Abstract
Colorectal cancer (CRC) is the third most common cancer in the world and its development is associated with oncogenic dysfunction. Therefore, the present study aimed to identify differentially expressed genes (DEGs) in CRC tissues and to determine the role of keratin 80 (KRT80) in CRC cell proliferation. DEGs were initially screened in 32 paired CRC tissues and matched adjacent normal tissues from RNA-Seq datasets in The Cancer Genome Atlas database using the limma package in R software. In total, 2,114 DEGs were identified, of which KRT80 was discovered to be the most upregulated in CRC tissues. Moreover, increased KRT80 expression levels were confirmed in tissues collected from 50 patients with CRC using reverse transcription-quantitative PCR, and its increased expression levels were significantly associated with increased lymph node and distant metastasis and a higher pathological stage. Furthermore, KRT80 knockdown using siRNA decreased the viability and proliferation of CRC cells. Finally, pathway analysis revealed that the proteins co-expressed with KRT80 in CRC were enriched in the cell cycle, DNA replication, immune system, metabolism of protein and RNA, signal transduction and other cellular processes. Among them, the cell cycle and DNA replication pathways contained the highest number of the proteins identified. In conclusion, the findings of the present study suggested that KRT80 may be overexpressed in CRC tissues. Furthermore, KRT80 may be involved in the proliferation of CRC cells, which is likely through its ability to regulate the cell cycle and DNA replication pathways, thus it may serve as a potential therapeutic target for patients with CRC.
Keywords: colorectal cancer, keratin 80, proliferation, cell cycle, DNA replication
Introduction
Colorectal cancer (CRC) is the most common malignancy of the digestive system and it is associated with high morbidity and mortality rates (1). According to the cancer statistics from 185 countries, ~800,977 new CRC cases were reported in 2018 and the mortality rate was as high as 47.8% (2). Although radical surgical resection combined with radiotherapy or chemotherapy is widely used for the treatment of CRC, its efficacy remains low; the 5-year survival rate of patients with CRC without metastasis is 40-90%, which drops to 10-15% in patients exhibiting metastasis (3). Thus, clinically effective therapeutic targets for CRC are still limited (4), and the identification of novel specific molecular targets is required to identify the mechanisms underlying CRC tumorigenesis and for the development of new drugs.
Gene expression profiling analysis based on large datasets serves an increasingly important role in determining potential molecular markers for different types of cancer (5,6). To investigate new CRC-related genes, the present study screened differentially expressed genes (DEGs) in 32 patients with CRC from RNA-Seq datasets in The Cancer Genome Atlas (TCGA) database and identified keratin 80 (KRT80) as the most upregulated gene. KRT80 is located on chromosome 12q13 and encodes a 452-amino-acid protein that is a type II keratin (7,8). Keratins are intermediate filament cytoskeletal proteins that maintain the structural integrity of epithelial cells and they have also been reported to be representative markers for epithelial cells (9,10). Keratin expression is tissue-specific and related to advanced tissue or cell differentiation (10). Previous studies have revealed that keratins were extensively expressed in human cancer and suggested that they may be used as molecular markers for the diagnosis of multiple types of tumor, such as basal cell carcinoma, oral squamous cell carcinoma, bladder cancer, breast cancer, hepatocellular carcinoma, cervical cancer and gastric adenocarcinoma (11). Furthermore, it has been reported that keratins served an important role in the regulation of cancer cell migration and invasion (12,13). In fact, the keratin family in humans consists of 28 type I keratins (KRT9-KRT40) and 26 type II keratins (KRT1-KRT8, KRT71-KRT86) (14,15). Among them, KRT7, KRT8 and KRT18-KRT20 were found to participate in the proliferation and differentiation of colon cells (16-18). However, to the best of our knowledge, there are few reports investigating the association between KRT80 expression levels and human cancer. Especially in CRC, only one previous study has demonstrated that KRT80 promoted the migration and invasion of CRC cells (19). Thus, the functional role of KRT80 in CRC cell proliferation remains relatively unclear.
In the present study, gene expression profiling analysis was performed to identify DEGs associated with CRC. Following the identification of KRT80, the study aimed to investigate the expression, function and underlying mechanisms of KRT80 in CRC cell proliferation. The findings supported the role of KRT80 in CRC development, and provided further experimental evidence and a theoretical basis for using KRT80 as a therapeutic target in patients with CRC.
Materials and methods
Data mining and analysis
The mRNA expression profiles of human CRC cases from RNA-Seq datasets in the TCGA database (TCGA_COADREAD_exp_HiSeqV2-2015-02-24) were downloaded from the UCSC Xena Browser (https://xenabrowser.net/datapages/). The DEGs between 32 CRC tissues and adjacent normal tissues were identified using the limma package (v3.34.9) of R software (20). DEGs with the cutoff criteria of false discovery rate (FDR) <0.001, |log2(fold-change)|≥1 and average expression value ≥5 were considered to be statistically significant. The mRNA expression levels of KRT80 in multiple forms of cancer (including bladder, brain, breast, cervical, colorectal, esophageal, gastric, head and neck, kidney, ovarian, liver, lung, pancreatic and prostate cancer and lymphoma, melanoma, myeloma, leukemia and sarcoma) were further analyzed using ONCOMINE database (access date: Januay 9th 2019; www.oncomine.org) (21), with cut-off values of P<0.001 and a fold-change of 2.0. The proteins co-expressed with KRT80 in TCGA-CRC cases (false discovery rate values <0.05 and absolute Spearman's rank correlation coefficient ≥0.4) were identified using the cBioPortal database (accessed March 2019; http://www.cbioportal.org) (22,23). The co-expressed proteins were subjected to pathway analysis using the Reactome database (accessed March 2019; https://reactome.org/PathwayBrowser) (24) and P<0.05 as the cut-off value.
Cell culture
Human CRC cell lines (HCT116, RKO, LoVo, HT29, SW480 and SW620), the normal colorectal epithelial cell line FHC and normal colorectal fibroblasts CCD18CO were obtained from the American Type Culture Collection. HCT 116 and HT-29 cells were cultured in McCoy's 5A medium (Gibco; Thermo Fisher Scientific, Inc.); RKO and CCD18CO cells were cultured in Eagle's MEM medium (Gibco; Thermo Fisher Scientific, Inc.); LoVo cells were cultured in Ham's F-12K medium (Gibco; Thermo Fisher Scientific, Inc.); SW480 and SW620 cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences); and FHC cells were cultured in DMEM/F12 medium (Gibco; Thermo Fisher Scientific, Inc.). All media were supplemented with 10% (v/v) FBS (Gibco; Thermo Fisher Scientific, Inc.), and maintained in a humidified incubator at 37˚C with 5% CO2.
Clinical tissue samples
The present study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by the Ethics Committee of Shantou University Medical College (approval no. SUMC-2015-42). All patients provided their oral informed consent prior to participation in the study. The need for additional written consent was waived by the Ethics Committee of Shantou University, as the patients had authorized the use of their samples in additional studies in a written consent form they signed when donating their tissues for use in an earlier study. None of the patients had received preoperative treatment or been diagnosed with other types of primary tumors. In total, 50 pairs of CRC tissues and matched adjacent normal tissues were collected from patients (age range, 30-86 years; mean age, 60.7 years; 31 males and 19 females) at the First Affiliated Hospital of Shantou University Medical College between October 2015 and January 2017. The samples were obtained during surgery following the removal of tissue for routine pathology examination. Postoperatively, the tumors were histologically classified and staged based on the AJCC 7th edition tumor node metastasis (TNM) system (25), and the clinical characteristics of these patients with CRC are presented in Table I. All the tissue samples were immediately flash frozen in liquid nitrogen and subsequently stored at -80˚C for reverse transcription-quantitative PCR (RT-qPCR) analysis.
Table I.
Clinicopathological characteristics of the patients with colorectal cancer (N=50).
| Variable | Cases n (%) |
|---|---|
| Age | |
| ≤60 years | 25(50) |
| >60 years | 25(50) |
| Sex | |
| Male | 31(62) |
| Female | 19(38) |
| Degree of differentiation | |
| Moderate to low | 9(18) |
| High | 38(76) |
| Unknown | 3(6) |
| Tumor invasion | |
| T1-2 | 6(12) |
| T3-4 | 44(88) |
| Distant metastasis | |
| No | 40(80) |
| Yes | 10(20) |
| Lymph node metastasis | |
| No | 32(64) |
| Yes | 18(36) |
RT-qPCR
Total RNA was extracted from CRC tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using a PrimeScript RT reagent kit with gDNA Eraser (Takara Bio, Inc.) according to the manufacturer's protocol. qPCR was subsequently performed using a QuantiNova™ SYBR Green PCR mix kit (Roche Diagnostics) and an ABI Prism 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The following primer pairs were used for the qPCR: KRT80 forward, 5'-CCTCCCTAATTGGCAAGGTG-3' and reverse, 5'-AGATGCCCGAGGTCGAAGAT-3'; and β-actin forward, 5'-CTGGAACGGTGAAGGTGACA-3' and reverse, 5'-AAGGGACTTCCTGTAACAATGCA-3'. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95˚C for 10 min; and 40 cycles of 95˚C for 10 sec and 60˚C for 30 sec. Expression levels of KRT80 were quantified using the 2-∆∆Cq method for cell experiments and the 2-∆Cq method for tissue samples (26), and normalized to the loading control β-actin.
Cell transfection
RNA interference was used to knock down KRT80 expression levels in SW480 and SW620 cells. Briefly, small interfering RNA (siRNA) against KRT80 (siRNA-KRT80; 5'-CCCTGGATGTCAAGTTGGA-3') and siRNA-negative control (NC: 5'-ACGUGACACGUUCGGAGAATT-3') were obtained from Guangzhou RiboBio Co., Ltd. A total of 2x105 cells/well were seeded into six-well plates in RPMI-1640 medium, supplemented with 10% FBS. Following overnight attachment in a humidified incubator at 37˚C with 5% CO2, SW480 and SW620 cells were transfected with 100 nM siRNA-KRT80 or siRNA-NC using Lipofectamine RNAiMAX reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C and 5% CO2. After 48 h, cells were collected and used for RNA extraction.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to determine cell viability, as previously described (27). A total of 4x103 SW480 and SW620 cells/well were plated into 96-well plates in RPMI-1640 medium, supplemented with 10% FBS. Following overnight attachment in a humidified incubator at 37˚C with 5% CO2, SW480 and SW620 cells were transfected with siRNA-KRT80 or siRNA-NC for 24, 48, 72 or 96 h. Subsequently, cell viability was detected using a CCK-8 assay kit (Dojindo Molecular Technologies, Inc.), according to the manufacturer's protocol. Optical density values were measured using a microplate reader (Bio-Rad Laboratories, Inc.) at 450 nm.
Colony formation assay
For the colony formation assay, SW480 and SW620 cells were transfected with siRNA-KRT80 or siRNA-NC for 24 h, plated at a density of 2x103 cells/well into six-well plates and incubated in 3 ml RPMI-1640 medium at 37˚C for 10 days. Subsequently, the cells were fixed with 4% paraformaldehyde for 30 min at 25˚C, air dried and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) for 20 min at 25˚C. The images of the colonies in the full well were captured with camera and colonies with >50 cells were counted using a light microscope [magnification, x200; (Nikon Corporation)]. To ensure all cells were counted only once the well surface area was divided using a 5x5 grid, the number of colonies in each segment of the grid were counted.
5-Ethynyl 5'-deoxyuridine (EdU) incorporation assay
EdU incorporation was performed using the Cell Light™ Edu Apollo 567 in vitro kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocol. A total of 5x103 SW480 and SW620 cells/well were plated into 96-well plates in RPMI-1640 medium, containing 10% FBS. Following overnight attachment in a humidified incubator at 37˚C with 5% CO2, SW480 and SW620 cells were transfected with siRNA-KRT80 or siRNA-NC for 48 h. Subsequently, the cells were incubated with 100 µl 50 µM EdU reagents at 37˚C. Following 2 h of incubation, the cells were fixed with 50 µl 4% paraformaldehyde for 30 min at 25˚C. Cells were washed three times in 100 µl PBS -0.1% Triton X-100 prior to incubation with 100 µl 1x Apollo solution (containing deionized water 93.8 µl, Apollo® reaction buffer 5 µl, Apollo® catalyst solution 1 µl, Apollo® fluorescent dye solution 0.3 µl and Apollo® buffer additive 0.9 mg) for 30 min at room temperature in the dark. The cells were subsequently stained with 100 µl 1X Hoechst 33342 nuclear dye for 30 min at room temperature. Stained cells were visualized using a DM IL LED fluorescence microscope (magnification, x200; Leica Microsystems GmbH), with an excitation wavelength of 567 nm. Image analysis was performed using ImageJ v1.8.0 software (National Institutes of Health).
Western blotting
SW480 cells were transfected with siRNA-KRT80 or siRNA-NC for 48 hand were lysed in radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck KGaA), supplemented with protease inhibitors (Roche Diagnostics). Total protein was quantified using a bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.) and 20 µg protein/lane was separated via 12% SDS-PAGE. The separated proteins were subsequently transferred to PVDF membranes (EMD Millipore) and blocked with 5% skimmed milk at room temperature for 1 h. The membranes were incubated with the following primary antibodies overnight at 4˚C: Anti-KRT80 (1:800; cat. no. 16835-1-AP; ProteinTech Group, Inc.), anti-protein phosphatase 1 catalytic subunit α (PPP1CA; 1:2,000; cat. no. 67070-1-Ig; ProteinTech Group, Inc.), anti-cyclin-dependent kinase inhibitor 1A (p21; 1:1,000; cat. no. 2947; Cell Signaling Technology, Inc.), anti-cyclin-dependent kinase inhibitor 1B (p27; 1:1,000; cat. no. 3686; Cell Signaling Technology, Inc.) and anti-β-actin (1:2,500; cat. no. A5441; Sigma-Aldrich; Merck KGaA). Following the primary antibody incubation, membranes were incubated with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG (H+L) secondary antibody (1:5,000; cat. no. SA00001-1; ProteinTech Group, Inc.) or an HRP-conjugated anti-rabbit IgG (H+L) secondary antibody (1:5,000; cat. no. SA00001-2; ProteinTech Group, Inc.) at room temperature for 2 h. Protein bands were visualized using the SuperSignal™ West Dura Extended Duration Substrate reagent (Thermo Fisher Scientific, Inc.) and an Amersham Imager 600 (GE Healthcare). Expression levels were quantified using ImageJ v1.8.0 software (National Institutes of Health) and normalized to β-actin, the loading control.
Statistical analysis
Statistical analysis was performed using SPSS 23.0 software (IBM Corp.). Statistical differences between groups were either determined using a two-tailed unpaired Student's t-test, a paired Student's t-test or one-way ANOVA, followed by Dunnett's multiple comparison test. All data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
Screening of DEGs identifies KRT80 as the most upregulated gene in CRC
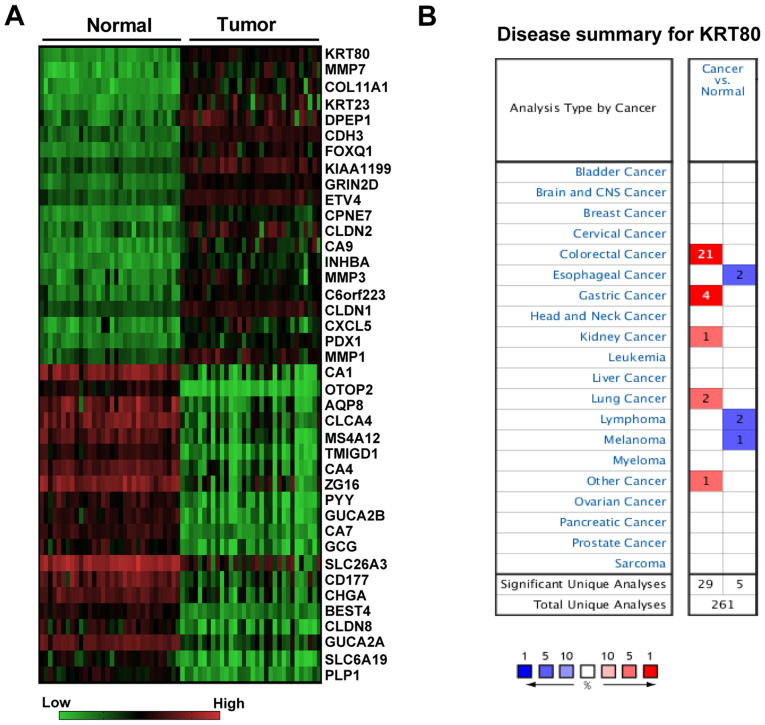
Among the CRC tissue samples from RNA-Seq datasets in the TCGA database, only 32 CRC specimens had matching adjacent normal tissues. Therefore, DEG analysis was only performed on these 32 pairs of tissue samples. A total of 2,114 DEGs were screened (consisting of 844 upregulated and 1,270 downregulated DEGs) between CRC and normal tissues, and KRT80 was discovered to be the most upregulated gene (Fig. 1A; Data S1). The ONCOMINE database was further used to compare the transcriptional levels of KRT80 in different types of cancer and the corresponding normal tissue; compared with the normal tissues, the mRNA expression levels of KRT80 were revealed to be significantly increased in 29 datasets (P<0.001), including CRC, gastric, lung and kidney cancer (Fig. 1B). Among these cancers, the CRC dataset accounted for 72.4% (21/29) of the cases (Fig. 1B).
Figure 1.
Identification of KRT80 as the most upregulated gene in CRC during the screening of DEGs. (A) Heat map of the top 20 DEGs that were upregulated and downregulated in 32 patients with CRC from the RNA-Seq datasets in The Cancer Genome Atlas database. (B) ONCOMINE database was used to determine the expression levels of KRT80 in various types of human cancer. CRC, colorectal cancer; DEGs, differentially expressed genes; KRT80, keratin 80; CNS, central nervous system.
To verify the above findings, KRT80 expression levels in CRC cell lines and cancerous tissues were further analyzed using RT-qPCR analysis. The expression levels of KRT80 mRNA were significantly increased in the CRC cell lines HCT116, RKO, LoVo, HT29, SW480 and SW620, compared with that noted in the normal colorectal cells FHC and CCD18CO (Fig. 2A). Consistent with the increased expression of KRT80 in CRC cell lines, increased expression levels of KRT80 was also observed in CRC tissues compared with the adjacent normal tissues (Fig. 2B). Furthermore, KRT80 expression was upregulated in 84% (42/50) of the patients with CRC (Fig. 2C). These findings are consistent with a previous study (19) and suggested that KRT80 expression levels may be increased in CRC tissues and cells.
Figure 2.
KRT80 expression levels are increased in CRC cells and tissues. (A) RT-qPCR analysis of KRT80 mRNA expression levels in colorectal normal cells (FHC and CCD18CO) and CRC cell lines (HCT116, RKO, LoVo, HT29, SW480 and SW620). β-actin was used as the internal loading control. Data are presented as the mean ± SD from 3 independent experimental repeats. *P<0.05, **P<0.01 vs. FHC. (B) RT-qPCR analysis of KRT80 mRNA expression levels in paired CRC tissues and adjacent normal tissues. Error bars represent 10-90 percentile (n=50). **P<0.01 vs. adjacent tissue. (C) Fold-change of KRT80 mRNA expression levels in each patient with CRC. The ratio of KRT80 expression levels in CRC tissue to corresponding tumor-adjacent normal tissue was measured to analyze the fold-change of each patient. n=50. KRT80, keratin 80; CRC, colorectal cancer; RT-qPCR, reverse transcription-quantitative PCR.
Increased KRT80 expression levels are associated with the pathological stage and metastasis of patients with CRC
To investigate the association between KRT80 expression levels and clinicopathological characteristics of patients with CRC, the clinical data of patients with CRC in the TCGA database were analyzed. The expression levels of KRT80 mRNA were significantly increased in patients with a high pathological stage, and both distant and lymph node metastasis compared with patients with a low pathological stage and metastasis, respectively (Fig. 3A-C). Moreover, clinical tissue sample analysis from 50 patients with CRC validated the above findings; compared with patients without distant metastasis, KRT80 expression levels were significantly increased in patients demonstrating metastasis (Fig. 3D).
Figure 3.
Increased expression levels of KRT80 are associated with the pathologic stage and metastasis of patients with CRC. Expression levels of KRT80 were analyzed in relation to the (A) pathologic stage, (B) distant metastasis and (C) lymph node metastasis using RNA-Seq datasets of CRC tissues in TCGA. (D) Reverse transcription-quantitative PCR analysis of KRT80 mRNA expression levels in tissues from patients with CRC with (n=10) or without (n=40) distant metastasis. β-actin was used as the internal loading control. Error bars represent the 10-90 percentile. **P<0.01 vs. I-II; #P<0.05, ##P<0.01 vs. No metastasis groups. KRT80, keratin 80; CRC, colorectal cancer; TCGA, The Cancer Genome Atlas.
Knockdown of KRT80 expression suppresses CRC cell proliferation
To determine the effects of KRT80 on the viability of CRC cells, cell viability was analyzed following KRT80 knockdown with siRNA. As hypothesized, the genetic knockdown of KRT80 using siRNA for 48 h significantly decreased KRT80 expression levels to ~40 and 30% of the control levels in SW480 and SW620 cells, respectively (Fig. 4A and B). The viability of SW480 cells was significantly decreased following 48 h in siRNA-KTR80-transfected cells compared with the siRNA-NC-transfected cells, whereas the cell viability in siRNA-KRT80-transfected cells SW620 cells was significantly decreased from 72 h compared with siRNA-NC-transfected cells (Fig. 4C and D). Moreover, the inhibitory effect of KRT80 knockdown on cell viability was more pronounced the longer the cells were cultured (Fig. 4C and D).
Figure 4.
Knockdown of KRT80 expression levels decreases the viability of colorectal cancer cells. SW480 and SW620 cells were transfected with siRNA-KRT80 or siRNA-NC for 48 h. Reverse transcription-quantitative PCR analysis was used to determine the expression levels of KRT80 in (A) SW480 and (B) SW620 cells. β-actin was the internal loading control. Cell Counting Kit-8 assay was used to determine the cell viability over 96 h in (C) SW480 and (D) SW620 cells transfected with siRNA-KRT80 or siRNA-NC. Data are presented as the mean ± SD from 3 independent experimental repeats. *P<0.05, **P<0.01 vs. siRNA-NC group. KRT80, keratin 80; siRNA, small interfering RNA; NC, negative control; OD, optical density.
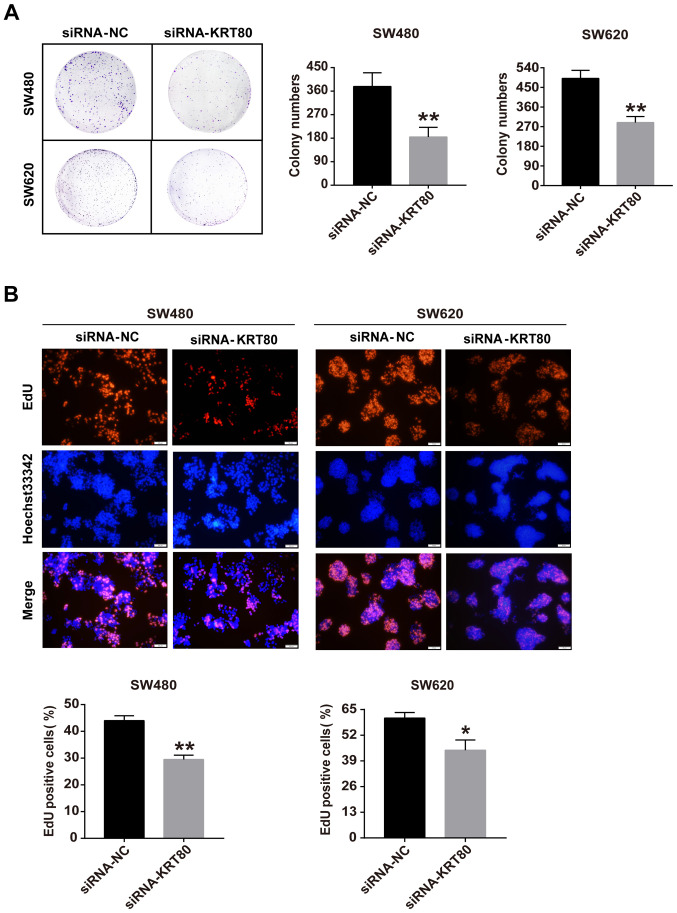
Furthermore, the effect of KRT80 on cell proliferation was determined using colony formation and EdU incorporation assays. The colony formation assay demonstrated that the genetic knockdown of KRT80 significantly decreased the number of colonies formed by ~50 and ~40% in SW480 and SW620 cells, respectively, compared with siRNA-NC-transfected cells (Fig. 5A). The EdU assay was conducted to assess the function of KRT80 with respect to cell proliferation. Compared with siRNA-NC-transfected cells, the number of EdU-positive cells was significantly reduced in both SW480 and SW620 cells transfected with the siRNA-KRT80 compared with the siRNA-NC-transfected cells (Fig. 5B). These data further supported the notion that KRT80 may be involved in cell proliferation in CRC.
Figure 5.
Knockdown of KRT80 expression levels suppresses colorectal cancer cell proliferation. SW480 and SW620 cells were transfected with siRNA-KRT80 or siRNA-NC for 48 h. (A) Colony formation and (B) EdU incorporation assays were used to determine the cell proliferation rate (magnification, x200). ImageJ v1.8.0 software was used for cell counting. Data are presented as the mean ± SD from 3 independent experimental repeats. *P<0.05, **P<0.01 vs. siRNA-NC group. KRT80, keratin 80; siRNA, small interfering RNA; NC, negative control; EdU, 5-ethynyl 2'-deoxyuridine.
Pathway analysis of KRT80 co-expressed proteins in CRC samples from TCGA
To predict the underlying mechanisms of KRT80 in CRC cell proliferation, data mining using the cBioPortal for TCGA was used to identify 354 proteins significantly co-expressed with KRT80 in CRC. In total, 112 co-expressed proteins with absolute Spearman's r≥0.4 were loaded into the Reactome database for pathway analysis (Data S2). The proteins were enriched in the following processes: Cell cycle, DNA replication, immune system, metabolism of RNA and proteins, transport of small molecules and signal transduction (Fig. 6). Among them, the pathways containing the most proteins were the cell cycle and DNA replication pathways.
Figure 6.
Pathway analysis of the proteins co-expressed with KRT80 in colorectal cancer. A total of 112 co-expressed proteins with absolute Spearman's r≥0.4 were loaded into the Reactome database for pathway analysis. The figure shows a genome-wide overview of the results of pathway analysis. Reactome pathways are arranged in a hierarchy. Each step away from the center represents the next level lower in the pathway hierarchy. The yellow color denotes that the proteins co-expressed with KRT80 were significantly enriched in that pathway (P<0.05), whereas light grey signifies pathways that were not significantly enriched. KRT80, Keratin 80.
Moreover, to verify the pathway analysis results, the expression levels of PPP1CA, which is co-expressed with KRT80 (Data S2) and involved in cell cycle and cell division (28), were analyzed in SW480 cells following the genetic knockdown of KRT80 expression. Western blotting discovered that the knockdown of KRT80 significantly decreased the protein expression levels of PPP1CA in SW480 cells compared with the siRNA-NC-transfected cells (Fig. 7). Additionally, KRT80 knockdown also significantly reduced the expression levels of cell cycle-related proteins, p21 and p27, compared with the siRNA-NC transfected cells (Fig. 7).
Figure 7.
Knockdown of KRT80 expression decreases the expression levels of cell cycle-related proteins. SW480 cells were transfected with siRNA-KRT80 or siRNA-NC for 48 h. (A) Western blotting was used to analyze the expression levels of KRT80, PPP1CA, p21 and p27. (B) Semi-quantification of the western blotting data in part A. β-actin was used as the internal loading control for the western blotting analysis. Data are presented as the mean ± SD from 3 independent experimental repeats. *P<0.05, **P<0.01 vs. siRNA-NC group. KRT80, keratin 80; PPP1CA, protein phosphatase 1 catalytic subunit α; p21, cyclin-dependent kinase inhibitor 1A; p27, cyclin-dependent kinase inhibitor 1B; siRNA, small interfering RNA; NC, negative control.
Discussion
Keratin, as a molecular marker of epithelial cells (10), serves an important role in maintaining the stability and integrity of epithelial cells, and it is also involved in various intracellular signal transduction processes, such as cell stress, proliferation and metabolism (29,30). Numerous studies have reported that several proteins of the keratin family were closely related to the development of CRC; for example, keratins were discovered to regulate colonic epithelial cell differentiation through the Notch 1 signaling pathway (18); keratin 8 deletion-induced colitis caused a predisposition to murine CRC through the inflammasome and IL-22 pathway (17); and increased expression levels of KRT7 were observed in CRCs with lymph node metastasis, which was associated with a poor prognosis (31). However, although KRT80 has been reported to mediate migration and invasion in CRC through activating the AKT signaling pathway (19), the role of KRT80 in CRC proliferation remains unknown.
In the present study, the TCGA and ONCOMINE databases were used to identify KRT80 as the most upregulated gene in CRC compared with the normal tissues. Furthermore, it was confirmed that KRT80 expression levels in clinical CRC tissues and CRC cell lines were significantly increased compared with the normal tissues and cells, and increased KRT80 expression levels were associated with the pathological stage and metastasis in patients with CRC. These results were consistent with the study by Li et al (19), which demonstrated that KRT80 was highly expressed in CRC and promoted the migration and invasion of CRC cells, suggesting that KRT80 may serve as an oncogene to promote the proliferation of CRC cells. To confirm this, KRT80 expression was knocked down and it was subsequently found that reduced KRT80 expression levels decreased the cell viability, reduced the number of colonies formed and suppressed cell proliferation. Together with previous reports (19), these findings validated that KRT80 may be a novel potential oncogene for CRC.
In addition, the results of the present study revealed that proteins co-expressed with KRT80 were enriched in the cell cycle and DNA replication processes, and the genetic knockdown of KRT80 significantly reduced the expression levels of cell cycle-related proteins, including PPP1CA, p21 and p27. PPP1CA, which is co-expressed with KRT80 in CRC, is reported to be one of three catalytic subunits of protein phosphatase 1(28); it has been demonstrated to be involved in prostate cancer and CRC tumorigenesis via the mitogen activated protein kinase signaling pathway (32,33). Although p21 and p27 mediate cell cycle arrest through binding to and inhibiting cyclin-dependent kinase/cyclin complexes, numerous studies over the past decade have revealed that p21 and p27 also serve an important role in carcinogenesis and tumor development (34-39). In fact, p21 and p27 have been reported to serve as oncogenic proteins or tumor suppressors, depending on their localization in the cytoplasm or the nucleus, respectively (40-43). In brief, cytoplasmic p21 and p27 have been found to favor antiapoptotic activities, whereas nuclear p21 and p27 have been associated with cell cycle arrest (36,39). Therefore, the findings of the present study provided a valuable reference for further study to investigate the function and underlying mechanisms of KRT80 in CRC tumorigenesis.
However, this study has some limitations. Firstly, only 50 pairs of tissue samples were used in this study, which is a small sample size. Additionally, some clinicopathological data related to CRC, such as patient survival, were not included. Finally, although pathway analysis indicated that proteins co-expressed with KRT80 were largely enriched in the cell cycle and DNA replication, the detailed underlying mechanisms have not been further explored. The above limitations should be resolved in future studies.
In conclusion, the findings of the present study suggested that KRT80 may be overexpressed in CRC tissues and it may function as an oncogene to promote the proliferation of CRC cells. KRT80 co-expressed proteins were discovered to be largely enriched in the cell cycle and DNA replication pathways, which are related to the process of tumorigenesis. Therefore, in-depth research into KRT80 to further reveal its functions and underlying mechanisms in CRC may provide novel insights for the early diagnosis and gene-targeted treatment of CRC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported by the Sanming Project of Medicine in Shenzhen (grant no. SZSM201612071) and the Shenzhen Science and Technology Project (grant no. JCYJ20180228163436705).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the UCSC Xena Browser (https://xenabrowser.net/datapages/), ONCOMINE gene expression array datasets (www.oncomine.org), cBioPortal database (http://www.cbioportal.org) and the Reactome database (https://reactome.org/PathwayBrowser).
Authors' contributions
HY and XX designed the study; JL, XF and JC performed the experiments and analyzed the data; XX contributed to obtaining the patient tissue samples; and JL and HY wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki and the study protocol was approved by the Ethics Committee of Shantou University Medical College (approval no. SUMC-2015-42).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mercier J, Voutsadakis IA. A systematic review and meta-analysis of retrospective series of regorafenib for treatment of metastatic colorectal cancer. Anticancer Res. 2017;37:5925–5934. doi: 10.21873/anticanres.12039. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Lin C, Liao G, Liu S, Ding J, Tang F, Wang Z, Liang X, Li B, Wei Y, et al. MicroRNA-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2. Oncotarget. 2015;6:32586–32601. doi: 10.18632/oncotarget.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Malapelle U, Mayo de-Las-Casas C, Rocco D, Garzon M, Pisapia P, Jordana-Ariza N, Russo M, Sgariglia R, De Luca C, Pepe F, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer. 2017;116:802–810. doi: 10.1038/bjc.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahjaman M, Kumar N, Ahmed MS, Begum A, Islam SMS, Mollah MNH. Robust feature selection approach for patient classification using gene expression data. Bioinformation. 2017;13:327–332. doi: 10.6026/97320630013327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strnad P, Paschke S, Jang KH, Ku NO. Keratins: Markers and modulators of liver disease. Curr Opin Gastroenterol. 2012;28:209–216. doi: 10.1097/MOG.0b013e3283525cb8. [DOI] [PubMed] [Google Scholar]
- 8.Rogers MA, Edler L, Winter H, Langbein L, Beckmann I, Schweizer J. Characterization of new members of the human type II keratin gene family and a general evaluation of the keratin gene domain on chromosome 12q13.13. J Invest Dermatol. 2005;124:536–544. doi: 10.1111/j.0022-202X.2004.23530.x. [DOI] [PubMed] [Google Scholar]
- 9.Uenishi T, Kubo S, Yamamoto T, Shuto T, Ogawa M, Tanaka H, Tanaka S, Kaneda K, Hirohashi K. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003;94:851–857. doi: 10.1111/j.1349-7006.2003.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurokawa I, Urakawa Y, Senba Y, Kawabata E, Nishimura K, Omoto Y, Tokime K, Mizutani H, Tsubura A. Keratin profiles may differ between intraepidermal and intradermal invasive eccrine porocarcinoma. Oncol Rep. 2006;16:473–477. [PubMed] [Google Scholar]
- 11.Toivola DM, Boor P, Alam C, Strnad P. Keratins in health and disease. Curr Opin Cell Biol. 2015;32:73–81. doi: 10.1016/j.ceb.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Omary MB, Ku NO, Strnad P, Hanada S. Toward unraveling the complexity of simple epithelial keratins in human disease. J Clin Invest. 2009;119:1794–1805. doi: 10.1172/JCI37762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F, Katoonizadeh A, Wouters J, van Kempen LC, Durnez A, et al. Keratin 19: A key role player in the invasion of human hepatocellular carcinomas. Gut. 2014;63:674–685. doi: 10.1136/gutjnl-2012-304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, Maltais L, Omary MB, Parry DA, Rogers MA, Wright MW. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob JT, Coulombe PA, Kwan R, Omary MB. Types I and II keratin intermediate filaments. Cold Spring Harb Perspect Biol. 2018;10(a018275) doi: 10.1101/cshperspect.a018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baribault H, Penner J, Iozzo RV, Wilson-Heiner M. Colorectal hyperplasia and inflammation in keratin 8-deficient FVB/N mice. Genes Dev. 1994;8:2964–2973. doi: 10.1101/gad.8.24.2964. [DOI] [PubMed] [Google Scholar]
- 17.Misiorek JO, Lähdeniemi IAK, Nyström JH, Paramonov VM, Gullmets JA, Saarento H, Rivero-Müller A, Husøy T, Taimen P, Toivola DM. Keratin 8-deletion induced colitis predisposes to murine colorectal cancer enforced by the inflammasome and IL-22 pathway. Carcinogenesis. 2016;37:777–786. doi: 10.1093/carcin/bgw063. [DOI] [PubMed] [Google Scholar]
- 18.Lähdeniemi IAK, Misiorek JO, Antila CJM, Landor SK, Stenvall CA, Fortelius LE, Bergström LK, Sahlgren C, Toivola DM. Keratins regulate colonic epithelial cell differentiation through the Notch1 signalling pathway. Cell Death Differ. 2017;24:984–996. doi: 10.1038/cdd.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Liu X, Liu Y, Liu X, Wang R, Liao J, Wu S, Fan J, Peng Z, Li B, Wang Z. Keratin 80 promotes migration and invasion of colorectal carcinoma by interacting with PRKDC via activating the AKT pathway. Cell Death Dis. 2018;9(1009) doi: 10.1038/s41419-018-1030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(e47) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(pl1) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48 (D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Li A, Yang S, Qiao R, Zhu X, Zhang J. CXCL5 promotes mitomycin C resistance in non-muscle invasive bladder cancer by activating EMT and NF-κB pathway. Biochem Biophys Res Commun. 2018;498:862–868. doi: 10.1016/j.bbrc.2018.03.071. [DOI] [PubMed] [Google Scholar]
- 28.Figueiredo J, da Cruz E Silva OA, Fardilha M. Protein phosphatase 1 and its complexes in carcinogenesis. Curr Cancer Drug Targets. 2014;14:2–29. doi: 10.2174/15680096113136660106. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Shan G, Cao P, He J, Lin Z, Huang Y, Ao N. Mechanical and biological properties of oxidized horn keratin. Mater Sci Eng C Mater Biol Appl. 2015;47:123–134. doi: 10.1016/j.msec.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, Qian X, Zhang F, Sun B. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5(e1549) doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czapiewski P, Bobowicz M, Pęksa R, Skrzypski M, Gorczyński A, Szczepańska-Michalska K, Korwat A, Jankowski M, Zegarski W, Szulgo-Paczkowska A, et al. Keratin 7 expression in lymph node metastases but not in the primary tumour correlates with distant metastases and poor prognosis in colon carcinoma. Pol J Pathol. 2016;67:228–234. doi: 10.5114/pjp.2016.63774. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Wan L, Zhang J, Zhang J, Mendez L, Clohessy JG, Berry K, Victor J, Yin Q, Zhu Y, et al. Deregulated PP1α phosphatase activity towards MAPK activation is antagonized by a tumor suppressive failsafe mechanism. Nat Commun. 2018;9(159) doi: 10.1038/s41467-017-02272-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun H, Ou B, Zhao S, Liu X, Song L, Liu X, Wang R, Peng Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine. 2019;48:236–247. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnit SS, Xie C, Yao M, Holst J, Bensoussan A, De Souza P, Li Z, Dong Q. p27(Kip1) signaling: Transcriptional and post-translational regulation. Int J Biochem Cell Biol. 2015;68:9–14. doi: 10.1016/j.biocel.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Sharma SS, Pledger WJ. The non-canonical functions of p27(Kip1) in normal and tumor biology. Cell Cycle. 2016;15:1189–1201. doi: 10.1080/15384101.2016.1157238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Currier AW, Kolb EA, Gorlick RG, Roth ME, Gopalakrishnan V, Sampson VB. p27/Kip1 functions as a tumor suppressor and oncoprotein in osteosarcoma. Sci Rep. 2019;9(6161) doi: 10.1038/s41598-019-42450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips AH, Ou L, Gay A, Besson A, Kriwacki RW. Mapping interactions between p27 and RhoA that stimulate cell migration. J Mol Biol. 2018;430:751–758. doi: 10.1016/j.jmb.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shamloo B, Usluer S. p21 in cancer research. Cancers (Basel) 2019;11(1178) doi: 10.3390/cancers11081178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Wang W, Chen Y, Huang Y, Zhang J, He S, Tan Y, Qiang F, Li A, Røe OD, et al. The opposite prognostic significance of nuclear and cytoplasmic p21 expression in resectable gastric cancer patients. J Gastroenterol. 2014;49:1441–1452. doi: 10.1007/s00535-013-0900-4. [DOI] [PubMed] [Google Scholar]
- 41.Koster R, di Pietro A, Timmer-Bosscha H, Gibcus JH, van den Berg A, Suurmeijer AJ, Bischoff R, Gietema JA, de Jong S. Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J Clin Invest. 2010;120:3594–3605. doi: 10.1172/JCI41939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georgakilas AG, Martin OA, Bonner WM. p21: A two-faced genome guardian. Trends Mol Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Denicourt C, Saenz CC, Datnow B, Cui XS, Dowdy SF. Relocalized p27Kip1 tumor suppressor functions as a cytoplasmic metastatic oncogene in melanoma. Cancer Res. 2007;67:9238–9243. doi: 10.1158/0008-5472.CAN-07-1375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the UCSC Xena Browser (https://xenabrowser.net/datapages/), ONCOMINE gene expression array datasets (www.oncomine.org), cBioPortal database (http://www.cbioportal.org) and the Reactome database (https://reactome.org/PathwayBrowser).