐containing proteins and Eukaryotic initiation factor 3 (eIF3)). 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 m6A modification has diverse biological functions such as nuclear RNA export, RNA splicing, protein translation regulation and RNA decay. In the YTHDF2‐mediated decay pathway, mRNAs with increased m6A abundance in 3’‐UTR are down‐regulated due to reduced RNA stability. 19
Several human diseases are associated with altered m6A modification. m6A levels of sperm RNA are elevated in patients with asthenozoospermina. 23 Increased m6A levels in the granulosa cells were reported in patients with premature ovarian insufficiency (POI), affecting apoptosis and cell proliferation in GCs. 24 Aberrant m6A modification, through the effects on RNA metabolism, plays critical roles in a variety of cancers. 25 However, whether m6A modification plays a role in the pathogenesis of PCOS is unknown.
Given that m6A modification affects the GC function in patients with POI, 24 we suggested alteration of the m6A profile in the luteinized GCs of PCOS patients, which may account for the dysregulation of certain key genes for PCOS. Here, we showed the differences of m6A distribution between the luteinized GCs of normovulatory women and PCOS patients following controlled ovarian hyperstimulation. We identified FOXO3 mRNA with differential m6A peaks, targeted for decay by YTHDF2. Our results indicated that hypomethylated FOXO3 mRNA caused the dysregulation of FOXO3 in luteinized GCs from PCOS patients following controlled ovarian hyperstimulation.
2. SUBJECTS AND METHODS
2.1. Subjects
Forty‐three control patients with tubal factor infertility or male infertility and 36 PCOS patients were recruited in the Reproductive Medicine Center at the First Affiliated Hospital of Wenzhou Medical University between December 2017 and August 2019. Inclusion criteria for control patients were aged between 25 and 35 years, having a regular menstrual cycle, serum testosterone (T) level < 2 nmol/L, 18.5 < body mass index (BMI) < 27, having a normal ovarian reserve and a normal uterus. PCOS was diagnosed according to the Rotterdam revised criteria. 26 The inclusion criteria for PCOS patients were aged between 25 and 35 years, having oligo‐ or anovulation, serum testosterone level > 2 nmol/L, 18.5 < BMI < 27, anti‐Müllerian hormone (AMH) > 7 ng/mL and having a normal uterus. The exclusion criteria were smoking, systemic diseases, endometriosis, abnormal serum level of prolactin, dysfunctional thyroid or having previous long‐term medication use.
This study was approved by the Ethics Committees of the First Affiliated Hospital of Wenzhou Medical (Approved number: YS2016‐063 and YS2019‐046). Written informed consent was obtained from all the participants.
2.2. Measurement of hormones
Plasma levels of AMH, luteinizing hormone (LH), follicle‐stimulating hormone (FSH), T and estradiol (E2) were collected and measured by chemiluminescence immunoassay (CLIA) between day 3 and day 5 of the menstrual cycle before the controlled ovarian stimulation. Serum fasting glucose levels and fasting insulin levels were measured by an oxidase‐peroxidase method and a CLIA method, respectively. The inter‐assay coefficients of variation were 6.1% for AMH, 7.2% for LH, 5.3% for FSH, 10.0% for T, 8.9% for E2, 4.3% for glucose and 5.4% for insulin. The intra‐assay coefficients of variation were 3.8% for AMH, 5.3% for LH, 4.6% for FSH, 8.1% for T, 6.8% for E2, 2.1% for glucose and 2.9% for insulin. Insulin in follicle fluids was detected by enzyme‐linked immunosorbent assay (ELISA, R&D Systems). HOMA‐IR was calculated by the formula (HOMA‐IR = fasting insulin (mIU/L) × fasting glucose (mmol/L)/ 22.5). 27
2.3. Transvaginal ultrasonography
Ultrasound examination was performed on the 3rd‐5th day of the menstrual cycle with a 7 MHz transvaginal transducer (LOGIC 400, General Electric Medical Systems) to calculate follicle number. The basal antral follicle count (AFC) was assessed as the sum of all follicles of 2‐10 mm in diameter. The polycystic ovary was defined as 12 or more AFC in each ovary.
2.4. Ovarian stimulation and GCs isolation
All participants underwent controlled ovarian stimulation with gonadotrophin‐releasing hormone (GnRH) agonist long protocol. Briefly, patients received 0.1 mg/day of GnRH agonist (Decapeptyl, Ferring, Germany) from day 20 of a spontaneous menstrual cycle until the day of human chorionic gonadotrophin (hCG) injection. When pituitary down‐regulation was achieved (usually after 14 days of GnRH agonist injection), ovarian stimulation was initiated with a minimum of 150 IU/day of recombinant human FSH (rhFSH, GonalF, Merck Serono). The dosages of rhFSH were adjusted according to serum E2 levels and follicle growth of the patients. 250 µg of recombinant hCG (Ovidrel, Merck Serono) was administered, when at least three follicles were >17 mm in diameter. Transvaginal aspiration was performed 34‐36 hours later to retrieve oocytes and follicular fluids. The follicular fluids from follicles >14 mm in diameter and without obvious blood contamination were collected. The follicular fluids were centrifuged at 340 g for 8 minutes to pellet the GCs. The cell pellets were resuspended in 1× PBS solution (Gibco), overlaid on 40%/80% gradient solution (PureCeption, SAGE) and centrifuged at 320 g for 20 minutes. GCs in the interface were collected and washed with 1× PBS solution.
2.5. RNA m6A quantification
Total RNA was isolated using the Total RNA Kit II Kit (Omega Bio‐Tek). Polyadenylated RNA was purified from total RNA using the GenElute mRNA Miniprep Kit (Sigma). 500 ng total RNA or polyadenylated RNA was used to determine the RNA m6A methylation levels using the EpiQuik m6A RNA Methylation Quantification Kit (Epigentek). Briefly, a standard curve was prepared according to the manufacturer's instruction. The RNA samples were coated on the strip wells, followed by incubation with capture antibody. After washes with washing buffer, detection antibody and enhance solution were added separately. Then, the signals were developed by the detection solution. The RNA m6A levels were quantified by the absorbance at 450 nm, and the m6A contents were calculated based on the standard curve.
2.6. Methylated RNA immunoprecipitation sequencing (MeRIP‐seq) and MeRIP‐qPCR
MeRIP was performed using Magna MeRIP m6A Kit (Millipore, Billerica, MA, USA) following the manufacturer's instruction. Briefly, 50 µg of total RNA was precipitated using 2.5 volumes of 100% ethanol, one‐tenth volumes of 3 mol/L sodium acetate and 1 mg/mL glycogen. Then, the RNA was resuspended, fragmented and immunoprecipitated with anti‐m6A antibody or normal mouse IgG. 10% of fragmented RNA was keep as input. The immunoprecipitated RNA was eluted by competition with m6A sodium salt and recovered using miRNeasy Mini Kit (QIAGEN, Hilden, Germany).
The recovered RNA was treated with DNase I (0.1 U/µL, Thermo Scientific, Waltham, MA, USA) and reverse‐transcribed by random hexamer primer and RevertAid Reverse Transcriptase (Thermo Scientific). Subsequently, second strand cDNA was synthesized using DNA Polymerase I and RNase H (Thermo Scientific). Sequencing library was constructed using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) and purified using AMPure XP system (Beckman Coulter). Library quality was assessed on a Bioanalyzer 2100 (Agilent). The clustering of the index‐coded samples was generated on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3‐cBot‐HS (Illumina). Library preparations were sequenced on a Hiseq 4000 Platform (Illumina).
Real‐time quantitative RT‐PCR analysis (qRT‐PCR) was carried out with the samples with anti‐m6A antibody, the samples with normal IgG, and input samples in triplicates. Ct of the samples with anti‐m6A antibody were normalized to input by subtracting the Ct of input from the Ct of IP samples: ∆Ct = CtIP‐(Ctinput‐Log2[Input Dlilution Factor]). Then, the per cent of input for each IP sample was calculated: % Input = 2−ΔCt(normalized IP). The primers for MeRIP‐qPCR were listed in Table S1.
2.7. MeRIP‐seq data analysis
Sequencing data were mapped to the reference genome (ftp://ftp.ensembl.org/pub/release‐94/fasta/homo_sapiens/dna/) using BWA mem (version 0.7.12). m6A peaks were identified by peak finding algorithm in MACS2 (version 2.1.0). The threshold of enrichment was set at q < 0.05. Differential peak analysis was based on the fold enrichment of peaks between the PCOS group and the control group. When the odds ratio > 2, a differential peak was determined. The statistics of pathway enrichment of genes with differential peaks in KEGG pathway was tested using KOBAS software.
2.8. Primary cell culture and RNA interference
The purified luteinized GCs were cultured in high‐glucose Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 10% foetal bovine serum (FBS, Gibco) and 1% penicillin‐streptomycin (Gibco) at 37°C in a humidified incubator under 5% CO2. The siRNAs against YTHDF2, METTL3, METTL14, FTO and ALKBH5 were obtained from GenePharma Corporation (China). When the confluence reached 80%, the cells were dissociated and seeded in 24‐well plates (105 cells/well). One day later, the cells were transfected with siRNA (GenePharma) targeting negative control, YTHDF2, METTL3, METTL14, FTO or ALKBH5 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. The sequences of siRNA were listed in Table S2.
2.9. Real‐time quantitative RT‐PCR analysis
For the GCs collected from follicular fluids, total RNA was extracted using the Total RNA Kit II Kit (Omega Bio‐Tek). For the GCs cultured in 24‐well plates, total RNA in each well was extracted using MicroElute Total RNA Kit (Omega Bio‐Tek). The total RNA was reverse‐transcribed by the PrimeScript RT reagent Kit with gDNA Eraser (Takara) according to the manufacturer's instruction. Real‐time quantitative RT‐PCR analysis (qRT‐PCR) was performed on a Corbett Rotor Gene 6000 Real‐time Cycler using TB Green Premix Ex Taq II (Takara). Each sample was analysed in triplicates to obtain the threshold cycle number. GAPDH mRNA was used as an internal control for normalization. Relative expression levels were quantified using comparative 2‐∆∆Ct method and expressed as fold changes relative to the controls. The sequences of the primers were listed in Table S1.
2.10. Western blot analysis
The total protein was extracted from the cultured GCs using RIPA buffer (Beyotime) containing 1mM PMSF (Beyotime). The concentration of the protein was measured using BCA protein assay kit (Solarbio). The cell extracts were separated on a 12% SDS‐PAGE Gel (ExpressPlus PAGE Gel, GenScript) and transferred to methanol‐activated polyvinylidenefluoride membranes. The membranes were blocked with 5% non‐fat milk powder for 1 hour and incubated with the primary antibody for 2 hour at room temperature. The mouse anti‐FOXO3 (Proteintech, 66428‐1‐Ig) or mouse anti‐Beta‐actin (Proteintech, 60008‐1‐Ig) antibody was used as primary antibody. Then, the membranes were incubated with HRP‐conjugated affinipure goat anti‐mouse IgG (H + L) (Biosharp, BL1001A) for 1 hour at room temperature. The signals were developed with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific).
2.11. Plasmid construction and dual‐luciferase reporter assay
The proximal 3’‐UTR of FOXO3 was amplified and inserted into 3’‐downstream of the Renilla luciferase gene of the psiCHECKTM‐2 Vector between XhoI and NotI restriction sites. The motif of m6A binding was mutated by fusion PCR with a pair of complementary primers containing the A‐to‐T mutation. The primary culture human GCs were transfected with wild‐type m6A motif reporter plasmid or mutant m6A motif reporter plasmid using Lipofectamine 2000. The Firefly and Renilla luciferase activities were measured using the Dual‐Luciferase Reporter Assay System (Promega) 48 hours later. The Renilla luciferase activities were normalized to Firefly luciferase activities.
2.12. Statistical analysis
Data are expressed as the means ± SD. The normality and homogeneity of variance of the data were assessed. Differences between groups were determined by one‐way analysis of variance (ANOVA) followed by the Tukey multiple comparison test using SPSS 16.0 software package. Significance was set at P < 0.05.
3. RESULTS
3.1. m6A levels in the luteinized GCs of the controls and PCOS patients
To investigate the role of m6A modification in the luteinized GCs of PCOS patients, we recruited normovulatory controls and PCOS patients in our reproductive centre (Table S3). The insulin‐glucose parameters indicated that the PCOS patients had insulin resistance in the present study. We examined m6A levels in the total RNA of the luteinized GCs collected following ovarian hyperstimulation by RNA m6A quantification. m6A levels were twofold higher in the luteinized GCs of PCOS patients compared with the controls (Figure 1A). We further isolated polyadenylated RNA from the total RNA and examined the m6A levels. Consistently, the m6A levels of polyadenylated RNA were increased in the luteinized GCs of PCOS patients (Figure 1B).
Figure 1.
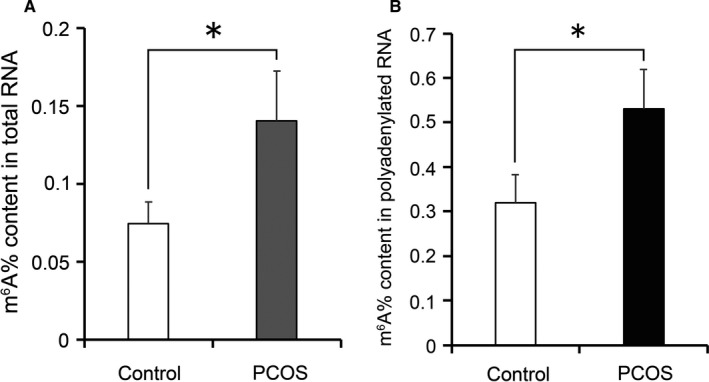
Quantification analysis of m6A modification in the luteinized GCs of the controls and PCOS patients. The m6A content in total RNA (A) and mRNA (B) of GCs were detected by colorimetric assay. Bars represent means ± SD, n = 5. *P < 0.05 vs the control
3.2. Differential m6A modification in the luteinized GCs of the controls and PCOS patients
We conducted MeRIP‐seq to analyse the transcriptome‐wide distribution of m6A modification in GCs. 2764 and 3405 m6A peaks were identified in the controls and PCOS patients, respectively (Table S4). We selected 8 transcripts with m6A peaks to verify the sequencing data by MeRIP‐qPCR. The qRT‐PCR results were in agreement with the sequencing data (Figure S1). We analysed the differential peaks based on the fold enrichment of peaks of the two groups. 1719 and 2195 peaks were distinct in the controls and PCOS patients, respectively, while 996 peaks were overlapped in both groups (Figure 2A and Table S5).
Figure 2.
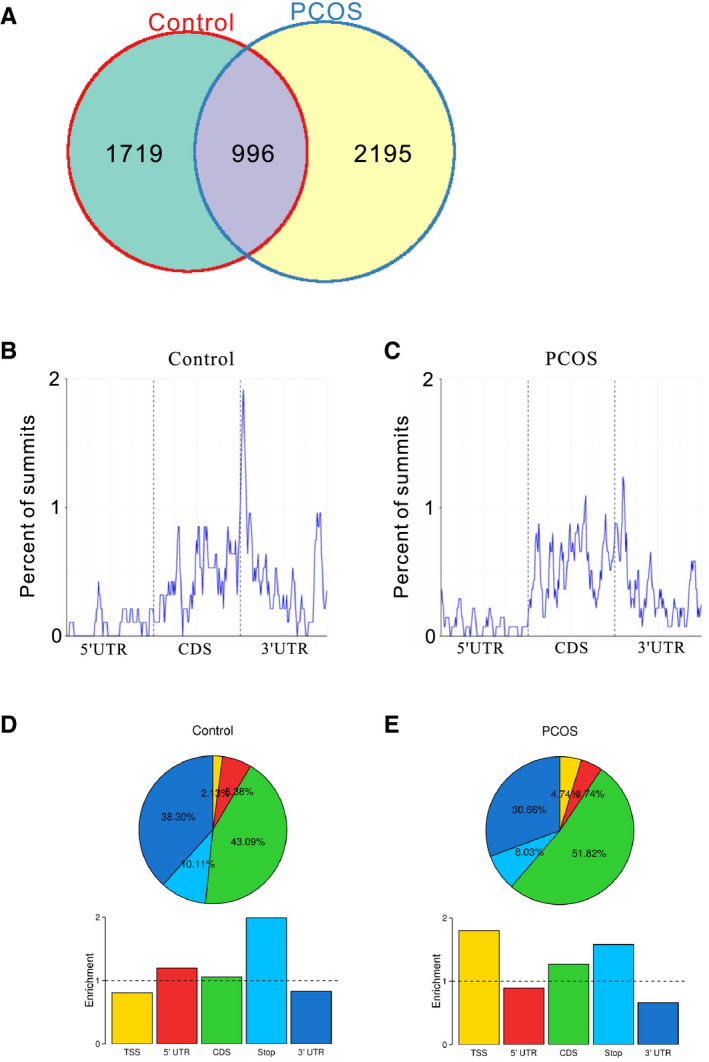
Distribution of m6A peaks in the luteinized GCs of the controls and PCOS patients. A, Comparison of the m6A peaks between the controls and PCOS patients. (B and C) Metagene profiles of enrichment of m6A peaks across mRNA transcriptome of the controls (B) and PCOS patients (C). (D and E) Distribution and enrichment of the m6A peaks within different gene regions in the GCs of the controls (D) and PCOS patients (E). TSS, transcription start sites; 5’‐UTR, 5’‐untranslated region; CDS, coding sequence; Stop, translation termination sites; 3’‐UTR, 3’‐untranslated region
Similar to the other studies, m6A peaks were strongly enriched around the stop codon in the controls (Figure 2B,D). 28 , 29 , 30 However, m6A peaks in PCOS patients showed a less prominent enrichment around the stop codon and increased locations to the CDS (coding sequence) and the TSS (transcription start sites) regions (Figure 2C,E).
3.3. m6A modification targets the FOXO3 transcript in the luteinized GCs of the controls but not PCOS patients
We suggested that m6A modification plays a role in dysregulation of PCOS‐associated genes. We examined the expression of AMH, AR, FOXO1 and FOXO3 in our study. Consistent with the previous studies, 2 , 3 , 31 the expression of AMH, AR, FOXO1 and FOXO3 were up‐regulated in the luteinized GCs of PCOS patients compared with the controls (Figure 3A). To screen potential candidates, we compared the expression of the m6A methyltransferases and demethylases in the luteinized GCs of the controls and PCOS patients. The expression of METTL3, METTL14, FTO and ALKBH5 were all elevated in PCOS patients compared with the controls (Figure 3B). We selectively knocked down the expression of METTL3, METTL14, FTO or ALKBH5 in the luteinized GCs of the controls and PCOS patients, respectively. The knockdown efficiency was confirmed by qRT‐PCR analysis (Figure S2). Due to differential m6A peaks identified in FOXO1 and FOXO3 transcripts, we examined the expression of FOXO1 and FOXO3 in these cells. In the controls, depletion of METTL3 or METTL14 increased the expression of FOXO3, and silencing of FTO suppressed the expression of FOXO3, suggesting m6A modification regulated FOXO3 mRNA degradation (Figure 3C,E,G). However, depletion of the m6A methyltransferases or demethylases did not affect the expression of FOXO3 in PCOS patients (Figure 3D,F,H), which indicated that effects of m6A modification on FOXO3 mRNA were cell‐specific. Our results also showed that knockdown of methyltransferases or demethylases did not affect the expression of FOXO1 in GCs (Figure 3C‐J). The common target gene SBK1 was used as a positive control to confirm the results. Collectively, these results demonstrated that selectively knocking down m6A methyltransferases or demethylases did not alter the expression of FOXO3 in the luteinized GCs of PCOS patients, but did so in the controls, suggesting an absence of m6A‐regulated transcription of FOXO3 in the luteinized GCs of PCOS patients.
Figure 3.
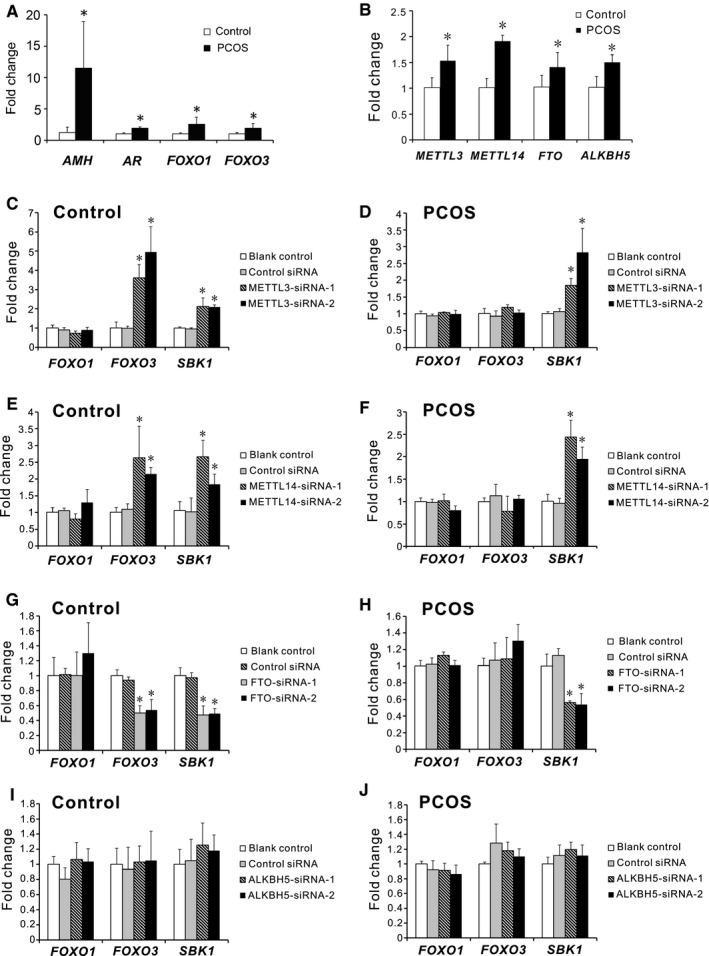
Involvement of m6A modification in regulation of FOXO3 mRNA in the luteinized GCs of the controls but not PCOS patients. A, mRNA levels of PCOS‐associated genes (AMH, AR, FOXO1 and FOXO3) in the luteinized GCs from the controls and PCOS patients. B, Expression of the methyltransferases (METTL3 and METTL14) and demethylases (FTO and ALKBH5) in the luteinized GCs of controls and PCOS patients. C‐J, The effects of selectively knockdown of the methyltransferases or demethylases on the expression of FOXO1, FOXO3 and SBK1 in the luteinized GCs of the controls and PCOS patients. GCs of the controls and PCOS patients were collected, cultured and subjected to METTL3‐siRNA (C,D), METTL14‐siRNA (E,F), FTO‐siRNA (G,H) or ALKBH5‐siRNA (I,J), respectively. qRT‐PCR analysis was performed to examine the mRNA levels of FOXO1, FOXO3 and SBK1. Data were expressed as fold changes relative to the blank controls. Bars represent means ± SD, n = 5. *P < 0.05 vs the blank controls
3.4. m6A modification regulates the stability of the FOXO3 transcript via the YTHDF2‐mediated decay pathway in the luteinized GCs of the controls
Our MeRIP‐seq showed a differential m6A peak in 3’‐UTR and near the stop codon of the FOXO3 transcript (Figure 4A). We performed MeRIP‐qPCR to confirm the differential m6A levels on the m6A target site (Figure 4B). To determine how m6A modification regulates the FOXO3 transcript, we analysed the sequence of the m6A peak. A putative m6A site was identified within the m6A peak of 3’‐UTR at position + 2279 (+1 relative to the translation start site) (Figure 4C). As YTHDF2 is the main binding protein that account for the decay of m6A‐modified mRNAs, we next examined the effects of YTHDF2‐knockdown on FOXO3 expression in human GCs. Depletion of YTHDF2 increased the amount of FOXO3 transcript, and total protein levels of FOXO3 (Figure 4D,E). The knockdown efficiency was checked by qRT‐PCR (Figure S2). To assess the functionalities of the m6A site in the 3’‐UTR of FOXO3 transcript, we constructed a reporter plasmid bearing FOXO3‐3’‐UTR with the putative m6A site mutated (Figure 4F). FTO‐siRNA suppress the luciferase activities of the reporter plasmid with the wild‐type m6A site (Figure 4G). However, mutation of the m6A site reversed the suppressive effects of FTO‐siRNA on the luciferase activities (Figure 4G). The presence of the m6A readers in human GCs was showed by a previous study. 32 To exclude the potential of lacking the m6A readers in GCs of PCOS patients, we examined the expression of the m6A readers in PCOS. The expression of YTHDF2 was significantly increased in the luteinized GCs of PCOS patients (Figure S3). These results indicated that m6A modification regulated FOXO3 mRNA decay through the m6A site in 3’‐UTR in the luteinized GCs of the controls.
Figure 4.
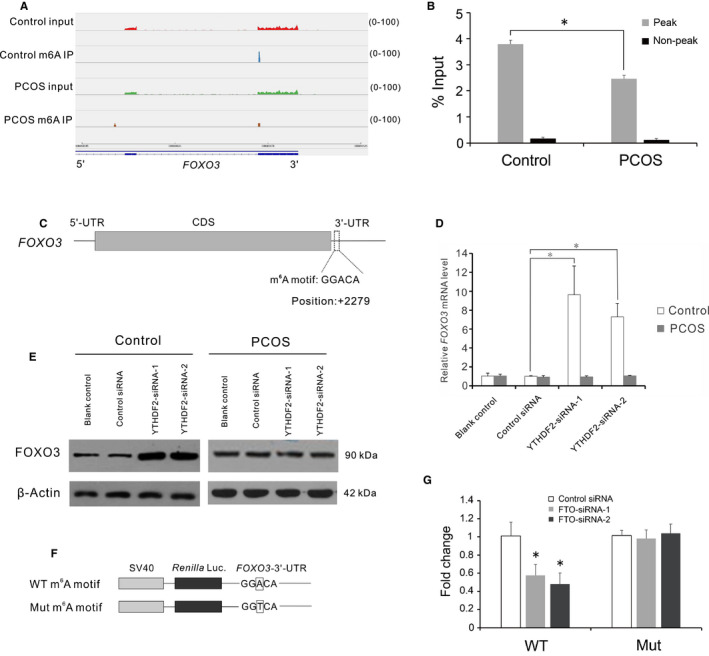
Differentially methylated FOXO3 transcript was regulated via the YTHDF2‐mediated decay pathway in the luteinized GCs of the controls. A, m6A peaks distribution across 3’‐UTR of the FOXO3 transcript. The Y‐axis shows read number. Blue boxes indicate exons, and blue line indicates introns. B, qRT‐PCR analysis of m6A peak region and non‐peak region in the 3’‐UTR of the FOXO3 transcript. Bars represent means ± SD, n = 5. *P < 0.05 for the differences between the indicated groups. C, Schematic representation of the position of m6A motif in the 3’‐UTR of the FOXO3 transcript. D, The mRNA level of FOXO3 in the YTHDF2‐knockdown cells. Bars represent means ± SD, n = 5. *P < 0.05 for the differences between the indicated groups. E, Western blot analysis of FOXO3 total protein level in the YTHDF2‐knockdown cells. F, Wild‐type or m6A motif mutant (A‐to‐T mutation) FOXO3‐3’‐UTR fused with Renilla luciferase reporter. G, Relative luciferase activity of the FOXO3‐3’‐UTR with wild‐type or mutant m6A motif after cotransfection with negative control siRNA or FTO‐siRNA Renilla luciferase activity was normalized to firefly luciferase activity. Bars represent means ± SD, n = 3. *P < 0.05 vs the control siRNA group
4. DISCUSSION
Increasing evidence showed that m6A modification play important roles in various physiological processes. 17 , 33 , 34 , 35 , 36 Elevated or decreased m6A level was associated with various human diseases. 23 , 24 , 25 We showed that the level of m6A modification was higher in the luteinized GCs of PCOS patients than that in the controls. Aberrant m6A levels affect gene expression and biological functions. However, whether human diseases alter m6A profiles is largely unknown. Here, we showed that not only elevated m6A level but also altered m6A profile are associated with PCOS in the luteinized GCs.
The site selection of m6A modification is largely unknown. miRNAs regulate m6A formation at corresponding target sites. 37 Altered miRNAs and other associated factors partially explain the differences of m6A profiles among cell types. 37 Furthermore, m6A profile was altered when cells exposed to heat shock, ultraviolet radiation or signalling molecules. 38 Acute stress regulates a fraction of genes with m6A modification in the cortex. 33 The GCs of PCOS patients present different cell characteristics. Cell survival, proliferation rate and responsiveness to FSH were altered in the GCs of PCOS patients. 39 , 40 Differentially expressed miRNAs were also identified in the GCs of PCOS patients. 41 , 42 Moreover, abnormal serum AMH, androgen, activin A and follistatin were observed in PCOS patients. 2 , 43 , 44 All these alterations may account for the altered m6A profile in PCOS. We identified 2195 differential m6A peaks in the luteinized GCs of PCOS patients compared to those of the controls. KEGG analysis showed that differential peaks were enriched in metabolic pathway (Figure S4). Whether m6A modification is involved in the regulation of these transcripts with the differential m6A modification needs further investigation.
Insulin resistance is prevalent in PCOS patients and plays a crucial role in metabolic abnormalities. Although obesity increases risks for insulin resistance, several studies have demonstrated that both obese and non‐obese PCOS patients had impaired insulin signalling. 45 , 46 In the present study, the non‐obese PCOS patients had significantly higher insulin 0, insulin 120 and HOMA‐IR compared with the controls. Moreover, the insulin levels in follicular fluid were notably higher in the non‐obese PCOS patients. These indicated that the PCOS patients in our study had insulin resistance. As FOXO proteins mediate the effects of insulin signalling on metabolism, 47 it is likely that altered expression of FOXO3 is involved in the defects of insulin signalling in the luteinized GCs of PCOS patients. Here, we demonstrated an absence of m6A‐mediated FOXO3 mRNA destabilization which altered expression of FOXO3 in the luteinized GCs of PCOS patients. Our findings have offered a potential mechanism for insulin resistance.
It is well known that insulin activates PI3K/Akt signalling to induce the nuclear exclusion of FOXO3. 9 However, the effects of PI3K/Akt signalling on GCs of PCOS patients are controversial. Zhao et al showed that WNT5a expression was elevated in GCs of PCOS patients. 10 Although they have demonstrated that WNT5a increased inflammation and oxidative stress via PI3K/Akt/NF‐κB signalling in human GCs, they did not confirm that WNT5a activated PI3K/Akt/NF‐κB signalling in GCs of PCOS patients. As insulin resistance is attributed to defects in PI3K/Akt signalling, metabolic insulin resistance in GCs of PCOS patients suggests impaired PI3K/Akt signalling. 9 , 11 Thus, it is possible that abnormalities in other signalling cascade rather than PI3K/Akt signalling contributed to chronic inflammation in GCs of PCOS patients. To date, the factors that up‐regulate and activate FOXO3 remain unclear in GCs of PCOS patients. Here, we demonstrated that YTHDF2 mediated FOXO3 mRNA decay via a putative m6A site in the 3’‐UTR in the luteinized GCs of the controls. In contrast to the findings in the controls, although the expression of YTHDF2 was elevated in the luteinized GCs of PCOS patients, FOXO3 mRNA was hypomethylated and irresponsive to m6A modification. Our results showed an absence of m6A‐based regulation of FOXO3 expression in the luteinized GCs of PCOS patients. Taken together, we inferred that altered m6A modification caused upregulation of FOXO3 in the luteinized GCs of PCOS patients following controlled ovarian hyperstimulation.
In summary, the present study provided m6A profiles of normovulatory women and PCOS patients in luteinized GCs following controlled ovarian hyperstimulation. We demonstrated that altered m6A modification disturbed the regulation of FOXO3 expression in the luteinized GCs of PCOS patients. However, the site selection mechanism of m6A in PCOS needs to be explored in future studies.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTION
Shen Zhang: Conceptualization (lead); Data curation (lead); Funding acquisition (equal); Investigation (lead); Methodology (lead); Writing‐original draft (lead); Writing‐review & editing (lead). Wenli Deng: Conceptualization (supporting); Data curation (supporting); Investigation (supporting); Methodology (supporting); Writing‐original draft (supporting); Writing‐review & editing (supporting). Qiongyou Liu: Investigation (supporting); Methodology (supporting); Writing‐review & editing (supporting). Peiyu Wang: Data curation (supporting); Investigation (supporting); Methodology (supporting). Wei Yang: Investigation (supporting); Methodology (supporting); Writing‐review & editing (supporting). Wuhua Ni: Funding acquisition (equal); Project administration (lead).
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Table S1
Table S2
Table S3
Table S4
Table S5
ACKNOWLEDGEMENTS
We thank our director Prof. Xuefeng Huang for supporting the project. We are indebted to Yan Li for help with the follicular fluid collection. The work was supported by grants from the National Natural Science Foundation of China (81903293 to WN), National Key R&D Program of China (2017YFC1001604) and Natural Science Foundation of Zhejiang Province (LQ19H040003 to SZ).
Zhang S, Deng W, Liu Q, Wang P, Yang W, Ni W. Altered m6A modification is involved in up‐regulated expression of FOXO3 in luteinized granulosa cells of non‐obese polycystic ovary syndrome patients. J Cell Mol Med. 2020;24:11874–11882. 10.1111/jcmm.15807
Contributor Information
Shen Zhang, Email: shenzhang@cqmu.edu.cn, Email: zhangshen09@126.com.
Wuhua Ni, Email: niwuhua228@163.com.
REFERENCES
- 1. Norman RJ, Dewailly D, Legro RS, et al. Polycystic ovary syndrome. Lancet. 2007;370:685‐697. [DOI] [PubMed] [Google Scholar]
- 2. Catteau‐Jonard S, Jamin SP, Leclerc A, et al. Anti‐Mullerian hormone, its receptor, FSH receptor, and androgen receptor genes are overexpressed by granulosa cells from stimulated follicles in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:4456‐4461. [DOI] [PubMed] [Google Scholar]
- 3. Haouzi D, Assou S, Monzo C, et al. Altered gene expression profile in cumulus cells of mature MII oocytes from patients with polycystic ovary syndrome. Hum Reprod. 2012;27:3523‐3530. [DOI] [PubMed] [Google Scholar]
- 4. Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421‐426. [DOI] [PubMed] [Google Scholar]
- 5. Sunayama J, Tsuruta F, Masuyama N, et al. JNK antagonizes Akt‐mediated survival signals by phosphorylating 14‐3‐3. J Cell Biol. 2005;170:295‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lehtinen MK, Yuan Z, Boag PR, et al. A conserved MST‐FOXO signaling pathway mediates oxidative‐stress responses and extends life span. Cell. 2006;125:987‐1001. [DOI] [PubMed] [Google Scholar]
- 7. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greer EL, Oskoui PR, Banko MR, et al. The energy sensor AMP‐activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107‐30119. [DOI] [PubMed] [Google Scholar]
- 9. Hopkins BD, Goncalves MD, Cantley LC. Insulin‐PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat Rev Endocrinol. 2020;16:276‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y, Zhang C, Huang Y, et al. Up‐regulated expression of WNT5a increases inflammation and oxidative stress via PI3K/AKT/NF‐kappaB signaling in the granulosa cells of PCOS patients. J Clin Endocrinol Metab. 2015;100:201‐211. [DOI] [PubMed] [Google Scholar]
- 11. Rice S, Christoforidis N, Gadd C, et al. Impaired insulin‐dependent glucose metabolism in granulosa‐lutein cells from anovulatory women with polycystic ovaries. Hum Reprod. 2005;20:373‐381. [DOI] [PubMed] [Google Scholar]
- 12. Mikaeili S, Rashidi BH, Safa M, et al. Altered FoxO3 expression and apoptosis in granulosa cells of women with polycystic ovary syndrome. Arch Gynecol Obstet. 2016;294:185‐192. [DOI] [PubMed] [Google Scholar]
- 13. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971‐3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jia G, Fu YE, Zhao XU, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7:885‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng G, Dahl J, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507‐519. [DOI] [PubMed] [Google Scholar]
- 17. Hsu PJ, Zhu Y, Ma H, et al. Ythdc2 is an N(6)‐methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li A, Chen Y‐S, Ping X‐L, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505:117‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Zhao B, Roundtree I, et al. N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meyer K, Patil D, Zhou J, et al. 5' UTR m(6)A promotes cap‐independent translation. Cell. 2015;163:999‐1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187‐1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Y, Huang W, Huang J‐T, et al. Increased N6‐methyladenosine in human sperm RNA as a risk factor for asthenozoospermia. Sci Rep. 2016;6:24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ding C, Zou Q, Ding J, et al. Increased N6‐methyladenosine causes infertility is associated with FTO expression. J Cell Physiol. 2018;233:7055‐7066. [DOI] [PubMed] [Google Scholar]
- 25. Liu J, Harada BT, He C. Regulation of gene expression by N(6)‐methyladenosine in cancer. Trends Cell Biol. 2019;29:487‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rotterdam EA‐SPcwg . Revised consensus on diagnostic criteria and long‐term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod. 2003;2004(19):41‐47. [DOI] [PubMed] [Google Scholar]
- 27. Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta‐cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412‐419. [DOI] [PubMed] [Google Scholar]
- 28. Zhou J, Wan JI, Gao X, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer K, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635‐1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li DA, You Y, Bi F‐F, et al. Autophagy is activated in the ovarian tissue of polycystic ovary syndrome. Reproduction. 2018;155:85‐92. [DOI] [PubMed] [Google Scholar]
- 32. Huang B, Ding C, Zou Q, et al. Cyclophosphamide regulates N6‐methyladenosine and m6A RNA enzyme levels in human granulosa cells and in ovaries of a premature ovarian aging mouse model. Front Endocrinol (Lausanne). 2019;10:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Engel M, Eggert C, Kaplick PM, et al. The role of m(6)A/m‐RNA methylation in stress response regulation. Neuron. 2018;99:389‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H‐B, Tong J, Zhu S, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL‐7/STAT5/SOCS pathways. Nature. 2017;548:338‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mendel M, Chen K‐M, Homolka D, et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol Cell. 2018;71(6):986‐1000.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tong J, Cao G, Zhang T, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28:253‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen T, Hao Y‐J, Zhang Y, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289‐301. [DOI] [PubMed] [Google Scholar]
- 38. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485:201‐206. [DOI] [PubMed] [Google Scholar]
- 39. Guedikian AA, Lee AY, Grogan TR, et al. Reproductive and metabolic determinants of granulosa cell dysfunction in normal‐weight women with polycystic ovary syndrome. Fertil Steril. 2018;109:508‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Das M, Djahanbakhch O, Hacihanefioglu B, et al. Granulosa cell survival and proliferation are altered in polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:881‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu BO, Zhang Y‐W, Tong X‐H, et al. Characterization of microRNA profile in human cumulus granulosa cells: Identification of microRNAs that regulate Notch signaling and are associated with PCOS. Mol Cell Endocrinol. 2015;404:26‐36. [DOI] [PubMed] [Google Scholar]
- 42. Sørensen AE, Wissing ML, Englund ALM, et al. MicroRNA species in follicular fluid associating with polycystic ovary syndrome and related intermediary phenotypes. J Clin Endocrinol Metab. 2016;101:1579‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Norman RJ, Milner CR, Groome NP, et al. Circulating follistatin concentrations are higher and activin concentrations are lower in polycystic ovarian syndrome. Hum Reprod. 2001;16:668‐672. [DOI] [PubMed] [Google Scholar]
- 44. Eldar‐Geva T, Spitz IM, Groome NP, et al. Follistatin and activin A serum concentrations in obese and non‐obese patients with polycystic ovary syndrome. Hum Reprod. 2001;16:2552‐2556. [DOI] [PubMed] [Google Scholar]
- 45. Dunaif A, Segal KR, Futterweit W, et al. Profound peripheral insulin resistance, independent of obesity, in polycystic ovary syndrome. Diabetes. 1989;38:1165‐1174. [DOI] [PubMed] [Google Scholar]
- 46. Dunaif A, Finegood DT. Beta‐cell dysfunction independent of obesity and glucose intolerance in the polycystic ovary syndrome. J Clin Endocrinol Metab. 1996;81:942‐947. [DOI] [PubMed] [Google Scholar]
- 47. Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183‐189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Table S1
Table S2
Table S3
Table S4
Table S5


