Abstract
BACKGROUND
Kinesin super family 23 (KIF23) is a member of the KIF family, and it plays an important role in mitosis and cytokinesis. Loss of expression can cause mitotic arrest. The Oncomine database is one of the largest oncogene chip databases in the world, and is an integrated data mining platform for cancer gene information. By querying the database, differences in expression between tumor tissue and normal tissue can be determined.
AIM
To study the expression and prognostic significance of KIF23 in gastric cancer (GC).
METHODS
We used immunohistochemistry to compare the expression of KIF23 in GC and normal gastric tissues. We mined the data on the expression and prognosis of KIF23 in GC using Oncomine and Kaplan–Meier plotter database.
RESULTS
Compared with normal gastric tissues, KIF23 expression was increased in GC tissues, and correlated with T, N, and tumor–node–metastasis stages. Survival analysis showed that patients with high expression of KIF23 had a poor overall survival. There were five studies in the Oncomine database in which expression of KIF23 was significantly higher in GC tissues than in normal gastric tissues (P < 0.05). Kaplan–Meier plotter database analysis showed that recurrence-free survival, overall survival, distant metastasis free survival, and post progression survival of patients with high expression of KIF23 were lower than those of patients with low expression. Further stratified analysis found that prognostic survival indicators worsened in patients with T2 and T3 poorly differentiated adenocarcinoma with high expression of KIF23.
CONCLUSION
KIF23 is highly expressed in GC and is associated with a poor prognosis of patients. It may be of great significance in the diagnosis, treatment, and prognostic evaluation of GC.
Keywords: Gastric cancer, Kinesin super family 23, Immunohistochemistry, Oncomine, Prognosis, Kaplan–Meier plotter
Core Tip: This study investigated the role of the kinesin super family 23 (KIF23) in the gastric cancer (GC), including its expression and prognosis significance in GC. We found that KIF23 is highly expressed in GC and is associated with a poor prognosis of patients. It may be of great significance in the diagnosis, treatment, and prognostic evaluation of GC. We believe that our study makes a significant contribution to the literature as it revealed a novel relationship between KIF23 and GC and may provide a new therapeutic target for treating GC.
INTRODUCTION
Gastric cancer (GC) is one of the most common malignant tumors, with a high mortality rate, ranking third among cancer-related deaths, and bringing a huge economic burden to society[1]. GC is characterized by insidious onset and lack of early clinical symptoms. However, most patients are diagnosed in the middle and advanced stages, which misses the best surgical treatment window and greatly affects prognosis. Surgical resection combined with radiotherapy and chemotherapy is the main treatment method for GC, but due to the characteristics of postoperative recurrence, high metastasis rate, and frequent drug resistance, the 5-year survival rate for GC is only 40%[2,3]. Therefore, for patients with GC, seeking effective diagnosis and treatment is the only way to improve prognosis. Exploration of specific and sensitive biomarkers may help clinicians predict prognosis and clarify the mechanism underlying gastric tumorigenesis.
The kinesin family members (KIFs) are microtubule-associated movement proteins that mediate multiple functions. Their abnormal expression plays an important role in tumorigenesis and development[4,5]. KIF23 is a member of the KIF family, and it plays an important role in mitosis and cytokinesis. Loss of KIF23 expression can cause mitotic arrest[6]. In recent years, research on the role of KIFs in tumors has received increasing attention[7,8]. Wang et al[9] found that KIF15 can promote the proliferation, invasion, and metastasis of pancreatic cancer cells through the MEK–ERK signaling pathway. Zhang et al[10] reported that KIF26B promoted the development of GC by activating the vascular endothelial growth factor pathway. Chen et al[10] confirmed that KIF4B is an independent prognostic factor for hepatocellular carcinoma. KIF23 is a candidate target gene for paclitaxel resistance in GC cells[12].
The Oncomine database is one of the largest oncogene chip databases in the world, and is an integrated data mining platform for cancer gene information. To date, the database has collected 715 gene expression data sets and sample data from 86733 cancerous and normal tissues. By querying the database, differences in expression between tumor tissue and normal tissue can be determined. Clinicopathological factors can also be analyzed to find differentially expressed genes in certain cancers and guide research.
In this study, we used immunohistochemistry to detect the expression level of KIF23 protein in GC and adjacent normal tissues, and analyzed the association between KIF23 protein expression and clinicopathological factors. The Oncomine database and high-throughput prognostic analysis (Kaplan–Meier plotter database) were used to analyze the association between expression of KIF23 mRNA and prognosis in GC, which may provide evidence for further research on the role of KIF23 in GC.
MATERIALS AND METHODS
Patients and specimens
We collected 174 GC specimen tissues and 174 paired non-tumor tissues (> 5 cm from the edge of the primary tumor) at the First Affiliated Hospital of China Medical University from January 2011 to December 2012. Written informed consent was obtained from all patients, and the study was approved by the institutional ethics committee of China Medical University. All specimens were obtained during surgical resection from patients who had not received chemotherapy or radiotherapy prior to surgery. All the patients were classified according to the 8th edition of the tumor–node–metastasis (TNM) classification of the International Union Against Cancer, and there were 18 patients with stage I, 53 with stage II, and 103 with stage III cancer. Among 174 patients, 94 had a tumor size > 4 cm. Patients were followed every 3 mo during the first year and every 6 mo thereafter. The patient or his relatives was called according to the follow-up guidelines of the National Comprehensive Cancer Network. The follow-up ended after December 2017. Overall survival (OS) was defined as the time from surgery to death from any cause or last follow-up.
Immunohistochemistry
All tissue pieces were cut into 4-μm slices, deparaffinized, rehydrated, stained using the Ultrasensitive TM S-P system (KIT-9710; MaiXin, Fuzhou, China), and incubated with anti-KIF23 antibody (1:300, #ab235955; Abcam, Cambridge, MA, USA) overnight at 4 °C. The tissue slices were incubated with secondary antibody labeled with biotin at room temperature for 30 min (Ultrasensitive TM S-P; MaiXin). Diaminobenzidine tetrahydrochloride substrate (MaiXin) was used as the chromogen. The intensity of KIF23 staining was scored as follows: 0 (no staining), 1 (weak), 2 (moderate), and 3 (high). Percentage scores were assigned as follows: 0 (< 10%), 1 (10%-25%), 2 (26%-50%), 3 (51%-75%), and 4 (76%-100%). The score of each tumor sample was multiplied to obtain a final score of 0-12, and the staining results were categorized into negative (0; −), low (1-4; +), moderate (5-8; ++), and high (9-12; +++). The images were viewed with a microscope (Nikon, Tokyo, Japan). The tissue sections from each patient were observed within eight fields of view, and two experienced pathologists read the sections without knowing the pathological grade and clinical data. The result was judged by the double blind method.
Oncomine analysis
Oncomine is a cancer microarray database and web-based data-mining platform that was used to analyze the mRNA levels of KIF23 in GC. We set the screening criteria as: (1) “Cancer Type: Gastric cancer”; (2) “Gene: KIF23”; (3) “Data Type: mRNA”; (4) “Analysis Type: Cancer vs Normal Analysis”; and (5) “Critical value setting condition: P < 10-4, fold change > 2, gene rank = top 10%”. The results are displayed by histogram.
Kaplan–Meier plotter analysis
Online survival analysis was performed using the GC data set of Kaplan Meier (KM) plotter database (http://kmplot.com/analysis/). The screening criteria were as follows: (1) “Cancer: Gastric Cancer”; (2) “Gene: KIF23”; (3) “Survival: OS”; and (4) “Exclude: GES62254”. This was because the KM database showed that GES62254 had distinct characteristics compared with other datasets (longer survival time and expression transformation). The database recommends excluding datasets when all samples are used together.
Statistical analysis
The χ2 test was used to evaluate the association between KIF23 expression and clinicopathological features in GC tissues. The relationship between KIF23 expression and prognosis of GC was analyzed using a Kaplan–Meier model. In order to determine the independent prognostic factors, we used Cox proportional hazards models for univariate and multivariate analyses. Statistical analyses were performed on all data using SPSS version 25.0. P < 0.05 showed statistical significance.
RESULTS
KIF23 expression in GC and relationship between KIF23 expression and clinicopathological characteristics
KIF23 protein expression was examined in GC and normal gastric tissues using immunohistochemistry. We detected KIF23 expression in 174 GC samples, and KIF23 was mainly expressed in the cytoplasm (Figure 1). For statistical analysis, - and + were considered low expression of KIF23, while ++ and +++ were considered high expression. We confirmed that the ratio of high KIF23 expression in GC was significantly higher than in normal gastric tissues (Table 1). We evaluated the association between KIF23 expression and the clinicopathological characteristics of GC using a chi-square test. As shown in (Table 2), high expression of KIF23 in GC was significantly related to lymph node metastasis (P = 0.047) and poor TNM stage (P = 0.022). However, there was no significant difference between KIF23 expression and other clinicopathological characteristics (such as age, gender, histological differentiation, and Borrmann type).
Figure 1.
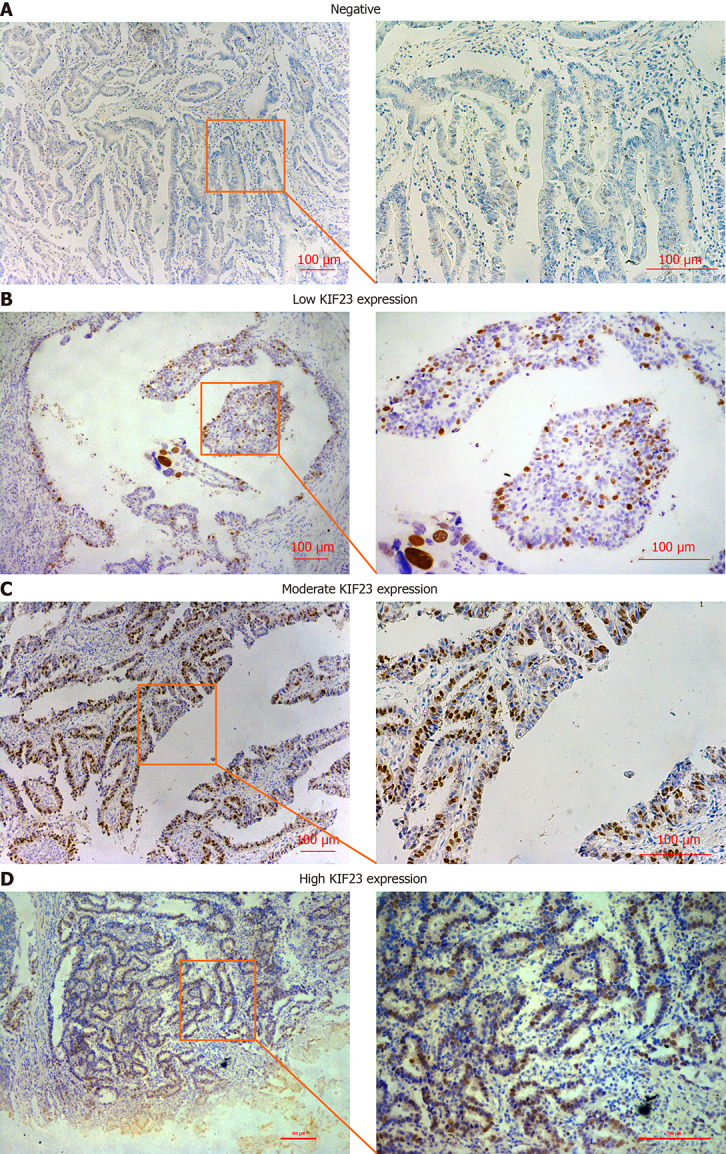
Immunohistochemical staining for KIF23 protein in gastric cancer. A: Negative; B: Low expression; C: Medium expression; D: High expression.
Table 1.
KIF 23 expression in gastric cancer and normal tissues, n (%)
| Cases |
KIF23 expression |
P value | ||
| Low | High | |||
| Cancer | 174 | 30 (17.2) | 144 (82.8) | < 0.001 |
| Normal | 174 | 169 (97.1) | 5 (2.9) | |
Table 2.
Correlation between KIF23 expression and clinicopathological features in gastric cancer
| n = 174 |
KIF23 expression |
P value | ||
| Low (n = 130) | High (n = 44) | |||
| Age (yr) | 0.539 | |||
| ≤ 60 | 90 | 69 | 21 | |
| > 60 | 84 | 61 | 23 | |
| Gender | 0.583 | |||
| Male | 129 | 95 | 34 | |
| Female | 45 | 35 | 10 | |
| Diameter (cm) | 0.667 | |||
| ≤ 4 | 80 | 61 | 19 | |
| > 4 | 94 | 69 | 25 | |
| Differentiation type | 0.188 | |||
| Poor | 104 | 74 | 30 | |
| Non-poor | 70 | 56 | 14 | |
| Borrmann type | 0.453 | |||
| I-II | 15 | 10 | 5 | |
| III-IV | 159 | 120 | 39 | |
| T stage | 0.051 | |||
| T1-T3 | 56 | 47 | 9 | |
| T4 | 118 | 83 | 35 | |
| N stage | 0.0471 | |||
| N0 | 61 | 51 | 10 | |
| N1-N3 | 113 | 79 | 34 | |
| TNM stage | 0.0221 | |||
| I-IIA | 37 | 33 | 4 | |
| IIB-IIIC | 137 | 97 | 40 | |
Significant difference.
Transcription levels of KIF23 in GC using Oncomine analysis
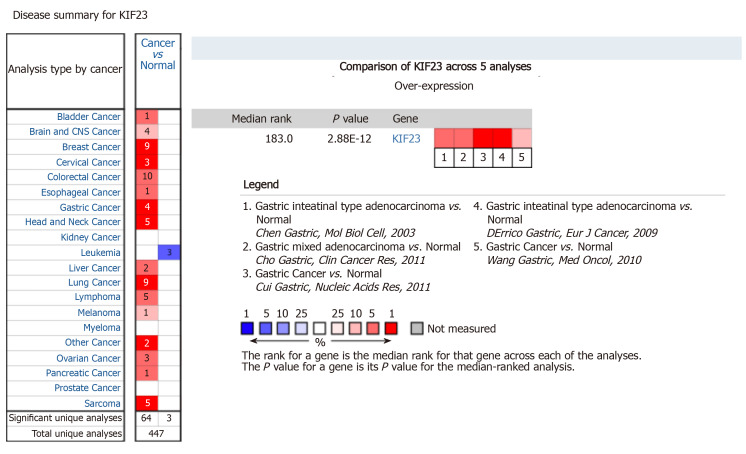
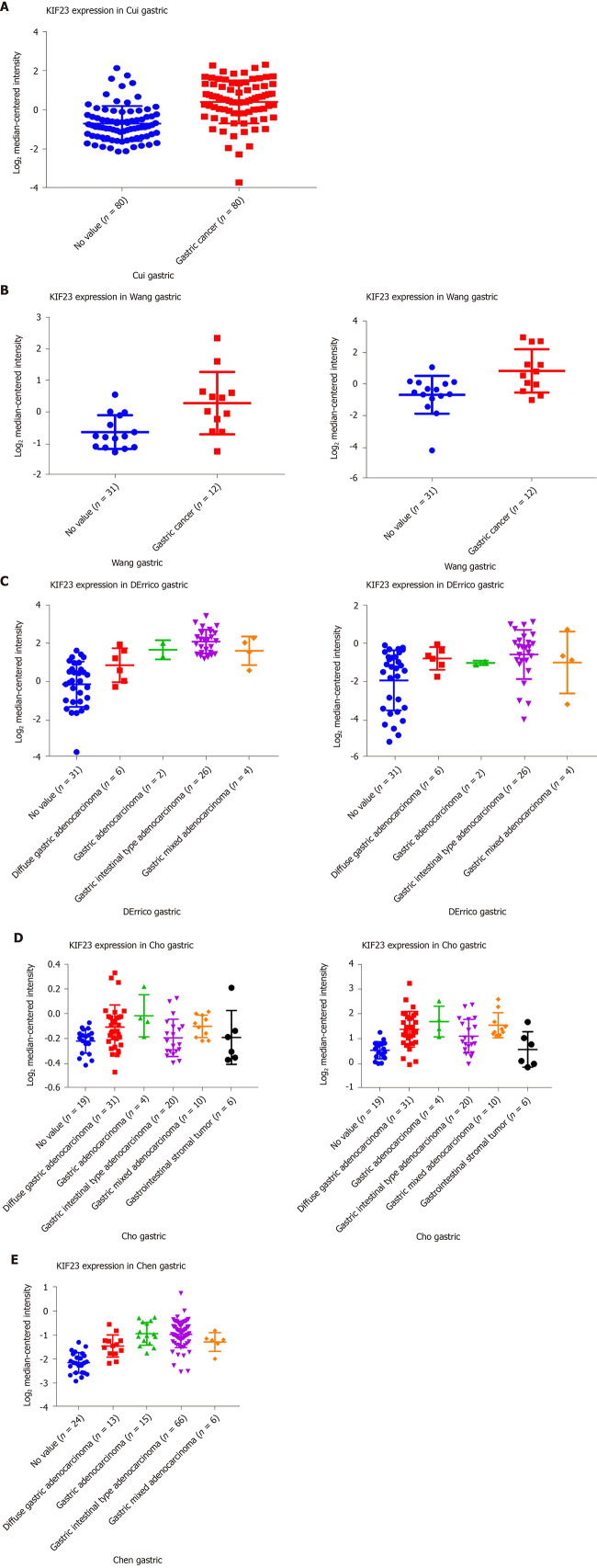
In Oncomine, we found that, since 2003, there were five studies involving expression of KIF23 in GC and normal tissues (Figure 2). A total of 478 samples including diffuse gastric adenocarcinoma, gastric adenocarcinoma, gastric intestinal type adenocarcinoma, gastric mixed adenocarcinoma, and gastrointestinal stromal tumors were compared with normal tissues[13-17]. Analysis of these studies showed that compared with the control group, the KIF23 gene ranked 183.0 among all differentially expressed genes (P = 2.88E-12), indicating that KIF23 was highly expressed in GC. Figure 3 shows the expression of KIF23 in the Oncomine database in different GC research chips. All these studies showed that expression of KIF23 in GC was higher than that in normal tissue (P < 0.05).
Figure 2.
KIF23 mRNA expression in tumors according to Oncomine database. A statistically significant dataset of high (red) or low (blue) KIF23 gene expression (cancer vs normal).
Figure 3.
Expression of KIF23 in gastric cancer in Oncomine database. A-E: The results of the five studies involved. The darker the red, the higher the expression of KIF23 in the chip.
Prognostic value of KIF23 in GC
Kaplan–Meier survival analysis showed that the OS rate of the KIF23-positive group was significantly lower than that of the KIF23-negative group, suggesting that the high level of KIF23 was associated with a poor prognosis (P = 0.0063) (Figure 4A). Univariate analysis revealed that differentiation type, Borrmann type, T stage, N stage, TNM stage, and KIF23 expression might be associated with the prognosis of GC patients (P < 0.05). Multivariate Cox analysis, using categorical variables, showed that TNM stage [hazard ratio (HR) = 1.869, 95%CI: 1.356-2.575, P = 0.000] and KIF23 expression (HR = 1.069, 95%CI: 1.006–1.135), P = 0.030) were independent prognostic factors for OS (Table 3). According to KIF23 expression and involved clinicopathological characteristics, we performed survival analysis in different subgroups. The OS of patients with high KIF23 expression was significantly lower than that of patients with T4, N1–N3, IIB–IIIC, or poorly differentiated GC with low KIF23 expression. However, there were no prognostic differences between high and low KIF expression in patients with T1–T3, N0, I–IIA, and well differentiated GC (Figure 4B-I).
Figure 4.
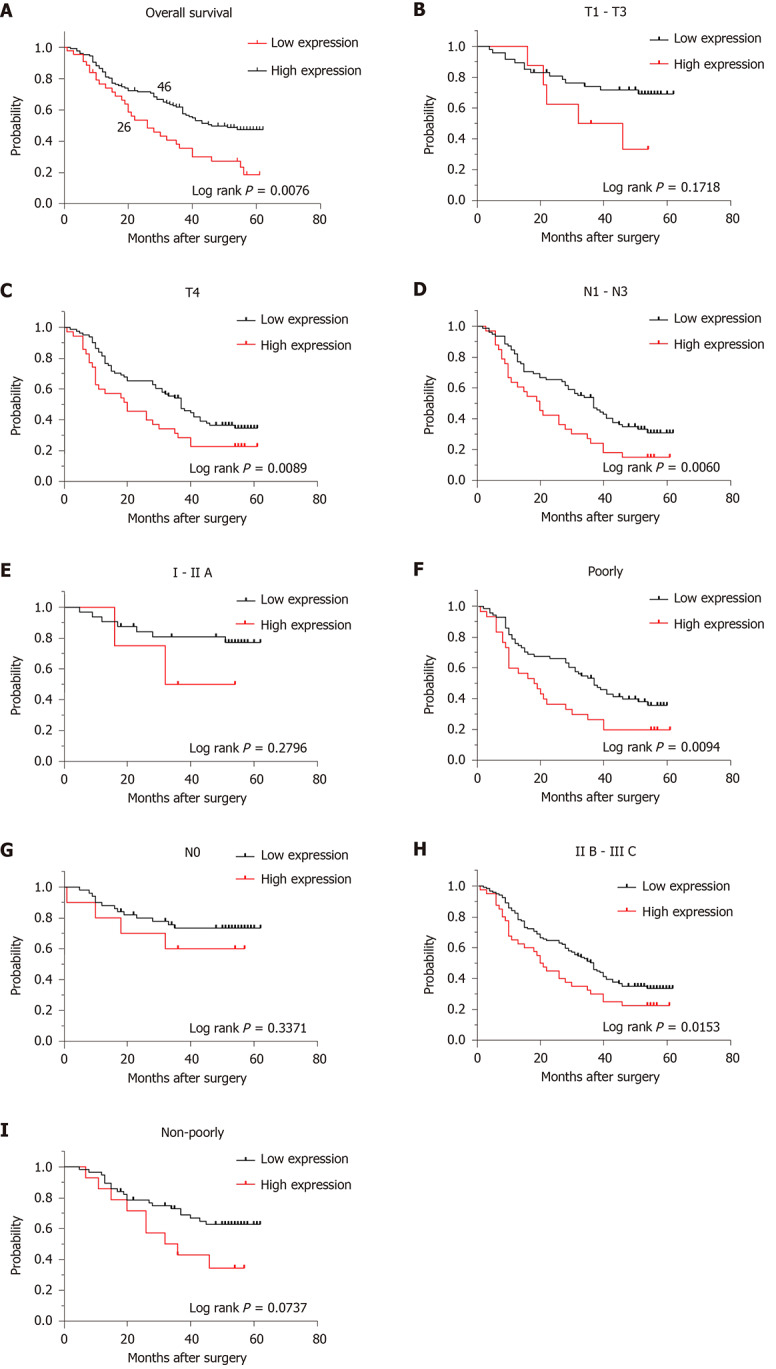
Prognostic effect of KIF23 expression in subgroups with different clinicopathological characteristics. A: Kaplan–Meier analysis of the overall survival (OS) rate between the low- and high-KIF23 expression groups. Median OS in the low- and high-KIF23 expression groups was 41 and 26 mo, respectively; B–I: Kaplan–Meier analysis of OS rate between the low- and high-KIF23 expression groups in GC patients with different T, N, and tumor–node–metastasis stages, and differentiation types.
Table 3.
Prognostic factors in Cox’s proportional hazards model in gastric cancer (n = 174)
| Variable |
Univariate |
Multivariate |
||||
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Age (yr): > 60 vs ≤ 60 | 0.822 | 0.555-1.217 | 0.328 | |||
| Gender: Male vs female | 1.297 | 0.838-2.008 | 0.243 | |||
| Diameter: > 4 cm vs ≤ 4 cm | 0.616 | 0.350-1.084 | 0.093 | |||
| Differentiation type: Poor vs non-poor | 0.552 | 0.336-0.906 | 0.0191 | 0.840 | 0.502-1.404 | 0.505 |
| Borrmann type: I-IV | 1.703 | 1.109-2.617 | 0.0151 | 1.410 | 0.916-2.169 | 0.119 |
| T stage: T1-T4 | 1.546 | 1.141-2.095 | 0.0051 | 1.310 | 0.858-2.000 | 0.210 |
| N stage: N0-N3 | 1.693 | 1.424-2.013 | 0.0001 | 1.869 | 1.356-2.575 | 0.0001 |
| TNM stage: I-IIIC | 2.410 | 1.637-3.547 | 0.0001 | 0.595 | 0.259-1.366 | 0.221 |
| KIF23 expression: High-level vs low-level | 1.064 | 1.006-1.126 | 0.0291 | 1.069 | 1.006-1.135 | 0.0301 |
Significant difference.
Prognostic analysis of KIF23 in GC using Kaplan–Meier survival information
Kaplan–Meier survival information for KIF23 can be found at https://www.kmplot.com. We examined the prognostic value of KIF23 mRNA expression in the database. Affymetrix ID is valid: 204709_s_at (KIF23). In all of the GC patients who were followed, KIF23 mRNA overexpression was found to be associated with worsening OS (HR = 1.64, 95%CI: 1.32-2.02, P = 3.9e-06). We then examined the correlation between KIF23 expression and OS in GC patients with different clinicopathological characteristics. High expression of KIF23 mRNA was associated with a poor OS in men, and patients with T3, M0, GDA, and poorly differentiated GC (Table 4). However, there were no significant differences in survival analysis in patients with other clinicopathological characteristics. As the sample size of patients with T1 and T4 GC was too small, no analysis was performed in these patients. Regarding the time span of involved KM survival, the guidelines for TNM staging were changed, so TNM stage was not included in the analysis[18-22].
Table 4.
Correlation between KIF23 mRNA expression and gastric cancer patients in Kaplan–Meier database
| Variable | Cases | HR (95%CI) | P value |
| Overall patients | 593 | 1.64 (1.32-2.02) | 3.9e-061 |
| T stage | |||
| T2 | 66 | 2.05 (0.98-4.26) | 0.0501 |
| T3 | 117 | 1.83 (1.14-2.94) | 0.0121 |
| N stage | |||
| N0 | 38 | 2.78 (0.88-8.73) | 0.069 |
| N1 + N2 + N3 | 175 | 1.33 (0.92-1.94) | 0.130 |
| M stage | |||
| M0 | 186 | 1.74 (1.17-2.59) | 0.00531 |
| M1 | 31 | 0.49 (0.19-1.23) | 0.120 |
| Gender | |||
| Male | 360 | 2.07 (1.60-2.69) | 1.9e-081 |
| Female | 138 | 1.50 (0.95-2.37) | 0.078 |
| Differentiation | |||
| Well | 32 | 0.41 (0.16-1.08) | 0.062 |
| Moderate | 67 | 0.75 (0.38-1.48) | 0.400 |
| Poor | 165 | 1.67 (1.12-2.49) | 0.0121 |
| Lauren classification | |||
| GITA | 179 | 0.76 (0.52-1.11) | 0.15 |
| GDA | 106 | 2.05 (1.23-3.39) | 0.00471 |
| GMA | 25 | 0.22 (0.03-1.68) | 0.11 |
Significant difference.
DISCUSSION
GC is one of the malignant tumors with the highest incidence and mortality worldwide. The prognosis of GC has been poor[23]. Therefore, it is of clinical importance to find the key molecules or targets for the treatment of GC. In this study, we discussed the prognostic value of KIF23 in GC. We examined the expression of KIF32 in GC cases from our hospital and analyzed the data from the Oncomine and Kaplan–Meier databases. Our results suggested that increased KIF23 expression in GC tissues predicted worse prognosis, and KIF23 might be a potential target for GC treatment.
Forty-five different functional KIFs have been found in humans[18-20]. They all share a highly conserved motor area, providing motion with microtubules. KIFs are divided into three types according to which area carries the sports field, including N-kinin, M-kinin, and C-kinin. KIF23 belongs to N-kinin, is located in the cytoplasm and nucleus, and is related to cell differentiation and proliferation[21]. KIF23 overexpression is a common event seen in various tumors, such as glioma[22], breast cancer[5], and lung cancer[24]. Overexpression of KIF23 in breast cancer is significantly associated with tumor grade, invasion, and prognosis[5]. In vivo and in vitro experiments have shown that knockdown of KIF23 significantly inhibits proliferation of glioma cells[22]. Murakami et al[12] found that KIF23 expression was significantly increased in GC cell lines. Previous evidence supported the role of KIF23 not in vesicular transport but in the bundling and transport of microtubules in specific locations and times in different cell types[25-28]. In our immunohistochemical study, KIF23 staining was observed mainly in the cytoplasm. It may be that overexpression of the kinesin family can affect normal mitosis, lead to aneuploidy and eventually tumor development.
We previously found that KIF23 is highly expressed in GC tissues by GSE63089 high-throughput microarray analysis, and its expression level is about 3.89 times that of adjacent tissues. The Cancer Genome Atlas database also confirmed that KIF23 is highly expressed in GC. Our study showed that KIF23 overexpression is a valuable independent prognostic factor in GC, particularly in T4, N1-N3, IIB-IIIC, and poorly differentiated cancer. Kaplan–Meier survival analysis suggested that high expression of KIF23 mRNA was associated with a poor OS in male patients, and patients with T3, M0, Lauren differentiation GDA, and poorly differentiated GC. Our data came from clinical cases and gene chips. The research methods were consistent and included a large sample size, which eliminated errors caused by sample size problems and increased the credibility of our conclusions. These findings lead us to believe that KIF23 overexpression represents an important transformation factor associated with a more malignant phenotype and hence poor prognosis, although further investigation is needed to validate this result.
The present study had some limitations. First, some information was obtained from open databases, and the medical parameters were not complete. Therefore, we were not able to perform a far-reaching survival analysis of KIF genes, considering each latent prognostic variable of GC in the multivariate Cox proportional hazards regression model. Second, because of the varied origin of GC patients, together with the number of elements affecting GC prognosis, we were not able to construct a comprehensive hazard score model. Third, for correlation with past research, the constraint of our present investigation suggested that it just studied the relationship between KIF23 expression and GC prognosis.
CONCLUSION
In summary, through in-depth exploration of KIF23-related information in GC tissues, we found that KIF23 overexpression in GC tissues is related to prognosis. Use of a database for large sample analysis can avoid errors caused by a small sample size and provide an important theoretical basis for clinical treatment. The specific mechanism of KIF23 in the development of GC needs further study.
ARTICLE HIGHLIGHTS
Research background
Kinesin super family 23 (KIF23) is a member of the KIF family, and it plays an important role in mitosis and cytokinesis. Loss of KIF23 expression can cause mitotic arrest. By querying the Oncomine database, differences in expression between tumor and normal tissues can be determined.
Research motivation
We detected the expression level of KIF23 protein in gastric cancer (GC) and adjacent normal tissues, and analyzed the association between KIF23 protein expression and clinicopathological factors.
Research objectives
This study aimed to study the expression and prognostic significance of KIF23 in GC.
Research methods
Immunohistochemistry was used to compare the expression of KIF23 in GC and normal gastric tissues. The data on the expression and prognosis of KIF23 in GC were mined using Oncomine and Kaplan–Meier plotter database.
Research results
Compared with normal gastric tissues, KIF23 expression was increased in GC tissues, and correlated with T, N, and tumor–node–metastasis stages. Survival analysis showed that patients with high expression of KIF23 had a poor overall survival. The prognostic survival indicators worsened in patients with T2 and T3 poorly differentiated adenocarcinoma with high expression of KIF23.
Research conclusions
KIF23 is highly expressed in GC, and it is associated with a poor prognosis of GC patients.
Research perspectives
We found that KIF23 overexpression in gastric cancer tissues is related to prognosis. Use of a database for large sample analysis can avoid errors caused by a small sample size and provide an important theoretical basis for clinical treatment. The specific mechanism of KIF23 in the development of gastric cancer needs further study.
Footnotes
Institutional review board statement: This study was approved by the Research Ethics Committee of China Medical University according to the Helsinki Declaration.
Conflict-of-interest statement: The authors declare no potential financial interests.
Manuscript source: Unsolicited manuscript
Peer-review started: April 14, 2020
First decision: May 15, 2020
Article in press: August 15, 2020
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aspichueta P, Jones D, Myers R S-Editor: Wang JL L-Editor: Wang TQ P-Editor: Li JH
Contributor Information
Wei-Tian Liang, Department of Surgical Oncology, the First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning Province, China.
Xiao-Fang Liu, Department of Surgical Oncology, the First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning Province, China.
Hai-Bo Huang, Department of Surgical Oncology, the First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning Province, China.
Zi-Ming Gao, Department of Surgical Oncology, the First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning Province, China.
Kai Li, Department of Surgical Oncology, the First Affiliated Hospital of China Medical University, Shenyang 110001, Liaoning Province, China. cmu_likai@126.com.
Data sharing statement
No additional data are available.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Hu J, Ge N, Wang S, Liu X, Guo J, Wang G, Sun S. The Role of Endoscopic Ultrasound and Endoscopic Resection for Gastric Glomus: A Case Series and Literature Review. J Transl Int Med. 2019;7:149–154. doi: 10.2478/jtim-2019-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirokawa N, Tanaka Y. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp Cell Res. 2015;334:16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Zou JX, Duan Z, Wang J, Sokolov A, Xu J, Chen CZ, Li JJ, Chen HW. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12:539–549. doi: 10.1158/1541-7786.MCR-13-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neef R, Klein UR, Kopajtich R, Barr FA. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol. 2006;16:301–307. doi: 10.1016/j.cub.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Gong H, Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013;104:651–656. doi: 10.1111/cas.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucanus AJ, Yip GW. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37:833–838. doi: 10.1038/onc.2017.406. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Guo X, Xie C, Jiang J. KIF15 promotes pancreatic cancer proliferation via the MEK-ERK signalling pathway. Br J Cancer. 2017;117:245–255. doi: 10.1038/bjc.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Ma RR, Wang XJ, Su ZX, Chen X, Shi DB, Guo XY, Liu HT, Gao P. KIF26B, a novel oncogene, promotes proliferation and metastasis by activating the VEGF pathway in gastric cancer. Oncogene. 2017;36:5609–5619. doi: 10.1038/onc.2017.163. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Li S, Zhou S, Cao S, Lou Y, Shen H, Yin J, Li G. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther. 2017;13:651–659. doi: 10.4103/jcrt.JCRT_491_17. [DOI] [PubMed] [Google Scholar]
- 12.Murakami H, Ito S, Tanaka H, Kondo E, Kodera Y, Nakanishi H. Establishment of new intraperitoneal paclitaxel-resistant gastric cancer cell lines and comprehensive gene expression analysis. Anticancer Res. 2013;33:4299–4307. [PubMed] [Google Scholar]
- 13.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, Ni Z, Zhang M, Kong X, Hoffman LL, Kang J, Su Y, Olman V, Johnson D, Tench DW, Amster IJ, Orlando R, Puett D, Li F, Xu Y. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–1207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, So S, Botstein D, Brown PO. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, Palombo F, Giuliani A, Dogliotti E. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, Noh SH, Park ES, Chu IS, Hong WK, Ajani JA, Lee JS. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 18.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 19.Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- 20.Vale RD, Schnapp BJ, Reese TS, Sheetz MP. Movement of organelles along filaments dissociated from the axoplasm of the squid giant axon. Cell. 1985;40:449–454. doi: 10.1016/0092-8674(85)90159-x. [DOI] [PubMed] [Google Scholar]
- 21.Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. Kinesin family in murine central nervous system. J Cell Biol. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, Liu X, Wang S, Ge N, Guo J, Sun S. Endoscopic Ultrasound-guided Gastroenterostomy: A Promising Alternative to Surgery. J Transl Int Med. 2019;7:93–99. doi: 10.2478/jtim-2019-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi S, Fusaki N, Ohta S, Iwahori Y, Iizuka Y, Inagawa K, Kawakami Y, Yoshida K, Toda M. Downregulation of KIF23 suppresses glioma proliferation. J Neurooncol. 2012;106:519–529. doi: 10.1007/s11060-011-0706-2. [DOI] [PubMed] [Google Scholar]
- 27.Kato T, Wada H, Patel P, Hu HP, Lee D, Ujiie H, Hirohashi K, Nakajima T, Sato M, Kaji M, Kaga K, Matsui Y, Tsao MS, Yasufuku K. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer. 2016;92:53–61. doi: 10.1016/j.lungcan.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Boman AL, Kuai J, Zhu X, Chen J, Kuriyama R, Kahn RA. Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil Cytoskeleton. 1999;44:119–132. doi: 10.1002/(SICI)1097-0169(199910)44:2<119::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.