Abstract
Regulator of G protein signaling 10 (RGS10) belongs to the superfamily of RGS proteins, defined by the presence of a conserved RGS domain that canonically binds and deactivates heterotrimeric G-proteins. RGS proteins act as GTPase activating proteins (GAPs), which accelerate GTP hydrolysis on the G-protein α subunits and result in termination of signaling pathways downstream of G protein-coupled receptors. RGS10 is the smallest protein of the D/R12 subfamily and selectively interacts with Gαi proteins. It is widely expressed in many cells and tissues, with the highest expression found in the brain and immune cells. RGS10 expression is transcriptionally regulated via epigenetic mechanisms. Although RGS10 lacks multiple of the defined regulatory domains found in other RGS proteins, RGS10 contains post-translational modification sites regulating its expression, localization, and function. Additionally, RGS10 is a critical protein in the regulation of physiological processes in multiple cells, where dysregulation of its expression has been implicated in various diseases including Parkinson’s disease, multiple sclerosis, osteopetrosis, chemoresistant ovarian cancer and cardiac hypertrophy. This review summarizes RGS10 features and its regulatory mechanisms, and discusses the known functions of RGS10 in cellular physiology and pathogenesis of several diseases.
Keywords: RGS10, RGS proteins, GPCR, G-protein, regulatory mechanisms, ovarian cancer
1-. Introduction
1.1. GPCR/G-protein signaling
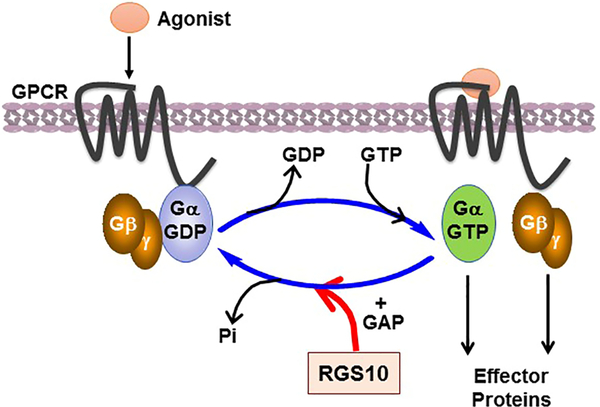
G protein-couple receptors (GPCRs) and the transduced heterotrimeric G-proteins, which modulate a cascade of intracellular effector proteins and chemical second messengers, are essential in the regulation of various physiological functions of cells and organ systems. They represent the largest family of FDA-approved targeted drugs for the treatment of a wide range of disorders caused by dysregulation of GPCR/G-protein signaling [1]. In the resting state, GPCRs are bound to inactive heterotrimeric G-proteins, consisting of the Gα-GDP and Gβγ dimer. In response to a variety of extracellular stimuli, GPCRs undergo conformational changes and act as guanine nucleotide exchange factors (GEFs) by promoting the exchange of GDP for GTP on Gα. As a result, activated GTP-Gα dissociates from Gβγ dimer, and thereby both can regulate downstream effector proteins [2] (Figure 1), such as enzymes, RhoGEFs, and ion channels that in turn initiate diverse signaling pathways mediating cellular responses [3–7]
Figure 1: G-protein activation and RGS protein-mediated deactivation cycle.
In response to various agonists, the seven transmembrane GPCR undergoes a conformational change and induces the exchange of GDP to GTP on Gα and dissociation from Gβγ dimer, which, in turn, both activated GTP-Gα and Gβγ dimer regulate downstream effectors. This cycle is terminated by the GAP activity of RGS proteins, which accelerates hydrolysis of GTP to GDP and thus terminates GPCR signaling.
The amplification and duration of signaling activity by Gα-GTP and Gβγ are tightly modulated by the intrinsic GTPase activity of Gα, in which the Gα subunit hydrolyzes GTP to GDP and promotes reassembly with the Gβγ dimer to reform the inactive Gαβγ protein complex. This, in turn, results in the deactivation of G-protein cycle. Even though Gα functions as a molecular switch and turns itself off by hydrolyzing its bound GTP to GDP, its intrinsic rate of GTP hydrolysis measured in vitro is very slow and does not account for the fast deactivation kinetic of G-proteins observed in vivo [8]. This observation has led to the discovery of proteins, named as regulators of G protein signaling that modulate the Gα-GTP hydrolysis rate, and thereby fine-tune the activation of G-proteins that mediate cellular signaling events.
1.2. RGS protein family
Regulator of G protein signaling (RGS) proteins that were initially discovered via genetic analysis in yeast and worm models [9–11], represent a diverse family of structural and multifunctional intracellular proteins that canonically regulate GPCRs and heterotrimeric G-protein signal transduction. RGS proteins control the lifetime of signaling pathways mediated by G-proteins. They act as a guanosine triphosphatase (GTPase) activating proteins (GAPs), which accelerate GTP hydrolysis of the active Gα-GTP form and, in turn, convert it back to the inactive Gα-GDP form. Consequently, this results in the termination of downstream G-protein signaling pathways (Figure 1).
To date, 20 canonical RGS proteins and 19 RGS-like proteins have been identified with various regulatory roles [12–14]. All canonical RGS proteins contain the conserved, approximately 120 amino acids long RGS domain, consisting of nine α helices that are subdivided into two sub-domains. The first subdomain forms a smaller helix bundle and is comprised of helices αI, II, III, VIII, and IX, while the larger bundle subdomain comprises helices αIV, V, VI, and VII [15]. The RGS domain selectively binds and stabilizes the Gα subunit in its transition state for GTP hydrolysis, mimicked by (GDP+AlF4−), resulting in GDP production and turning off signaling cascades regulated by G-proteins [15, 16].
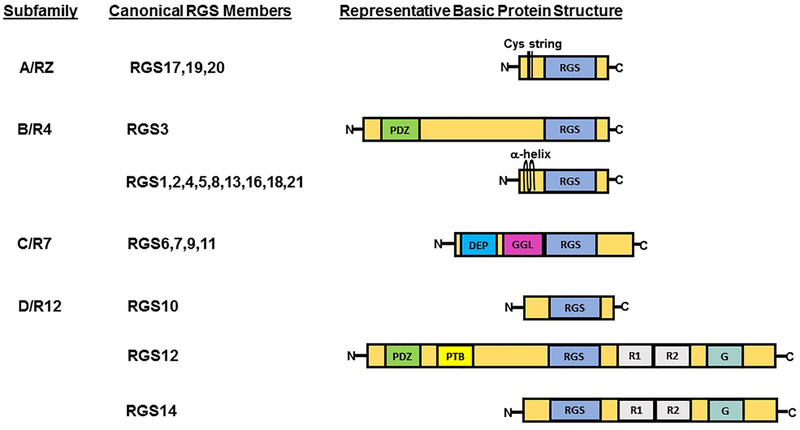
The structure of canonical RGS proteins ranges from small proteins comprised solely of an RGS domain responsible for GAP activity to more complex proteins containing multiple motifs and domains with functions in subcellular localization, protein stability, and protein-protein interactions. Based on the homology of the RGS domain and the presence of other common structural domains, canonical RGS proteins are categorized in four main subfamilies, consisting of A/RZ, B/R4, C/R7, and D/R12 (Figure 2). As negative regulators of G-protein signaling, RGS proteins mainly exert their GAP function on α subunits of the Gi/o and Gq families of G-proteins. Although there are no confirmed reports on GAP activity of any RGS domain toward Gαs, RGS proteins indirectly regulate Gαs downstream signaling through interaction with subtypes of adenylate cyclase (AC) [17, 18].
Figure 2: Classification and illustration of canonical RGS proteins based on the homology of the RGS domain and the presence of other common structural domains.
Protein domain and motif abbreviations are indicated as following: Cys string, cysteine string; α-helix, an amphipathic alpha–helix; RGS, regulator of G-protein signaling domain; DEP, disheveled EGL10-Pleckstrin homology domain; GGL, G protein gamma subunit-like domain; R1&R2, Ras/Rap-binding domain; G, G protein regulator motif; PTB, phosphotyrosine binding domain; PDZ, PSD95/Dlg/Z0-1/2 domain.
1.3. RGS D/R12 proteins subfamily
RGS D/12 subfamily is composed of three distinct structural proteins (Figure 2), containing RGS10, RGS12, and RGS14, which share high sequence identity within a conserved RGS domain (Figure 3A) and act as GAP for the Gi family Gα subunits. Whereas RGS10, at 20 kDa, is a relatively simple RGS protein, RGS12 and RGS14 are much larger and more complex than RGS10, and have multiple common structural and regulatory motifs and functional domains, including a pair of tandem Ras-binding domains (RBDs) and a C-terminal G-protein regulatory (GPR) motif, also known as the GoLoco motif. Through their first RBD domain, both RGS12 and RGS14 interact with activated small G-proteins, such as H-Ras-GTP and Rap-2-GTP [19–21], while the two tandem RBDs of RGS14 mediate interactions with Raf kinases [21] and calcium dependent-calmodulin (CaM) [22]. The GPR motif plays a role in subcellular localization and selectively binds inactive Gα proteins-GDP, thereby inhibiting the exchange of GDP toward GTP through its guanine nucleotide dissociation inhibitor (GDI) activity [23].
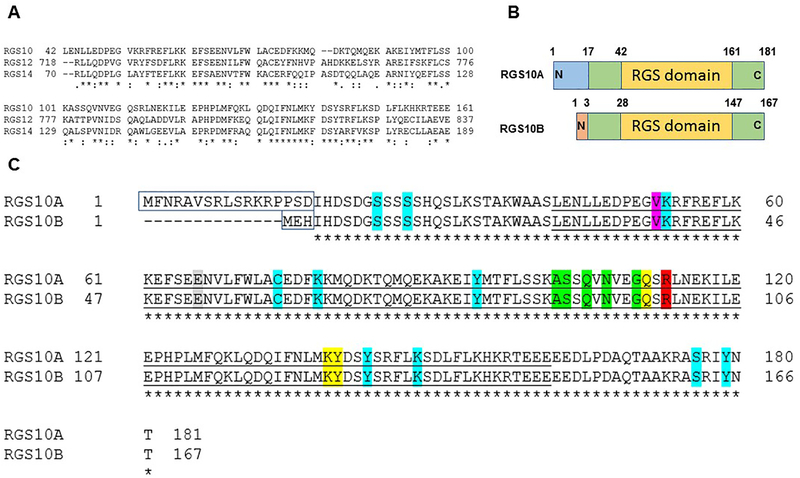
Figure 3: RGS10 isoforms: structure and analysis.
(A) Clustal Omega as a multiple sequence alignment program was used to align the RGS domain sequences within the D/R12 subfamily of human RGS proteins. (B) Schematic of the RGS10 isoform structures. (C) Alignment of human RGS10 isoform sequences using Clustal Omega program. Unfilled boxed amino acids represent N-terminal splice variants. Conserved catalytic RGS domains are underlined. The bright green highlighted residues, including (A102(88), S103(89), Q105(91), N107(93), G110(96)) are critical residues of RGS10 for the interaction with Gαi3 according to RGS10:Gαi3-AlF4− crystal structure (Protein Data Bank: 2IHB). The yellow-highlighted residues are putative disruptor residues, including Q111(97), K139(125), and Y140(126) that are predicted to impair the interaction with the Gα helical domain. The red-highlighted residue R113(99) is a conserved arginine residue within the RGS domain that does not contact the Gαi3 switch III region. The pink highlighted residue is a putative SNP that has been linked to schizophrenia. The gray-highlighted residue is the GAP dead mutation site based on the characterized GAP-dead mutation site in the RGS domain of RGS12 protein. Residue positions highlighted with turquoise are reported human RGS10 PTMs sites, including phosphorylation residues (S24(10), S27(13), Y94(80), Y143(129), S176(162), Y179(165)), ubiquitination residues (K53(39), K148(134)), palmitoylation C74(60) residue, and acetylation K78(64) residue.
Unlike RGS14, RGS12 is expressed as multiple isoforms. The longest RGS12 isoform is called trans-spliced-RGS12TS-L, is a 156 kDa protein that possesses two additional domains, including PTB and PDZ domains, making RGS12 the largest member of the RGS protein family. The PTB domain is involved in interacting with neuronal N-type calcium channels [24, 25], while the PDZ domain binds the C-terminal of the CXCR2 receptor [26], and both PTB/PDZ domains significantly attenuate ERK phosphorylation downstream of PDGFβ receptor activation [27]. RGS12 is abundant in the brain [28], heart [29] and osteoclasts [30], and is implicated in neuronal differentiation [31], bone disorders [32], cardiac hypertrophy [33] and lung and prostate cancers [34, 35].
RGS14, at 61 kDa, is expressed in the brain with a high enrichment in CA2 hippocampal neurons, where RGS14 naturally suppresses synaptic plasticity and limits learning and spatial memory [36]. Further, RGS14 is associated with cardiac remodeling [37], Parkinson’s disease (PD) [38], and kidney diseases [39–41]. This review primarily focuses on the small RGS protein RGS10, and discusses, in-depth, its characterization, regulatory mechanisms, and the physiological and pathological roles of RGS10 in several cells and mouse models.
2-. Characterization of RGS10 protein
2.1. Gene and protein organization
RGS10 is a small RGS protein that resembles the structure of R4 subfamily members. However, based on RGS domain sequence similarity, RGS10 is classified as a third member of the D/R12 subfamily which also includes the proteins RGS12 and RGS14 (Figure 3A) [42–44]. Despite the homology within their RGS domains, RGS10 is one of the smallest proteins of the entire RGS protein family and shares only a single conserved RGS domain in common with both RGS12 and RGS14 that contain more accessory domains. In addition to its conserved and functional RGS domain, RGS10 contains short disordered amino and carboxy-terminal extensions, harboring regulatory modification sites (Figure 3B).
RGS10 is highly conserved in humans and rodents and is encoded by a single gene (Rgs10) located at the position 10q26 on chromosome 11 in humans and on murine chromosome 7 [45, 46]. Alternative splicing of the first exon of the Rgs10 mouse gene results in two transcripts isoforms that share the last four exons and correspond to human transcripts with conserved exon structures. These two transcript variants yield two proteins, differing only by a few amino acids in their N-terminal sequences (Figure 3C). The long isoform, called RGS10A (RGS10–1/RGS10L), is a 181 amino-acid peptide, while the short isoform, termed RGS10B (RGS10–2/RGS10S), consists of 167 amino acids (Figure 3B). The third human isoform (173 aa), arising from an alternative start site upstream of the first shared exon, has been reported and shown to have GAP activity, but its homolog in mouse has not found yet [42, 45].
RGS10A (RGS10–1/RGS10L) is the predominant and functional isoform that is naturally expressed in osteoclasts [47], microglia [48], and ovarian cancer cells [49], while RGS10B (RGS10–2/RGS10S) lacking 14 amino acids at the N-terminus has impaired GAP function [50].
2.2. Tissue distribution
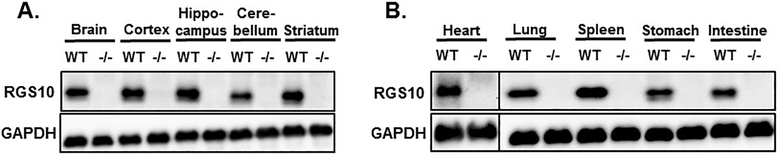
RGS10 is ubiquitously expressed; it has been detected in several tissues of humans and other mammals. Our results shown in Figure 4 confirm the wide tissue distribution of RGS10 protein expression and its lack in RGS10−/− mice. In human, the RGS10 mRNA is predominately expressed in the brain [51, 52], as well as subsets of immune cells, including CD4+ T cells [53], and monocyte-derived dendritic cells [54]. Also, the RGS10 transcript is highly expressed throughout the mouse and rat brains, with specific enrichment in the hippocampus, dorsal raphe, and striatum, regions of the brain that are implicated in mood disorders and anti-depressant treatment response [43, 55]. Further, RGS10 is found in peripheral tissues, including the cornea [56], heart, lung, testis and immune organs including the bone marrow, lymph nodes, and spleen, but its expression is not detectable in liver, kidney, and muscles [45]. Using an antibody ([C-20, sc-6206], Santa Cruz Biotechnology) raised against a synthetic peptide identical to the last 20 residues (EEDLPDAQTAAKRASRIYNT) at the C-terminus of RGS10 [55], protein expression of RGS10 was confirmed in our laboratory by western blotting in multiple parts of the brain including the cortex, hippocampus, cerebellum, and striatum (Figure 4A), and in several peripheral tissues including the heart, lung, spleen, stomach and intestine (Figure 4B). RGS10 expression is absent in the liver and the kidney (data not shown).
Figure 4: RGS10 protein expression in various tissues.
RGS10 protein expression was determined and compared by immunoblot analysis in (A) indicated brain regions, or (B) peripheral tissues (heart, lung, spleen, stomach, and intestine) of WT and RGS10-deficient mice. GAPDH was used as a loading control.
2.3. Subcellular localization
Instead of localizing to the plasma membrane, where the canonical G protein targets reside, numerous RGS proteins are primarily accumulated in the nucleus [57–60]. The nucleus as a cellular storage compartment may serve to restrain RGS proteins’ availability from regulating G-protein signaling or may allow them independently of their canonical function to regulate nuclear signal transduction through undefined nuclear interacting proteins [61]. Several reports (summarized in Table 1 and described below) demonstrated the localization of RGS10 to both the nucleus and the cytoplasm, with no significant plasma membrane localization. Chatterjee and Fisher (2000) first reported that RGS10 that is ectopically expressed in COS-7 cells and endogenously is expressed in H4-neuroglioma cells, is mainly localized to the nucleus [62]. In contrast to this finding, Burgon et al. (2001) demonstrated a predominant distribution in the cytoplasm of HEK-293 cells expressing RGS10-GFP fusion protein [63], while Lee and her colleagues observed endogenous RGS10 throughout cytoplasmic and nuclear cellular compartments in primary microglia [48]. Collectively, these findings suggest that the subcellular localization of RGS10 is cell type-specific.
Table 1:
Subcellular localization of RGS10
| Cells | Endogenous/Ectopic expression | Predominant localization under resting condition | Stimulus/modification | Predominant localization after cellular stimulation | Reference |
|---|---|---|---|---|---|
| COS-7 | Ectopic | Nucleus | ND | ND | [57] |
| Neurogliom a (H4) | Endogenous | Nucleus | ND | ND | |
| HEK293 | Ectopic | Cytoplasm | Forskolin/ phosphorylation by PKA at (Ser 168) | Nucleus | [63] |
| PC3-AR | Ectopic | Nucleus | Melatonin or 8-bromo-cGMP | Cytoplasm | [64] |
| Primary microglia | Endogenous | Cytoplasm=Nucleus | LPS | Nucleus | [48] |
ND indicates not determined.
Indeed, the cytoplasmic/nuclear translocation of RGS10 is a highly dynamic process. RGS10 can shuttle between the cytoplasm and the nucleus in response to cellular stimuli or signal-induced covalent modifications (Table 1). In microglia, RGS10 appears to be evenly distributed between the cytoplasm and nucleus under resting conditions. However, following lipopolysaccharide (LPS) stimulation, RGS10 is robustly enriched in the nucleus [48]. Furthermore, activation of Gαi signaling by melatonin promotes RGS10 translocation from the nucleus to the cytoplasm in PC3-AR prostate cancer cells [64]. Importantly, nuclear localization of RGS10 is induced through the cyclic AMP-dependent protein kinase A (PKA)-mediated phosphorylation of RGS10 on serine 168 [63], a residue located outside of its common RGS domain that contains a defined putative nuclear localization sequence [62]. Therefore, further sequence and deletion analyses are required to identify the potential nuclear localization sequences outside of the RGS domain and to elucidate the molecular mechanism underlying RGS10 nuclear localization in response to phosphorylation by PKA.
2.4. Gα binding and GAP activity
Many RGS proteins selectively target and negatively regulate a particular type of activated Gα protein signaling, while others have an affinity to interact with different Gα subtypes. RGS10 is selective to bind and terminate Gi family Gα subunits, including Gαi, Gαo, and Gαz [42, 44, 65]. Previous studies have defined and described the structure of RGS10 in an uncomplexed form compared to the structure of RGS10 complexed with Gαi3 [66, 67]. Soundararajan et al. (2008) demonstrated that RGS10 possesses a shorter flexible αVI helix and an extended αV-αVI loop containing 18 residues compared to typical 14 residues of αV- αVI loop for RGS domains of R4, R7, and Rz subfamilies [66].
Generally, the interaction between RGS10 and Gαi3 (critical residues highlighted in Figure 3C) is similar and consistent with R4 subfamily members-Gαi complexes with observed differences in the αVI helix. Due to the disorder of the entire RGS10 αVI helix in its complex with Gαi3, a conserved arginine residue in the αVI helix (R113(99), a putative modulatory residue in RGS domain highlighted in Figure 3C) does not make a direct interaction with Gαi3 switch III region as R4 subfamily members do interact with Gαi complexes through this conserved residue [66]. The RGS10 RGS domain exhibits a higher GAP activity toward Gαi1 and Gαz compared to RGS domains of RGS4 and RGS19 (GAIP) [44] and approximately the same GAP activity similar to RGS domains of RGS4 and RGS16 against Gαo [68]. However, the GAP activity of RGS10 is lower toward Gαi1 and Gαo compared to higher GAP RGS proteins, such as RGS4 and RGS16 [69], where they localize to the plasma membrane through their N-terminal amphipathic domain, efficiently contributing to their GAP activity with Gαz. Interestingly, replacement of the N-terminal domain of RGS10 with the N-terminal domain of RGS4 enhances its GAP activity toward Gαi and Gαz [50, 70], suggesting that plasma membrane targeting is essential for its GAP activity. Moreover, unlike RGS4 and RGS16, RGS10 contains putative disruptor residues (highlighted in Figure 3C) including Q111(97), K139(125), and Y140(126) that disrupt the contact with both Gαi1 and Gαo and consequently result in reduced GAP activity [69].
RGS10 has been shown to modulate Gαi-mediated receptor signaling. Adenoviral gene delivery-mediated RGS10 expression in CHOK1 cells stably expressing the human serotonin 5-HT1A receptor results in a significant reduction of Gαi-mediated inhibition of AC and inhibition of forskolin-stimulated cAMP production [71]. Consistent with its effect on 5-HT1A signaling, RGS10 overexpression attenuates the inhibition of AC activity mediated by μ (mu)-opioid receptor [72]. Further, RGS10 blocks CXCL12- stimulated chemokine CXCR4 receptor-mediated signaling in T cells [53]. Together, these findings support the selectivity of RGS10 in regulating GPCR-coupled Gαi signaling.
3-. Regulation of RGS10 function and expression
3.1. Epigenetic mechanisms
Numerous studies (summarized in Table 2) have shown the regulation of RGS10 expression in response to various stimuli in several cell types and disease models. Unlike other RGS proteins, RGS10 is regulated at the transcriptional level via epigenetic mechanisms. Typically, the transcription of genes is epigenetically regulated through alteration of chromatin structure, containing nucleosomes formed by DNA coiled around histones proteins [73]. Modification of DNA and histones leads to chromatin remodeling, which either facilitates or impedes the transcriptional machinery’s access to DNA. This results in the initiation or repression of gene expression [73]. Common mechanisms underlying chromatin modifications are DNA methylation and histone acetylation/deacetylation. DNA methylation is carried out by DNA methyltransferases (DNMTs) that add a methyl group directly to the fifth carbons of cytosine residues at CpG dinucleotides and mainly leads to gene silencing [74]. Histone acetylation is generally associated with transcriptional activation, and is mediated by adding acetyl groups on lysine residues of histone tails by histone acetyltransferases (HATs). In contrast, histone deacetylation involves the removal of acetyl groups from histone residues by histone deacetylases (HDACs) and is linked to transcriptional repression [75].
Table 2:
Regulation of RGS10 expression
| Cell/Tissue | Stimuli/Disease model | mRNA/Protein | Expression | Reference(s) |
|---|---|---|---|---|
| BMDM cell | FSL-1 100 nM (2&6 h) | mRNA | Decrease | [132] |
| LPS 10 ng/ml (3 h) | mRNA | |||
| LPS 100 ng/ml (48 h) | Protein | Decrease | [101] | |
| Primary microglia cell | LPS 10 ng/ml (6 h) | mRNA | Decrease | [78] |
| LPS 10 ng/ml (24,48&72 h) | Protein | Decrease | [48] | |
| BV2 cell | LPS 10 ng/ml (4–72 h) | mRNA | Decrease | [78] |
| LPS 10 ng/ml (48 h) | Protein | [48] | ||
| TNF-α 10 ng/ml (24,48&72 h) | Protein | |||
| TSA 100,250,500 nM (24 h) | mRNA | Increase | [78] | |
| S1P 10 μM (48 h) | mRNA | |||
| Spinal dorsal horn (L4-L5) tissue | pSNL model of inflammatory neuropathic pain (72 h) | Protein | Decrease | |
| MN9D cell | TNF-α 10 ng/ml (6&24 h) | mRNA/ Protein | Decrease | [99] |
| Phagocytic (MGdNΦ) microglia cell | Injection of apoptotic neurons (dN) into the cortex and hippocampus of WT mice | Protein | Decrease | [133] |
| Chondrocytes | Differentiation (9,12&15 days) | mRNA | Increase | [113] |
| Hippocampus | Acute electroconvulsive seizures ECS (24 h) | mRNA | Decrease | [134] |
| Ventral tegmental | Amphetamine (AMPH) self-administration | mRNA | Decrease | [135] |
| SKOV-3 cell | Cisplatin 100 μM (48 h) | mRNA | Decrease | [124] |
| 5-Aza (3,5,7&9 days) | mRNA | Increase | [49] | |
| A2780-AD cell | TSA 500 nM (48&36 h) | mRNA | Increase | [76] |
| 5-Aza 10 μM&20 μM (3,5&7 days) | mRNA | |||
| Caco-2 cell | Black tea polyphenol (Theaflavins TF-2) 50 μM (4–24 h) | mRNA | Increase | [136] |
| Striatum | Reserpine Acute treatment (30 min) | mRNA | Increase | [137] |
| Daily reserpine (5 days) | Decrease | |||
| Neonatal rat cardiomyocytes | Angiotensin II 50 μmol/L (24&48 h) | protein | Decrease | [108] |
| Human neural progenitor (hNP) | 5-Aza 5 μM (5 days) | mRNA | Increase | [77] |
| Molt-4 cell | CXCL-12 (30&60 min) | protein | Increase | [53] |
| Osteoclast derived BMM | RANKL+M-CSF (10 ng/ml) (30 min-96 h) | mRNA | Increase | [111] |
| protein | ||||
| RAW264.7 cell | RANKL+M-CSF (10 ng/ml) (30 min-96 h) | mRNA | Increase | [47] |
| Paw and spinal cord | Trimethylamine N-oxide (TMAO) (24 h) | Protein | Decrease | [138] |
BMDM, bone marrow derived macrophage; FSL-1, Pam2CGDPKHPKSF, synthetic diacylated lipoprotein; LPS, lipopolysaccharide; MN9D, mesencephalon neuroblastoma cell line; RANKL, receptor activator of nuclear factor-κB-ligand; TNF-α, tumor necrosis factor-alpha; TSA, trichostatin A; 5-Aza, 5-Azacytidine; M-CSF, Macrophage colony-stimulating factor; S1P, sphingosine-1-phosphate; pSNL, partial sciatic nerve ligation.
Several studies have demonstrated that DNA hypermethylation and histone deacetylation are critical epigenetic mechanisms mediating the regulation of RGS10 transcription in cancer cells, as well as neurons and macrophages. RGS10 expression is suppressed in chemoresistant ovarian cancer cells due to an increase in DNA methylation and histone deacetylation at its promoter compared to chemosensitive counterparts [49]. Suppression of either DNA methyltransferase 1 (DNMT1) or histone deacetylase 1 (HDAC1) using siRNA knockdown significantly increases the expression of RGS10 in chemoresistant A2780-AD ovarian cancer cells [76]. In addition, pharmacological inhibition of the activity of DNA methyltransferase with 5-Aza deoxycytidine (5-Aza) enhances the RGS10 transcript level in human neural progenitor cells [77]. In contrast, LPS-mediated activation of microglia, where RGS10 is highly enriched in the central nervous system [55], suppresses RGS10 expression by histone deacetylation with no significant effect in DNA methylation [78]. The suppression of RGS10 expression following microglia activation involving HDAC recruitment to the RGS10 promoter is blocked by Trichostatin A (TSA), a pharmacological inhibitor of HDAC enzyme activity [78]. Collectively, these studies provide evidence that RGS10 is regulated in a cell type-specific manner in response to distinct epigenetic mechanisms.
The previous studies have demonstrated a dynamic regulation of RGS10 expression by epigenetic mechanisms, suggesting that modified RGS10 could be used as a biomarker. Wen et al. (2015) use a method called Methylated CpG Tandems Amplification and Sequencing (MCTA-Seq) to detect DNA methylation in freely circulating DNA in the blood of hepatocellular carcinoma (HCC) patients and their control subjects. Strikingly, the study finds that RGS10 is among only four biomarkers that have strongly hypermethylated CpG islands in the blood of hepatocellular carcinoma patients compared to healthy individuals. Therefore, hypermethylated RGS10 may potentially aid in detecting the early stage of hepatocellular carcinoma [79].
In addition to chromatin-modifying mechanisms regulating RGS10 expression, miRNAs, which are short non-coding RNAs, control the expression of genes via binding and degrading their transcripts and thus subsequently resulting in repression in protein translation [80]. They have been implicated in the pathogenesis of numerous diseases through the dysregulation of their target genes including RGS genes [81]. A microarray study in multidrug-resistant (MDR) laryngeal cancer followed by miRNA Target Prediction Analysis has revealed that RGS10 is a putative target of has-miR-93 [82]. Although RGS10 is suppressed in various types of chemoresistant ovarian cancer, its expression is upregulated and inversely correlated with down-regulation of has-miR-93 expression in MDR laryngeal cancer Hep-2/v resistant cells compared to chemosensitive Hep-2 cells [82]. Taken together, these findings suggest that dysregulation of RGS10 expression could contribute to an acquired chemoresistant phenotype depending on the cancer cell type and its regulation by epigenetic mechanisms.
3.2. Post-translational modifications
Several RGS proteins are subjected to a wide range of post-translational modifications (PTM), which have profound impacts on their stability, GAP activity, interaction with binding partners, and subcellular localization [81, 83]. Despite the lack of multiple defined regulatory domains found in RGS12 and RGS14, RGS10 has various defined regulatory PTM sites, including phosphorylation, palmitoylation and ubiquitination (Figure 3C).
Phosphorylation of RGS10 on serine 168 at its C terminus by cAMP-dependent PKA triggers RGS10 localization from the cytosol to the nucleus. It thus results in attenuation of RGS10 availability at the plasma membrane needed to regulate G-protein-dependent activation of the G-protein-coupled inwardly-rectifying potassium (GIRK) channels without affecting its GTPase activity against the Gα protein [63]. Furthermore, a proteomic approach has identified RGS10 phosphorylation in human platelets following thrombin receptor activating peptide (TRAP) stimulation [84]. However, the site of RGS10 phosphorylation and the kinase activated by TRAP was not determined. The effects of RGS10 phosphorylation on its GAP activity and platelet activation are also unclear.
Unlike other RGS subfamily members that have the N-terminal amphipathic helix or cysteine string to localize to the cell membrane [85], RGS10 targets the plasma membrane by palmitoylation, which is a reversible reaction involving the attachment of a 16-carbon fatty acids palmitate to cysteine residues via a thioester linkage [86]. RGS10 is palmitoylated on a conserved cysteine 66 residue buried inside helix 4 within the RGS domain, which influences its GAP activity based on the assay used. Palmitoylation of this residue inhibits RGS10 GTPase activity in the single turnover-GTP hydrolysis assay in detergent solution but substantially potentiates steady-state GAP activity in receptor/G-protein reconstituted proteoliposomes [70, 87, 88]. Consistent with the previous positive impact of palmitoylation on RGS10 GAP activity in Spodoptera frugiperda (Sf9) insect cells, Castro-Fernandez et al. (2002) show constitutive palmitoylation of RGS10 on a conserved cysteine 60 in mammalian GH3 cells stably expressing the GnRH receptor (GGH3). This constitutive palmitoylation results in the inhibition of GnRH agonist-induced inositol phosphate (I.P.) and cAMP production, while the elimination of the palmitoylation effect by substitution of cysteine 60 by alanine abolishes RGS10 GAP activity on GnRH signaling [89].
In addition to phosphorylation and palmitoylation, a recent study using quantitative proteomics has identified RGS10 as a substrate for ubiquitination by the E3 ubiquitin ligase tripartite motif protein 32 (TRIM32). Degraded RGS10 is detected in lateral/medial ganglionic eminence (L/MGE) progenitors and results in promoting Rheb-GTP/mTOR hyperactivity that is required for GABAergic interneuron generation via enhancing L/MGE proliferation. On the other hand, the accumulation of RGS10 in response to TRIM32 deficiency or the MG-132 proteasome inhibitor inhibits mTOR signaling and thus causes L/MGE autophagy, a feature of autism-like behaviors [90]. Since RGS10 contains undefined functional domains outside of its RGS domain, it will be interesting to determine if the RGS domain mediates the TRIM32-RGS10 interaction and whether TRIM32 also interacts with other RGS proteins.
In addition to the defined and functional regulatory modification sites mentioned above, Squires et al. (2018) have identified and reported a list of PTM sites found on human RGS10 (highlighted in Figure 3C), including K53(39) and K148(134) (Ubiquitination), K78(64) (Acetylation), and S24(10), S27(13), Y94(80), Y143(129), and Y179(165) (Phosphorylation). The majority of these regulatory sites are found within the canonical RGS domain, and half of these sites overlap with reported human variants in RGS10 [91]. Therefore, further studies are needed to characterize the functional roles of these sites in regulating RGS10 GAP activity, subcellular localization, protein stability, and their implications in disease models.
4-. RGS10 functions in physiology and pathological diseases
Given its broad expression in diverse cells and tissues, RGS10 has emerged as an essential regulator involving cellular processes, physiological responses, and pathological conditions. This section reviews and discusses the results from biochemical, cellular, and in vivo studies that have revealed the physiological roles of RGS10 and its implications in pathological states. The overview of phenotypes and disease links arising from loss of RGS10 expression is present in Table 3.
Table 3:
RGS10-associated loss of function phenotypes and disease links
| Phenotype(s) | Proposed disease links | Year | Reference(s) |
|---|---|---|---|
| Impaired osteoclasts differentiation | Osteopetrosis | 2007 | [47, 111] |
| Neuroinflammation and neurodegeneration | Parkinson’s disease and a possible link to schizophrenia | 2008 | [48, 94, 96] |
| Reduced chemotherapy-induced cell death | Ovarian cancer chemoresistance | 2010 | [124, 125] |
| Hypertrophy following pressure overload | Heart failure | 2015 | [108] |
| Platelets aggregation and thrombogenesis | Aspirin resistance | 2016 | [119, 121] |
| Increased body weight with insulin resistance and glucose intolerance | Obesity and related metabolic syndromes | 2019 | [131] |
4.1. Neuroinflammation
RGS10 is normally expressed at high levels in the central nervous system, with particular enrichment in microglia [55, 92]. Microglia are the brain’s resident macrophages, which generally engulf pathogens and cell debris. However, chronic microglia activation leads to an amplified release of proinflammatory and neurotoxic mediators that eventually enhance inflammation-induced neurotoxicity. Injured neurons, in turn, produce danger signals that ultimately augment inflammatory microglial responses. This interplay between microglia and neurons results in an uncontrolled and persistent neuroinflammation, which is known to drive the progression of neurodegenerative pathogeneses, such as Alzheimer’s disease and PD [93, 94]. Microglia are mainly activated in response to bacterial LPS, tumor necrosis factor α (TNF-α), neurotoxins, extracellular ATP and 6-OHDA.
A polymorphism in the Rgs10 gene has been associated with age-related maculopathy [95] and schizophrenia [96]. Interestingly, activation of microglia by LPS or TNF-α triggers suppression of RGS10, which could directly amplify microglial inflammatory responses mediating neurotoxicity. Furthermore, RGS10-deficient mice exhibit enhanced CNS microglia burden, and activation. Interestingly, these mice are hypersensitive to systemic exposure of LPS-mediated substantia nigra pars compacta (SNpc) DA neuron degeneration, a phenotype of PD [48]. Activated primary microglia isolated from RGS10-deficient mice produce higher levels of inflammatory mediators, which consequently elevate media toxicity toward MN9D (mesencephalon DA neuroblastoma) cells, compared to WT mice [48, 97]. Similarly, dwon-regulation of RGS10 via siRNA in BV2 mouse microglia cells upregulates inflammation-related gene expression following LPS activation of TLR4, including TNF-α and IL-1β. Also, knockdown of RGS10 enhances MN9D cell death in response to activated BV2 cultural media incubation, which could be prevented by etanercept-mediated neutralization of TNF-α neurotoxic effects [48]. In primary microglia exposed to TNF-α or LPS, RG10 deficiency leads to enhanced expression and transcriptional activity of NF-κB, a major transcription factor mediating inflammatory signaling. Reciprocally, re-expression of RGS10 in RGS10-deficient primary microglia suppresses microglia activation, NF-κB activity, proinflammatory cytokines release, and inflammatory neurotoxicity. In addition, virus-mediated gene delivery of RGS10 into SNpc of rats significantly decreases 6-OHDA neurotoxin-induced microglia activation and DA neuron degeneration [97]. Consistent with these data, knockdown of RGS10 in BV2 cells enhances LPS-induced cyclooxygenase-2 (COX-2) expression and the release of its downstream effector, PGE2. Interestingly, amplification of LPS-stimulated TNF-α and COX-2 following microglial RGS10 suppression does not require Gαi activity [98]. Despite the importance of these results, the molecular mechanism underlying RGS10 regulation of microglial inflammatory signaling has not been clearly defined.
In addition to its higher microglial expression, RGS10 is also high and detectable within the nuclei of neurons [55]. MN9D cells transfected with direct siRNA-targeting RGS10 are more sensitive to the toxic effects of activated BV2 cultural media [48]. Due to the direct role of RGS10 in promoting DA neuron survival and regulation of microglial inflammatory responses, specifically TNF-α, and the fact that TNF-α/TNFR1 signaling is involved in DA neurons loss, the prosurvival function of RGS10 in DA neurons against TNF-α-induced cytotoxicity has been addressed. Treatment of MN9D cells with TNF-α suppresses RGS10 protein expression. As discussed RGS10 subcellular localization in the above sections, phosphorylation of RGS10 at serine 168 triggers its translocation to the nucleus. The overexpression of WT RGS10, but not of RGS10 S168A mutant which is resistant to PKA phosphorylation, reduces the neurotoxic effects of TNF-α in MN9D cells, through the inhibition of PARP-1 and caspase 3 cleavages and potentiation of PKA/CREB-mediated Bcl-2 anti-apoptotic expression. Consistent with these data, blocking PKA-induced RGS10 phosphorylation and the subsequent nuclear translocation limit the protective effect of RGS10 against TNF-α toxicity [99]. Together, these results suggest that phosphorylation of RGS10 by PKA is required for RGS10’s neuroprotective effect against cytotoxicity induced by TNF-α. In the context of aging, as a major risk factor for neurodegenerative diseases, RGS10 expression is decreased within Iba1+ cells in the brains of aged mice, which possibly causes changes in inflammatory responses or age-related cellular functions that could predispose to neurodegeneration [100]. Collectively, these findings provide evidence of the anti-inflammatory and neuroprotective roles of RGS10 in the CNS.
4.2. Immune system
Various types of leukocytes express RGS10. Within the immune system, RGS10 is strongly expressed in monocytes/macrophages [101], T lymphocytes [53], dendritic cells [54], and mast cells [102], with relatively low expression in B lymphocytes [103] and neutrophils [104].
In peripheral macrophages, RGS10 is the most highly expressed RGS protein and has a central role in macrophage polarization. RGS10 suppresses classical M1 activation through the downregulation of NF-κB activation and inflammatory cytokines release, and promotes markers of the alternatively activated M2 phenotype [101]. RGS10 also modulates NLRP3 and NLRC4 inflammasome activity in activated BMDM [105]. Interestingly, long-term, but not short-term, Gαi inhibition by PTX treatment during macrophage growth and differentiation suppresses LPS-induced M1 markers and upregulates IL-4-induced M2 markers [105]. Thus, it will be interesting to test whether amplification of inflammatory M1-related genes seen in RGS10-deficient, activated BMDMs is affected by long-term PTX treatment. Therefore, RGS10 acts as an anti-inflammatory protein and a critical modulator in the regulation of peripheral macrophage activation and differentiation.
In addition to macrophages, high expression of RGS10 has been detected in human monocyte-derived dendritic cells and murine BM-derived dendritic cells [54]. Although RGS10 expression is downregulated following LPS treatment in macrophages [101], LPS-mediated dendritic cell activation does not affect RGS10 expression [54]. Dendritic cells are professional antigen-presenting cells, mainly inducing T cell activation. Similar to dendritic cells of wild-type mice, antigen presentation and induction of CD4+ T cell proliferation are also observed in dendritic cells derived from RGS10-deficient mice [106].
A previous study has shown that RGS10 is found in human and mouse T cells, where its expression is upregulated in response to the activation of the chemokine receptor CXCR4 [53]. Functionally, loss of RGS10 enhances chemokine-induced Vav1–Rac1 activation mediating α4β1 and αLβ1 integrin-dependent T cell adhesion, while overexpression of RGS10 reduces this adhesion response triggered by chemokine treatment. In addition, the silencing of RGS10 amplifies Cdc42 activation mediating T cell migration following chemokine stimulation [53]. Collectively, these findings demonstrate that RGS10 is an efficient regulator that desensitizes signaling pathways controlling cellular immune responses.
4.3. Cardiac hypertrophy
Numerous recent studies have emerged that describe novel functions of RGS proteins in cardiovascular physiology and pathology [107]. Among various RGS proteins, RGS10 is expressed in normal human and mouse hearts, whereas RGS10 protein is significantly expressed at lower levels in failing human hearts and hypertrophic murine hearts. In addition, RGS10 protein expression is downregulated following Angiotensin II (AngII) treatment in neonatal rat cardiomyocytes [108]. However, the precise mechanism for the downregulation of RGS10 remains undefined.
Furthermore, the biological function of RGS10 in the heart has been proposed in the murine model of cardiac hypertrophy. Aortic banding mediated-pressure overload in RGS10-deficient mice displays cardiac hypertrophy, while overexpression of RGS10 alleviates this cardiac hypertrophy response [108]. Moreover, in RGS10-deficient neonatal cardiomyocytes, AngII induces hypertrophy, potentially through amplification of the MEK/ERK signaling pathway [108].
In addition, RGS10 has been reported to mediate the effect of β-adrenergic activation on endogenous G-protein-gated (GIRK) current deactivation in rat atrial myocytes [109]. Together, these findings strongly suggest that RGS10 is a cardioprotective protein through inhibition of the signaling downstream of AngII receptor, while its suppression may contribute to cardiac remodeling pathogenesis.
4.4. Osteoclastogenesis
Substantial evidences have revealed critical roles of RGS proteins in modulating functions of osteoclasts, bone-resorbing cells that tightly regulate and contribute to the process of bone remodeling and related pathological diseases [110]. RGS10, like other RGS proteins, is highly expressed in human osteoclasts and murine osteoclast like-cells, whereas RGS10 expression is induced in osteoclasts precursors by RANKL stimulation [47, 111], a control step required for differentiation of osteoclasts precursors into mature osteoclasts.
More importantly, the silencing of RGS10 in preosteoclasts or BMMs treated with RANKL impairs osteoclast differentiation through blocking intracellular Ca2+ oscillation and loss of expression of NFATc1, a master switch transcription factor in the regulation of terminal differentiation of osteoclasts [47, 111]. In contrast, overexpression of RGS10 results in enhanced RANKL sensitivity leading osteoclastogenesis by regulating Ca2+ oscillation-NFATc1 signaling pathway. Consistent with its role in modulating osteoclast cell differentiation, RGS10-deficient mice exhibit severe osteopetrosis and osteoclast differentiation defects [111]. Interestingly, the reintroduction of either RGS10 or NFATc1 in BMMs derived from RGS10-deficient mice rescues osteoclastogenesis defects. Further elucidation of the mechanism reveals that RGS10 interacts in a competitive fashion with calmodulin (CaM) and phosphatidylinositol 3,4,5-triphosphate (PIP3) in a calcium-dependent manner leading to PLCγ activation and Ca2+ oscillation, which subsequently activate NFATc1 signaling required for osteoclast terminal differentiation [47, 111]. Therefore, RGS10 is a crucial factor of osteoclastogenesis through potential regulation of the crosstalk between G-protein signaling and RANKL-dependent signaling pathway.
Moreover, the inhibition of RGS10 mediated by adeno-associated virus (AAV) reduces osteoclast markers and regulates the immune response in a murine model of periodontal disease, which is associated with bone loss [112]. In addition to its role in osteoclasts differentiation, RGS10 is expressed in chondrocytes, the resident cells of the cartilage. Similar to RGS5 and RGS7, RGS10 promotes chondrocyte differentiation [113], suggesting that RGS10 is essential for chondrogenesis involved in the normal function and development of cartilage.
4.5. Platelet functions
Many GPCR agonists, such as thrombin, adenosine diphosphate (ADP), and thromboxane A2 (TxA2), regulate platelet activation, which is essential for physiological hemostasis or thrombosis [114]. In addition to RGS18, RGS10 is predominately expressed in quiescent platelets [115] and modulates platelet activation via binding to the spinophilin (SPL) scaffolding protein forming a complex with the SHP-1 protein tyrosine phosphatase [116]. Following exposure to platelet activators, SHP-1 is phosphorylated, which subsequently triggers the dephosphorylation of SPL and, in turn, the decay of this complex [116]. Dissociation of the complex increases free RGS10 levels and its availability to limit G-protein signaling and allows SPL to interact with PP1 phosphatase and inhibits its activity [117], both of which regulate platelet activation. In addition to SPL/RGS10/SHP-1 complex, RGS10 interacts with 14–3-3γ scaffolding protein in resting platelets, whereas agonists of platelets induce RGS10 release from 14–3-3γ binding [118].
Furthermore, although RGS10 is a selective GAP for Gi family Gα subunits, platelets isolated from mice lacking RGS10 are hyperresponsive to stimuli that act via receptors coupled to Gi, Gq, and G12/13 proteins [118, 119]. In response to these stimuli, RGS10-deficient platelets exhibit aggregation, α-granule secretion, and integrin activation and enhance hemostasis and thrombogenesis [118, 119], without affecting platelet count and survival [120]. Along with more thrombosis formation, RGS10-deficient mice are more susceptible to ischemia [119]. Interestingly, RGS10 expression is higher in platelets of patients with aspirin resistance and metabolic syndrome compared to aspirin-sensitive patients [121], suggesting an indirect role for RGS10 in regulating platelet functions. Collectively, these studies indicate that RGS10 serves as a molecular brake on excessive activation of platelets and the process of pathological thrombosis.
4.6. Cancers
Given the enormous implications of GPCRs and cognate G-protein signaling in cancer initiation and progression [122], emerging evidences suggest a direct contribution of RGS proteins in tumor evolution, where dysregulation of RGS protein expression is observed and linked to many types of cancers [123]. RGS10 has been studied in ovarian cancer cells, where its expression is lower than normal ovary cells [49]. Interestingly, datasets comparing gene expression between multiple models of chemoresistant ovarian cancer cells and their parental, chemosensitive counterparts have demonstrated that the RGS10 transcript level is suppressed during the development of chemoresistance [124]. Further, RGS10 has been identified as a key modulator of ovarian cancer survival and chemoresistance, in which RGS10 knockdown via siRNA leads to enhanced ovarian cell viability and promotes chemotherapeutic drug resistance [124], potentially through activation of Rheb-GTP/mTOR signaling mediated ovarian cancer cell survival. Altman et al. (2015) have shown that RGS10 interacts with the monomeric GTP-binding Rheb protein, which is known to bind and activate the mechanistic target of rapamycin complex 1 (mTORC1). Elevated levels of active GTP-bound Rheb are observed in RGS10-deficient cells, which consequently amplify phosphorylation levels of AKT, mTOR, 4EB-BP1, p70S6 kinase, eIF2a, and ribosomal protein S6 [125].
A recent study has demonstrated RGS10-mediated regulation of Inflammatory signaling in SKOV-3 ovarian cancer cells, where the expression of TNF-α and COX-2 is robustly enhanced following RGS10 knockdown compared to control cells. Interestingly, the effects of RGS10 suppression on TNF-α and COX-2 expressions are independent of amplified Gαi signaling [98]. Given the finding that RGS10 modulates survival and chemoresistance of ovarian cancer and regulates inflammatory mediators’ expressions that have recently linked to ovarian cancer chemoresistance [126–128], further work is needed to investigate whether amplification of inflammatory signaling accounts for the enhanced survival and chemoresistance observed in RGS10-deficient ovarian cancer cells.
In addition to the functional studies of RGS10 in ovarian cancer, there is a couple of correlative studies linking the change in RGS10 expression to poor prognosis of multiple cancers, including laryngeal cancer [82], hepatocellular carcinoma [79], and pediatric acute myeloid leukemia [122]. Therefore, these studies suggest that RGS10 may serve as a biomarker for cancer diagnosis and detection.
4.7. Multiple sclerosis
Multiple sclerosis (MS) is a chronic inflammatory and demyelinating autoimmune disorder affecting the CNS. Even though the exact etiology of this disease remains unknown, several hypotheses and a variety of factors have been proposed, involving the pathological role of autoreactive infiltrating T cells attacking myelin and initiating inflammatory processes mediated by proinflammatory cytokines release, which in turn exacerbate MS disease severity [129]. The global lack of RGS10 does not alter the distribution and baseline numbers of immune cells, including B cells, CD4+ and CD8+ T cells, and monocytes in the brain, blood, and lymphoid tissues [100]. In comparison to WT cells, naïve splenic CD4+ T cells derived from RGS10-deficient mice exhibit intact TCR-induced proliferation, cytokine release, and in vitro Th1 or Th17 effector cell differentiation following activation by polarizing stimuli [106]. In contrast, in experimental autoimmune encephalomyelitis (EAE), a murine model of MS induced by myelin oligodendrocyte glycoprotein peptide fragment 35–55 (MOG35–55) immunization that mimics the immunopathological features of human MS, infiltration of both Th1 and Th17 cells in the CNS of RGS10-deficient mice (C57BL/6 background) is significantly reduced compared to WT counterparts (C57BL/6 background) [106].
Further, immunized RGS10-deficient mice have dramatically less clinical EAE symptoms associated with a significant reduction in T cell proliferation and cytokine production in response to in vitro MOG35–55 immunogen reexposure. Injection of in vitro differentiated RGS10-deficient Th1 cells (but not Th17 cells) into WT naïve recipient mice results in less EAE clinical phenotype compared to mice receiving in vitro Th1 cells from WT mice [106]. Due to its role in regulating T cell chemotaxis and its high expression in other immune cells, such as macrophages, further studies involving conditional knockout mice should aid in our understanding how RGS10 augments MS. Interestingly, in addition to the RGS1 protein, the transcript level of RGS10 is also higher in peripheral blood mononuclear cells (PBMCs) from MS patients compared to healthy individuals [130]. Therefore, this finding suggests that RGS10 modulates T cell functions that could contribute to this autoimmune disease.
4.8. Metabolic disorders
A high-fat diet (HFD), as one of the highly impactful environmental factors, induces body weight gain and central obesity, with various metabolic abnormalities. The role of RGS10 in metabolic changes related to HFD consumption has been studied in mice, where RGS10 expression is downregulated in the liver and upregulated in adipose tissues of WT mice fed with HFD compared to low-fat diet (LFD)-fed mice [131]. In comparison to WT, HFD-fed RGS10-deficient mice are more susceptible to the body weight gain associated with larger white adipose and liver tissue masses. More importantly, HFD-fed RGS10-deficient mice exhibit insulin resistance, while glucose intolerance and higher leptin levels are observed in RGS10-deficient mice following either LFD or HFD feeding compared to WT mice [131]. Furthermore, since RGS10 serves as an anti-inflammatory protein in macrophage activation, RGS10-deficient mice fed with HFD are more susceptible to chronic inflammation, as evidenced by higher M1 inflammatory gene transcripts in the liver and adipose tissues and lower mRNAs of Fizz1 and YM1 anti-inflammatory M2 markers [131]. Thus, this study sheds light on the importance of RGS10 in managing HFD-induced body weight gain and related metabolic disorders.
5-. Targeting RGS10 expression
Compelling evidence from several studies has shown that suppression of RGS10 in various cell types is linked to the pathogenesis of diverse diseases models, such as PD [48], cardiac hypertrophy [108], ovarian cancer chemoresistance [124], and osteopetrosis [47, 111]. More importantly, RGS10 expression is silenced in many of these cells, where the impact of RGS10 loss leads to these pathologies. For example, RGS10 expression is suppressed in microglia and neurons by inflammatory stimuli and in cardiomyocytes in response to AngII treatment. Likewise, RGS10 expression is significantly downregulated in multiple chemoresistant ovarian cancer cells compared to chemosensitive ones. As discussed above, both DNA methylation by DNMTs and histone deacetylation by HDACs mediate epigenetic silencing of RGS10 that could consequently contribute to the development of resistance to chemotherapy in ovarian cancer. Pharmacological inhibition of both enzymatic activities of DNMTs by 5-Aza and HDACs by TSA restore RGS10 expression and enhance the sensitivity of chemoresistant ovarian cancer cells to chemotherapy. Indeed, these inhibitors are not selective and have more side effects; thus, more work is needed to elucidate the molecular mechanisms and further protein targets that control RGS10 expression and function. Identification of these molecular mechanisms will provide valuable insight into the function of RGS10 and will facilitate strategies to target RGS10 in various disorders in which its function is implicated. Altogether, stabilizing RGS10 expression could potentially be a useful and promising strategy in the treatment of various ailments.
6-. Conclusions and future perspective
Since its initial discovery, growing knowledge and considerable progress have been achieved in defining the canonical GAP functions, as well as regulatory mechanisms of RGS10 and its potential roles in a number of pathophysiological states. Despite its simple structure, the studies discussed herein suggest that RGS10 is a critical modulator of G-protein and inflammatory signaling and a wide range of physiological cellular processes, including survival, polarization, adhesion, chemotaxis, and differentiation in multiple types of cells. Together, these findings implicate RGS10 as an attractive and promising therapeutic target for the treatment of pathological conditions in various target cells, in which alteration in its expression and function is involved. Although RGS10 has many functions in these systems, the molecular mechanisms governing its functions have not been fully defined. Therefore, further research should focus on identifying binding partners of RGS10, which will be fundamental in understating both the mechanism underlying RGS10 function and the mechanism controlling RGS10 expression in different disease states. As discussed above, RGS10 expression is suppressed following different signals in several cellular and animal models, which in turn impacts their pathophysiology. Thus, defining regulatory factors in a specific cellular environment will facilitate the development of therapeutic strategies to target RGS10 in all its implicated diseases. In addition, it will help future efforts to design and identify selective compounds that stabilize RGS10 expression using high-throughput screening libraries. Given the high expression of RGS10 in immune cells, particularly macrophages, and its role in the regulation of inflammatory responses in these cells, future studies could explore the role of RGS10 in infectious disease models in which immune and inflammatory responses have a significant influence.
Highlights.
RGS10 is dysregulated in response to multiple stimuli and various disease models.
Epigenetic and post-translational mechanisms regulate RGS10 expression and function.
RGS10 has emerged functions in cellular physiology and pathology.
Stabilizing RGS10 expression represents a promising therapeutic strategy.
Acknowledgments
Funding: This work was supported by the University of Georgia research Foundation startup grant to B.R. and the National Institutes of Health (to B.R., R01AI146857-01A1).
Abbreviations
- AC
adenylate cyclase
- AlF4
aluminum fluoride
- AngII
angiotensin II
- AR
androgen receptor
- ATP
adenosine triphosphate
- BMDMs
bone marrow-derived macrophages
- CaM
calmodulin
- CNS
central nervous system
- COX-2
cyclooxygenase-2
- DNMTs
DNA methyltransferases
- ERK
extracellular signal-regulated kinase
- GAP
GTPase-activating protein
- GDP
guanosine diphosphate
- GEFs
guanine nucleotide exchange factors
- GFP
green fluorescent protein
- GIRK
G protein-coupled inwardly-rectifying potassium channel
- GPCR
G protein–coupled receptor
- GTP
guanosine triphosphate
- HATs
histone acetyltransferases
- HDACs
histone deacetylases
- 6-OHDA
6-hydroxydopamine
- HFT
high fat diet
- kDa
kilodalton
- KD
knockdown
- KO
knockout
- LFD
low-fat diet
- L/MGE
lateral/medial ganglionic
- LPS
lipopolysaccharide
- MN9D
mesencephalon neuroblastoma cell
- MS
multiple sclerosis
- NFATc1
nuclear factor of activated T cells 1
- NF-κB
nuclear factor kappa B
- PGE2
prostaglandin E2
- PD
Parkinson’s disease
- PKA
protein kinase A
- PTM
post-translational modifications
- PTX
pertussis toxin
- RANKL
receptor activator of nuclear factor (NF)-kB-ligand
- RGS
regulator of G protein signaling
- siRNA
small interfering RNA
- SNP
single nucleotide polymorphism
- SNpc
substantia nigra pars compacta
- TLR
toll-like receptor
- TNF-α
tumor necrosis factor-alpha
- TSA
trichostatin A
- WT
wild-type
Footnotes
Competing Interests
The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lagerstrom MC, Schioth HB, Structural diversity of G protein-coupled receptors and significance for drug discovery, Nat Rev Drug Discov 7(4) (2008) 339–57. [DOI] [PubMed] [Google Scholar]
- [2].Gilman AG, G proteins: transducers of receptor-generated signals, Annu Rev Biochem 56 (1987) 615–49. [DOI] [PubMed] [Google Scholar]
- [3].Clapham DE, Neer EJ, G protein beta gamma subunits, Annu Rev Pharmacol Toxicol 37 (1997) 167–203. [DOI] [PubMed] [Google Scholar]
- [4].Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC, p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13, Science 280(5372) (1998) 2109–11. [DOI] [PubMed] [Google Scholar]
- [5].Simonds WF, G protein regulation of adenylate cyclase, Trends Pharmacol Sci 20(2) (1999) 66–73. [DOI] [PubMed] [Google Scholar]
- [6].Kammermeier PJ, Ruiz-Velasco V, Ikeda SR, A voltage-independent calcium current inhibitory pathway activated by muscarinic agonists in rat sympathetic neurons requires both Galpha q/11 and Gbeta gamma, J Neurosci 20(15) (2000) 5623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rhee SG, Regulation of phosphoinositide-specific phospholipase C, Annu Rev Biochem 70 (2001) 281–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tsang SH, Burns ME, Calvert PD, Gouras P, Baylor DA, Goff SP, Arshavsky VY, Role for the target enzyme in deactivation of photoreceptor G protein in vivo, Science 282(5386) (1998) 117–21. [DOI] [PubMed] [Google Scholar]
- [9].Chan RK, Otte CA, Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones, Mol Cell Biol 2(1) (1982) 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J, Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit), Mol Cell Biol 16(9) (1996) 5194–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Koelle MR, Horvitz HR, EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins, Cell 84(1) (1996) 115–25. [DOI] [PubMed] [Google Scholar]
- [12].Ross EM, Wilkie TM, GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins, Annu Rev Biochem 69 (2000) 795–827. [DOI] [PubMed] [Google Scholar]
- [13].Hollinger S, Hepler JR, Cellular regulation of RGS proteins: modulators and integrators of G protein signaling, Pharmacol Rev 54(3) (2002) 527–59. [DOI] [PubMed] [Google Scholar]
- [14].Willars GB, Mammalian RGS proteins: multifunctional regulators of cellular signalling, Semin Cell Dev Biol 17(3) (2006) 363–76. [DOI] [PubMed] [Google Scholar]
- [15].Tesmer JJ, Berman DM, Gilman AG, Sprang SR, Structure of RGS4 bound to AlF4−-activated G(i alpha1): stabilization of the transition state for GTP hydrolysis, Cell 89(2) (1997) 251–61. [DOI] [PubMed] [Google Scholar]
- [16].Berman DM, Kozasa T, Gilman AG, The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis, J Biol Chem 271(44) (1996) 27209–12. [DOI] [PubMed] [Google Scholar]
- [17].Roy AA, Baragli A, Bernstein LS, Hepler JR, Hebert TE, Chidiac P, RGS2 interacts with Gs and adenylyl cyclase in living cells, Cell Signal 18(3) (2006) 336–48. [DOI] [PubMed] [Google Scholar]
- [18].Talbot JN, Roman DL, Clark MJ, Roof RA, Tesmer JJ, Neubig RR, Traynor JR, Differential modulation of mu-opioid receptor signaling to adenylyl cyclase by regulators of G protein signaling proteins 4 or 8 and 7 in permeabilised C6 cells is Galpha subtype dependent, J Neurochem 112(4) (2010) 1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Traver S, Bidot C, Spassky N, Baltauss T, De Tand MF, Thomas JL, Zalc B, Janoueix-Lerosey I, Gunzburg JD, RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of galphao, Biochem J 350 Pt 1 (2000) 19–29. [PMC free article] [PubMed] [Google Scholar]
- [20].Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP, Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector, PLoS One 4(3) (2009) e4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shu FJ, Ramineni S, Hepler JR, RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways, Cell Signal 22(3) (2010) 366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Evans PR, Gerber KJ, Dammer EB, Duong DM, Goswami D, Lustberg DJ, Zou J, Yang JJ, Dudek SM, Griffin PR, Seyfried NT, Hepler JR, Interactome Analysis Reveals Regulator of G Protein Signaling 14 (RGS14) is a Novel Calcium/Calmodulin (Ca(2+)/CaM) and CaM Kinase II (CaMKII) Binding Partner, J Proteome Res 17(4) (2018) 1700–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kimple RJ, De Vries L, Tronchere H, Behe CI, Morris RA, Gist Farquhar M, Siderovski DP, RGS12 and RGS14 GoLoco motifs are G alpha(i) interaction sites with guanine nucleotide dissociation inhibitor Activity, J Biol Chem 276(31) (2001) 29275–81. [DOI] [PubMed] [Google Scholar]
- [24].Schiff ML, Siderovski DP, Jordan JD, Brothers G, Snow B, De Vries L, Ortiz DF, Diverse-Pierluissi M, Tyrosine-kinase-dependent recruitment of RGS12 to the N-type calcium channel, Nature 408(6813) (2000) 723–7. [DOI] [PubMed] [Google Scholar]
- [25].Richman RW, Strock J, Hains MD, Cabanilla NJ, Lau KK, Siderovski DP, Diverse-Pierluissi M, RGS12 interacts with the SNARE-binding region of the Cav2.2 calcium channel, J Biol Chem 280(2) (2005) 1521–8. [DOI] [PubMed] [Google Scholar]
- [26].Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, Siderovski DP, GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain, J Biol Chem 273(28) (1998) 17749–55. [DOI] [PubMed] [Google Scholar]
- [27].Sambi BS, Hains MD, Waters CM, Connell MC, Willard FS, Kimple AJ, Pyne S, Siderovski DP, Pyne NJ, The effect of RGS12 on PDGFbeta receptor signalling to p42/p44 mitogen activated protein kinase in mammalian cells, Cell Signal 18(7) (2006) 971–81. [DOI] [PubMed] [Google Scholar]
- [28].Snow BE, Antonio L, Suggs S, Gutstein HB, Siderovski DP, Molecular cloning and expression analysis of rat Rgs12 and Rgs14, Biochem Biophys Res Commun 233(3) (1997) 770–7. [DOI] [PubMed] [Google Scholar]
- [29].Doupnik CA, Xu T, Shinaman JM, Profile of RGS expression in single rat atrial myocytes, Biochim Biophys Acta 1522(2) (2001) 97–107. [DOI] [PubMed] [Google Scholar]
- [30].Yang S, Li YP, RGS12 is essential for RANKL-evoked signaling for terminal differentiation of osteoclasts in vitro, J Bone Miner Res 22(1) (2007) 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Willard MD, Willard FS, Li X, Cappell SD, Snider WD, Siderovski DP, Selective role for RGS12 as a Ras/Raf/MEK scaffold in nerve growth factor-mediated differentiation, EMBO J 26(8) (2007) 2029–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yuan X, Cao J, Liu T, Li YP, Scannapieco F, He X, Oursler MJ, Zhang X, Vacher J, Li C, Olson D, Yang S, Regulators of G protein signaling 12 promotes osteoclastogenesis in bone remodeling and pathological bone loss, Cell Death Differ 22(12) (2015) 2046–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Huang J, Chen L, Yao Y, Tang C, Ding J, Fu C, Li H, Ma G, Pivotal Role of Regulator of G-protein Signaling 12 in Pathological Cardiac Hypertrophy, Hypertension 67(6) (2016) 1228–36. [DOI] [PubMed] [Google Scholar]
- [34].Dai J, Gu J, Lu C, Lin J, Stewart D, Chang D, Roth JA, Wu X, Genetic variations in the regulator of G-protein signaling genes are associated with survival in late-stage non-small cell lung cancer, PLoS One 6(6) (2011) e21120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang Y, Wang J, Zhang L, Karatas OF, Shao L, Zhang Y, Castro P, Creighton CJ, Ittmann M, RGS12 Is a Novel Tumor-Suppressor Gene in African American Prostate Cancer That Represses AKT and MNX1 Expression, Cancer Res 77(16) (2017) 4247–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Evans PR, Dudek SM, Hepler JR, Regulator of G Protein Signaling 14: A Molecular Brake on Synaptic Plasticity Linked to Learning and Memory, Prog Mol Biol Transl Sci 133 (2015) 169–206. [DOI] [PubMed] [Google Scholar]
- [37].Li Y, Tang XH, Li XH, Dai HJ, Miao RJ, Cai JJ, Huang ZJ, Chen AF, Xing XW, Lu Y, Yuan H, Regulator of G protein signalling 14 attenuates cardiac remodelling through the MEK-ERK1/2 signalling pathway, Basic Res Cardiol 111(4) (2016) 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vogt IR, Lees AJ, Evert BO, Klockgether T, Bonin M, Wullner U, Transcriptional changes in multiple system atrophy and Parkinson’s disease putamen, Exp Neurol 199(2) (2006) 465–78. [DOI] [PubMed] [Google Scholar]
- [39].Urabe Y, Tanikawa C, Takahashi A, Okada Y, Morizono T, Tsunoda T, Kamatani N, Kohri K, Chayama K, Kubo M, Nakamura Y, Matsuda K, A genome-wide association study of nephrolithiasis in the Japanese population identifies novel susceptible Loci at 5q35.3, 7p14.3, and 13q14.1, PLoS Genet 8(3) (2012) e1002541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yasui T, Okada A, Urabe Y, Usami M, Mizuno K, Kubota Y, Tozawa K, Sasaki S, Higashi Y, Sato Y, Kubo M, Nakamura Y, Matsuda K, Kohri K, A replication study for three nephrolithiasis loci at 5q35.3, 7p14.3 and 13q14.1 in the Japanese population, J Hum Genet 58(9) (2013) 588–93. [DOI] [PubMed] [Google Scholar]
- [41].Mahajan A, Rodan AR, Le TH, Gaulton KJ, Haessler J, Stilp AM, Kamatani Y, Zhu G, Sofer T, Puri S, Schellinger JN, Chu PL, Cechova S, van Zuydam N, Consortium S, BioBank Japan P, Arnlov J, Flessner MF, Giedraitis V, Heath AC, Kubo M, Larsson A, Lindgren CM, Madden PAF, Montgomery GW, Papanicolaou GJ, Reiner AP, Sundstrom J, Thornton TA, Lind L, Ingelsson E, Cai J, Martin NG, Kooperberg C, Matsuda K, Whitfield JB, Okada Y, Laurie CC, Morris AP, Franceschini N, Trans-ethnic Fine Mapping Highlights Kidney-Function Genes Linked to Salt Sensitivity, Am J Hum Genet 99(3) (2016) 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hunt TW, Fields TA, Casey PJ, Peralta EG, RGS10 is a selective activator of G alpha i GTPase activity, Nature 383(6596) (1996) 175–7. [DOI] [PubMed] [Google Scholar]
- [43].Gold SJ, Ni YG, Dohlman HG, Nestler EJ, Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain, J Neurosci 17(20) (1997) 8024–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Popov S, Yu K, Kozasa T, Wilkie TM, The regulators of G protein signaling (RGS) domains of RGS4, RGS10, and GAIP retain GTPase activating protein activity in vitro, Proc Natl Acad Sci U S A 94(14) (1997) 7216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Haller C, Fillatreau S, Hoffmann R, Agenes F, Structure, chromosomal localization and expression of the mouse regulator of G-protein signaling10 gene (mRGS10), Gene 297(1–2) (2002) 39–49. [DOI] [PubMed] [Google Scholar]
- [46].Sierra DA, Gilbert DJ, Householder D, Grishin NV, Yu K, Ukidwe P, Barker SA, He W, Wensel TG, Otero G, Brown G, Copeland NG, Jenkins NA, Wilkie TM, Evolution of the regulators of G-protein signaling multigene family in mouse and human, Genomics 79(2) (2002) 177–85. [DOI] [PubMed] [Google Scholar]
- [47].Yang S, Chen W, Stashenko P, Li YP, Specificity of RGS10A as a key component in the RANKL signaling mechanism for osteoclast differentiation, J Cell Sci 120(Pt 19) (2007) 3362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee JK, McCoy MK, Harms AS, Ruhn KA, Gold SJ, Tansey MG, Regulator of G-protein signaling 10 promotes dopaminergic neuron survival via regulation of the microglial inflammatory response, J Neurosci 28(34) (2008) 8517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ali MW, Cacan E, Liu Y, Pierce JY, Creasman WT, Murph MM, Govindarajan R, Eblen ST, Greer SF, Hooks SB, Transcriptional suppression, DNA methylation, and histone deacetylation of the regulator of G-protein signaling 10 (RGS10) gene in ovarian cancer cells, PLoS One 8(3) (2013) e60185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ajit SK, Young KH, Analysis of chimeric RGS proteins in yeast for the functional evaluation of protein domains and their potential use in drug target validation, Cell Signal 17(7) (2005) 817–25. [DOI] [PubMed] [Google Scholar]
- [51].Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M, Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system, Brain Res Mol Brain Res 122(1) (2004) 24–34. [DOI] [PubMed] [Google Scholar]
- [52].Taymans JM, Leysen JE, Langlois X, Striatal gene expression of RGS2 and RGS4 is specifically mediated by dopamine D1 and D2 receptors: clues for RGS2 and RGS4 functions, J Neurochem 84(5) (2003) 1118–27. [DOI] [PubMed] [Google Scholar]
- [53].Garcia-Bernal D, Dios-Esponera A, Sotillo-Mallo E, Garcia-Verdugo R, Arellano-Sanchez N, Teixido J, RGS10 restricts upregulation by chemokines of T cell adhesion mediated by alpha4beta1 and alphaLbeta2 integrins, J Immunol 187(3) (2011) 1264–72. [DOI] [PubMed] [Google Scholar]
- [54].Shi GX, Harrison K, Han SB, Moratz C, Kehrl JH, Toll-like receptor signaling alters the expression of regulator of G protein signaling proteins in dendritic cells: implications for G protein-coupled receptor signaling, J Immunol 172(9) (2004) 5175–84. [DOI] [PubMed] [Google Scholar]
- [55].Waugh JL, Lou AC, Eisch AJ, Monteggia LM, Muly EC, Gold SJ, Regional, cellular, and subcellular localization of RGS10 in rodent brain, J Comp Neurol 481(3) (2005) 299–313. [DOI] [PubMed] [Google Scholar]
- [56].Wu F, Lee S, Schumacher M, Jun A, Chakravarti S, Differential gene expression patterns of the developing and adult mouse cornea compared to the lens and tendon, Exp Eye Res 87(3) (2008) 214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chatterjee TK, Fisher RA, Novel alternative splicing and nuclear localization of human RGS12 gene products, J Biol Chem 275(38) (2000) 29660–71. [DOI] [PubMed] [Google Scholar]
- [58].Dulin NO, Pratt P, Tiruppathi C, Niu J, Voyno-Yasenetskaya T, Dunn MJ, Regulator of G protein signaling RGS3T is localized to the nucleus and induces apoptosis, J Biol Chem 275(28) (2000) 21317–23. [DOI] [PubMed] [Google Scholar]
- [59].Zmijewski JW, Song L, Harkins L, Cobbs CS, Jope RS, Second messengers regulate RGS2 expression which is targeted to the nucleus, Biochim Biophys Acta 1541(3) (2001) 201–11. [DOI] [PubMed] [Google Scholar]
- [60].Chatterjee TK, Liu Z, Fisher RA, Human RGS6 gene structure, complex alternative splicing, and role of N terminus and G protein gamma-subunit-like (GGL) domain in subcellular localization of RGS6 splice variants, J Biol Chem 278(32) (2003) 30261–71. [DOI] [PubMed] [Google Scholar]
- [61].Sethakorn N, Yau DM, Dulin NO, Non-canonical functions of RGS proteins, Cell Signal 22(9) (2010) 1274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chatterjee TK, Fisher RA, Cytoplasmic, nuclear, and golgi localization of RGS proteins. Evidence for N-terminal and RGS domain sequences as intracellular targeting motifs, J Biol Chem 275(31) (2000) 24013–21. [DOI] [PubMed] [Google Scholar]
- [63].Burgon PG, Lee WL, Nixon AB, Peralta EG, Casey PJ, Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10), J Biol Chem 276(35) (2001) 32828–34. [DOI] [PubMed] [Google Scholar]
- [64].Rimler A, Jockers R, Lupowitz Z, Sampson SR, Zisapel N, Differential effects of melatonin and its downstream effector PKCalpha on subcellular localization of RGS proteins, J Pineal Res 40(2) (2006) 144–52. [DOI] [PubMed] [Google Scholar]
- [65].Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ, RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits, Nature 383(6596) (1996) 172–5. [DOI] [PubMed] [Google Scholar]
- [66].Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundstrom M, Doyle DA, Siderovski DP, Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits, Proc Natl Acad Sci U S A 105(17) (2008) 6457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Taylor VG, Bommarito PA, Tesmer JJ, Structure of the Regulator of G Protein Signaling 8 (RGS8)-Galphaq Complex: MOLECULAR BASIS FOR Galpha SELECTIVITY, J Biol Chem 291(10) (2016) 5138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY, Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions, Nat Struct Mol Biol 18(7) (2011) 846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Asli A, Sadiya I, Avital-Shacham M, Kosloff M, “Disruptor” residues in the regulator of G protein signaling (RGS) R12 subfamily attenuate the inactivation of Galpha subunits, Sci Signal 11(534) (2018). [DOI] [PubMed] [Google Scholar]
- [70].Tu Y, Woodson J, Ross EM, Binding of regulator of G protein signaling (RGS) proteins to phospholipid bilayers. Contribution of location and/or orientation to Gtpase-activating protein activity, J Biol Chem 276(23) (2001) 20160–6. [DOI] [PubMed] [Google Scholar]
- [71].Ghavami A, Hunt RA, Olsen MA, Zhang J, Smith DL, Kalgaonkar S, Rahman Z, Young KH, Differential effects of regulator of G protein signaling (RGS) proteins on serotonin 5HT1A, 5-HT2A, and dopamine D2 receptor-mediated signaling and adenylyl cyclase activity, Cell Signal 16(6) (2004) 711–21. [DOI] [PubMed] [Google Scholar]
- [72].Xie Z, Li Z, Guo L, Ye C, Li J, Yu X, Yang H, Wang Y, Chen C, Zhang D, Liu-Chen LY, Regulator of G protein signaling proteins differentially modulate signaling of mu and delta opioid receptors, Eur J Pharmacol 565(1–3) (2007) 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Venkatesh S, Workman JL, Histone exchange, chromatin structure and the regulation of transcription, Nat Rev Mol Cell Biol 16(3) (2015) 178–89. [DOI] [PubMed] [Google Scholar]
- [74].Smith ZD, Meissner A, DNA methylation: roles in mammalian development, Nat Rev Genet 14(3) (2013) 204–20. [DOI] [PubMed] [Google Scholar]
- [75].Bannister AJ, Kouzarides T, Regulation of chromatin by histone modifications, Cell Res 21(3) (2011) 381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cacan E, Ali MW, Boyd NH, Hooks SB, Greer SF, Inhibition of HDAC1 and DNMT1 modulate RGS10 expression and decrease ovarian cancer chemoresistance, PLoS One 9(1) (2014) e87455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Tuggle K, Ali MW, Salazar H, Hooks SB, Regulator of G protein signaling transcript expression in human neural progenitor differentiation: R7 subfamily regulation by DNA methylation, Neurosignals 22(1) (2014) 43–51. [DOI] [PubMed] [Google Scholar]
- [78].Alqinyah M, Maganti N, Ali MW, Yadav R, Gao M, Cacan E, Weng HR, Greer SF, Hooks SB, Regulator of G Protein Signaling 10 (Rgs10) Expression Is Transcriptionally Silenced in Activated Microglia by Histone Deacetylase Activity, Mol Pharmacol 91(3) (2017) 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wen L, Li J, Guo H, Liu X, Zheng S, Zhang D, Zhu W, Qu J, Guo L, Du D, Jin X, Zhang Y, Gao Y, Shen J, Ge H, Tang F, Huang Y, Peng J, Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients, Cell Res 25(12) (2015) 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z, The microRNA world: small is mighty, Trends Biochem Sci 28(10) (2003) 534–40. [DOI] [PubMed] [Google Scholar]
- [81].Alqinyah M, Hooks SB, Regulating the regulators: Epigenetic, transcriptional, and post-translational regulation of RGS proteins, Cell Signal 42 (2018) 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yin W, Wang P, Wang X, Song W, Cui X, Yu H, Zhu W, Identification of microRNAs and mRNAs associated with multidrug resistance of human laryngeal cancer Hep-2 cells, Braz J Med Biol Res 46(6) (2013) 546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kach J, Sethakorn N, Dulin NO, A finer tuning of G-protein signaling through regulated control of RGS proteins, Am J Physiol Heart Circ Physiol 303(1) (2012) H19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Garcia A, Prabhakar S, Hughan S, Anderson TW, Brock CJ, Pearce AC, Dwek RA, Watson SP, Hebestreit HF, Zitzmann N, Differential proteome analysis of TRAP-activated platelets: involvement of DOK-2 and phosphorylation of RGS proteins, Blood 103(6) (2004) 2088–95. [DOI] [PubMed] [Google Scholar]
- [85].Bernstein LS, Grillo AA, Loranger SS, Linder ME, RGS4 binds to membranes through an amphipathic alpha -helix, J Biol Chem 275(24) (2000) 18520–6. [DOI] [PubMed] [Google Scholar]
- [86].Jones TL, Role of palmitoylation in RGS protein function, Methods Enzymol 389 (2004) 33–55. [DOI] [PubMed] [Google Scholar]
- [87].Tu Y, Wang J, Ross EM, Inhibition of brain Gz GAP and other RGS proteins by palmitoylation of G protein alpha subunits, Science 278(5340) (1997) 1132–5. [DOI] [PubMed] [Google Scholar]
- [88].Tu Y, Popov S, Slaughter C, Ross EM, Palmitoylation of a conserved cysteine in the regulator of G protein signaling (RGS) domain modulates the GTPase-activating activity of RGS4 and RGS10, J Biol Chem 274(53) (1999) 38260–7. [DOI] [PubMed] [Google Scholar]
- [89].Castro-Fernandez C, Janovick JA, Brothers SP, Fisher RA, Ji TH, Conn PM, Regulation of RGS3 and RGS10 palmitoylation by GnRH, Endocrinology 143(4) (2002) 1310–7. [DOI] [PubMed] [Google Scholar]
- [90].Zhu JW, Zou MM, Li YF, Chen WJ, Liu JC, Chen H, Fang LP, Zhang Y, Wang ZT, Chen JB, Huang W, Li S, Jia WQ, Wang QQ, Zhen XC, Liu CF, Li S, Xiao ZC, Xu GQ, Schwamborn JC, Schachner M, Ma QH, Xu RX, Absence of TRIM32 Leads to Reduced GABAergic Interneuron Generation and Autism-like Behaviors in Mice via Suppressing mTOR Signaling, Cereb Cortex (2019). [DOI] [PubMed] [Google Scholar]
- [91].Squires KE, Montanez-Miranda C, Pandya RR, Torres MP, Hepler JR, Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease, Pharmacol Rev 70(3) (2018) 446–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL, Identification of a unique TGF-beta-dependent molecular and functional signature in microglia, Nat Neurosci 17(1) (2014) 131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Subhramanyam CS, Wang C, Hu Q, Dheen ST, Microglia-mediated neuroinflammation in neurodegenerative diseases, Semin Cell Dev Biol 94 (2019) 112–120. [DOI] [PubMed] [Google Scholar]
- [94].Tansey MG, Goldberg MS, Neuroinflammation in Parkinson’s disease: its role in neuronal death and implications for therapeutic intervention, Neurobiol Dis 37(3) (2010) 510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB, Susceptibility genes for age-related maculopathy on chromosome 10q26, Am J Hum Genet 77(3) (2005) 389–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hishimoto A, Shirakawa O, Nishiguchi N, Aoyama S, Ono H, Hashimoto T, Maeda K, Novel missense polymorphism in the regulator of G-protein signaling 10 gene: analysis of association with schizophrenia, Psychiatry Clin Neurosci 58(5) (2004) 579–81. [DOI] [PubMed] [Google Scholar]
- [97].Lee JK, Chung J, McAlpine FE, Tansey MG, Regulator of G-protein signaling-10 negatively regulates NF-kappaB in microglia and neuroprotects dopaminergic neurons in hemiparkinsonian rats, J Neurosci 31(33) (2011) 11879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Alqinyah M, Almutairi F, Wendimu MY, Hooks SB, RGS10 Regulates the Expression of Cyclooxygenase-2 and Tumor Necrosis Factor Alpha through a G Protein-Independent Mechanism, Mol Pharmacol 94(4) (2018) 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Lee JK, Chung J, Druey KM, Tansey MG, RGS10 exerts a neuroprotective role through the PKA/c-AMP response-element (CREB) pathway in dopaminergic neuron-like cells, J Neurochem 122(2) (2012) 333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kannarkat GT, Lee JK, Ramsey CP, Chung J, Chang J, Porter I, Oliver D, Shepherd K, Tansey MG, Age-related changes in regulator of G-protein signaling (RGS)-10 expression in peripheral and central immune cells may influence the risk for age-related degeneration, Neurobiol Aging 36(5) (2015) 1982–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lee JK, Chung J, Kannarkat GT, Tansey MG, Critical role of regulator G-protein signaling 10 (RGS10) in modulating macrophage M1/M2 activation, PLoS One 8(11) (2013) e81785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bansal G, DiVietro JA, Kuehn HS, Rao S, Nocka KH, Gilfillan AM, Druey KM, RGS13 controls g protein-coupled receptor-evoked responses of human mast cells, J Immunol 181(11) (2008) 7882–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Moratz C, Harrison K, Kehrl JH, Regulation of chemokine-induced lymphocyte migration by RGS proteins, Methods Enzymol 389 (2004) 15–32. [DOI] [PubMed] [Google Scholar]
- [104].Cho H, Kamenyeva O, Yung S, Gao JL, Hwang IY, Park C, Murphy PM, Neubig RR, Kehrl JH, The loss of RGS protein-Galpha(i2) interactions results in markedly impaired mouse neutrophil trafficking to inflammatory sites, Mol Cell Biol 32(22) (2012) 4561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Vural A, Nabar NR, Hwang IY, Sohn S, Park C, Karlsson MCI, Blumer JB, Kehrl JH, Galphai2 Signaling Regulates Inflammasome Priming and Cytokine Production by Biasing Macrophage Phenotype Determination, J Immunol 202(5) (2019) 1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lee JK, Kannarkat GT, Chung J, Joon Lee H, Graham KL, Tansey MG, RGS10 deficiency ameliorates the severity of disease in experimental autoimmune encephalomyelitis, J Neuroinflammation 13 (2016) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zhang P, Mende U, Regulators of G-protein signaling in the heart and their potential as therapeutic targets, Circ Res 109(3) (2011) 320–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Miao R, Lu Y, Xing X, Li Y, Huang Z, Zhong H, Huang Y, Chen AF, Tang X, Li H, Cai J, Yuan H, Regulator of G-Protein Signaling 10 Negatively Regulates Cardiac Remodeling by Blocking Mitogen-Activated Protein Kinase-Extracellular Signal-Regulated Protein Kinase 1/2 Signaling, Hypertension 67(1) (2016) 86–98. [DOI] [PubMed] [Google Scholar]
- [109].Bender K, Nasrollahzadeh P, Timpert M, Liu B, Pott L, Kienitz MC, A role for RGS10 in beta-adrenergic modulation of G-protein-activated K+ (GIRK) channel current in rat atrial myocytes, J Physiol 586(8) (2008) 2049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Jules J, Yang S, Chen W, Li YP, Role of Regulators of G Protein Signaling Proteins in Bone Physiology and Pathophysiology, Prog Mol Biol Transl Sci 133 (2015) 47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yang S, Li YP, RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation, Genes Dev 21(14) (2007) 1803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Yang S, Hao L, McConnell M, Zhou X, Wang M, Zhang Y, Mountz JD, Reddy M, Eleazer PD, Li YP, Chen W, Inhibition of Rgs10 Expression Prevents Immune Cell Infiltration in Bacteria-induced Inflammatory Lesions and Osteoclast-mediated Bone Destruction, Bone Res 1(3) (2013) 267–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Appleton CT, James CG, Beier F, Regulator of G-protein signaling (RGS) proteins differentially control chondrocyte differentiation, J Cell Physiol 207(3) (2006) 735–45. [DOI] [PubMed] [Google Scholar]
- [114].Offermanns S, Activation of platelet function through G protein-coupled receptors, Circ Res 99(12) (2006) 1293–304. [DOI] [PubMed] [Google Scholar]
- [115].Kim SD, Sung HJ, Park SK, Kim TW, Park SC, Kim SK, Cho JY, Rhee MH, The expression patterns of RGS transcripts in platelets, Platelets 17(7) (2006) 493–7. [DOI] [PubMed] [Google Scholar]
- [116].Ma P, Cierniewska A, Signarvic R, Cieslak M, Kong H, Sinnamon AJ, Neubig RR, Newman DK, Stalker TJ, Brass LF, A newly identified complex of spinophilin and the tyrosine phosphatase, SHP-1, modulates platelet activation by regulating G protein-dependent signaling, Blood 119(8) (2012) 1935–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ma P, Foote DC, Sinnamon AJ, Brass LF, Dissociation of SHP-1 from spinophilin during platelet activation exposes an inhibitory binding site for protein phosphatase-1 (PP1), PLoS One 10(3) (2015) e0119496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ma P, Gupta S, Sampietro S, DeHelian D, Tutwiler V, Tang A, Stalker TJ, Brass LF, RGS10 shapes the hemostatic response to injury through its differential effects on intracellular signaling by platelet agonists, Blood Adv 2(16) (2018) 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hensch NR, Karim ZA, Druey KM, Tansey MG, Khasawneh FT, RGS10 Negatively Regulates Platelet Activation and Thrombogenesis, PLoS One 11(11) (2016) e0165984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].DeHelian DJ, Gupta S, Wu J, Thorsheim CL, Estevez B, Cooper M, Litts K, Lee-Sundlov MM, Hoffmeister K, Poncz M, Ma P, Brass LF, RGS10 and RGS18 differentially limit platelet activation, promote platelet production, and prolong platelet survival, Blood (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Mao Y, Lei L, Su J, Yu Y, Liu Z, Huo Y, Regulators of G protein signaling are upregulated in aspirin-resistant platelets from patients with metabolic syndrome, Pharmazie 69(5) (2014) 371–3. [PubMed] [Google Scholar]
- [122].Chaudhury S, O’Connor C, Canete A, Bittencourt-Silvestre J, Sarrou E, Prendergast A, Choi J, Johnston P, Wells CA, Gibson B, Keeshan K, Age-specific biological and molecular profiling distinguishes paediatric from adult acute myeloid leukaemias, Nat Commun 9(1) (2018) 5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Hurst JH, Hooks SB, Regulator of G-protein signaling (RGS) proteins in cancer biology, Biochem Pharmacol 78(10) (2009) 1289–97. [DOI] [PubMed] [Google Scholar]
- [124].Hooks SB, Callihan P, Altman MK, Hurst JH, Ali MW, Murph MM, Regulators of G-Protein signaling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells, Mol Cancer 9 (2010) 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Altman MK, Alshamrani AA, Jia W, Nguyen HT, Fambrough JM, Tran SK, Patel MB, Hoseinzadeh P, Beedle AM, Murph MM, Suppression of the GTPase-activating protein RGS10 increases Rheb-GTP and mTOR signaling in ovarian cancer cells, Cancer Lett 369(1) (2015) 175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Spinella F, Rosano L, Di Castro V, Nicotra MR, Natali PG, Bagnato A, Inhibition of cyclooxygenase-1 and −2 expression by targeting the endothelin a receptor in human ovarian carcinoma cells, Clin Cancer Res 10(14) (2004) 4670–9. [DOI] [PubMed] [Google Scholar]
- [127].Mantovani A, Allavena P, Sica A, Balkwill F, Cancer-related inflammation, Nature 454(7203) (2008) 436–44. [DOI] [PubMed] [Google Scholar]
- [128].Maccio A, Madeddu C, Inflammation and ovarian cancer, Cytokine 58(2) (2012) 133–47. [DOI] [PubMed] [Google Scholar]
- [129].Filippi M, Bar-Or A, Piehl F, Preziosa P, Solari A, Vukusic S, Rocca MA, Author Correction: Multiple sclerosis, Nat Rev Dis Primers 4(1) (2018) 49. [DOI] [PubMed] [Google Scholar]
- [130].Kemppinen AK, Kaprio J, Palotie A, Saarela J, Systematic review of genome-wide expression studies in multiple sclerosis, BMJ Open 1(1) (2011) e000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Fang X, Chung J, Olsen E, Snider I, Earls RH, Jeon J, Park HJ, Lee JK, Depletion of regulator-of-G-protein signaling-10 in mice exaggerates high-fat diet-induced insulin resistance and inflammation, and this effect is mitigated by dietary green tea extract, Nutr Res 70 (2019) 50–59. [DOI] [PubMed] [Google Scholar]
- [132].Riekenberg S, Farhat K, Debarry J, Heine H, Jung G, Wiesmuller KH, Ulmer AJ, Regulators of G-protein signalling are modulated by bacterial lipopeptides and lipopolysaccharide, FEBS J 276(3) (2009) 649–59. [DOI] [PubMed] [Google Scholar]
- [133].Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, Greco DJ, Smith ST, Tweet G, Humulock Z, Zrzavy T, Conde-Sanroman P, Gacias M, Weng Z, Chen H, Tjon E, Mazaheri F, Hartmann K, Madi A, Ulrich JD, Glatzel M, Worthmann A, Heeren J, Budnik B, Lemere C, Ikezu T, Heppner FL, Litvak V, Holtzman DM, Lassmann H, Weiner HL, Ochando J, Haass C, Butovsky O, The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases, Immunity 47(3) (2017) 566–581 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Gold SJ, Heifets BD, Pudiak CM, Potts BW, Nestler EJ, Regulation of regulators of G protein signaling mRNA expression in rat brain by acute and chronic electroconvulsive seizures, J Neurochem 82(4) (2002) 828–38. [DOI] [PubMed] [Google Scholar]
- [135].Sun H, Calipari ES, Beveridge TJ, Jones SR, Chen R, The brain gene expression profile of dopamine D2/D3 receptors and associated signaling proteins following amphetamine self-administration, Neuroscience 307 (2015) 253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lu J, Gosslau A, Liu AY, Chen KY, PCR differential display-based identification of regulator of G protein signaling 10 as the target gene in human colon cancer cells induced by black tea polyphenol theaflavin monogallate, Eur J Pharmacol 601(1–3) (2008) 66–72. [DOI] [PubMed] [Google Scholar]
- [137].Geurts M, Maloteaux JM, Hermans E, Altered expression of regulators of G-protein signaling (RGS) mRNAs in the striatum of rats undergoing dopamine depletion, Biochem Pharmacol 66(7) (2003) 1163–70. [DOI] [PubMed] [Google Scholar]
- [138].Zhang Y, Zhang C, Li H, Hou J, The Presence of High Levels of Circulating Trimethylamine N-Oxide Exacerbates Central and Peripheral Inflammation and Inflammatory Hyperalgesia in Rats Following Carrageenan Injection, Inflammation 42(6) (2019) 2257–2266. [DOI] [PubMed] [Google Scholar]