Abstract
Background
Sexual reproduction of scleractinians has captured the attention of researchers and the general public for decades. Although extensive ecological data has been acquired, underlying molecular and cellular mechanisms remain largely unknown. In this study, to better understand mechanisms underlying gametogenesis, we isolated ovaries and testes at different developmental phases from a gonochoric coral, Euphyllia ancora, and adopted a transcriptomic approach to reveal sex- and phase-specific gene expression profiles. In particular, we explored genes associated with oocyte development and maturation, spermiogenesis, sperm motility / capacitation, and fertilization.
Results
1.6 billion raw reads were obtained from 24 gonadal samples. De novo assembly of trimmed reads, and elimination of contigs derived from symbiotic dinoflagellates (Symbiodiniaceae) and other organisms yielded a reference E. ancora gonadal transcriptome of 35,802 contigs. Analysis of 4 developmental phases identified 2023 genes that were differentially expressed during oogenesis and 678 during spermatogenesis. In premature/mature ovaries, 631 genes were specifically upregulated, with 538 in mature testes. Upregulated genes included those involved in gametogenesis, gamete maturation, sperm motility / capacitation, and fertilization in other metazoans, including humans. Meanwhile, a large number of genes without homology to sequences in the SWISS-PROT database were also observed among upregulated genes in premature / mature ovaries and mature testes.
Conclusions
Our findings show that scleractinian gametogenesis shares many molecular characteristics with that of other metazoans, but it also possesses unique characteristics developed during cnidarian and/or scleractinian evolution. To the best of our knowledge, this study is the first to create a gonadal transcriptome assembly from any scleractinian. This study and associated datasets provide a foundation for future studies regarding gametogenesis and differences between male and female colonies from molecular and cellular perspectives. Furthermore, our transcriptome assembly will be a useful reference for future development of sex-specific and/or stage-specific germ cell markers that can be used in coral aquaculture and ecological studies.
Keywords: Scleractinian corals, Euphyllia ancora, Ovary, Testis, Gonads, RNA-seq, Transcriptome assembly, Sex-specific, Phase-specific, Oogenesis, Spermatogenesis
Background
Since the discovery of scleractinian mass spawning events in the Great Barrier Reef in the 1980s [1–3], sexual reproduction of scleractinians has captured the attention of researchers and the general public. Studies on various aspects of sexual reproduction, such as the timing of broadcast spawning or brooding, general cellular processes of gametogenesis, and sexuality (hermaphroditic or gonochoric), have been undertaken mainly from an ecological perspective in many scleractinian species in many locations during the past 3 decades [4–6]. Although large amounts of data are now available from more than four hundred species [7], our current understanding of intrinsic mechanisms underlying key processes of sexual reproduction, such as sex determination/differentiation, gametogenesis, and ovulation/spawning, is quite limited.
Gametogenesis is a highly organized process whereby genetically diverse haploid gametes are created from diploid germ cells through meiosis with recombination. Generally, scleractinian germ cells are developed in endodermal mesenteries of polyps [4, 8, 9]. Sites of germ cell development are often observed as swellings in polyps during active gametogenesis, and are termed gonads [4]. Oogenesis begins with mitotic division of a small number of oogonia along the gonadal mesoglea, a thin layer composed of extracellular matrix. After oogonia differentiate into oocytes by entering a meiotic phase, oocytes increase in size and migrate into the mesoglea layer [4, 9–11]. There, oocytes further increase in size until maturation by accumulating yolk proteins, lipids, and other essential materials for embryonic development [12, 13]. Spermatogenesis begins with the active mitotic division of spermatogonia in the gonadal mesogleal layer. After spermatogonia form many small clusters comprising dozens of spermatogonia, they migrate into the mesoglea layer and form many spermatogenic compartments called spermaries. Further proliferation of spermatogonia, meiotic differentiation into spermatocytes, and spermiogenesis take place within each spermary [4, 14].
Studies of molecular and cellular aspects of scleractinian gametogenesis have just recently begun. Only several reports are available describing genes related to oogenesis, including vitellogenesis [12–20] and spermatogenesis [21, 22]. Currently, in order to cope with recent declines of coral reefs, reef restoration efforts via aquaculture are being initiated worldwide [23–25]. A comprehensive understanding of intrinsic mechanisms of gametogenesis will enable us to approach coral reef restoration from a new perspective. For instance, hormonal induction of gametogenesis and spawning under artificial rearing systems would allow more efficient propagation of target species [14]. Sex-and stage-specific molecular markers for germ cells would also enable us to monitor and to evaluate the developmental status of germ cells in corals cultured in captivity [21]. Moreover, because scleractinians belong to the phylum Cnidaria (e.g., corals, sea anemones, hydras, and jellyfish), which are regarded as evolutionarily basal in the animal kingdom, studies highlighting common mechanisms of sexual reproduction between scleractinians and advanced animals (e.g., vertebrates) should provide insights into the evolution of sexual reproduction in metazoans [14].
Transcriptome analysis using high-throughput sequencing has greatly enhanced identification of transcripts involved in sexual reproduction in various taxa [26–30]. This study performed gonadal transcriptome sequencing of a scleractinian coral, Euphyllia ancora, commonly known as the anchor or hammer coral (Fig. 1 a-c). E. ancora was selected for the following reasons: (i) These corals are common in the Indo-Pacific region. (ii) They are gonochoric, and their annual gametogenic cycle in reefs along southern Taiwan has been studied histologically in both male and female colonies [8, 9]. For instance, a single oogenic or spermatogenic cycle in this region takes approximately a year in females and half a year in males. Annual spawning occurs within a week after a full moon in April or May, or occasionally in June. Finally, (iii) They have large polyps (3–5 cm in diameter) that allow us to isolate ovaries and testes with relative ease [12]. This transcriptomic analysis of isolated gonads was undertaken in order to discover genes participating in gametogenesis.
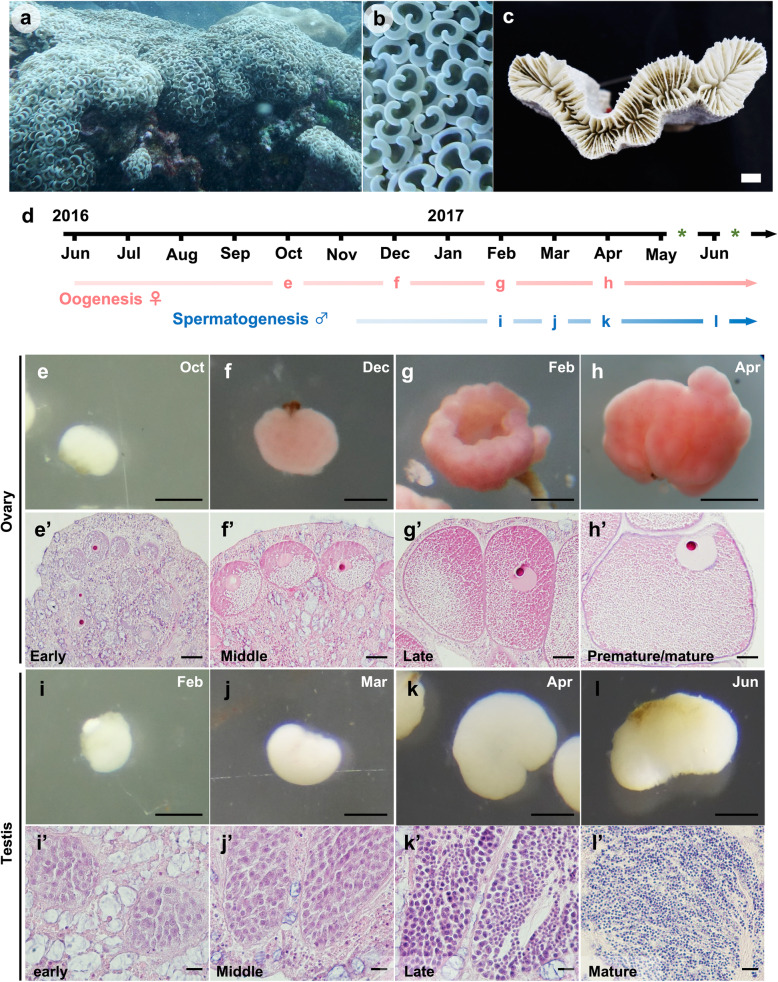
Fig. 1.
Euphyllia ancora and its germ cells observed histologically in isolated gonads at different sampling times. a External appearance of an E. ancora colony. b External appearance of tentacles of an E. ancora colony. Anchor-like tentacles and the flabello-meandroid skeleton typify E. ancora. The pictures were taken at Nanwan Bay, Kenting National Park, in southern Taiwan in October 2016. c A top view of an E. ancora skeleton. The picture was taken after removal of polyp tissue in the laboratory. d Periods of oogenesis (pink arrow) and spermatogenesis (blue arrow) and predicated spawning timing (*). Letters (e-l) on the arrows correspond to Figure 1 (e-l) below, and indicate the timing (month) of sampling for ovaries and testes. e-h The external appearance of isolated ovaries in October and December 2016 and February and April 2017. e’-h’ Histological observation of the isolated ovaries. e, e’ The early phase of ovaries. f, f’ The middle phase of an ovary. g, g’ The late phase of an ovary. h, h’ The premature/mature phase of an ovary. i-l The external appearance of isolated testes in February, March, April, and June 2017. i’-l’ Histological observation of isolated testes. i, i’ The early phase of a testis with spermatogonia. j, j’ The middle phase of a testis having spermatogonia and primary spermatocytes. k, k’ The late phase of a testis with secondary spermatocytes and spermatids. l, l’ The mature phase of a testis with mature sperm. Sections were stained with hematoxylin and eosin. Scale bars = 1 cm (c); 500 μm (e-l); 50 μm (e’-h’); 10 μm (i’-l’)
The present study isolated ovaries and testes at different developmental phases from wild E. ancora colonies in order to reveal sex- and phase-specific gene expression profiles. In particular, we focused on premature and mature phases of gonads to identify candidate genes associated with oocyte development and maturation, spermiogenesis, sperm motility and capacitation, and fertilization, because of their importance for coral aquaculture (e.g., induction of sexual maturation) and ecological studies (e.g., monitoring germline development or predicting spawning time). These findings may highlight conserved molecular mechanisms of gametogenesis between scleractinians and other animals, including humans.
Results
Histological analysis of E. ancora gonads collected at different times
Ovaries and testes were isolated from wild colonies at different times during a period of 9 months in 2016–2017 (Fig. 1d). Progress of gametogenesis was histologically confirmed as the spawning season approached (April–June, 2017). Gametogenesis is generally synchronized among polyps in a colony. Histological analysis of isolated ovaries showed that oocytes grew steadily during the 9-month investigation, and that ovaries isolated at 4 sampling dates generally displayed different oocyte developmental stages: October 2016 (oocytes with cytoplasmic polarization, < 125 μm in diameter), December 2016 (oocytes with accumulation of yolk and other components, 126–200 μm in diameter), February 2017 (oocytes with accumulation of yolk and other components, 201–275 μm in diameter), and April 2017 (oocytes with ‘U’-like germinal vesicles or GVBD, > 276 μm in diameter) (Fig. 1d, Table 1). Notably, in the April 2017 samples, most oocyte nuclei had translocated to the peripheral membrane (Fig. 1 e-h), and some oocyte nuclei had disappeared (Additional file 1), indicating that germinal vesicle breakdown (GVBD) had occurred in those oocytes. These ovarian samples were then classified into 4 phases, early, middle, late, and premature/mature, and were used for RNA-seq (Fig. 1 e-h, Table 1).
Table 1.
Criteria for classification of gonadal phases
| Gonad | Phase | The most represented germ cells observed in the gonads | Approximate timings of collection in 2016 to 2017 |
|---|---|---|---|
| Ovary | Early | Oocytes with cytoplasmic polarization (<125 μm in diameter) | October, 2016 |
| Middle | Oocytes with accumulation of yolk and other components (126-200 μm in diameter) | December, 2016 | |
| Late | Oocytes with accumulation of yolk and other components (201-275 μm in diameter) | February, 2017 | |
| Premature/ mature | Oocytes with ‘U’-like germinal vesicles and GVBD (>276 μm in diameter) | April, 2017 | |
| Testis | Early | Spermatogonia | February, 2017 |
| Middle | Spermatogonia and primary spermatocytes | March, 2017 | |
| Late | Secondary spermatocytes and spermatids | April, 2017 | |
| Mature | Sperm | June, 2017 |
Similarly, testes isolated at the following 4 sampling dates in 2017 possessed germ cells in different developmental stages: February (spermatogonia), March (spermatogonia and primary spermatocytes), April (secondary spermatocytes and spermatids), and June (mature sperm) (Fig. 1 d, Table 1) (Fig. 1 i-l). In the June samples, although a small number of spermaries with both round spermatids and mature sperm were observed in some testes, cytological observation confirmed the presence of morphologically mature sperm (Additional file 1). Testis samples were then classified into 4 phases, early, middle, late, and mature, and were subjected to RNA-seq (Fig. 1 i-l, Table 1).
De novo transcriptome assembly of E. ancora gonads, identification of coral contigs, and functional annotation
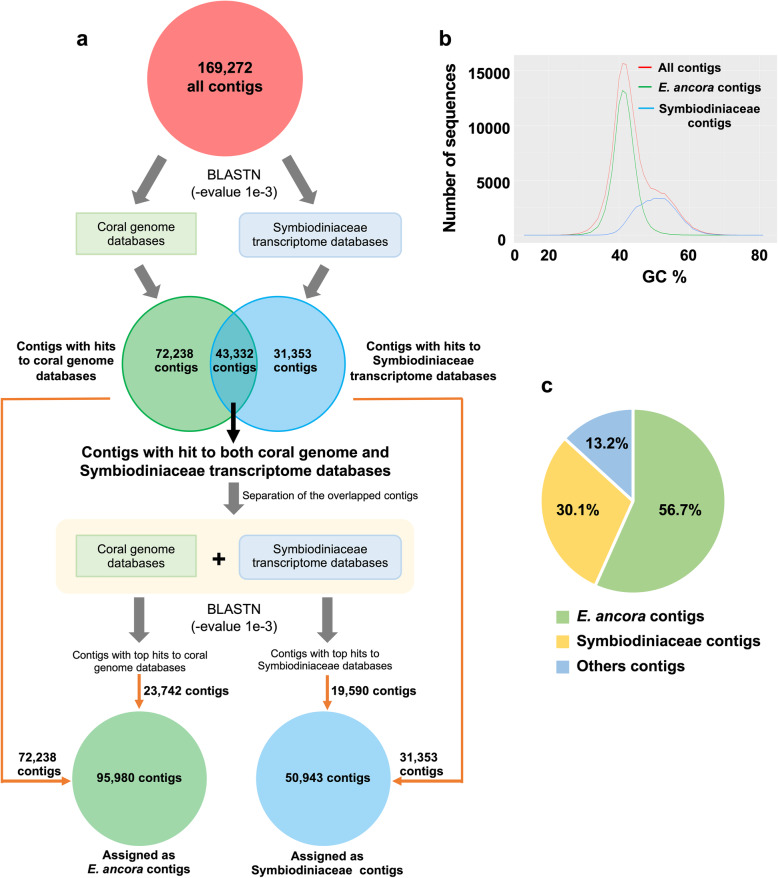
1.6 billion raw reads comprising approximately 240 Gb of clean transcriptomic sequencing data were obtained by Illumina paired-end sequencing from the selected 12 testis (3 colonies, 4 time points) and 12 ovary (3 colonies, 4 time points) samples. Clean reads were deposited in the Sequence Read Archive (SRA) of DDBJ under BioProject number PRJDB9831 (Additional file 2). De novo assembly of all clean reads produced 169,272 initial contigs with an average size of 2321 bp and an N50 of 4610 bp. Maximum contig length reached 52,720 bp (Table 2). The assembled transcriptome sequences were also deposited in DDBJ under accession number ICQS01000001-ICQS01169272. In addition, we provided the accession numbers that are included in the reference gonadal transcriptome, E. ancora contigs and Symbiodiniaceae as Additional files 3, 4, 5 respectively.
Table 2.
Summary of transcriptome assemblies in this study
| E. ancora holobiont (all contigs) | Assigned as E. ancora contigs | Assigned as Symbiodiniaceae contigs | Reference E. ancora gonadal transcriptome contigs used for this study | |
|---|---|---|---|---|
| Number of contigs sequences | 169,272 | 95,980 (56.7%) | 50,943 (30.1%) | 35,802 |
| Total basepair (bp) | 392,911,637 | 328,250,055 | 53,222,313 | 125,288,259 |
| Minimum length (bp) | 200 | 200 | 200 | 297 |
| Average (bp) | 2,321 | 3,420 | 1,045 | 3,500 |
| Maximum unigene length (bp) | 52,720 | 52,720 | 16,557 | 43,419 |
| N50 size (bp) | 4,610 | 5,384 | 1,318 | 5,019 |
| GC (%) | 44.2 | 41.5 | 50.6 | 42.1 |
| BUSCO completeness (%) | 99.7 | 98 | 65 | 92.1 |
Since the initial transcriptome assembly contained contigs from E. ancora gonads, symbiotic dinoflagellates (Symbiodiniaceae), and other organisms (e.g., bacteria), we first bioinformatically identified possible E. ancora contigs prior to detailed analyses (Fig. 2a). All assembled contigs were aligned to available genome databases of 4 scleractinian species (Acropora digitifera, Pocillopora damicornis, Stylophora pistillata, and Orbicella faveolata) and transcriptomic databases of 6 Symbiodiniaceae (Symbiodinium sp. A1, Symbiodinium sp. A2, Breviolum sp. B2, Breviolum muscatinei, Uncultured Cladocopium sp. and Uncultured Durusdinium sp.) (For more details, see Additional file 6), and contigs unambiguously matched to coral genomic databases (72,238 contigs) and to Symbiodiniaceae transcriptomic databases (31,353 contigs) were separated (Fig. 2a). Contigs matching both databases (43,332 contigs) were further aligned to the combined databases of coral genomes and Symbiodiniaceae transcriptomes, and were separated into coral contigs (23,742 contigs) and symbiotic dinoflagellate contigs (19,590 contigs) based on top hit results of BLASTN (−evalue 1e-3). Eventually, 95,980 contigs were assigned as E. ancora, and 50,943 to Symbiodiniaceae (Fig. 2a). E. ancora contigs had a GC peak at 41.5%, while symbiotic Symbiodiniaceae peaked at 50.6% (Fig. 2b). These GC contents correspond well to previous genomic studies of corals and Breviolum minutum [31–33].
Fig. 2.
Identification of E. ancora contigs from the transcriptome assembly of an E. ancora holobiont. a A flow chart for identification of E. ancora contigs from the transcriptome assembly (all contigs) that contains contigs from the host coral, symbiotic (dinoflagellates), and other organisms (bacteria). b Distribution of GC percentages of assembled contigs. Red line: all contigs, green line: E. ancora contigs, blue line: extracted Symbiodiniaceae contigs. c Proportions of contigs from E. ancora, Symbiodiniaceae, and other symbiotic organisms (other contigs) in the initial whole holobiont transcriptome assembly. Only extracted E. ancora contigs (56.7%) were used for further analysis
In order to remove sequence heterogeneity originating from different individuals or different haplotypes in the same individual, translated sequences of the extracted 95,980 E. ancora contigs were further clustered using CD-HIT with 95% amino acid sequence identity. Finally, 35,802 contigs totaling approximately 125 Mbp (N50, 5019 bp) were used as the reference E. ancora gonadal transcriptome with bench-marking universal single-copy orthologs (BUSCO) of more than 90% (Table 2), which covers all E. ancora candidate genes involved in gametogenesis. BLAST search (BLASTP, −evalue 1e-5) revealed that 21,569 of 35,802 (60.2%) contigs had significant similarities to sequences in the SWISS-PROT database (Fig. 3a). Moreover, 23,686 of 35,802 (66.2%) contigs matched conserved protein domains in the Pfam database (Fig. 3b).
Fig. 3.
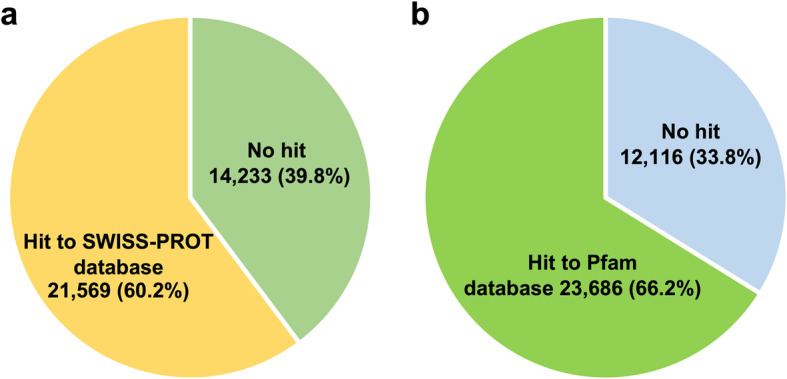
Contig numbers in the reference E. ancora gonadal transcriptome that matched SWISS-PROT and Pfam databases. a Results of BLAST searches against the SWISS-PROT database (cut-off -evalue le-5). Note that 21,569 out of 35,802 contigs (60.2%) had significant similarities with database sequences. bIdentification of protein domains using the Pfam database (cut-off -evalue le-5) for contigs from the reference E. ancora gonadal transcriptome. Note that 23,686 out of 35,802contigs (66.2%) had conserved protein domains
The reference E. ancora gonadal transcriptome contained reproduction-related genes identified in our previous studies using degenerate PCR or cDNA libraries (Additional file 7). Furthermore, evolutionarily conserved genes associated with germline development (Gcl, Mago, Boule, and Pum1) were identified. Genes involved in meiotic processes, such as invasion and pairing of the homologous strand (Msh4, Msh5, Mlh1), formation of a synaptonemal complex (Sycp1, Sycp3), and maintenance of chromosome structure integrity (Rad21) were also identified (Additional file 8).
Differential gene expression analysis among different developmental stages of ovaries and testes
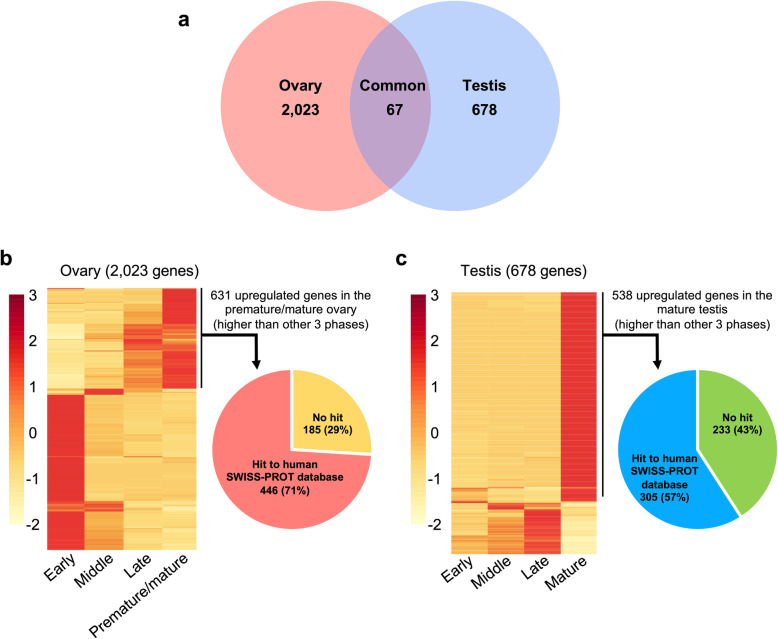
Hierarchical cluster analysis of 24 selected samples (12 testes and 12 ovaries) determined that 2 samples (Oct-female-1 and Feb-male-1) differed from all others (Additional file 9). These were assigned as outliers, possibly resulting from accidental collection of allospecific samples adjacent to the labeled colonies. To minimize data variation, the foregoing 2 samples were removed, and the remaining 22 samples were used for downstream analysis. Differential gene expression analysis of 4 developmental phases of ovaries and testes identified 2023 and 678 differentially expressed genes during oogenesis and spermatogenesis, respectively, and 67 differentially expressed genes in both ovaries and testes during gametogenesis (q-value< 0.05, Fig. 4a). There were 1165, 89, 138, and 631 upregulated genes specific to the early, middle, late, and premature/mature ovarian phases, respectively (Fig. 4b). In the testis, there were 6, 19, 115, and 538 upregulated genes specific to the early, middle, late, and mature phases, respectively (Fig. 4c).
Fig. 4.
Differentially expressed genes in oogenesis and spermatogenesis of E. ancora. a 2023 and 678 genes were differentially expressed during oogenesis and spermatogenesis, respectively, and 67 of those genes were differentially expressed in both oogenesis and spermatogenesis (q-value< 0.05, ANOVA). b Relative gene expression levels of differentially expressed genes (2023 genes) at different phases of ovaries. CPM values were scaled to row Z-scores for each of the genes. In premature/mature ovaries, 631 genes were expressed at higher levels than in the other 3 phases. Among the 631 genes, 446 genes (71%) matched the SWISS-PROT human database, as shown in the pie chart. c Relative gene expression levels of differentially expressed genes (678 genes) at different phase of testes. CPM values were scaled to row Z-scores for each of the genes. In mature testes, 538 genes were expressed more highly than in the other 3 phases. Among the 538 genes, 305 (57%) matched the SWISS-PROT human database, as shown in the pie chart. In the heatmaps, each row represents a differentially expressed gene and the columns represent time points. The color bar on the left indicates expression levels
Upregulated genes of premature/mature ovaries
The 631 genes specifically upregulated in premature/mature ovaries were further analyzed. Four hundred forty six of those genes (71%) matched the human SWISS-PROT database (Fig. 4b). Analysis of enriched functional terms revealed that 18 GO terms were enriched in premature/mature ovaries (P < 0.05 and enrichment > 4-fold; Additional file 10): 16 biological processes (BP) and 2 molecular functions (MF). Of the enriched BP terms, terms related to neuronal activity such as positive regulation of synaptic transmission, GABAergic (GO: 0050806), calcium ion-regulated exocytosis of neurotransmitter (GO: 0048791), neurotransmitter transport (GO:0006836) and neuronal action potential (GO: 0019228) were highly enriched. Among enriched MF terms, extracellular ligand-gated ion channel activity (GO: 0005230) was most enriched (Additional file 10).
In premature/mature ovaries, genes upregulated > 5-fold more than in the other three phases (log2, FDR < 0.05), 9 genes, including those encoding GFP-like fluorescent chromoprotein, neurogenic locus notch homolog protein 3, carbonic anhydrase 2, octopamine receptor beta-1R, beta-1,4-galactosyltransferase galt-1 were identified. One of the 9 genes could not be annotated (Table 3).
Table 3.
Genes highly upregulated in premature/mature ovaries and mature testes compared to the other 3 phases
| Gonad | Annotation | Assembly ID | E-value |
|---|---|---|---|
| Ovaries (log2>5) | GFP-like fluorescent chromoprotein | CL9557.Contig3_All | 4.00E-101 |
| Neurogenic locus notch homolog protein 3 | CL2173.Contig10_All | 1.00E-63 | |
| Carbonic anhydrase 2 | CL8194.Contig1_All | 3.00E-50 | |
| Octopamine receptor beta-1R | CL4990.Contig1_All | 1.00E-22 | |
| Beta-1,4-galactosyltransferase galt-1 | Unigene16679_All | 1.00E-13 | |
| Polcalcin Juno 2 | CL7546.Contig1_All | 2.00E-12 | |
| Transmembrane protein 26 | Unigene179326_All | 7.00E-10 | |
| Integrin-linked protein kinase | Unigene80860_All | 1.00E-08 | |
| Testes (log2>8) | Creatine kinase S-type, mitochondrial | CL3023.Contig3_All | 0 |
| Creatine kinase, flagellar | CL12240.Contig1_All | 0 | |
| Omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1 | Unigene27219_All | 4.00E-114 | |
| Glutamate receptor ionotropic, kainate 2 | Unigene8664_All | 1.00E-105 | |
| Testis-specific serine/threonine-protein kinase 4 | CL327.Contig1_All | 2.00E-91 | |
| Fibrillin-2 | CL1782.Contig1_All | 8.00E-71 | |
| Monocarboxylate transporter 10 | CL1025.Contig1_All | 4.00E-66 | |
| Latent-transforming growth factor beta-binding protein 1 | CL10775.Contig3_All | 4.00E-54 | |
| Uncharacterized protein KIAA0895-like | CL11131.Contig2_All | 1.00E-44 | |
| Disheveled-associated activator of morphogenesis 1 | CL7734.Contig1_All | 2.00E-39 | |
| BTB/POZ domain-containing protein 8 | Unigene35893_All | 1.00E-33 | |
| Cyclic nucleotide-binding domain-containing protein 2 | CL4865.Contig1_All | 8.00E-23 | |
| F-box/LRR-repeat protein 14 | Unigene33934_All | 7.00E-13 | |
| Testis-expressed protein 11 | CL11981.Contig40_All | 1.00E-12 | |
| Nuclear receptor corepressor 2 | CL7171.Contig3_All | 8.00E-10 | |
| Netrin receptor UNC5C | CL47.Contig4_All | 2.00E-09 | |
| cAMP-dependent protein kinase regulatory subunit | Unigene2733_All | 7.00E-09 | |
| Polysialoglycoprotein | Unigene16171_All | 3.00E-07 |
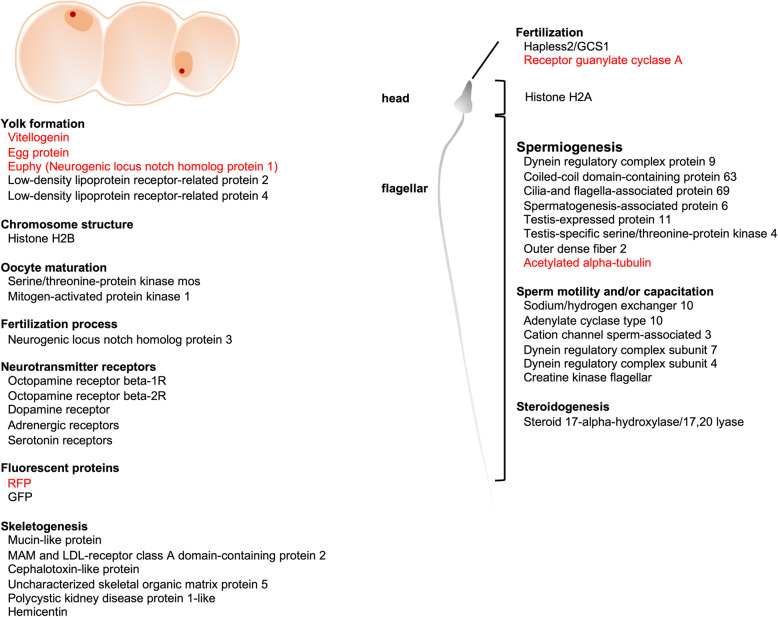
Evolutionarily conserved genes associated with oocyte development (vitellogenin-A2, low-density lipoprotein receptor-related proteins), formation of chromosome structure (histone H2B), and oocyte maturation (serine/threonine-protein kinase mos, mitogen-activated protein kinase 1) were identified (Table 4). Additionally, several sequences similar to components of skeletal organic matrix proteins of scleractinians (mucin-like protein, MAM and LDL-receptor class A domain-containing protein 2, cephalotoxin-like protein, uncharacterized skeletal organic matrix protein 5, polycystic kidney disease protein 1-like, and hemicentin) were also identified (Table 4, Fig. 5).
Table 4.
Upregulated genes of interest in premature/mature ovaries showing similarities to oocyte development/maturation-related genes in other animals
| Category | Annotation | Assembly ID | Reference |
|---|---|---|---|
| Oocyte development | Vitellogenin-A2 | CL4556.Contig6_All | (E. ancora Vitellogenin) [12] |
| Uncharacterized skeletal organic matrix protein 5 | Unigene22577_All | (E. ancora Egg protein) [12] | |
| Neurogenic locus notch homolog protein 1 | Unigene28293_All | (E. ancora Euphy)[13] | |
| Low-density lipoprotein receptor-related protein 2 | Unigene39647_All | [39, 40] | |
| Low-density lipoprotein receptor-related protein 4 | Unigene20580 All | [39, 40] | |
| Chromosome structure | Histone H2B | Unigene6592_All | [119] |
| Oocyte maturation | Serine/threonine-protein kinase mos | Unigene58091_All | [44] |
| Mitogen-activated protein kinase 1 | CL13032.Contig3_All | [44] | |
| Neurotransmitter receptors | Octopamine receptor beta-1R | CL4990.Contig1_All | [57, 58] |
| Octopamine receptor beta-2R | Unigene7363_All | [57, 58] | |
| Dopamine receptor | Unigene35882_All | [59] | |
| Skeletogenesis | Mucin-like protein | CL7569.Contig3_All | [41] |
| MAM and LDL-receptor class A domain-containing protein 2 | CL3169.Contig2_All | [41] | |
| Cephalotoxin-like protein | Unigene22179_All | [41] | |
| Uncharacterized skeletal organic matrix protein 5 | Unigene22577_All | [41] | |
| Polycystic kidney disease protein 1-like | CL5263.Contig3_All | [42] | |
| Hemicentin | CL1601.Contig3_All | [43] |
Fig. 5.
Genes potentially involved in oocyte development and maturation, and in sperm motility/capacitation, and fertilization in E. ancora. Genes indicated in red have been reported in our previous studies
Upregulated genes of mature testes
There were 538 specifically upregulated genes in mature testes. Of those, 305 (57%) matched human genes in the SWISS-PROT database (Fig. 4c). GO functional enrichment analysis showed that 32 GO terms were enriched (P < 0.05 and > 4-fold enrichment; Additional file 11): 21 biological processes (BP), 8 cellular components (CC), and 3 molecular functions (MF). Of the enriched BP terms, response to corticosteroid (GO: 0031960), sequestering of TGF beta in extracellular matrix (GO:0035583), and regulation of cellular response to growth factor stimulus (GO: 0090287) were highly enriched. The term spermatid development (GO: 0007286) was also identified, and further queries of genes representative of the term identified genes encoding testis-specific serine/threonine-protein kinases, outer dense fiber protein 2, alstrom syndrome protein 1, radial spoke head 1 homolog, and dynein regulatory complex protein 9. Of the enriched CC terms, microfibril (GO: 0001527) was among the most enriched. Of the 3 MF terms identified, the term extracellular matrix structural constituent (GO: 0030021) was most enriched (Additional file 11).
Among significantly upregulated genes in mature testes (log2 > 8-fold change compared to the other 3 phases, FDR < 0.05), we identified 28 genes, including those encoding creatine kinase, S-type mitochondrial, creatine kinase flagellar, omega-6 fatty acid desaturase, glutamate receptor ionotropic, kainate 2, testis-specific serine/threonine-protein kinase 4, and fibrillin-2 (Table 3). Ten of the 28 genes could not be annotated.
Evolutionarily conserved genes involved in spermiogenesis and fertilization were further explored among upregulated genes in mature testes (Table 5). We identified a number of important genes encoding proteins associated with spermiogenesis (spermatogenesis-associated protein 6, cilia- and flagella-associated protein 69), sperm motility and/or capacitation (dynein regulatory complex subunit 7, sodium/hydrogen exchanger 10, creatine kinase, flagellar, adenylate cyclase type 10, and cation channel sperm-associated protein 3), and fertilization process (hapless 2/generative cell specific 1 and receptor guanylate cyclase). We also identified a gene encoding steroid 17α-hydroxylase/17,20-lyase (Cyp17a), a key enzyme in sex steroid and cortisol production (Fig. 5, Table 5).
Table 5.
Upregulated genes of interest in mature testes showing similarities to sperm-related genes in other animals
| Category | Annotation | Assembly ID | Reference |
|---|---|---|---|
| Spermiogenesis | Spermatogenesis-associated protein 6 | Unigene40770_All | [76] |
| Cilia- and flagella-associated protein 69 | Unigene30111_All | [77] | |
| Testis-specific serine/threonine-protein kinase 1 | CL2440.Contig1_All | [78, 79] | |
| Testis-specific serine/threonine-protein kinase 2 | Unigene8363_All | [78, 79] | |
| Outer dense fiber protein 2 | CL11766.Contig4_All | [80] | |
| Alstrom syndrome protein 1 | CL2440.Contig1_All | [81] | |
| Radial spoke head 1 homolog | CL2440.Contig1_All | [82] | |
| Dynein regulatory complex protein 9 | CL4871.Contig6_All | [83] | |
| Sperm motility/capacitation | Dynein regulatory complex subunit 7 | CL8060.Contig33_All | [86] |
| Sodium/hydrogen exchanger 10 | CL11505.Contig1_All | [87] | |
| Creatine kinase, flagellar | CL12240.Contig1_All | [88] | |
| Adenylate cyclase type 10 | CL1089.Contig3_All | [87] | |
| Cation channel sperm-associated protein 3 | Unigene27629_All | [87] | |
| Cyclin-F | Unigene55093_All | [89] | |
| Dynein regulatory complex subunit 4 | Unigene7541_All | [90] | |
| Chromosome structure | Histone H2A | CL4659.Contig4_All | [120] |
| Fertilization process | Hapless 2 | CL1879.Contig3_All | [109, 110] |
| Receptor guanylate cyclases | CL4659.Contig4_All | [22] | |
| Cyclic nucleotide-gated channel cone photoreceptor subunit alpha | CL3457.Contig2_All | [153] | |
| Steroidogenesis | Steroid 17-alpha-hydroxylase/17,20 lyase | Unigene38823_All | [104] |
Discussion
Scleractinian gonadal transcriptome assembly
Since scleractinian gametogenesis occurs exclusively in gonads, isolated gonads (but not whole polyps) are useful to explore genes associated with gametogenesis. However, gonad isolation is technically difficult in many scleractinians due to small polyp sizes. Gonad isolation not only requires an understanding of polyp anatomy, but also technical skill. The present study applied previously established techniques for gonad isolation from E. ancora polyps [12] to the current transcriptomic study. Bioinformatics methods to eliminate contigs from symbiotic dinoflagellates or other contaminants were also employed [33]. 60.2% of the contigs in the E. ancora gonadal transcriptome assembly showed similarities to entries in the SWISS-PROT database (Fig. 3). Specifically, 68% were similar to Stylophora pistilata gene models [34] and 59.7% to Pocillpora damicornis gene models [35] in SWISS-PROT. In addition, conserved Pfam protein domains were detected in 66.2% of contigs in the E. ancora gonadal transcriptome (Fig. 3). Conserved Pfam protein domains were detected in 54% of sequences in the Heliopora coerulea transcriptome assembly [36]. Other transcriptome assemblies showed similar percentages: Dendrophyllia sp. (48.8%), Eguchipsammia fistula (45.4%), and Rhizotrochus typus (51.3%) [37]. The E. ancora gonadal transcriptome is clearly comparable to other coral genomic or transcriptomic datasets. The present transcriptome assembly allowed us to identify sex-specific and gonadal phase-specific upregulated genes as well as evolutionarily conserved genes associated with germ cell development. The resulting dataset will provide a foundation for future research investigating molecular and cellular mechanisms of gametogenesis in scleractinians.
Characteristics of premature/mature ovaries as assessed by anatomical and histological analyses
The observed growth of oocytes and the loss of germinal vesicles in oocytes of premature/mature ovaries suggest that oocytes were still actively accumulating essential materials (e.g., yolk and other components) for survival and development of embryos until just before maturation. Also, the oocyte maturation process, including germinal vesicle breakdown (GVBD) and resumption of meiosis occurred in some oocytes.
Upregulated genes in premature/mature ovaries
Yolk formation and accumulation is one of the most important aspects of oogenesis for oviparous animals. In scleractinian eggs, several major yolk proteins, including vitellogenin (Vg), a female-specific phosphoglycolipoprotein, and large amounts of lipids (e.g., wax esters, fatty acids, phosphatidylethanolamines, and phosphatidylcholines) have been identified to date [12, 13, 19, 20, 38]. The present study found that transcripts encoding 3 major yolk proteins were upregulated (Vg, Egg protein, and Euphy, Fig. 5, Table 4), in agreement with histological observations, indicating that oocytes were actively accumulating yolk materials. Those yolk proteins are produced by ovarian somatic cells adjacent to oocytes [12, 13]. However, little is known about the uptake mechanisms of yolk proteins by oocytes. Although receptor-mediated endocytosis has been hypothesized, related receptor molecules have not been identified yet [12]. The present study also identified transcripts encoding two types of low-density lipoprotein receptor-related proteins (Lrps) as upregulated genes in premature/mature ovaries (Fig. 5, Table 4). In some teleosts, a member of Lrps, Lrp13, serves as one of the Vg receptors expressed on oocyte membranes [39, 40]. Thus, the Lrps identified here may be involved in uptake mechanisms of yolk materials in scleractinians, and are promising candidate receptors for Vg and/or other lipoproteins in future studies.
In addition to the major yolk materials, eggs of scleractinians are assumed to accumulate materials essential for larval development. Among the upregulated genes in premature/mature ovaries, we identified several sequences similar to components of skeletal organic matrix proteins found in A. digitifera [41], A. millepora [42], and S. pistillata [43]. Since no skeleton formation occurs in ovaries, it is likely that these gene products (mRNA and proteins) are stored in oocytes during oogenesis to be used for skeleton formation during larval development. We cannot rule out the possibility that the identified genes may have other functions in oocyte development/maturation.
The occurrence of GVBD in some oocytes of ovaries collected in April 2017 was an unexpected finding, because mature gametes were not observed in testes collected at the same time. It is possible that timing of oocyte maturation was split among oocytes and/or ovaries over April and May (or June) for unknown reasons. We cannot completely rule out the possibility that the GVBD was partially induced by isolation of ovaries from polyps, i.e., mechanical stress. Nevertheless, this study successfully identified two sequences similar to the serine/threonine-protein kinase mos (Mos) gene and the mitogen-activated protein kinase 1 (Mapk1) gene, which contribute to signaling pathways of oocyte maturation in a variety of animals, including cnidarians [44] (Fig. 5). Upregulation of these two genes in premature/mature ovaries implies that they may also function in oocyte maturation in scleractinians. Previous studies regarding oocyte maturation in scleractinians were limited to histological observations and focused on the presence and timing of GVBD [45, 46]. To the best of our knowledge, this is the first study to identify these candidate molecules in oocyte maturation of scleractinians.
In a variety of animals, hormones (i.e., steroids, growth factors, peptides, and other substances) are involved in the reproduction [47–54]. In Acropora species, transcriptomic studies suggest that melanopsin-like homolog and /or neuropeptides [55] and Rhodopsin-like receptors [56] are involved in the signaling pathway for spawning in scleractinians. Enriched BP terms in E. ancora premature/mature ovaries imply that neuronal activity is significantly higher than during other phases. Upregulation of transcripts similar to genes encoding monoamine receptors (e.g., octopamine receptors, dopamine receptors, adrenergic receptors, and serotonin receptors, [57, 58] Fig. 5, Table 4) also support this assumption. Recent studies show that some neurotransmitters (dopamine and serotonin) are also involved in regulation of scleractinian spawning. Treatment of Acropora tenuis with dopamine during the final phase of gametogenesis inhibited spawning [59]. By contrast, treatments with serotonin and its precursor, L-5-hydroxytryptophan (5-HTP) induced spawning of Acropora cervicornis [60]. Taking all these lines of evidence into account, the identified monoamine receptors may also be essential during the premature/mature phase of E. ancora oogenesis. It will be of interest to investigate whether treatment of female E. ancora with these neurotransmitters induces or inhibits oocyte maturation and spawning.
Of particular interest is the upregulation of three genes encoding neurogenic locus notch homolog proteins in premature/mature ovaries (Additional file 12). The Notch signaling pathway is conserved across animal taxa, and regulates cell-cell interactions and cell fate determination [61]. One of the identified genes, neurogenic locus notch homolog protein 1, encodes Euphy, a novel major yolk protein in E. ancora oocytes identified in our previous study [13]. The remaining 2 genes have not been previously reported. Although both sequences possess EGF-like domain repeats typifying notch homolog proteins, they are structurally distinct from Notch1 identified in vertebrates (e.g., human Notch1). These may be novel genes that emerged after gene duplication, domain shuffling, and rapid molecular evolution in cnidarian/scleractinian lineages [41, 42]. Interestingly, one of them, neurogenic locus notch homolog protein 3, was highly and significantly upregulated, and contains a zona pellucida (ZP) protein and transmembrane domains (Fig. 5, Additional file 12). The ZP is the extracellular matrix (ECM) surrounding mammalian oocytes, composed of four glycoproteins (ZP1-ZP4). ZP functions during oogenesis, fertilization, and preimplantation development in mammals [62]. In jellyfish, a ZP domain-containing protein called mesoglein, which resembles mammalian ZP, was identified in the contact plate of oocytes [63]. Although scleractinian oocytes have neither a protective coat nor a membrane surrounding them, this finding implies that the identified ZP domain-containing protein probably participates in oogenesis and subsequent fertilization processes.
GFP is one of the natural pigments of corals [64–67]. Although the natural functions of GFP remain obscure, proposed functions include photoprotection from high UVA/blue irradiation, photosynthetic enhancement, phototaxis of zooxanthellae [68–72], and antioxidant activity [73, 74]. We previously showed that E. ancora oocytes express an endogenous RFP with H2O2 degradation activity from early to mature stages of oocytes, and suggested a possible role of RFP in protecting oocytes from oxidative stress during oogenesis [15]. Our finding implies that not only RFP, but also GFP may serve in oogenesis, particularly during the premature/mature phase (Fig. 5).
Characteristics of mature testes as assessed by histological and cytological analyses
Spermiogenesis is a process by which haploid spermatids undergo a complex series of morphological changes, and eventually become elongated functional sperm. The presence of spermaries having both round spermatids and mature sperm in testes collected in June 2017 suggested that spermiogenesis was occurring in the testes at the time of collection, and that genes involved in regulation of spermiogenesis were being expressed in testes.
Upregulated genes in mature testes
Morphological changes of male germ cells during spermiogenesis include flagellum formation, nuclear DNA condensation, and elimination of organelles and cytoplasm. Scleractinian spermiogenesis is generally morphologically similar to that of vertebrates, except that male germ cells possess long flagella from early to late stages of development [21]. Nevertheless, scleractinian male germ cells possess typical flagellar axonemes, characterized by a“9 + 2” arrangement of microtubules [21, 75]. In this study, further queries of genes associated with spermatid development (GO term:0007286), together with literature-based gene identification, allowed us to identify various candidate genes encoding proteins of flagellar components [76–83]. The presence of a conserved molecular toolkit for spermiogenesis suggests that scleractinians and vertebrates share similar characteristics at both morphological and molecular levels.
Sperm motility is important for most scleractinians, which fertilize externally in seawater. Sperm of acroporid corals remain completely immotile in seawater until they come close to eggs, whereupon they acquire motility [84]. The presence of chemoattractants and involvement of intracellular pH elevation and Ca2+-dependent signal transduction in sperm motility have been experimentally demonstrated [84, 85]. Molecules regulating flagellar motility still remain largely unexplored in scleractinians. This study identified a number of important genes encoding proteins involved in sperm motility and/or capacitation in mammals and sea urchins, such as cation channel sperm-associated protein 3 (CatSper3), sodium/hydrogen exchanger (sNHE), and adenylate cyclase type 10 (sAC) [86–90] (Fig. 5, Table 5). These findings support the hypothesis of Romero and Nishigaki that CatSper3, sNHE, and sAC form prototypical machinery for sperm flagellar beating in metazoans [87]. This study further identified the gene encoding creatine kinase, flagellar, which was first identified from flagella of sea urchin sperm, participating in energy transport from sperm heads to the flagella during sperm motility [89]. Genes associated with sperm motility and/or capacitation in scleractinians suggest that these features were most likely present in the common ancestor prior to divergence of the cnidarian and bilaterian lineages.
Sex steroids are critical for sex differentiation, gametogenesis, and gamete maturation in vertebrates [91–95]. Sex steroids (e.g., estrogen, testosterone, and progesterone) have been demonstrated in several scleractinians, including E. ancora [96–99]. Additionally, the correlation between sex steroid levels and gametogenic cycles has led to the hypothesis that sex steroids may be involved in regulation of scleractinian reproduction [97, 99]. Steroid biosynthesis is catalyzed by various steroidogenic enzymes. Although steroid biosynthetic activities are known from extracts of some scleractinian tissues [97, 98, 100–102], only one gene encoding a steroidogenic enzyme, 17β-hydroxysteroid dehydrogenase type 14 (17β-hsd 14), has been identified and characterized so far [103]. In the present study, a gene encoding steroid 17α-hydroxylase/17,20-lyase (Cyp17a) (Fig. 5, Table 5), a key enzyme in production of sex steroids and cortisol [104], was upregulated in mature testes. Although further analysis is required to clarify its activity, the presence of this enzyme implies that steroid biosynthesis occurs in mature testes, and the produced sex steroids/cortisol could be associated with maturation of male germ cells in scleractinians.
Molecules involved in fertilization remain largely unknown in scleractinians. We found that a gene similar to Hapless 2/Generative Cell Specific 1 (Hap2/Gcs1) was upregulated in mature testes (Fig. 5, Table 5). Hap2/Gcs1 was first identified as a male gamete-specific transmembrane protein in lilies [105]. The coding gene is found in genomes of most major eukaryotic taxa (e.g., protozoa, plants, and animals) except fungi [106, 107]. Functional analysis with the mutant/gene targeting system showed that Hap2/Gcs1 are essential for gamete fusion in Arabidopsis [105], the protozoan parasite, Plasmodium [106], and the green alga, Chlamydomonas [108]. Expression of Hap2/Gcs1 was also confirmed in male germ cells of some cnidarians, such as Hydra [109] and the starlet sea anemone, Nematostella vectensis [110], and its involvement in fertilization has been demonstrated in sea anemones [110]. Upregulation of Hap2/Gcs1 in E. ancora mature testes suggests that Hap2/Gcs1 participates in scleractinian sperm-egg fusion. Most recently, we reported that receptor guanylate cyclase A (rGC-a) (also known as atrial natriuretic peptide receptor 1 in mammals) is expressed in E. ancora sperm flagella [22] (Fig. 5, Table 5). rGCs are expressed on sperm and serve as receptors for egg-derived sperm-activating and sperm-attracting factors in some echinoderms and mammals [111–114]. Taken together, evolutionarily conserved proteins underlie fertilization mechanisms of scleractinians.
Other major findings and potential applications
Genes encoding Histone H2B and Histone H2A were upregulated in premature/mature ovaries and mature testes, respectively (Fig. 5, Table 4, 5). Histones are the major protein components of chromatins in eukaryote cell nuclei. Five histone protein families exist: the core histone families (H2A, H2B, H3, and H4) and the linker histone family (H1) [115]. Core histones are components of the nucleosome core, whereas linker histones are present in adjacent nucleosomes, where they bind to nucleosomal core particles, and stabilize both nucleosome structure and higher-order chromatin architecture [115, 116]. Various isoforms of each family have been identified as histone variants, and their importance in diverse cellular processes (e.g., transcriptional control, chromosome segregation, DNA repair and recombination, and germline specific translational regulation) have been revealed [117–120]. In scleractinians, although sequences of the histone gene cluster have been identified in Acropora formosa (H3, H4, H2A, and H2B) [121] and Acropora gemmifera (H3, and H2B) [122], differences in gene expression levels between ovaries and testes have not been reported so far. This study revealed the existence of histone variants showing sexually dimorphic expression in scleractinians. In the cnidarian model organism, Hydractinia echinata, 19 genes encoding histones were identified, and some of them, such as histone H2A.X and five H2B variants, are specifically expressed in female and male germ cells, respectively [123]. Our findings imply that the identified histone may control gene expression in female and male germ cells during scleractinian gametogenesis.
Studies of a variety of animals have revealed that a set of specialized and highly conserved genes govern germline specification, development, meiosis, and/or maintenance in metazoans [124–136]. In the gonadal transcriptome, we could identify many genes associated with germline specification and meiotic processes (Additional file 8). Although further spatiotemporal expression analyses and functional assays are required to clarify their functions, their expression in gonads implies that these genes participate in scleractinian germline development and meiosis.
The E. ancora gonadal transcriptome assembly includes a large number of genes without homology to sequences in the SWISS-PROT database. These findings suggest that although scleractinian gametogenesis shares many common molecular characteristics with gametogenesis in other metazoans, it also possesses characteristics that developed in evolutionarily unique ways. Further characterization and functional studies of these unannotated genes will clarify unique features in scleractinian gametogenesis, and this will eventually lead to comprehensive understanding of scleractinian gametogenesis.
The knowledge obtained in the present study will be useful for ecological studies and coral aquaculture. For instance, since scleractinian corals have no secondary sexual characteristics, histological analysis has traditionally been used to investigate polyp or colony sex, as well as to determine the status of germ cell development. However, histological analysis of scleractinians is time consuming. It generally requires decalcification steps, and the whole histological process sometimes takes 1–2 weeks. Identification of molecular markers for determining colony sex and germ cell development status offers a useful alternative process. Colony sex and germ-cell type could be determined faster using PCR with markers, than by histological means. Sex- and gonad phase-specific genes identified in this study would be candidates.
Conclusions
Analysis of upregulated genes in premature/mature gonads allowed us to identify many genes potentially involved in oocyte development, oocyte maturation, spermiogenesis, sperm motility/capacitation, and fertilization processes (Fig. 5). We identified a large number of sex-biased or sex-specific genes and shed light on possible molecular mechanisms of scleractinian gametogenesis, which appear to be coordinated by both conserved and novel genes. This study and its generated datasets thus provide a foundation for future studies regarding gametogenesis and differences between sexes from molecular and cellular perspectives. Furthermore, our transcriptome assembly will be a useful reference for future development of sex-specific and/or stage-specific markers for germ cells for use in coral aquaculture and ecological studies.
Methods
Sample collection
E. ancora specimens were collected by scuba divers at Nanwan Bay, Kenting National Park, in southern Taiwan (21°57′N, 120°46′E). Approximately 10 colonies were labeled, and gonads (> 20 gonads) of labeled colonies were microscopically isolated at different times during a 9-month period from October 2016 (non-spawning period) to June 2017 (spawning period) (Fig. 1d). Collection was approved by the administrative office of Kenting National Park (issue number: 1010006545). For RNA-seq, collected samples were snap frozen in liquid nitrogen, and stored at − 80 °C until use. Some of the isolated gonads were also fixed with 20% Zinc-Formal-Fixx (Thermo Fisher Scientific, Pittsburgh, PA, USA) for histological analysis. Experiments were performed in accordance with principles and procedures approved by the Institutional Animal Care and Use Committee, National Taiwan Ocean University, Taiwan.
Histological analysis for sample selection
Histological analyses were performed to determine developmental phases of gonads, and to select samples for RNA-seq. Isolated gonads (> 10 gonads/colony/time point) were analyzed according to the methodology in our previous studies [8, 9]. Developmental stages of germ cells were determined according to previous criteria [8, 9] with some modifications (see Table 1).
RNA extraction and RNA-seq library construction
In total, 12 testes and 12 ovaries (3 colonies × 4 time points) were selected based on the results of histological analyses. Total RNA of the 24 samples was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. DNase I treated-RNA samples were sent to Beijing Genomics Institute (BGI, Shenzhen). RNAs were qualified using a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA) with an RNA 6000 labchip kit (Agilent Technologies) and all samples were confirmed as high-quality RNA (RIN > 8). Twenty four RNA-seq libraries were constructed using TruSeq mRNA Library Prep Kits v2 (Illumina, San Diego, CA, USA), and sequenced with 150-bp paired-ends (150PE) on an Illumina HiSeq X Ten. Illumina adaptors, low-quality sequences (Phred value Q < 20), and reads with a high proportion of N (> 5%) were removed. Cleaned sequencing data were used for subsequent analyses.
De novo assembly and annotation of the E. ancora transcriptome
The transcriptome assembly of the E. ancora “holobiont”, the host and its symbiotic organisms, was created by BGI as follows. Clean reads of 24 individual samples were assembled de novo using Trinity v2.0.6 software [137] (parameter settings: –min_contig_length 150 –CPU 8 –min_kmer_cov 3 –min_glue 3 –bfly_opts ‘-V 5 –edge-thr = 0.1) and assembled sequences were clustered using Tgicl v2.0.6 software [138] (parameter settings: -l 40 -c 10 -v 25 -O ‘-repeat_stringency 0.95 -minmatch 35 -minscore 35′). Since gonadal samples contained substantial numbers of symbiotic dinoflagellate cells, we bioinformatically separated sequences originating from E. ancora, algal symbionts (Symbiodiniaceae), or microbes as follows. All assembled sequences were aligned to available genomic databases of 4 scleractinian corals and 6 Symbiodiniaceae transcriptomic databases using BLASTN (−evalue 1e-3). These databases included A. digitifera [31, 139], P. damicornis [35, 140], S. pistillata [34], and O. faveolata [141], Symbiodinium sp. A1 [142], Symbiodinium sp. A2 [143], Breviolum sp. B2 [143], Breviolum muscatinei [144], Uncultured Cladocopium sp. [145] and uncultured Durusdinium sp. [145] (For more detailed information on the databases, see Additional file 6). Contigs aligned exclusively to the coral genome database were annotated as “E. ancora contigs”, while those that aligned only to Symbiodiniaceae transcriptome databases were annotated as “Symbiodiniaceae contigs”. To separate contigs aligned to both the coral genome and Symbiodiniaceae transcriptomic databases, contigs were re-aligned BLASTN (−evalue 1e-3) using a combined database of coral genomes and Symbiodiniaceae transcriptomes. Based on the top hit results of BLASTN (corals or Symbiodiniaceae), contigs were annotated as “E. ancora contigs” or “Symbiodiniaceae contigs” (Fig. 2). All databases used in the present study were downloaded on 3/18/2019. Nucleotide sequences were again clustered using CD-HIT [146] with 97% identity for removing sequences possibly originating from different individuals or haplotypes in a single individual. Finally, contigs were translated into amino acid sequences using the longorf script [147] and clustered using CD-HIT with 95% identity. Completeness using the assembled sequences was assessed using BUSCO (bench-marking universal single-copy orthologs) version 3 [148, 149] in transcriptome mode. Reference E. ancora gonadal transcriptome contigs were annotated as follows: 1) BLAST searches against public protein databases: SWISS-PROT database (−evalue 1e-5) (Consortium 2011) (3/18/2019), 2) Identification of conserved protein domains with the Pfam database (−evalue 1e-5) [150, 151].
Identification of reproduction-related genes in ovaries and testes
Genes important in metazoan reproduction were searched in the reference E. ancora gonadal transcriptome, based on the literature [8, 9, 12, 13, 15, 16, 21, 22, 72, 126–136, 152, 153]. Two strategies were adopted. 1) Full-length cDNA sequences of genes in vertebrates and invertebrates were retrieved from Genbank (NCBI), and local BLAST searches were conducted (BLASTP, cut-off -evalue 1e-5) against translated sequences from the reference E. ancora gonadal transcriptome. 2) Gene names or keyword searches for target categories were performed in SWISS-PROT annotation results.
Differential gene expression analysis
First, possible outlier RNA-seq samples were examined by mapping raw reads to assembled sequences with Bowtie2 v2.2.6 software [154] (parameter setting: -q –phred33 –sensitive –dpad 0 –gbar 99,999,999 –mp 1,1 –np 1 –score-min L,0,-0.1 -I 1 -X 1000 –no- mixed –no-discordant -p 1 -k 200) and the mapping coverage of contigs was determined with RSEM v1.2.12 software [154] under default settings. The hclust package in R was used to perform a hierarchical cluster analysis of RNA-seq samples [155]. The above analyses were performed by BGI. Illumina adaptors and low-quality sequences (quality score > Q20, reads length > 25 bp) were removed from raw RNA sequences of the remaining samples using CUTADAPT v1.16 [156] . Using SALMON v0.13.1 [157], clean reads were mapped to the reference E. ancora transcriptome contigs. Further statistical analyses based on mapping counts were done using edgeR v3.24.3 [158, 159] in R software. Mapping counts were normalized by the trimmed mean of M values (TMM) method, and then converted to counts per million (CPM). Differentially expressed genes in each phase of ovaries and testes were identified using the likelihood ratio test (glmLRT). All P-values obtained by likelihood ratio test were adjusted with the Benjamini-Hochberg method. Genes (or transcripts) representing FDR < 0.05 were considered as differentially expressed genes. CPM values were used to identify genes that were differentially expressed in each phase of ovaries and testes, respectively (ANOVA with q-value< 0.05). For heatmap generation, CPM values were scaled to row Z-scores for each of the genes that were highly expressed in each phase of gonads.
Gene enrichment analysis
UniProt IDs were assigned for each reference E. ancora gonadal transcriptome contig based on best matches against the human SWISS-PROT database with BLASTP (cutoff of -evalue 1e-5) [160]. Gene enrichment analysis of Gene Ontology (GO) was performed with the assigned UniProt ID using DAVID Bioinformatics Resources 6.8 (> 4-fold enrichment and P < 0.05) [161, 162]. UniPort IDs of the reference E. ancora gonadal transcriptome were used as a background for the DAVID analysis.
Supplementary information
Additional file 1 Microscopic observation of ovaries and testes isolated in April and June 2017, respectively. (a) External appearances of oocytes having germinal vesicles in isolated ovaries collected in April 2017. (b) The external appearance of oocytes without germinal vesicles collected at the same times as the samples shown in (a). Only one oocyte has a germinal vesicle (arrow). (c) Cytological appearance of an isolated testis collected in June 2017. Morphologically mature sperm with triangular head shapes were observed (arrows).
Additional file 2. (Table) Summary of clean read data for 24 samples used in de novo assembly.
Additional file 3. DDBJ accession numbers for the reference gonadal transcriptome
Additional file 4 DDBJ accession numbers for the E. ancora contigs
Additional file 5. DDBJ accession numbers for the Symbiodiniaceae contigs of DDBJ accession number
Additional file 6 (Table) Reference databases used for identifying E. ancora-originated contigs from the E. ancora holobiont transcriptome assembly
Additional file 7 (Table) Reproduction-related genes of E. ancora identified in our previous studies
Additional file 8 (Table) Evolutionarily conserved genes in metazoan reproduction identified in the E. ancora gonadal transcriptome assembly
Additional file 9 Hierarchical Clustering analysis of E. ancora gonadal samples used in this study. Twelve testis samples (3 colonies, 4 time points) and 12 ovary samples (3 colonies, 4 time points) were subjected to analysis. The cluster dendrogram showed that 2 samples (Oct-female-1 and Feb-male-1) are outliers, while others belong to a similar group. The 2 outliers were removed and the 22 remaining samples were used for gene expression analysis.
Additional file 10 GO functional analysis of upregulated genes in premature/mature ovaries. Significantly (> 4-fold change, P < 0.05) enriched GO terms in biological processes (blue bar), and molecular function (yellow bar). The X-axis represents the magnitude of change. The Y-axis represents the GO functional category.
Additional file 11. GO functional analysis of upregulated genes in mature testes. Significantly (> 4-fold change, P < 0.05) enriched GO terms in biological processes (blue bar), cellular component (green bar), and molecular function (yellow bar). The X-axis represents the magnitude of change. The Y-axis represents the GO functional category.
Additional file 12. Schematic figures depicting domain structures of neurogenic locus notch homolog protein. (a) Neurogenic locus notch homolog proteins 1. (b) Neurogenic locus notch homolog proteins 2. (c) Neurogenic locus notch homolog proteins 3.
Acknowledgements
We thank our colleagues and divers who helped us collect samples. We especially thank Dr. Pung-Pung Hwang from Academia Sinica, Taiwan, who served as a scientific advisor.
Abbreviations
- GVBD
Germinal vesicle breakdown
- SRA
Sequence read archive
- DDBJ
DNA Data Bank of Japan
- bp
Base pair
- GC content
Guanine-Cytosine content
- Pfam
Protein family
- Gcl
Germ cell-less
- Mago
Mago nashi
- Boule
Boule-like
- Pum1
Pumilio homolog 1
- Dmc1
DNA meiotic recombinase 1
- Muts4
MutS protein homolog 4
- Muts5
MutS protein homolog 5
- Mlh1
DNA mismatch repair protein Mlh1
- Sycp1
Synaptonemal complex protein 1
- Sycp2
Synaptonemal complex protein 2
- Sycp3
Synaptonemal complex protein 3
- Rad21
Double-strand-break repair protein rad21
- FDR
False discovery rate
- GO
Gene ontology
- BP
Biological processes
- MF
Molecular functions
- CC
Cellular components
- SOMPs
Skeletal organic matrix proteins
- Lrps
Low-density lipoprotein receptor related proteins
- Lrp13
Low-density lipoprotein receptor-related-protein 13
- Vg
Vitellogenin
- Mos
Serine/threonine-protein kinase mos
- Map k1
Mitogen-activated protein kinase 1
- 5-HTP
5-Hydroxytryptophan
- Notch1
Neurogenic locus notch homolog protein 1
- ZP
Zona pellucida
- ECM
Extracellular matrix
- GFP
Green fluorescent protein
- UVA
Ultraviolet A
- RFP
Red fluorescent protein
- CatSper3
Cation channel sperm-associated protein 3
- sNHE
Sodium/hydrogen exchange
- sAC
Adenylate cyclase type 10
- 17beta-hsd 14
17beta-hydroxysteroid dehydrogenase type 14
- Cyp17a
17α-hydroxylase/17,20-lyase
- Hap2/Gcs1
Hapless 2/Generative Cell Specific 1
- rGC
Receptor guanylate cyclase
- PCR
Polymerase chain reaction
- BGI
Beijing Genomics Institute
- RIN
RNA integrity number
- PE
Paired-end
- BUSCO
Bench-marking universal single-copy orthologs
- CPM
Count per million
- ANOVA
Analysis of variance
- DAVID
Database for annotation, visualization and integrated discovery
Authors’ contributions
SS conceived the idea and designed the experiments. YLC and SS performed the sampling and experiments. CS, YY, YLC, and SS analyzed the data. YLC, SS, CFC, and CS wrote the manuscript. The authors read and approved the final manuscript. The authors read and approved the final manuscript.
Funding
This research was funded by grants from the Ministry of Science and Technology, Taiwan (MOST) (104–2313-B019-MY3 to CFC), and partially supported by (MOST 103–2621-B-019-006-MY3, MOST 108–2628-B-019-001 to SS) and Japan Society for the Promotion of Science (JSPS) KAKENHI (17KT0027 and 17 K07949 to CS).
Availability of data and materials
Datasets created and/or analyzed in this study are available in the Sequence Read Archive (SRA) of DNA DataBank of Japan (DDBJ) under BioProject accession PRJDB9831. Transcriptome assemblies have been deposited at the DDBJ/European Nucleotide Archive/GenBank Transcriptome Shotgun Assembly (TSA) under accession numbers ICQS01000001-ICQS01169272. The datasets supporting the conclusions of this article are included within the article and its additional files (Additional files 6, 7, 8).
Ethics approval and consent to participate
Collection of E. ancora specimens was approved by the administrative office of Kenting National Park (issue number: 1010006545).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shinya Shikina, Email: shikina@mail.ntou.edu.tw.
Chuya Shinzato, Email: c.shinzato@aori.u-tokyo.ac.jp.
Ching-Fong Chang, b0044@email.ntou.edu.tw.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-07113-9.
References
- 1.Harrison PL, Babcock RC, Bull GD, Oliver JK, Wallace CC, Willis BL. Mass spawning in tropical reef corals. Science. 1984;223:1186–9. [DOI] [PubMed]
- 2.Willis BL, Babcock RC, Harrison PL, Oliver JK, Wallace CC. Patterns in the mass spawning of corals on the great barrier reef from 1981 to 1984. In: Proceedings of the 5th international coral reef congress. French: Polynesia; 1985. p. 343–8.
- 3.Babcock RC, Bull GD, Harrison PL, Heyward AJ, Oliver JK, Wallace CC, et al. Synchronous spawnings of 105 scleractinian coral species on the great barrier reef. Mar Biol. 1986;90:379–94.
- 4.Harrison PL, Wallace CC. Reproduction, dispersal and recruitment of scleractinian corals. In: Dubinsky Z, editor. Ecosystems of the world 25, coral reefs. Amsterdam: Elsevier; 1990. p. 133–207.
- 5.Richmond RH, Hunter CL. Reproduction and recruitment of corals: comparisons among the Caribbean, the tropical Pacific, and the Red Sea. Mar Ecol Prog Ser. 1990;60:185–203.
- 6.Baird AH, Guest JR, Willis BL. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu Rev Ecol Evol Syst. 2009;40:551–71.
- 7.Harrison PL. Sexual reproduction in scleractinian corals. In: Dubinsky Z, Stambler N, editors. Coral reefs: ecosystem in transition. Netherlands: Springer; 2011. p. 59–85.
- 8.Shikina S, Chen CJ, Liou JY, Shao ZF, Chung YJ, Lee YH, et al. Germ cell development in the scleractinian coral Euphyllia ancora (Cnidaria, Anthozoa). POLS One. 2012;7:e41569. [DOI] [PMC free article] [PubMed]
- 9.Shikina S, Chung YJ, Wang HM, Chiu YL, Shao ZF, Lee YH, et al. Localization of early germ cells in a stony coral, Euphyllia ancora: potential implications for a germline stem cell system in coral gametogenesis. Coral Reefs. 2015a;34:639–53.
- 10.Szmant-Froelich A, Yevich P, Pilson MEQ. Gametogenesis and early development of the temperate coral Astrangia danae (Anthozoa: scleractinia). Biol Bull. 1980;158:257–69.
- 11.Szmant-Froelich A, Reutter M, Riggs L. Sexual reproduction of Favia fragum (Esper): lunar patterns of gametogenesis, embryogenesis and planulation in Puerto Rico. Bull Mar Sci. 1985;37:880–92.
- 12.Shikina S, Chen CJ, Chung YJ, Shao ZF, Liou JY, Tseng HP, et al. Yolk formation in a stony coral Euphyllia ancora Cnidaria, Anthozoa: insight into the evolution of Vitellogenesis in Nonbilaterian animals. Endocrinology. 2013;154:3447–59. [DOI] [PubMed]
- 13.Shikina S, Chiu YL, Lee YH, Chang CF. From somatic cells to oocytes: a novel yolk protein produced by ovarian somatic cells in a stony coral, Euphyllia ancora. Biol Reprod. 2015b;93:1–10. [DOI] [PubMed]
- 14.Shikina S, Chang CF. Sexual reproduction in stony corals and insight into the evolution of oogenesis in Cnidaria. In: Goffredo S, Dubinsky Z, editors. The Cnidaria, past, present and future. The world of medusa and her sisters. Switzerland: Springer; 2016. p. 249–68.
- 15.Shikina S, Chiu YL, Chung YJ, Chen CJ, Lee YH, Chang CF. Oocytes express an endogenous red fluorescent protein in a stony coral, Euphyllia ancora: a potential involvement in coral oogenesis. Sci Rep. 2016b;6:25868. [DOI] [PMC free article] [PubMed]
- 16.Chen CJ, Shikina S, Chen WJ, Chung YJ, Chiu YL, Bertrand JA, et al. A novel female-specific and sexual reproduction-associated Dmrt gene discovered in the stony coral, Euphyllia ancora. Biol Reprod. 2016;94:40. [DOI] [PubMed]
- 17.Imagawa S, Nakano Y, Watanabe T. Molecular analysis of a major soluble egg protein in the scleractinian coral Favites chinensis. Comp Biochem Physiol B Biochem Mol Biol. 2004;137:11–9. [DOI] [PubMed]
- 18.Hayakawa H, Nakano Y, Andoh T, Watanabe T. Sex-dependent expression of mRNA encoding a major egg protein in the gonochoric coral Galaxea fascicularis. Coral Reefs. 2005;24:488–94.
- 19.Hayakawa H, Andoh T, Watanabe T. Precursor structure of egg proteins in the coral Galaxea fascicularis. Biochem Biophys Res Commun. 2006;344:173–80. [DOI] [PubMed]
- 20.Hayakawa H, Andoh T, Watanabe T. Identification of a novel yolk protein in the hermatypic coral Galaxea fascicularis. Zoolog Sci. 2007;24:249–55. [DOI] [PubMed]
- 21.Shikina S, Chiu YL, Chen CJ, Yang SH, Yao JI, Chen CC, et al. Immunodetection of acetylated alpha-tubulin in stony corals: evidence for the existence of flagella in coral male germ cells. Mol Reprod Dev. 2017;84:1285–95. [DOI] [PubMed]
- 22.Zhang Y, Chiu YL, Chen CJ, Ho YY, Shinzato C, Shikina S, et al. Discovery of a receptor guanylate cyclase expressed in the sperm flagella of stony corals. Sci Rep. 2019;9:1–9. [DOI] [PMC free article] [PubMed]
- 23.Epstein N, Bak RPM, Rinkevich B. Applying forest restoration principles to coral reef rehabilitation. Aquat Conserv. 2003;13:387–95.
- 24.Rinkevich B. Management of coral reefs: we have gone wrong when neglecting active reef restoration. Mar Pollut Bull. 2008;56:1821–4. [DOI] [PubMed]
- 25.Omori M. Coral restoration research and technical developments: what we have learned so far. Mar Biol Res. 2019;15:377–409.
- 26.Stewart MJ, Stewart P, Rivera-Posada J. De novo assembly of the transcriptome of Acanthaster planci testes. Mol Ecol Resour. 2015;15:953–66. [DOI] [PubMed]
- 27.Zhao C, Wang P, Qiu L. RNA-Seq-based transcriptome analysis of reproduction- and growth-related genes in Lateolabrax japonicus ovaries at four different ages. Mol Biol Rep. 2018;45:2213–25. [DOI] [PubMed]
- 28.González-Castellano I, Manfrin C, Pallavicini A, Martínez-Lage A. De novo gonad transcriptome analysis of the common littoral shrimp Palaemon serratus: novel insights into sex-related genes. BMC Genomics. 2019;20:757. [DOI] [PMC free article] [PubMed]
- 29.Piprek RP, Damulewicz M, Tassan JP, Kloc M, Kubiak JZ. Transcriptome profiling reveals male- and female-specific gene expression pattern and novel gene candidates for the control of sex determination and gonad development in Xenopus laevis. Dev Genes Evol. 2019;229:53–72. [DOI] [PMC free article] [PubMed]
- 30.Esteve-Codina A, Kofler R, Palmieri N, Bussotti G, Notredame C, Pérez-Enciso M. Exploring the gonad transcriptome of two extreme male pigs with RNA-seq. BMC Genomics. 2011;12:552. [DOI] [PMC free article] [PubMed]
- 31.Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–3. [DOI] [PubMed]
- 32.Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23:1399–408. [DOI] [PubMed]
- 33.Shinzato C, Inoue M, Kusakabe M. A snapshot of a coral "holobiont": a transcriptome assembly of the scleractinian coral, porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS One. 2014;9:e85182. [DOI] [PMC free article] [PubMed]
- 34.Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot JF, et al. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep. 2017;7:17583. [DOI] [PMC free article] [PubMed]
- 35.Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci Rep. 2018;8:16134. [DOI] [PMC free article] [PubMed]
- 36.Guzman C, Shinzato C, Lu TM, Conaco C. Transcriptome analysis of the reef-building octocoral, Heliopora coerulea. Sci Rep. 2018;8:8397. [DOI] [PMC free article] [PubMed]
- 37.Yum LK, Baumgarten S, Röthig T, Roder C, Roik A, Michell C, et al. Transcriptomes and expression profiling of deep-sea corals from the Red Sea provide insight into the biology of azooxanthellate corals. Sci Rep. 2017;7:6442. [DOI] [PMC free article] [PubMed]
- 38.Arai I, Kato M, Heyward A, Ikeda Y, Iizuka T, Maruyama T. Lipid composition of positively buoyant eggs of reef building corals. Coral Reefs. 1993;12:71–5.
- 39.Reading BJ, Hiramatsu N, Schilling J, Molloy KT, Glassbrook N, Mizuta H. Lrp13 is a novel vertebrate lipoprotein receptor that binds vitellogenins in teleost fishes. J Lipid Res. 2014;55:2287–95. [DOI] [PMC free article] [PubMed]
- 40.Mushirobira Y, Mizuta H, Luo W, Todo T, Hara A, Reading BJ, et al. Molecular cloning and partial characterization of a low-density lipoprotein receptor-related protein 13 (Lrp13) involved in vitellogenin uptake in the cutthroat trout (Oncorhynchus clarki). Mol Reprod Dev. 2015;82:986–1000. [DOI] [PubMed]
- 41.Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and Transcriptomics. PLoS One. 2016;11:e0156424. [DOI] [PMC free article] [PubMed]
- 42.Ramos-Silva P, Kaandorp J, Huisman L, Marie B, Zanella-Cleon I, Guichard N, et al. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 2013;30:2099–112. [DOI] [PMC free article] [PubMed]
- 43.Drake JL, Mass T, Haramaty L, Zelzion E, Bhattacharya D, Falkowski PG. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci U S A. 2013;110:3788–93. [DOI] [PMC free article] [PubMed]
- 44.Amiel A, Leclère L, Robert L, Chevalier S, Houliston E. Conserved functions for Mos in eumetazoan oocyte maturation revealed by studies in a cnidarian. Curr Biol. 2009;19:305–11. [DOI] [PubMed]
- 45.Okubo N, Isomura N, Motokawa T, Hidaka M. Possible self-fertilization in the brooding coral Acropora (Isopora) brueggemanni. Zoolog Sci. 2007;24:277–80. [DOI] [PubMed]
- 46.Suwa R, Nakamura M. A precise comparison of developmental series of oocyte growth and oocyte maturation between real-oocytes and pseudo-oocytes in the coral Galaxea fascicularis. Galaxea (Tokyo). 2018;20:1–7.
- 47.Kanatani H, Hiramoto Y. Site of action of 1-methyladenine in inducing oocyte maturation in starfish. Exp Cell Res. 1970;61:280–4. [DOI] [PubMed]
- 48.Testart J, Lefevre B, Gougeon A. Effects of gonadotrophin-releasing hormone agonists (GnRHa) on follicle and oocyte quality. Hum Reprod. 1993;8:511–8. [DOI] [PubMed]
- 49.Okumura T. Perspectives on hormonal manipulation of shrimp reproduction. Japan Agric Res Quarterly. 2004;38:49–54.
- 50.Nagahama Y, Yamashita M. Regulation of oocyte maturation in fish. Dev Growth Differ. 2008;50:S195–219. [DOI] [PubMed]
- 51.Mita M, Yoshikuni M, Ohno K, Shibata Y, Paul-Prasanth B, Pitchayawasin S, et al. A relaxin-like peptide purified from radial nerves induces oocyte maturation and ovulation in the starfish, Asterina pectinifera. Proc Natl Acad Sci U S A. 2009;106:9507–12. [DOI] [PMC free article] [PubMed]
- 52.Kato S, Tsurumaru S, Taga M, Yamane T, Shibata Y, Ohno K, et al. Neuronal peptides induce oocyte maturation and gamete spawning of sea cucumber, Apostichopus japonicus. Dev Biol. 2009;326:169–76. [DOI] [PubMed]
- 53.Takeda N, Kon Y, Quiroga Artigas G, Lapébie P, Barreau C, Koizumi O, et al. Identification of jellyfish neuropeptides that act directly as oocyte maturation-inducing hormones. Development. 2018;145:dev156786. [DOI] [PubMed]
- 54.Quiroga Artigas G, Lapébie P, Leclère L, Bauknecht P, Uveira J, Chevalier S, et al. A G protein-coupled receptor mediates neuropeptide-induced oocyte maturation in the jellyfish Clytia. PLoS Biol. 2020;18:e3000614. [DOI] [PMC free article] [PubMed]
- 55.Kaniewska P, Alon S, Karako-Lampert S, Hoegh-Guldberg O, Levy O. Signaling cascades and the importance of moonlight in coral broadcast mass spawning. Elife. 2015;4:e09991. [DOI] [PMC free article] [PubMed]
- 56.Rosenberg Y, Doniger T, Harii S, Sinniger F, Levy O. Canonical and cellular pathways timing gamete release in Acropora digitifera, Okinawa, Japan. Mol Ecol. 2017;26:2698–710. [DOI] [PubMed]
- 57.Rubinstein CD, Wolfner MF. Drosophila seminal protein ovulin mediates ovulation through female octopamine neuronal signaling. Proc Natl Acad Sci U S A. 2013;110:17420-5. [DOI] [PMC free article] [PubMed]
- 58.Lim J, Sabandal PR, Fernandez A, Sabandal JM, Lee HG, Evans P, et al. The octopamine receptor Octβ2R regulates ovulation in Drosophila melanogaster. PLOS One. 2014;9:e104441. [DOI] [PMC free article] [PubMed]
- 59.Isomura N, Yamauchi C, Takeuchi Y, Takemura A. Does dopamine block the spawning of the acroporid coralAcropora tenuis? Sci Rep. 2013;3:2649. [DOI] [PMC free article] [PubMed]
- 60.Flint M, Than JT. Potential spawn induction and suppression agents in Caribbean Acropora cervicornis corals of the Florida Keys. PeerJ. 2016;4:e1982. [DOI] [PMC free article] [PubMed]
- 61.Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129-38. [PubMed]
- 62.Wassarman PM, Litscher ES. Mammalian fertilization: the egg's multifunctional zona pellucida. Int J Dev Biol. 2008;52:665-76. [DOI] [PubMed]
- 63.Adonin LS, Shaposhnikova TG, Podgornaya OI. Aurelia aurita (Cnidaria) Oocytes contact plate structure and development. PLOS One. 2012;7:e46542. [DOI] [PMC free article] [PubMed]
- 64.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969-73. [DOI] [PubMed]
- 65.Labas YA, Gurskaya NG, Yanushevich YG, Fradkov AF, Lukyanov KA, Lukyanov SA, et al. Diversity and evolution of the green fluorescent protein family. Proc Natl Acad Sci U S A. 2002;99:4256-61. [DOI] [PMC free article] [PubMed]
- 66.Field SF, Bulina MY, Kelmanson IV, Bielawski JP, Matz MV. Adaptive evolution of multicolored fluorescent proteins in reef- building corals. J Mol Evol. 2006;62:332-9. [DOI] [PubMed]
- 67.Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, et al. Diversity and evolution of coral fluorescent proteins. PLOS One. 2008;3:e2680. [DOI] [PMC free article] [PubMed]
- 68.Salih A, Larkum A, Cox G, Kühl M, Hoegh Guldberg O. Fluorescent pigments in corals are photoprotective. Nature. 2000;408:850-53. . [DOI] [PubMed]
- 69.Takahashi S, Whitney SM, Badger MR. Different thermal sensitivity of the repair of photodamaged photosynthetic machinery in cultured Symbiodinium species. Proc Natl Acad Sci U S A. 2009;106:3237-42. [DOI] [PMC free article] [PubMed]
- 70.Roth MS, Latz MI, Goericke R, Dehyn DD. Green fluorescent protein regulation in the coral Acropora yongei during photoacclimation. J Exp Biol. 2010;213:3644-55. [DOI] [PubMed]
- 71.Ben‐Zvi O, Eyal G, Loya Y. Light‐dependent fluorescence in the coral Galaxea fascicularis. Hydrobiologia. 2015;759:15-26.
- 72.Chiu YL, Shikina S, Chang CF. Testicular somatic cells in the stony coral Euphyllia ancora express an endogenous green fluorescent protein. Mol Reprod Dev. 2019;86:798-811. [DOI] [PubMed]
- 73.Bou‐Abdallah F, Chasteen ND, Lesser MP. Quenching of superoxide radicals by green fluorescent protein. Biochim Biophys Acta. 2006;1760:1690-5. [DOI] [PMC free article] [PubMed]
- 74.Palmer CV, Modi CK, Mydlarz LD. Coral fluorescent proteins as antioxidants. PLOS One. 2009;4:e7298. [DOI] [PMC free article] [PubMed]
- 75.Padilla-Gamin ~o JL, Weatherby TM, Waller RG, Gates RD. Formation and structural organization of the egg-sperm bundle of the scleractinian coral Montipora capitata. Coral Reefs. 2011;30:371-80.
- 76.Yuan S, Stratton CJ, Bao J, Zheng H, Bhetwal BP, Yanagimachi R, et al. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc Natl Acad Sci U S A. 2015;112:E430-9. [DOI] [PMC free article] [PubMed]
- 77.Dong FN, Amiri-Yekta A, Martinez G, Saut A, Tek J, Stouvenel L, Lorès P, et al. Absence of CFAP69 Causes Male Infertility due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am J Hum Genet. 2018;102:636-48. [DOI] [PMC free article] [PubMed]
- 78.Xu B, Hao Z, Jha KN, Zhang Z, Urekar C, Digilio L, et al. Targeted deletion of Tssk1 and 2 causes male infertility due to haploinsufficiency. Dev Biol. 2008;319:211-22. [DOI] [PMC free article] [PubMed]
- 79.Hao Z, Jha KN, Kim YH, Vemuganti S, Westbrook VA, Chertihin O, et al. Expression analysis of the human testis-specific serine/threonine kinase (TSSK) homologues. A TSSK member is present in the equatorial segment of human sperm. Mol Hum Reprod. 2004;10:433-44. [DOI] [PubMed]
- 80.Tarnasky H, Cheng M, Ou Y, Thundathil JC, Oko R, van der Hoorn FA. Gene trap mutation of murine outer dense fiber protein-2 gene can result in sperm tail abnormalities in mice with high percentage chimaerism. BMC Dev Biol. 2010;10:67. [DOI] [PMC free article] [PubMed]
- 81.Arsov T, Silva DG, O'Bryan MK, Sainsbury A, Lee NJ, Kennedy C, et al. Fat aussie-a new Alström syndrome mouse showing a critical role for ALMS1 in obesity, diabetes, and spermatogenesis. Mol Endocrinol. 2006;20:1610-22. [DOI] [PubMed]
- 82.Abbasi F, Miyata H, Shimada K, Morohoshi A, Nozawa K, Matsumura T, et al. RSPH6A is required for sperm flagellum formation and male fertility in mice. J Cell Sci. 2018;131:jcs221648. [DOI] [PMC free article] [PubMed]
- 83.Li RK, Tan JL, Chen LT, Feng JS, Liang WX, Guo XJ, et al. Iqcg is essential for sperm flagellum formation in mice. PLOS One. 2014;9:e98053. [DOI] [PMC free article] [PubMed]
- 84.Morita M, Nishikawa A, Nakajima A, Iguchi A, Sakai K, Takemura A, et al. Eggs regulate sperm flagellar motility initiation, chemotaxis and inhibition in the coral Acropora digitifera, A. gemmifera and A. tenuis. J Exp Biol. 2006;209:4574-9. [DOI] [PubMed]
- 85.Morita M, Iguchi A, Takemura A. Roles of calmodulin and calcium/calmodulin-dependent protein kinase in flagellar motility regulation in the coral Acropora digitifera. Mar Biotechnol (NY). 2009;11:118-23. [DOI] [PubMed]
- 86.Morohoshi A, Miyata H, Shimada K, Nozawa K, Matsumura T, Yanase R, et al. Nexin-Dynein regulatory complex component DRC7 but not FBXL13 is required for sperm flagellum formation and male fertility in mice. PLOS Genet. 2020;16:e1008585. [DOI] [PMC free article] [PubMed]
- 87.Romero F, Nishigaki T. Comparative genomic analysis suggests that the sperm-specific sodium/proton exchanger and soluble adenylyl cyclase are key regulators of CatSper among the Metazoa. Zoological Lett. 2019;5:25. [DOI] [PMC free article] [PubMed]
- 88.Wothe DD, Charbonneau H, Shapiro BM. The phosphocreatine shuttle of sea urchin sperm: flagellar creatine kinase resulted from a gene triplication. Proc Natl Acad Sci U S A. 1990;87:5203-7. [DOI] [PMC free article] [PubMed]
- 89.Sun Y, Fu L, Xue F, Li Y, Xu H, Chen J. Digital gene expression profiling and validation study highlight Cyclin F as an important regulator for sperm motility of chickens. Poult Sci. 2019;98:5118-26. [DOI] [PubMed]
- 90.Yeh SD, Chen YJ, Chang AC, Ray R, She BR, Lee WS, et al. Isolation and properties of Gas8, a growth arrest-specific gene regulated during male gametogenesis to produce a protein associated with the sperm motility apparatus. J Biol Chem. 2002;277:6311-7. [DOI] [PubMed]
- 91.Slater CH, Schreck CB, Swanson P. Plasma profiles of the sex steroids and gonadotropins in maturing female spring chinook salmon (Oncorhynchus tshawytscha). Comp Biochem Physiol A Mol Integr Physiol. 1990;109:167-75.
- 92.Tobet SA, Hanna IK. Ontogeny of sex differences in the mammalian hypothalamus and preoptic area. Cell Mol Neurobiol. 1997;17:565-601. [DOI] [PMC free article] [PubMed]
- 93.Vanderhyden B. Molecular basis of ovarian development and function. Front Biosci. 2002;7:d2006-22. [DOI] [PubMed]
- 94.Papadaki M, Piferrer F, Zanuy S, Maingot E, Divanach P, Mylonas CC. Growth, sex differentiation and gonad and plasma levels of sex steroids in male- and female-dominant populations of Dicentrarchus labrax obtained through repeated size grading. J Fish Biol. 2005;66:938-56.
- 95.Lahnsteiner F, Berger B, Kletzl M, Weismann T. Effect of 17beta-estradiol on gamete quality and maturation in two salmonid species. Aquat Toxicol. 2006;79:124-31. [DOI] [PubMed]
- 96.Atkinson S, Atkinson MJ. Detection of estradiol 17b during mass coral spawn. Coral Reefs. 1992;11:33-35.
- 97.Tarrant AM, Atkinson S, Atkinson MJ. Estrone and estradiol-17 beta concentration in tissue of the scleractinian coral, Montipora verrucosa. Comp Biochem Physiol A Mol Integr Physiol. 1999;122:85-92. [DOI] [PubMed]
- 98.Slattery M, Hines GA, Starmer J, Paul VJ. Chemical signals in gametogenesis, spawning, and larval settlement and defense of the soft coral Sinularia polydactyla. Coral Reefs. 1999;18:75-84.
- 99.Twan WH, Hwang JS, Chang CF. Sex steroids in scleractinian coral, Euphyllia ancora: implication in mass spawning. Biol Reprod. 2003;68:2255-60. [DOI] [PubMed]
- 100.Gassman NJ, Kennedy CJ. Cytochrome P-450 content and xenobiotic metabolizing enzyme activities in the scleractinian coral, Favia fragum (Esper). Bull Mar Sci. 1992;50:320-30.
- 101.Twan WH, Hwang JS, Lee YH, Tung YH, Wu HF, Tung YH, et al. Hormones and reproduction in scleractinian corals. Comp Biochem Physiol A Mol Integr Physiol. 2006;144:247-53. [DOI] [PubMed]
- 102.Blomquist CH, Lima PH, Tarrant AM, Atkinson MJ, Atkinson S. 17Beta-hydroxysteroid dehydrogenase (17beta-HSD) in scleractinian corals and zooxanthellae. Comp Biochem Physiol B Biochem Mol Biol. 2006;143:397-403. [DOI] [PubMed]
- 103.Shikina S, Chung YJ, Chiu YL, Huang YJ, Lee YH, Chang CF. Molecular cloning and characterization of a steroidogenic enzyme, 17β-hydroxysteroid dehydrogenase type 14, from the stony coral Euphyllia ancora (Cnidaria, Anthozoa). Gen Comp Endocrinol. 2016a;228:95-104. [DOI] [PubMed]
- 104.Di Cerbo A, Biason-Lauber A, Savino M, Piemontese MR, Di Giorgio A, Perona M, et al. Combined 17alpha-Hydroxylase/17, 20-lyase deficiency caused by Phe93Cys mutation in the CYP17 gene. J Clin Endocrinol Metab. 2002;87:898-905. [DOI] [PubMed]
- 105.Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol. 2006;8:64-71. [DOI] [PubMed]
- 106.Hirai M, Arai M, Mori T, Miyagishima SY, Kawai S, Kita K, et al. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol. 2008;18:607-13. [DOI] [PubMed]
- 107.Wong JL, Leydon AR, Johnson MA. HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. PLOS Genet. 2010;6:e1000882. [DOI] [PMC free article] [PubMed]
- 108.Liu Y, Tewari R, Ning J, Blagborough AM, Garbom S, Pei J, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;22:1051-68. [DOI] [PMC free article] [PubMed]
- 109.Steele RE, Dana CE. Evolutionary History of the HAP2/GCS1 Gene and Sexual Reproduction in Metazoans. PLOS One. 2009;4:e7680. [DOI] [PMC free article] [PubMed]
- 110.Ebchuqin E, Yokota N, Yamada Y, Yasuoka Y, Akasaka M, Arakawa M, et al. Evidence for participation of GCS1 in fertilization of the starlet sea anemone Nematostella vectensis: implication of a common mechanism of sperm-egg fusion in plants and animals. Biochem Biophys Res Commun. 2014;451:522-8. [DOI] [PubMed]
- 111.Nishigaki T, Chiba K, Hoshi M. A 130-kDa membrane protein of sperm flagella is the receptor for asterosaps, sperm-activating peptides of starfish Asterias amurensis. Dev Biol. 2000;219:154-62. [DOI] [PubMed]
- 112.Kaupp UB, Hildebrand E, Weyand I. Sperm chemotaxis in marine invertebrates-molecules and mechanisms. J Cell Physiol. 2006;208:487-94. [DOI] [PubMed]
- 113.Bian F, Mao G, Guo M, Mao G, Wang J, Li J, et al. Gradients of natriuretic peptide precursor A (NPPA) in oviduct and of natriuretic peptide receptor 1 (NPR1) in spermatozoon are involved in mouse sperm chemotaxis and fertilization. J Cell Physiol. 2012;227:2230-9. [DOI] [PubMed]
- 114.Kong N, Xu X, Zhang Y, Wang Y, Hao X, Zhao Y, et al. Natriuretic peptide type C induces sperm attraction for fertilization in mouse. Sci Rep. 2017;7:39711. [DOI] [PMC free article] [PubMed]
- 115.Fyodorov DV, Zhou BR, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol. 2018;19:192-206. [DOI] [PMC free article] [PubMed]
- 116.Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439-53. [DOI] [PMC free article] [PubMed]
- 117.Henikoff S, Smith MM. Histone variants and epigenetics. Cold Spring Harb Perspect Biol. 2015;7:a019364. [DOI] [PMC free article] [PubMed]
- 118.Pérez-Montero S, Carbonell A, Azorín F. Germline-specific H1 variants: the "sexy" linker histones. Chromosoma. 2016;125:1-13. [DOI] [PubMed]
- 119.Hinkley C, Perry M. Histone H2B gene transcription during Xenopus early development requires functional cooperation between proteins bound to the CCAAT and octamer motifs. Mol Cell Biol. 1992;12:4400-11. [DOI] [PMC free article] [PubMed]
- 120.Zhang ZH, Mu SM, Guo MS, Wu JL, Li YQ, Zhang H, et al. Dynamics of histone H2A, H4 and HS1ph during spermatogenesis with a focus on chromatin condensation and maturity of spermatozoa. Sci Rep. 2016;6:25089. [DOI] [PMC free article] [PubMed]
- 121.Miller DJ, Harrison PL, Mahony TJ, McMillan JP, Miles A, Odorico DM, et al. Nucleotide sequence of the histone gene cluster in the coral Acropora formosa (Cnidaria; Scleractinia): features of histone gene structure and organization are common to diploblastic and triploblastic metazoans. J Mol Evol. 1993;37:245-53. [DOI] [PubMed]
- 122.Oldach MJ, Workentine M, Matz MV, Fan TY, Vize PD. Transcriptome dynamics over a lunar month in a broadcast spawning acroporid coral. Mol Ecol. 2017;26:2514-26. [DOI] [PubMed]
- 123.Török A, Schiffer PH, Schnitzler CE, Ford K, Mullikin JC, Baxevanis AD, et al. The cnidarian Hydractinia echinataemploys canonical and highly adapted histones to pack its DNA. Epigenetics Chromatin. 2016;9:36. [DOI] [PMC free article] [PubMed]
- 124.Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869-84. [DOI] [PubMed]
- 125.Gustafson EA, Wessel GM. Vasa genes: emerging roles in the germ line and in multipotent cells. Bioessays. 2010;32:626-37. [DOI] [PMC free article] [PubMed]
- 126.Jongens TA, Hay B, Jan LY, Jan YN. The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell. 1992;70:569-84. [DOI] [PubMed]
- 127.Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, et al. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468-78. [DOI] [PubMed]
- 128.Kuales G, De Mulder K, Glashauser J, Salvenmoser W, Takashima S, Hartenstein V et al. Boule-like genes regulate male and female gametogenesis in the flatworm Macrostomum lignano. Dev Biol. 2011;357:117-32. . [DOI] [PMC free article] [PubMed]
- 129.Mak W, Fang C, Holden T, Dratver MB, Lin H. An Important Role of Pumilio 1 in Regulating the Development of the Mammalian Female Germline. Biol Reprod. 2016;94:134. [DOI] [PMC free article] [PubMed]
- 130.Neyton S, Lespinasse F, Moens PB, Paul R, Gaudray P, Paquis-Flucklinger V, et al. Association between MSH4 (MutS homologue 4) and the DNA strand-exchange RAD51 and DMC1 proteins during mammalian meiosis. Mol Hum Reprod. 2004;10:917-24. [DOI] [PubMed]
- 131.Santucci-Darmanin S, Neyton S, Lespinasse F, Saunières A, Gaudray P, Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11:1697-706. [DOI] [PubMed]
- 132.Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 2000;14:1539-47. [DOI] [PubMed]
- 133.Ollinger R, Alsheimer M, Benavente R. Mammalian protein SCP1 forms synaptonemal complex-like structures in the absence of meiotic chromosomes. Mol Biol Cell. 2005;16:212-7. [DOI] [PMC free article] [PubMed]
- 134.Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Höög C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell. 2000;5:73-83. [DOI] [PubMed]
- 135.Parra MT, Viera A, Gómez R, Page J, Benavente R, Santos JL, et al. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J Cell Sci. 2004;117:1221-34. [DOI] [PubMed]
- 136.Urban E, Nagarkar-Jaiswal S, Lehner CF, Heidmann SK. The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLOS Genet. 2014;10:e1004540. [DOI] [PMC free article] [PubMed]
- 137.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644-52. [DOI] [PMC free article] [PubMed]
- 138.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20;1453-4. [DOI] [PubMed]
- 139.Shinzato C, Mungpakdee S, Arakaki N, Satoh N. Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Sci Rep. 2015;5:18211. [DOI] [PMC free article] [PubMed]
- 140.Flot JF, Tillier S. The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: the putative D-loop and a novel ORF of unknown function. Gene. 2007;401:80-7. [DOI] [PubMed]
- 141.Prada C, Medina M, Knowlton N. BioProject: PRJNA378841 biology-Naos. Panama City, Washington, DC: Smithsonian Tropical Research Institute; 2017.
- 142.Baumgarten S, Bayer T, Aranda M, Liew YJ, Carr A, Micklem G, et al. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics. 2013;14:704. [DOI] [PMC free article] [PubMed]
- 143.Rosic N, Ling EY, Chan CK, Lee HC, Kaniewska P, Edwards D, et al. Unfolding the secrets of coral-algal symbiosis. ISME J. 2015;9:844-56. . [DOI] [PMC free article] [PubMed]
- 144.Macrander JC, Dimond JL, Bingham BL, Reitzel AM. Transcriptome sequencing and characterization of Symbiodinium muscatinei and Elliptochloris marina, symbionts found within the aggregating sea anemone Anthopleura elegantissima. Mar Genomics. 2018;37:82-91. [DOI] [PubMed]
- 145.Yuyama I, Ishikawa M, Nozawa M, Yoshida MA, Ikeo K. Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci Rep. 2018;8:16802. [DOI] [PMC free article] [PubMed]
- 146.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658-9. [DOI] [PubMed]
- 147.Kortschak RD, Samuel G, Saint R, Miller DJ. EST Analysis of the Cnidarian Acropora millepora Reveals Extensive Gene Loss and Rapid Sequence Divergence in the Model Invertebrates. Curr Biol. 2003;13:2190-5. [DOI] [PubMed]
- 148.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. Bioinformatics. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210-2. [DOI] [PubMed]
- 149.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol Biol Evol. 2018;35:543-8. [DOI] [PMC free article] [PubMed]
- 150.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247-51. [DOI] [PMC free article] [PubMed]
- 151.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427-32. [DOI] [PMC free article] [PubMed]
- 152.Shikina S, Chiu YL, Zhang Y, Yao YC, Liu TY, Tsai PH, et al. Involvement of GLWamide neuropeptides in polyp contraction of the adult stony coral Euphyllia ancora. Sci Rep. 2020;10:9427. [DOI] [PMC free article] [PubMed]
- 153.Wiesner B, Weiner, Middendorff R, Hagen V, Kaupp UB, Weyand I. Cyclic nucleotide-gated channels on the flagellum control Ca2+ entry into sperm. J Cell Biol. 1998;142:473-84. [DOI] [PMC free article] [PubMed]
- 154.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA- Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed]
- 155.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863-8. [DOI] [PMC free article] [PubMed]
- 156.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10-2.
- 157.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417-9. [DOI] [PMC free article] [PubMed]
- 158.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [DOI] [PMC free article] [PubMed]
- 159.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288-97. [DOI] [PMC free article] [PubMed]
- 160.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-9. [DOI] [PMC free article] [PubMed]
- 161.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1-13. [DOI] [PMC free article] [PubMed]
- 162.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009b;4:44-57. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Microscopic observation of ovaries and testes isolated in April and June 2017, respectively. (a) External appearances of oocytes having germinal vesicles in isolated ovaries collected in April 2017. (b) The external appearance of oocytes without germinal vesicles collected at the same times as the samples shown in (a). Only one oocyte has a germinal vesicle (arrow). (c) Cytological appearance of an isolated testis collected in June 2017. Morphologically mature sperm with triangular head shapes were observed (arrows).
Additional file 2. (Table) Summary of clean read data for 24 samples used in de novo assembly.
Additional file 3. DDBJ accession numbers for the reference gonadal transcriptome
Additional file 4 DDBJ accession numbers for the E. ancora contigs
Additional file 5. DDBJ accession numbers for the Symbiodiniaceae contigs of DDBJ accession number
Additional file 6 (Table) Reference databases used for identifying E. ancora-originated contigs from the E. ancora holobiont transcriptome assembly
Additional file 7 (Table) Reproduction-related genes of E. ancora identified in our previous studies
Additional file 8 (Table) Evolutionarily conserved genes in metazoan reproduction identified in the E. ancora gonadal transcriptome assembly
Additional file 9 Hierarchical Clustering analysis of E. ancora gonadal samples used in this study. Twelve testis samples (3 colonies, 4 time points) and 12 ovary samples (3 colonies, 4 time points) were subjected to analysis. The cluster dendrogram showed that 2 samples (Oct-female-1 and Feb-male-1) are outliers, while others belong to a similar group. The 2 outliers were removed and the 22 remaining samples were used for gene expression analysis.
Additional file 10 GO functional analysis of upregulated genes in premature/mature ovaries. Significantly (> 4-fold change, P < 0.05) enriched GO terms in biological processes (blue bar), and molecular function (yellow bar). The X-axis represents the magnitude of change. The Y-axis represents the GO functional category.
Additional file 11. GO functional analysis of upregulated genes in mature testes. Significantly (> 4-fold change, P < 0.05) enriched GO terms in biological processes (blue bar), cellular component (green bar), and molecular function (yellow bar). The X-axis represents the magnitude of change. The Y-axis represents the GO functional category.
Additional file 12. Schematic figures depicting domain structures of neurogenic locus notch homolog protein. (a) Neurogenic locus notch homolog proteins 1. (b) Neurogenic locus notch homolog proteins 2. (c) Neurogenic locus notch homolog proteins 3.
Data Availability Statement
Datasets created and/or analyzed in this study are available in the Sequence Read Archive (SRA) of DNA DataBank of Japan (DDBJ) under BioProject accession PRJDB9831. Transcriptome assemblies have been deposited at the DDBJ/European Nucleotide Archive/GenBank Transcriptome Shotgun Assembly (TSA) under accession numbers ICQS01000001-ICQS01169272. The datasets supporting the conclusions of this article are included within the article and its additional files (Additional files 6, 7, 8).