Abstract
BACKGROUND
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related deaths worldwide, but there is a shortage of effective biomarkers for its diagnosis.
AIM
To explore blood exosomal micro ribonucleic acids (miRNAs) as potential biomarkers for HCC diagnosis.
METHODS
T
RESULTS
The principal component analysis suggested that daily alcohol consumption could alter the blood exosomal miRNA profiles of hepatitis B virus positive non-HCC patients through miR-3168 and miR-223-3p. The miRNA profiles also revealed the tumor stages of HCC patients. High expression of miR-455-5p and miR-30c-5p, which significantly correlated with better overall survival in tumor tissues, could also be detected in blood exosomes. Two pairs of miRNAs (miR-584-5p/miR-106-3p and miR-628-3p/miR-941) showed a 94.1% sensitivity and 68.4% specificity to differentiate HCC patients from non-HCC patients. The specificity of the combination was substantially influenced by alcohol consumption habits.
CONCLUSION
This study suggested that blood exosomal miRNAs can be used as new non-invasive diagnostic tools for HCC. However, their accuracy could be affected by tumor stage and alcohol consumption habits.
Keywords: Blood exosomal micro ribonucleic acids, Biomarker, Differential diagnosis, Alcohol consumption habit, Hepatocellular carcinoma, Bioinformatics
Core Tip: To identify the potential biomarkers for hepatocellular carcinoma (HCC) diagnosis, we focused on exosomal micro ribonucleic acids (miRNAs) using miRNA sequencing. To our knowledge, this is the first study to explore the exosomal miRNAs for differential diagnosis of HCC using miRNA sequencing. This study provided a hint that blood exosomal miRNAs can be used as new non-invasive diagnostic tools for HCC in clinical implementation.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-associated mortality worldwide, especially in East and South Asia[1,2]. In China, more than 460000 new cases of HCC were diagnosed with 422100 deaths in 2015[3,4]. Due to its insidious onset without specific early symptoms, most of the HCC patients are diagnosed at an advanced stage. The lack of effective early diagnosis methods for HCC is a major challenge to improving the outcomes of these patients, who therefore lose the opportunity for therapeutic treatments, such as resection and sorafenib[5-7]. Hence, there is an urgent demand for the identification of potential biomarkers for early detection to cut down the high mortality of HCC.
It was reported that some tumor biomarkers in serum had been discovered and might play an important role in HCC diagnosis[8-10]. At present, the serum αfetoprotein (AFP) test is a common and important early diagnostic test for liver cancer, but this biomarker has a low specificity[11]. In addition, carbohydrate antigen (CA) 19-9 (CA19-9) and carcinoembryonic antigen have been widely used as serum tumor markers for the clinical diagnosis of HCC[12]. However, the efficacy of these tumor markers in the clinical diagnosis of HCC remains unsatisfactory. In addition, candidate tumor biomarkers, such as circulating tumor cells, serum cell-free deoxyribonucleic acid (DNA), non-coding ribonucleic acids (RNAs), and microRNAs (miRNAs), show limited progress in advanced HCC to date[7,9,13,14]. In the face of the difficult problem of exploring biomarkers for diagnosis of HCC, it is particularly important to adopt new techniques and study related interference factors.
Exosomes, which are 50-150 nm membrane microvesicles secreted by a number of cell types, have been found in many types of body fluids, including saliva, plasma, breast milk, urine, and malignant effusions[15]. It has been reported that exosomes serve as a key regulator of the tumor microenvironment by promoting HCC occurrence and progression[16]. The transfer of exosomes from primary tumors to the circulatory system has been demonstrated in various models[17], and many studies have indicated that cancer-associated exosomal miRNAs participate in the regulation of cancer cell growth, metastasis, and drug resistance[18,19]. This rapidly expanding field includes studies investigating opportunities for developing serum-derived exosomal contents as new biomarkers in cancer diagnosis, treatment, and drug resistance.
Considering the frequency of late diagnosis in HCC, biomarkers as potential prognostic indicators and their complexity are worthy of further investigation. Several studies have reported the application of exosomal miRNAs as biomarkers in early detection, diagnosis, and therapy of HCC[7,20,21]. However, the previous research was on a small scale and showed differences in candidate biomarkers using real-time polymerase chain reaction (PCR) or microarrays, which are classic methods for miRNA expression analysis and can detect only known and limited miRNAs. In previous studies, there was a lack of comprehensive miRNA expression profiling. Such comprehensive profiling has proven useful in diagnosing and monitoring the development and progression of tumors. Recently, with the rapid development of next-generation sequencing, miRNA sequencing (miRNA-seq) has begun to offer increased specificity and sensitivity in miRNA profiling[22]. In particular, it possesses the ability to identify novel miRNAs, which has significantly enabled rapid profiling and deep investigation of miRNAs. Therefore, we adopted two relatively large population cohorts [including hepatitis B virus (HBV)-infected individuals, liver cirrhosis patients, and HCC patients] for exosomal miRNA profiling using miRNA-seq. Cohort analysis of blood exosomal miRNA profiling was performed and revealed the complexity caused by drinking alcohol and potential biomarkers for differential diagnosis in HCC. This work was the first study revealing how exosomal miRNA biomarkers are made more complex by drinking alcohol in the differential diagnosis of HCC using exosomal miRNA-seq.
MATERIALS AND METHODS
Patient clinical information and clinical sample collection
This study was approved by the Ethics Committee of Xiangya Hospital of Central South University (No. 201803818). There were 136 patients enrolled for this study between August 2017 and May 2018. After excluding patients with non-HCC malignant tumors and hemolysis, this study included 89 patients, comprising 51 patients with HCC confirmed by surgical pathology, 20 liver cirrhosis patients, and 18 HBV-infected individuals. These patients were divided into two cohorts (cohorts 1 and 2). Detailed clinical characteristics of these patients are shown in Table 1. We collected data about the patients’ age, sex, cirrhosis, tumor stage, alcohol consumption, and levels of various tumor biomarkers (AFP, CA19-9, and carcinoembryonic antigen) (Table 1). Daily consumption of alcohol under 12 g was defined as “No daily drinking,” while daily drinking greater than or equal to 12 g of alcohol was defined as “Daily drinking” in this study. All of the individuals at the Xiangya Hospital of Central South University gave their written consent for their plasma samples and pathology information to be used in this research.
Table 1.
Clinical characteristics of patients and control individuals
|
|
Cohort 1 (n = 44)
|
Cohort 2 (n = 45)
|
| Age (yr), average | 46.0 ± 10.24 | 47.2 ± 11.02 |
| Gender | ||
| Male | 36 | 41 |
| Female | 8 | 4 |
| Category | ||
| HBV infected group | 10 | 8 |
| Liver cirrhosis group | 10 | 10 |
| HCC tumor group | 24 | 27 |
| BCLC stage1 | ||
| Stage A | 8 | 9 |
| Stage B | 8 | 8 |
| Stage C | 8 | 10 |
| Drinking history | ||
| No daily drinking2 | 29 | 21 |
| Daily drinking3 | 15 | 24 |
| Intake of alcohol (g/d), average | 97 ± 143 | 121 ± 121 |
| AFP (ng/mL) of HCC, average | 350.75 ± 361.98 | 221.86 ± 274.84 |
| CA19-9 (KU/L) of HCC, average | 44.63 ± 59.17 | 28.96 ± 59.2 |
| CEA (ng/mL) of HCC, average | 2.98 ± 1.98 | 3.94 ± 7.32 |
Barcelona Clinic Liver Cancer stage.
Daily consumption of alcohol under 12 g was defined as “No daily drinking”.
Daily drinking greater than or equal to 12 g of alcohol was defined as “Daily drinking”. HBV: Hepatitis B virus; HCC: Hepatocellular carcinoma; AFP: αfetoprotein.
The blood samples of these patients were collected in vacutainers with anticoagulant (REF367863, BD, United States) and then shipped at 4 °C after collection. When we received the blood samples, we first centrifuged them at 4 °C (1600 × g, 10 min). After this, we determined and recorded the hemolysis level. Plasma samples with a hemolysis level beyond four were not used in this study. Followed by a second centrifugation at 4 °C (16000 × g, 10 min), we finally transferred each 1 mL of fraction of the supernatant to a 1.5 mL fresh tube and stored them at -80 °C for further study.
Plasma exosome isolation
As previously described[23], plasma exosome isolation was performed using 3D Medicine isolation reagent (L3525; 3DMed, Shanghai, China). After plasma samples were incubated in a water bath and then centrifuged at 4 °C (12000 × g, 10 min), we transferred the supernatants into a 0.45 µm tube filter (Costar, CLS8163-100EA, Corning, United States), centrifuged them at 4 °C (12000 × g, 5 min), and then filtered them with 0.22 µm tube filters (Costar, CLS8161-100EA, United States). The supernatants were transferred into fresh 1.5 mL tubes. We then added one-quarter volume of exosome isolation reagent (L3525, 3DMed, Shanghai, China). We gently vortexed the mixtures and incubated them for 30 min at 4 °C, and then centrifuged them at 4 °C (4700 × g, 30 min). The supernatants were discarded, and the precipitates, containing the exosomes, were resuspended in 200 μL of phosphate-buffered saline.
Characterization of exosomes by scanning electron microscopy
Scanning electron microscopy (SEM) was performed as previously described[24]. Briefly, exosomes were suspended in phosphate-buffered saline before being fixed in 5% glutaraldehyde. Then, exosomes were immobilized in 1% OsO4 and were dehydrated with various concentrations of ethanol. Subsequently, samples underwent drying at room temperature for 24 h and were finally analyzed by SEM (SU8020, Hitachi High-Technologies, Japan).
Quantification of exosomal proteins
Exosomal protein extraction was done with exosome isolation reagent (N3525, 3DMed). Then, exosomes were lysed in 200 μL of lysis buffer (P0013B, Beyotime, Shanghai, China). Equal amounts of proteins were run on a 4%–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel (No. 4561095, Bio-Rad, United States). Proteins were transferred onto a polyvinylidene fluoride membrane (Millipore), and then incubated at room temperature for 2 h with primary antibodies anti-CD9 (1:500 dilution, ab92726, Abcam, England) and anti-CD63 (1:2000 dilution, ab68418, Abcam). Anti-rabbit immunoglobulin G (sc-2004, Santa Cruz, United States) was added and blots were incubated at room temperature for 40 min. Blots were visualized using an enhanced chemiluminescence system according to the manufacturer’s protocol (Tanon-5200Multi, Shanghai, China).
MiRNA isolation from exosomes
Exosomal miRNAs were extracted with the miRNeasy Serum/Plasma Kit according to the manufacturer’s instructions (217184, QIAGEN, Shanghai, China). The yield, distribution, and quality of miRNAs were analyzed with the Agilent 2100 bioanalyzer using Small RNA Chips (5067-1548, Agilent, United States).
MiRNA sequencing library preparation
A total of 100 ng RNA per sample was used to construct the miRNA library using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (E7300L, NEB) according to the manufacturer’s protocol. Eighteen cycles of PCR were performed after first strand cDNA synthesis. The DNA libraries were purified by NucleoSpin Gel and PCR Clean-up (740609.50, QIAGEN, Shanghai, China). The quality and distribution of libraries were analyzed using the LabChip GX Touch 24 Nucleic Acid Analyzer. The pooled libraries were sequenced using an Illumina HiSeq PE150 analyzer.
Bioinformatics analysis of miRNA-seq data
The miRNA-seq data were quantified and tested for differential expression with DESeq2 package in R/Bioconductor[25]. After raw data cleaning, sequences with a length less than 30 nt were mapped to hg19 by BWA[26] 0.7.12-r1039. The raw expression of a miRNA was defined by the number of reads mapped to the locus of the mature miRNA, which was obtained from miRBase[27] v21. Due to the limited sequencing depth, only those miRNAs which had at least one read in each sample were considered in the following analysis. Before comparing the expression between samples, the raw expression was normalized. The 75th percentile expression of samples was calculated as the size factor. The miRNA profiling was normalized using counts per million mappable miRNA sequences. The method is described in DESeq2[25].
Statistical analysis
To scale for miRNA expression, principal component analysis (PCA) was performed, wherein the normalized expression levels of each miRNA were scaled by dividing the expression levels by the maximum expression levels of the miRNAs. PCA was then performed on the matrix constructed by the scaled expression levels, using the prcomp function in R 3.3.3.
To test the performance of biomarkers, the area under the curve (AUC) was calaulated using r = /span >/Bioconductor pROC (v1.12.1) package. Clinicopathologic diagnoses were used as the gold standard to assess the diagnostic accuracy of a group of exosomal miRNAs by AUC. Kaplan-Meier plot analysis of The Cancer Genome Atlas data was performed using the online bioinformatics tool, OncoLnc (http://www.oncolnc.org/). P < 0.05 was considered statistically significant.
RESULTS
Blood exosomal miRNA profiling in HCC and non-HCC patients
To study the blood exosomal miRNA profiles, 89 HBV positive patients were enrolled and their blood was collected between August 2017 and May 2018. Among these patients, there were 51 patients diagnosed with HCC, 20 liver cirrhosis patients, and 18 HBV-infected individuals. All of the patients were assigned to two cohorts (cohorts 1 and 2). The clinical information and statistics of the two cohorts are listed in Table 1.
To characterize the exosomes from plasma obtained from these patients, Western blot and SEM analysis were performed. SEM images of two representative samples are shown in Supplementary Figure 1A. Two characteristic proteins of exosomes are CD9 and CD63. The protein levels in four representative samples are shown in Supplementary Figure 1B. These results suggested that exosomes from the plasma of these patients were successfully obtained by this method.
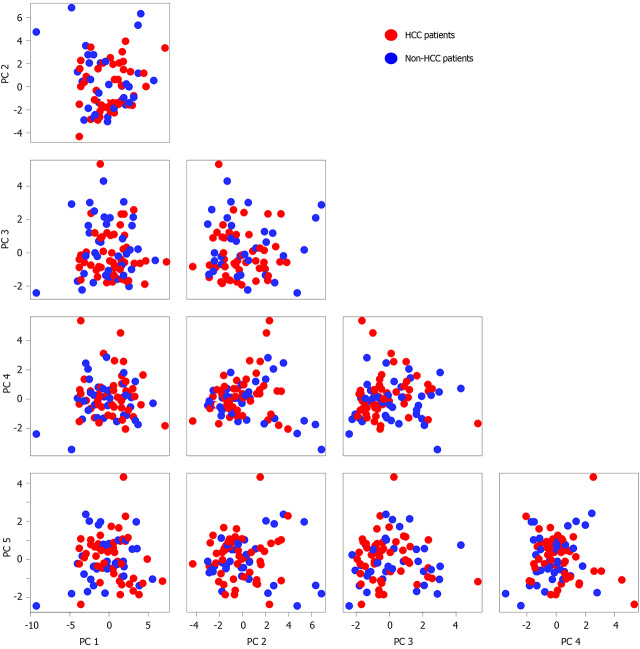
All of the patients were pooled to explore the potential differential features between cancer patients and non-cancer patients. PCA was performed to identify the major variations, which might reveal their biological nature (Figure 1). The distribution of cancer patients and non-cancer patients generally overlapped in the top five principal components (PC) and their combinations. The PCA result suggested that multiple other factors could more significantly affect the blood exosomal miRNA profiles than the biological difference between HBV infected HCC and non-HCC patients.
Figure 1.
Blood exosomal microRNA profiling in hepatocellular carcinoma patients and non-hepatocellular carcinoma patients. The top 5 principal components of blood exosomal micro RNA profiles from hepatocellular carcinoma (HCC) patients and non-HCC patients are shown. The red represents HCC patients, and the green represents non-HCC patients. HCC: Hepatocellular carcinoma.
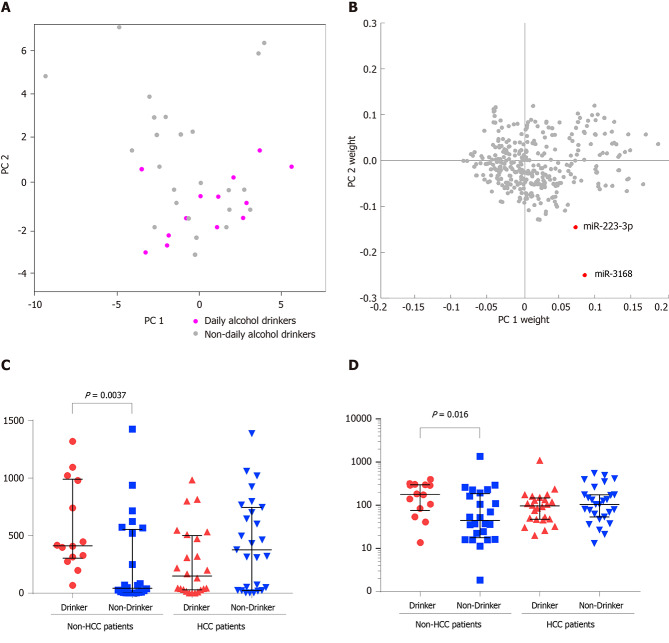
Daily alcohol consumption could alter the blood exosomal miRNA profiles of HBV positive non-HCC patients
On the coordinates of PC1 and PC2, non-HCC patients who had a daily alcohol consumption habit tended to distribute in the bottom right corner (Figure 2A), suggesting that alcohol consumption might shift the blood exosomal miRNA profile. To identify the key miRNAs that are expressed in response to alcohol consumption, a two-dimensional coordinate was constructed by the weights of each miRNA in PC1 and PC2 (Figure 2B). In this coordinate, miR-3168 and miR-223-3p were spotted as outliers in the bottom right corner. Both miRNAs showed significantly higher levels in non-HCC patients with daily alcohol consumption (Figure 2C and D). However, non-HCC patients who had a daily alcohol consumption habit and HCC patients (drinking or non-drinking) showed comparable blood exosomal miRNA levels.
Figure 2.
Daily alcohol consumption alters the blood exosomal microribonucleic acid profiles of hepatitis B virus positive non-hepatocellular carcinoma patients. A: Distribution of daily alcohol drinkers and non-daily alcohol drinkers on principal components 1 (PC1) and PC2; B: Distribution of PC1 weight and PC2 weight of each microRNA; C and D: Expression level comparisons of miR-3168 (C) and miR-223-3p (D) from daily alcohol drinkers and non-daily alcohol drinkers between hepatocellular carcinoma patients and non-hepatocellular carcinoma patients are demonstrated. HCC: Hepatocellular carcinoma.
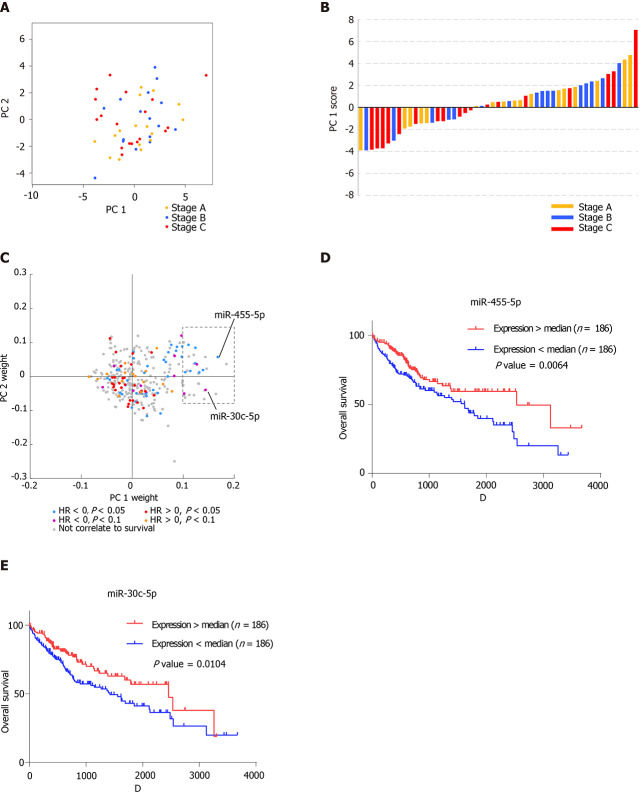
Blood exosomal miRNA profiles reveal the stage of tumor tissues
On the coordinates constructed from PC1 and PC2, the HCC patients are colored according to the Barcelona Clinic Liver Cancer tumor staging system[28] (Figure 3A). Interestingly, patients with stage C HCC tumors were mainly distributed in the negative direction on the PC1 coordinate, indicating that the vital miRNAs that contributed to the variation of PC1 might reveal tumor stage (Figure 3B). Since blood exosomes could derive from various types of cells, the clinical and miRNA-seq data of 372 HCC tissue samples from The Cancer Genome Atlas were analyzed to discover the connection between tumor tissues and blood exosomes in further detail. A Cox regression model was performed on miRNAs that were expressed in all of the tissue samples. Then, the Cox regression results from the tumor tissues were integrated with the PC weights of the blood exosomal miRNAs (Figure 3C). We found that 29 miRNAs possessed a positive weight of PC1 larger than 0.1 (in the grey rectangle), while none of the miRNAs possessed a negative weight less than 0.05, suggesting that the 29 miRNAs mainly contributed to the variation of PC1. Among the 29 miRNAs, 11 showed significant or marginal P values in the Cox regression model, and all of those miRNAs possessed negative hazard ratios. MiR-455-5p and miR-30c-5p ranked among the top five miRNAs with positive weights. Both miRNAs were suggested to be tumor suppressors in various types of cancers[29,30], and patients with higher expression levels showed significantly better overall survival (Figure 3D and E). Blood exosomal miRNAs were related to the stage of the tumor tissues, which might predict the survival of HCC patients. However, miRNAs from other tissues or cells might also influence the signals from tumor tissues.
Figure 3.
Blood exosomal micro ribonucleic acid profiles reveal the stage of tumor tissues. A: Distribution of hepatocellular carcinoma patients with stage A, stage B, and stage C on principal components 1 (PC1) and PC2. The yellow represents the patients with stage A, the blue represents the patients with stage B, and the red represents the patients with stage C; B: Histogram displaying PC1 scores of patients with stage A, stage B, and stage C. The histogram color is labeled as in A; C: Distribution of PC1 weight and PC2 weight of each micro ribonucleic acid. The colorindicates the hazardous levels based on The Cancer Genome Atlas data; D: Kaplan–Meier analysis of overall survival of two groups (high expression and low expression of miR-455-5p); E: Kaplan–Meier analysis of overall survival of the two groups (high expression and low expression of miR-30c-5p).
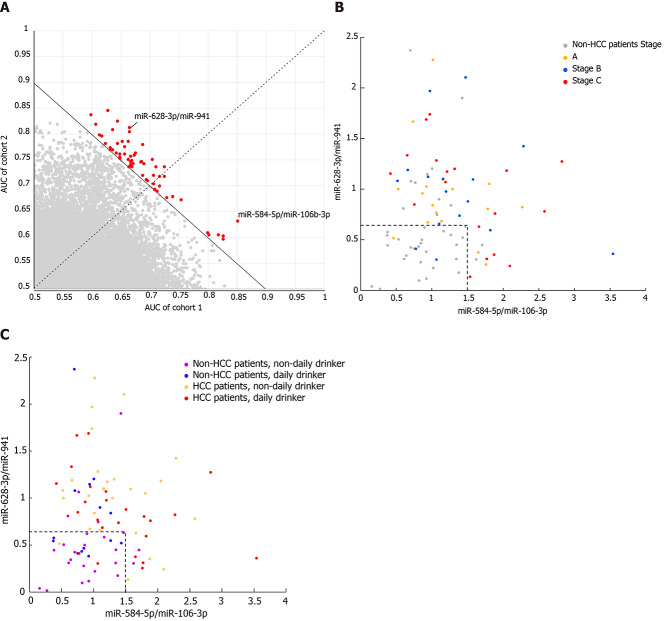
Accuracy of biomarkers from blood exosome miRNAs to classify HCC and non-HCC patients is affected by tumor stage and alcohol consumption habits
Since blood exosomal miRNAs might be derived from various sources, normalization of all miRNAs with the same size factor (or the same housekeeping miRNA) might not reveal the real difference of each miRNA between HCC and non-HCC patients. Similar to the normalization concept of qPCR, the quotient between a pair of miRNAs, where one is considered the target gene and the other is considered the reference gene, was calculated. This exhaustive method was used to find the best reference gene to normalize each target gene, and their quotients were considered the candidate biomarkers. To avoid potential batch effects, the patients were allocated to two cohorts according to their experimental date. The accuracy of each pair of miRNAs was measured by AUC analysis. The pairs of miRNAs were ranked according to the average AUC between the two cohorts, where the average AUC should be larger than 0.7 (Figure 4A). The top two pairs of miRNAs were miR-628-3p/miR-941 and miR-584-5/miR-106b-3p, and their average AUCs were 0.7423 and 0.7421, respectively. To distinguish HCC patients from non-HCC patients, miR-584-5p/miR-106b-3p showed a 94.7% specificity under a cutoff of 1.5, but the sensitivity was relatively low (37.2%). Under a cutoff of 0.65, miR-628-3p/miR-941 showed a 76.5% sensitivity and 73.7% specificity. When we considered a sample which had either of the quotients higher than their cutoffs as HCC patients, the combination of two quotients raised the sensitivity to 94.1%, while the specificity was maintained at 68.4% (Figure 4B). Combining the two quotients increased the overall accuracy to 83.1%, which was higher than that for miR-584-5p/miR-106b-3p (61.8%) or miR-628-3p/miR-941 (75.3%).
Figure 4.
Accuracy of biomarkers from blood exosomal micro ribonucleic acid to classify hepatocellular carcinoma and non-hepatocellular carcinoma patients is affected by tumor stage and alcohol consumption habits. A: The area under the curve of paired exosomal micro ribonucleic acid in cohort 1 and cohort 2; B: Distribution of the two paired blood exosomal miRNAs (miR-628-3p/miR-941 and miR-584-5p/miR-106-3p) colored according to tumor stage; and C: Alcohol consumption habits. HCC: Hepatocellular carcinoma.
While miR-584-5p/miR-106b-3p did not show any bias toward distinguishing HCC patients from non-HCC patients at any stages, among the HCC patients who could not be identified by miR-584-5p/miR-106b-3p, the HCC patients in stages A and B showed lower overall levels of miR-628-3p/miR-941 than those in stage C, and thus they were more difficultly distinguished from non-HCC patients (Figure 4B and Supplementary Figure 2). Additionally, non-HCC patients with different alcohol consumption habits also showed effects on their levels of miR-628-3p/miR-941 (Figure 4C). Non-HCC patients with daily alcohol consumption presented significantly higher levels of miR-628-3p/miR-941 than those without daily alcohol consumption and were more likely to be identified as false-positive HCC patients (Supplementary Figure 3).
DISCUSSION
It has been reported that miRNAs in blood could distinguish HBV-infected HCC patients from HBV-infected non-HCC patients[31], which suggests that the signals released from HCC can be observed. Mohamed et al[32] found that circulating miR-23a showed an accuracy of 79.3% in diagnosing HCC patients with a sensitivity of 89.47% and specificity of about 64.91%. Zhu et al[33] adopted a diagnostic 2-miRNA panel to differentiate HCC patients from healthy controls (AUC = 0.823, P < 0.0001) and cirrhosis patients (AUC = 0.859, P < 0.0001). Huang et al[34] established a score comprising five blood miRNAs and a binary etiology variable that was capable of differentiating between cirrhotic patients and HCC patients (AUC = 72.5%, P < 0.001)[34]. These results bring some hope for the differential diagnosis of HCC, but they are still not satisfactory for the clinical demand. However, blood miRNAs can be derived from various sources. Exosomes are small extracellular vesicles released from living cells for intercellular communications, and they also reveal the metabolic events occurring inside the cells. Hence, studying the miRNA profiles from HCC exosomes could provide insights for research and clinical diagnosis.
The exosomal miRNA profiles showed that in HBV-positive patients, the signals from tumors or triggered by tumors might not be major factors able to shift the whole profiles to be significantly distinct from those of non-HCC patients. However, other clinical features could be revealed through exosomal miRNA profiling. Among these clinical features, alcohol consumption is known to cause several liver diseases, including alcoholic liver disease, alcoholic fatty liver, alcoholic hepatitis, and alcoholic liver cirrhosis[35]. The liver is the main organ responsible for metabolizing ethanol, and thus it has been considered for a long time a major organ damaged by the harmful use of alcohol[36]. The expression of a wide variety of miRNAs is potentially regulated by many factors, such as alcohol, diet, cigarette smoking, and other drugs[37]. To date, there has only been one previous study focusing on the role of alcohol-regulated miRNAs in HCC pathogenesis and progression. That study reported that alcohol consumption in patients with HBV positivity and those with HCC regulates miRNAs (miR-944 and miR-223-3p) from tumor tissue. These miRNAs likely play previously uncharacterized roles in the alcohol-associated carcinogenesis of HCC[38]. Our study showed that miR-3168 and miR-223-3p had significantly higher levels in non-HCC patients with daily alcohol consumption than in those without, suggesting that daily alcohol consumption could alter the blood exosomal miRNA profiles of HBV positive non-HCC patients. MiR-223-3p, which was reported to play a role in the alcohol-associated carcinogenesis of HCC, was discovered as a blood exosomal biomarker for alcohol consumption in our study. In addition, it has also been reported that when treated by alcohol, the miR-223-3p level in tumor cell lines and in the immortalized liver cell line L02 increased[38]. A similar trend was also revealed in blood exosomal miRNA. These results led to the idea that the function of miR-223-3p in the alcohol-associated carcinogenesis of HCC deserves further study.
The Barcelona clinic liver cancer staging system classifies HCC patients into four stages that are highly correlated with prognosis[28]. Patients in early stage A or B are usually asymptomatic without vascular invasion or extrahepatic spread, while patients in advanced stage C have either symptomatic tumors, vascular invasion, or extrahepatic spread, which causes poorer survival than early stages. Intriguingly, tumor stages are major contributors in shifting the miRNA profiles among tumor patients, especially between stage C and early stages. We found that 29 miRNAs conferred the major differences between stage C and the early stages in blood exosomes, and 37.9% of them predicted prognosis in the same direction in HCC tissues, suggesting that these miRNAs were mainly released from tumors. It is possible that miR-455-5p and miR-30c-5p could function as tumor suppressors[29,30], with decreased levels in stage C tumors compared with the early stages. Hence, profiles of blood exosomal miRNAs could be harnessed to monitor the development of tumors.
Affected by multiple factors, accurately differentiating HBV-positive HCC patients and non-HCC patients by blood miRNA levels is difficult. Zhou et al[31] constructed a 7-miRNA model to distinguish HCC patients from non-HCC patients with chronic hepatitis B. The sensitivity was 79.1% and the specificity was 76.4% in a clinical trial of 934 patients. However, this method is not significantly more accurate than current protein biomarkers, such as AFP, HSP, and GPC3[39,40]. In this study, because the exosomes could be derived from a vast range of cells, conventional normalization methods for each miRNA might not be appropriate. Instead, the biomarkers were searched in pairs to find the best reference sequence for each miRNA. More biomarkers included in a model could increase the accuracy, but also could increase the risk of over-fitting. Due to the limitation of sample size, only the top two pairs of miRNAs were included in the model, and their performance was comparable to that of other miRNA models or protein-based biomarkers. The sensitivity of this model was 94.1%, and advanced HCC patients were more easily detected than those in early stages. However, the specificity of this model was relatively low compared with other models or methods. One of the critical factors causing false positive results is alcohol consumption. Hence, to further improve the diagnostic accuracy, a more complex model constituted of biomarkers from multiple categories is necessary to eliminate the influence of various factors. Such a complex model will need to be validated in a large cohort of patients.
CONCLUSION
This study performed a cohort analysis of blood exosomal miRNA profiling to discover the complexity of differential diagnostic biomarkers for HCC and to confirm that exosomal miRNAs could be used as new biomarkers for HCC and liver cirrhosis. In addition, we showed that the accuracy of biomarkers to classify HCC and non-HCC patients from blood exosomal miRNAs could be affected by tumor stage and alcohol consumption habits. This study laid a foundation for understanding the complexity of blood exosomal miRNA profiles and for promoting the clinical application of blood exosomal miRNAs as biomarkers in the differential diagnosis of HCC.
ARTICLE HIGHLIGHTS
Research background
Hepatocellular carcinoma (HCC) is common malignancy with high morbidity and mortality. The differential diagnosis of HCC from non-HCC is an urgent demand in clinical practice. However, there is a lack of effective non-invasive diagnosis methods for the differential diagnosis of HCC. Blood exosomal micro ribonucleic acids (miRNAs) have been reported as promising biomarkers in various types of cancer, when biopsy or resection is not the first choice.
Research motivation
To better distinguish between HCC patients and non-HCC patients, a convenient, non-invasive method with high accuracy is needed in clinical practice.
Research objectives
In this study, we aimed to characterize the blood exosomal miRNA profiling between HCC patients and non-HCC patients, and investigate the feasibility of blood exosomal miRNAs as potential biomarkers for differential diagnosis.
Research methods
Eighty-nine patients from Xiangya Hospital of Central South University were enrolled in this study between August 2017 and May 2018. The expression of blood exosomal miRNAs in these patients was assessed using miRNA sequencing.
Research results
We explored the characteristics of blood exosomal miRNAs in 89 patients with HCC and non-HCC patients, who were enrolled for cohort 1 (n = 44) and cohort 2 (n = 45). The analysis result of principal component analysis suggested that daily alcohol consumption could alter the blood exosomal miRNA profiles of non-HCC patients through miR-3168 and miR-223-3p. The blood exosomal miRNA profiles also revealed that some miRNAs are related to the stage of the tumor tissues. Based on the exosomal miRNA expression profiles of 89 patients, two pairs of miRNAs (miR-584-5p/miR-106-3p and miR-628-3p/miR-941) were established to identify HCC. The pairs of miRNAs showed a 94.1% sensitivity and 68.4% specificity to differentiate HCC patients from non-HCC patients. In addition, alcohol consumption habit was found to influence the specificity.
Research conclusions
Exosomal miRNAs miR-584-5p/miR-106-3p combined with miR-628-3p/miR-941 could distinguish HCC patients and non-HCC patients. However, tumor stage and alcohol consumption habits could affect the accuracy of the two pairs of blood exosomal miRNAs.
Research perspectives
This study suggested the potentiality of blood exosomal miRNAs as novel non-invasive diagnosis biomarkers to differentiate between HCC patients and non-HCC patients.
ACKNOWLEDGEMENTS
The authors would like to thank all of the patients and their families for their support of this study.
Footnotes
Institutional review board statement: The study was approved by the ethics committee of Xiangya Hospital and informed consent was obtained from all patients or their relatives.
Conflict-of-interest statement: The authors declare no potential financial interests.
STROBE statement: The authors have read the STROBE Statement—checklist of items, and the manuscript was prepared and revised according to the STROBE Statement—checklist of items.
Manuscript source: Unsolicited manuscript
Peer-review started: June 2, 2020
First decision: July 21, 2020
Article in press: August 31, 2020
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Watanabe T S-Editor: Zhang L L-Editor: Wang TQ P-Editor: Wang LL
Contributor Information
Lang-Qing Sheng, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Jia-Rong Li, Department of Biliopancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Hao Qin, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Ling Liu, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Da-Dong Zhang, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Qi Zhang, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Meng-Li Huang, The Medical Department, 3D Medicines Inc., Shanghai 201114, China.
Xiao-Li Li, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Xiao-Ya Xu, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Yang-Nian Wei, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Zi-Shuo Chen, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Hui Luo, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Ji-Yang Zhang, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Cheng-Hui Zhou, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Hao Chen, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Ze-Guo Chen, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China.
Fu-Gen Li, Research and Development Institute of Precision Medicine, 3D Medicines Inc., Shanghai 201114, Hunan Province, China.
Nian-Feng Li, Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Changsha 410008, Hunan Province, China. 1535909540@qq.com.
Data sharing statement
The data used to support the findings of this study are available from the corresponding author upon request.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139–152. doi: 10.1038/s41575-019-0229-4. [DOI] [PubMed] [Google Scholar]
- 3.Chen W. Cancer statistics: updated cancer burden in China. Chin J Cancer Res. 2015;27:1. doi: 10.3978/j.issn.1000-9604.2015.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, Negri E. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302–309. doi: 10.1016/j.jhep.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peck-Radosavljevic M. Drug therapy for advanced-stage liver cancer. Liver Cancer. 2014;3:125–131. doi: 10.1159/000343868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:135–142. doi: 10.1002/jcb.27165. [DOI] [PubMed] [Google Scholar]
- 8.Liu XN, Cui DN, Li YF, Liu YH, Liu G, Liu L. Multiple "Omics" data-based biomarker screening for hepatocellular carcinoma diagnosis. World J Gastroenterol. 2019;25:4199–4212. doi: 10.3748/wjg.v25.i30.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Han X, Yu X, Xu Z, Yang G, Liu B, Xiu P. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res. 2018;37:213. doi: 10.1186/s13046-018-0893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, Yin Z, Xie X, Wang X, Chen W, Zhou L, Wang X, Li Y, Chen H, Gao L, Lu X, Wu T, Wang H, Niu J, Xu G. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67:662–675. doi: 10.1002/hep.29561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon Y, Jang ES, Choi YS, Kim JW, Jeong SH. Glypican-3 level assessed by the enzyme-linked immunosorbent assay is inferior to alpha-fetoprotein level for hepatocellular carcinoma diagnosis. Clin Mol Hepatol. 2016;22:359–365. doi: 10.3350/cmh.2016.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banales JM, Iñarrairaegui M, Arbelaiz A, Milkiewicz P, Muntané J, Muñoz-Bellvis L, La Casta A, Gonzalez LM, Arretxe E, Alonso C, Martínez-Arranz I, Lapitz A, Santos-Laso A, Avila MA, Martínez-Chantar ML, Bujanda L, Marin JJG, Sangro B, Macias RIR. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatology. 2019;70:547–562. doi: 10.1002/hep.30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444–1452. doi: 10.1002/ijc.31931. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, Jiang J, Huang X, Tong H, Tian Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018;7:1670–1679. doi: 10.1002/cam4.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun F, Wang JZ, Luo JJ, Wang YQ, Pan Q. Exosomes in the Oncobiology, Diagnosis, and Therapy of Hepatic Carcinoma: A New Player of an Old Game. Biomed Res Int. 2018;2018:2747461. doi: 10.1155/2018/2747461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383–390. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu LX, Zhang BL, Yang Y, Wang MC, Lei GL, Gao Y, Liu H, Xiao CH, Xu JJ, Qin H, Xu XY, Chen ZS, Zhang DD, Li FG, Zhang SG, Liu R. Exosomal microRNAs as potential biomarkers for cancer cell migration and prognosis in hepatocellular carcinoma patient-derived cell models. Oncol Rep. 2019;41:257–269. doi: 10.3892/or.2018.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suehiro T, Miyaaki H, Kanda Y, Shibata H, Honda T, Ozawa E, Miuma S, Taura N, Nakao K. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett. 2018;16:3267–3273. doi: 10.3892/ol.2018.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan JH, Zhou H, Zhao XX, Ding H, Li W, Qin L, Pan YL. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: Potential in diagnosis and antitumour treatments (Review) Int J Mol Med. 2018;41:1809–1816. doi: 10.3892/ijmm.2018.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buitrago DH, Patnaik SK, Kadota K, Kannisto E, Jones DR, Adusumilli PS. Small RNA sequencing for profiling microRNAs in long-term preserved formalin-fixed and paraffin-embedded non-small cell lung cancer tumor specimens. PLoS One. 2015;10:e0121521. doi: 10.1371/journal.pone.0121521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JT, Qin H, Man Cheung FK, Su J, Zhang DD, Liu SY, Li XF, Qin J, Lin JT, Jiang BY, Song Dong, Liao RQ, Qiang N, Yang XN, Tu HY, Zhou Q, Yang JJ, Zhang XC, Zhang YN, Wu YL, Zhong WZ. Plasma extracellular vesicle microRNAs for pulmonary ground-glass nodules. J Extracell Vesicles. 2019;8:1663666. doi: 10.1080/20013078.2019.1663666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manri C, Yokoi T, Nishida H. Size-Selective Harvesting of Extracellular Vesicles for Strategic Analyses towards Tumor Diagnoses. Appl Biochem Biotechnol. 2017;182:609–623. doi: 10.1007/s12010-016-2348-5. [DOI] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 29.Lai YH, Chen J, Wang XP, Wu YQ, Peng HT, Lin XH, Wang WJ. Collagen triple helix repeat containing-1 negatively regulated by microRNA-30c promotes cell proliferation and metastasis and indicates poor prognosis in breast cancer. J Exp Clin Cancer Res. 2017;36:92. doi: 10.1186/s13046-017-0564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandoval-Bórquez A, Polakovicova I, Carrasco-Véliz N, Lobos-González L, Riquelme I, Carrasco-Avino G, Bizama C, Norero E, Owen GI, Roa JC, Corvalán AH. MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin Epigenetics. 2017;9:114. doi: 10.1186/s13148-017-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed AA, Ali-Eldin ZA, Elbedewy TA, El-Serafy M, Ali-Eldin FA, AbdelAziz H. MicroRNAs and clinical implications in hepatocellular carcinoma. World J Hepatol. 2017;9:1001–1007. doi: 10.4254/wjh.v9.i23.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu HT, Liu RB, Liang YY, Hasan AME, Wang HY, Shao Q, Zhang ZC, Wang J, He CY, Wang F, Shao JY. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37:888–896. doi: 10.1111/liv.13356. [DOI] [PubMed] [Google Scholar]
- 34.Huang YH, Liang KH, Chien RN, Hu TH, Lin KH, Hsu CW, Lin CL, Pan TL, Ke PY, Yeh CT. A Circulating MicroRNA Signature Capable of Assessing the Risk of Hepatocellular Carcinoma in Cirrhotic Patients. Sci Rep. 2017;7:523. doi: 10.1038/s41598-017-00631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos SGRD, Mattos AA, Guimarães MM, Boger BS, Coral GP. Alcohol Consumption Influences Clinical Outcome in Patients Admitted to a Referral Center for Liver Disease. Ann Hepatol. 2018;17:470–475. doi: 10.5604/01.3001.0011.7391. [DOI] [PubMed] [Google Scholar]
- 36.Rocco A, Compare D, Angrisani D, Sanduzzi Zamparelli M, Nardone G. Alcoholic disease: liver and beyond. World J Gastroenterol. 2014;20:14652–14659. doi: 10.3748/wjg.v20.i40.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu T, Li L, Hu HQ, Meng XM, Huang C, Zhang L, Qin J, Li J. MicroRNAs in alcoholic liver disease: Recent advances and future applications. J Cell Physiol. 2018;234:382–394. doi: 10.1002/jcp.26938. [DOI] [PubMed] [Google Scholar]
- 38.Zheng H, Zou AE, Saad MA, Wang XQ, Kwok JG, Korrapati A, Li P, Kisseleva T, Wang-Rodriguez J, Ongkeko WM. Alcohol-dysregulated microRNAs in hepatitis B virus-related hepatocellular carcinoma. PLoS One. 2017;12:e0178547. doi: 10.1371/journal.pone.0178547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1:593–598. doi: 10.3892/mco.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou F, Shang W, Yu X, Tian J. Glypican-3: A promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med Res Rev. 2018;38:741–767. doi: 10.1002/med.21455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.