Abstract
Protein CK2 has gained much interest as an anti-cancer drug target in the last decade. We had previously described the identification of a new allosteric site on the catalytic α-subunit, along with first small molecule ligands based on the 4-(4-phenylthiazol-2-ylamino) benzoic acid scaffold. In the present work, structure optimizations guided by a binding model led to the identification of the lead compound 2-hydroxy-4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (27), showing a submicromolar potency against purified CK2α (IC50 = 0.6 μM). Furthermore, 27 induced apoptosis and cell death in 786-O renal cell carcinoma cells (EC50 = 5 μM) and inhibited STAT3 activation even more potently than the ATP-competitive drug candidate CX-4945 (EC50s: 1.6 μM vs. 5.3 μM). Notably, the potencies of our allosteric ligands to inhibit CK2 varied depending on the individual substrate. Altogether, the novel allosteric pocket was proved a druggable site, offering an excellent perspective to develop efficient and selective allosteric CK2 inhibitors.
Keywords: CK2, kinase inhibitors, allosteric inhibition, kinase selectivity, anti-cancer
Graphical Abstract

INTRODUCTION
The protein kinase CK2 has an exceptional position within the human kinome due to the significant proportion of the cellular phosphoproteome that can be attributed to this kinase.1 This ubiquitously expressed protein kinase is involved in the activation of several pro-oncogenic pathways that are critical for cell proliferation, differentiation and survival.2 CK2 overexpression has been widely correlated with a poor prognosis for patients with prostate cancer, breast carcinoma, glioblastoma, leukemias, gastric carcinoma or renal cell carcinoma (see ref.3 for a recent review). Indeed, CK2 promotes prosurvival and antiapoptotic signals by interfering with NF-κB, Wnt and Akt pathways.4 The crucial role of CK2 as a cancer driver makes it an interesting target in cancer therapy, and CK2 inhibition proved to be an effective method to induce tumor regression.5
Optimal activity toward most protein substrates is reached when CK2 is assembled as a tetramer consisting of a dimer of regulatory subunits (CK2β) and two catalytic subunits, CK2α and/or CK2α’.6 The catalytic subunit of CK2 is believed to be constitutively active, while the regulatory subunit modulates the selectivity toward a subset of substrate proteins.7, 8 However, there is clear evidence that the catalytic CK2α subunit also exists as a population independent from CK2β in cells.8, 9
Most of the known CK2 inhibitors bind to the ATP pocket and show an ATP-competitive behavior in enzymatic assays.10 CX-4945 (5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid, silmitasertib) is one of the most potent CK2 inhibitors (IC50 = 14 nM) and is currently in phase II clinical trials.11 X-ray crystallographic analysis revealed that CX-4945 binds to the ATP-binding pocket of CK2α in the active conformation.12 Looking for splicing regulation through phosphorylation of serine/arginine rich (SR) proteins, Kim et al. discovered a CK2-independent implication of CX-4945 in cellular regulation of SR (serine/arginine–rich) protein phosphorylation.13 Indeed, this compound was found to be a strong inhibitor of Cdc2-like kinase-2 (Clk-2) (IC50 = 4 nM), almost fourfold more potent than toward CK2α (IC50 = 14 nM).13 In addition, CX-4945 was identified as a potent inhibitor of the dual-specificity tyrosine-phosphorylation regulated kinases (Dyrk) 1A and 1B with IC50s of 6.8 and 6.4 nM, respectively.14 These off-target activities of silmitasertib, potentially causing adverse effects, underline the need for potent and more selective CK2 inhibitors.
Therefore, continuous efforts are spent to identify alternative druggable sites on CK2. Given the large conformational flexibility of CK2α in the glycine-rich loop, the hinge/helix αD region, and the β4β5 loop at the CK2β interface,15–17 it is likely that several catalytically inactive conformations exist in the dynamic equilibrium, some of which can be stabilized by the binding of small molecule ligands to unique allosteric pockets. Indeed, one such small pocket, termed αD pocket, was identified on the large C-terminal lobe of the catalytic subunit.18 The most elaborated αD pocket ligand, CAM4712 (MW 453 g/mol), exhibited a moderate potency in the cell-free assay (IC50 = 7 μM) and good efficacy in cells, while the selectivity over other kinases required further optimization.19
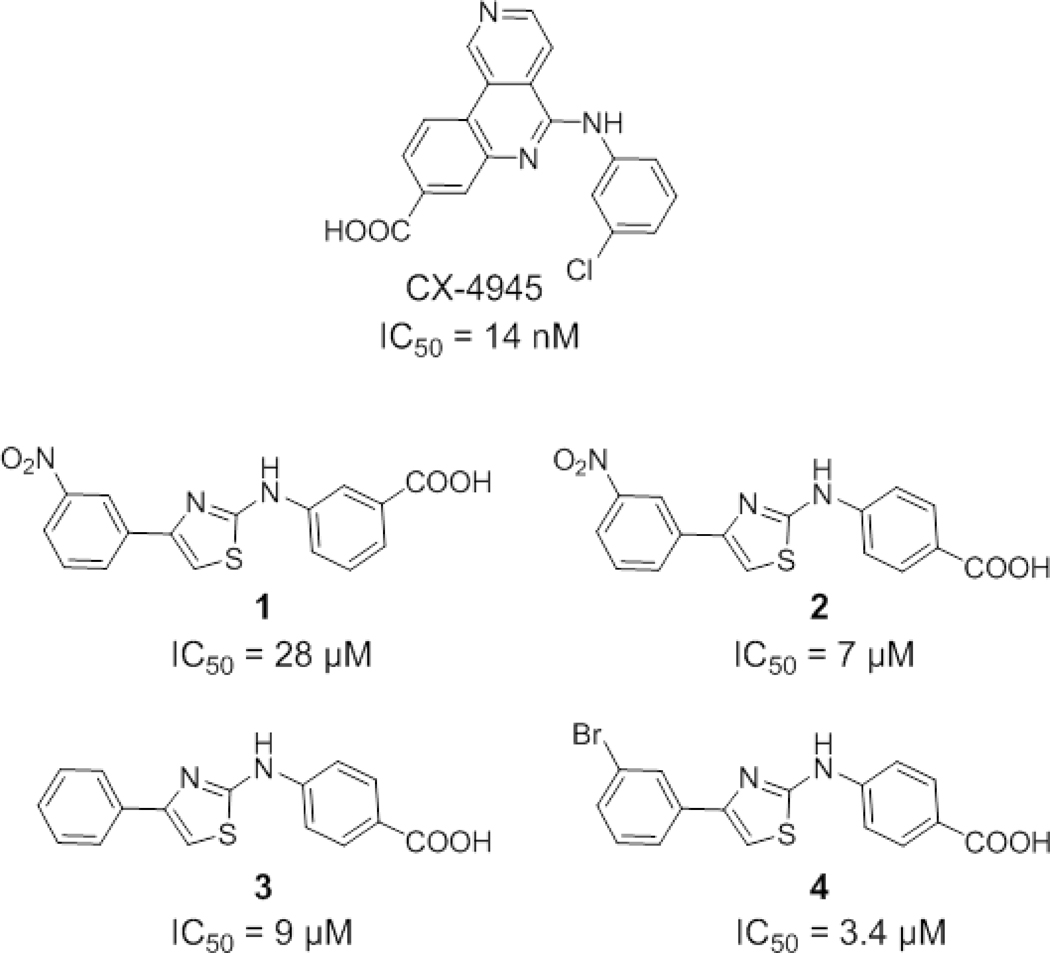
Recently, we identified 2-aminothiazole derivatives as novel allosteric inhibitors of CK2α;20 exemplarily shown are compounds 1–4 (Figure 1). Using complementary methods, it was demonstrated that compound 3 binds in an allosteric pocket adjacent to the ATP binding site, between the glycine-rich loop and the αC-helix. A preliminary hit optimization led to compound 4, exhibiting an IC50 of 3.4 μM. However, it had yet to be shown that the new CK2 modulator class can be developed further into more potent drugs, which is often a limitation with allosteric target sites that were not evolutionary designed for high-affinity interactions with small molecules.21, 22 Therefore, we carried out a compound optimization guided by a binding model, as will be described below. The cellular effects of the allosteric CK2 modulators were investigated and compared to that of the ATP-competitive reference inhibitor CX-4945. Our results revealed a distinct modulation of several signal transduction pathways by our new compounds, attributable to their novel mode of action.
Figure 1.
Chemical structures of CX-4945 and previously described allosteric inhibitors (1-4).20 The IC50s against human CK2α are indicated.
RESULTS AND DISCUSSION
Chemistry.
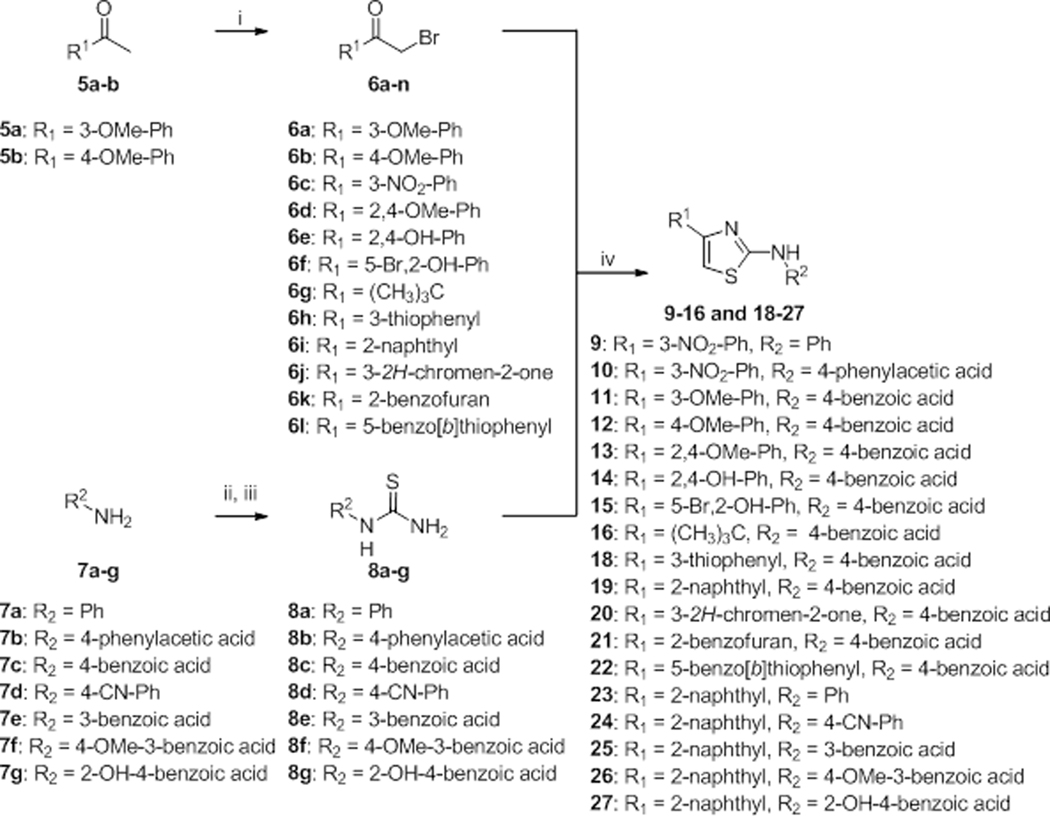
The aminothiazole core was prepared according to Hantzsch-type syntheses from 2-bromoacetophenones and thiourea intermediates (Scheme 1). Most of the 2-bromoacetophenones 6a-n were commercially available or easily prepared 6a-b from commercially available acetophenones 5a-b through a bromination step, as described in the literature.23, 24 Thioureas 8a-g were obtained after reaction of isothiocyanate intermediates with ammonia solution. The latter were synthesized after reaction of the primary amine of aniline derivatives 7a-g with carbon disulfide, in presence of triethylamine, followed by iodine oxidation.25 Hantzsch syntheses were carried out in ethanol and the resulting products were directly obtained as solids, after their precipitation with water. Semi-preparative HPLC, recrystallization, or washing procedures led to the final products with a LC-UV (254 nm) purity higher than 95%. In one case, the ethyl ester derivative was obtained instead of the free carboxylic acid. A saponification step with NaOH was then carried out to obtain the desired product 10. In this way, aminothiazoles (9–16 and 18–27) were obtained in varying yield, with the exception of compound 17 which was commercially available.
Scheme 1.
Hantzsch-type synthesis. Reagents and conditions: i) Br2, CHCl3, 40 °C, 5 min or Br2, HBr 32% in AcOH, MeOH, 60 °C, 4h. ii) CS2, Et3N in THF/H2O 1/1, RT, 24 h, then I2 in THF, 0 °C to RT, 3 h. iii) NH4OH 30%, RT, 6 h. iv) EtOH, reflux, 3–12 h.
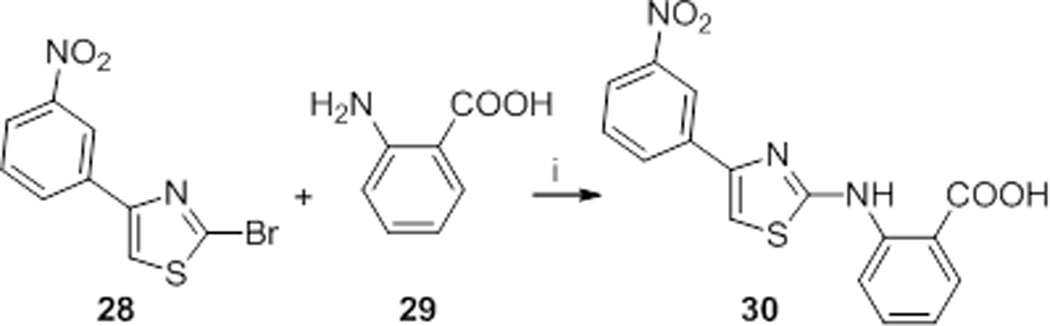
Compound 30 could not be obtained through Hantzsch-type synthesis, maybe due to the spatial proximity between the primary amine and the carboxylic acid of 2-aminobenzoic acid, which prevents the formation of the isothiocyanate intermediate. Consequently, a nucleophilic substitution between 2-bromo-4-(3-nitrophenyl)thiazole (28) and 2-aminobenzoic acid (29) in hydrochloric acid was performed to obtain the desired 2-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoic acid (30) albeit in a low 2 % yield (Scheme 2).
Scheme 2.
Preparation of compound 30 through aromatic nucleophilic substitution. Reagents and conditions: i) HCl(aq) 37%, 1,4-dioxane, 90 °C, 48 h.
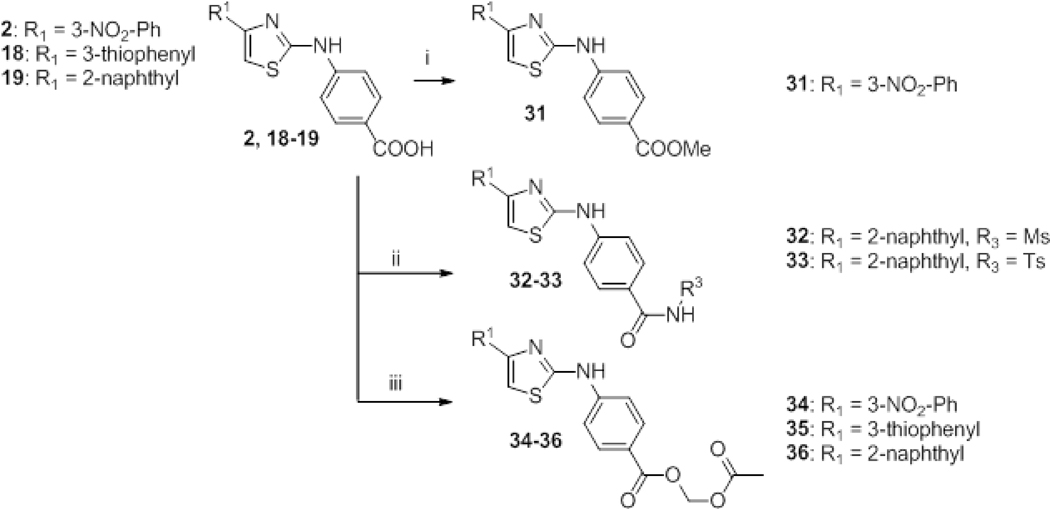
Several modifications were considered on the carboxylic acid function (Scheme 3). The methyl ester derivative 31 was easily obtained through a classical acidic esterification step. Sulfonamide derivatives 32–33 were obtained according to peptide coupling procedures whereas acetoxymethyl prodrugs 34–36 were accessed from carboxylic acid derivatives (2 or 18–19) and bromomethyl acetate through a previously described method.26
Scheme 3.
Syntheses of esters and amides 31–36. Reagents and conditions: i) H2SO4 95% 3 drops, MeOH, reflux, 2 h. ii) 32: HATU, DIPEA, dry DMF, 30 min; NaH, RSO2NH2, dry THF, RT, 30 min; then RT, 4 h. 33: EDC·HCl, DMAP, RSO2NH2, CH2Cl2, RT, 36 h. iii) Bromomethyl acetate, Et3N, dry DMF, RT, 12 h.
Optimization of potency.
The preliminary Structure-Activity Relationships (SAR) obtained from a small series of analogs of the original hit compound 1 (Figure 1) had suggested that the three aryl ring system as well as the carboxylic acid function are needed for CK2α inhibition.20
In order to gain further insight into essential key features and to optimize the inhibitory potency, we synthesized a targeted set of derivatives to systematically explore the SAR around the new scaffold. First, we varied the position and number of the essential carboxyl function (compounds 9 and 30, Table 1). Although this did not further improve the activity, placing the carboxylic acid group in the ortho-position gave compound 30, exhibiting a potency similar to that of the para-regioisomer 2. This suggested that different protein side chains might be available for interaction with the carboxylate, which, however, cannot easily reach the carboxylate when it is in the meta-position, since compound 1 (Figure 1) was four times less potent than 2 or 30. Changing the benzoic acid to a phenyl acetic acid moiety (derivative 10) was detrimental to the activity, indicative of a specific interaction of the carboxylate that is strictly dependent on the position. Altogether, the p-position appeared most suitable for the placement of the essential acidic function.
Table 1.
Evaluation of the benzoic acid moiety in the allosteric inhibitors.
IC50 values are the mean of ≥ two experiments with S.D. ≤ 20%, except for 1 and 30:
S.D. ≤ 40%.
Kd values determined by at least three runs, S.D.<20%; n.d., not determined.
Included for comparison; previous hit compounds from Ref. 20
Next, we modified the second phenyl ring in the thiazole 4-position while keeping the p-benzoic acid moiety constant (Table 2). Various substituents were introduced to modulate the electron density of the phenyl ring: 3-methoxy (compound 11), 4-methoxy (compound 12), 2,4-dimethoxy (compound 13), 2,4-dihydroxy (compound 14) and 5-bromo-2-hydroxy (compound 15). The rather flat SAR observed with these and previous derivatives indicated that the electron density of the phenyl ring was not relevant to the binding affinity. For example, compounds 12 and 14 (Table 2), carrying electron–donating substituents, exhibited the same potency as the 3-nitrophenyl derivative 1 (cf. Figure 1).20 We also synthesized and tested the pyridinyl derivative 17 in order to explore more polar heterocycles with a ring dipole moment similar to the nitrophenyl compound. However, the measured IC50 value of 19 μM was unfavorable, suggesting that the polar H-bond acceptor nitrogen in the ring is not complementary to the electrostatic potential of the binding pocket. In the previously synthesized compound 4 (Figure 1), bromine as a large hydrophobic substituent had a positive impact on the potency, suggesting that an extension of the aromatic ring (cf. compounds 19, 20, 21, 22) or exchange by a bulky substituent such as tert-butyl (compound 16) could be tolerated by the binding pocket and further increase the potency. The bulky tert-butyl moiety in 16 could not successfully replace the aromatic ring, as indicated by the strong decrease of the CK2α inhibition. On the other hand, some of the bicyclic aromatic systems were good substitutes for the potentially unstable or toxic 3-bromophenyl moiety, with the naphthalenyl derivative 19 being the most potent (Table 2). More polar bicycles, such as chromene (20) and 2-benzofuran (21), were less active than 19, underlining the preference of a hydrophobic aromatic moiety by this part of the binding pocket. Interestingly, a plain thiophene ring (18, IC50 = 5 μM) led to an almost twofold increase of potency compared with the original phenyl congener (3, Figure 1). This promising result prompted us to introduce a 5-benzothiophene substituent, however, no significant improvement of the inhibitory activity was noted (22, IC50 = 4 μM).
Table 2.
Optimization of the hydrophobic moiety in the thiazolyl-4 position.
 |
|||
|---|---|---|---|
| Cpd | R1 | IC50 (μM)a | Kd (μM)b |
| 3c |  |
9 | 6 |
| 4c |  |
3.4 | n.d. |
| 11 |  |
12 | 14 |
| 12 |  |
6 | 7 |
| 13 |  |
13 | n.d. |
| 14 |  |
7 | n.d. |
| 15 |  |
5 | n.d. |
| 16 |  |
>50 | n.d. |
| 17 |  |
19 | 20 |
| 18 |  |
5 | 6 |
| 19 |  |
3 | 0.6 |
| 20 |  |
8 | n.d. |
| 21 |  |
14 | n.d. |
| 22 |  |
4 | n.d. |
IC50 values are the mean of ≥ two experiments with S.D. ≤ 20%.
Kd values determined by at least three runs, S.D.<20%; n.d., not determined.
Included for comparison; previous hit compounds from Ref. 20
So far, the naphthalenyl derivative had shown the highest potency among the aryl aminothiazole compounds. In order to obtain insight in the potential binding mode with CK2α, we started docking simulations using the available crystal structures of human CK2α and the CK2α2/β2 holoenzyme. A prior inspection of the different PDB entries revealed a very high mobility of the P-loop, with the tip residue Tyr50 spanning a distance of more than 20 Å (Tyr-OH group) between its extreme locations in 4UB727 and 1JWH6, with many intermediate positions showing up in the other CK2 crystal structures. In X-ray structures displaying active CK2α, Tyr50 occludes the lipophilic allosteric pocket (e. g., in the active symmetric CK2α2/β2 tetramer in 4MD728 and in the CK2α/AMPPNP complex in 3NSZ12). Contrastingly, the allosteric pocket was accessible and gave reasonable poses in our preliminary docking runs (data not shown) in the inactive conformations of CK2α, 3FWQ29 and 4UB727, and the holoenzyme structures 1JWH6 and 4DGL30, displaying distorted asymmetric CK2α2/β2 tetramers organized in inactive trimers.28 Because it was more logic to correlate the alanine scanning mutational analysis (cf. Figure 4) with docking simulations both carried out with the isolated CK2α subunit, we selected PDB entry 3FWQ for further docking studies. The docking poses with the ten highest scores were evaluated for plausibility taking into account all previous results20: i) the binding pocket is located close to the ATP binding site without or with only marginal overlap; ii) the binding pocket is delimited by residues Val73, Lys74 and Lys77; iii) the observed pose fits to the STD-NMR–based binding epitope mapping as previously determined with 4. The most consistent binding model of 19 in the 3FWQ structure (Chain B) displayed a tight fitting of the naphthalene ring in the hydrophobic pocket, with an additionally fixed orientation due to a double CH–π interaction of Gly177 with the naphthalene cycle (Figure 5A).
Figure 4.
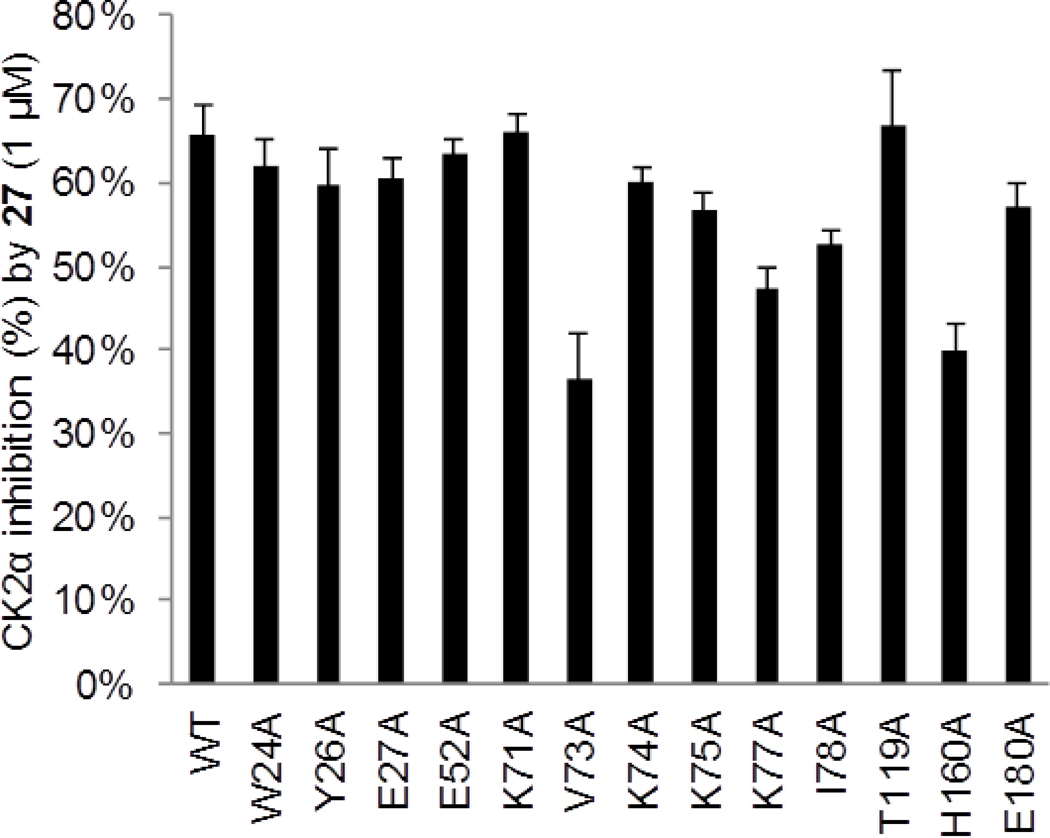
Percentage of inhibition of GST-CK2α (wild type (WT) or single alanine mutant) in the presence of compound 27 (1 μM); see Table S1 (Supporting Information) for explicite values. The error bars indicate ± S.D.
Figure 5.
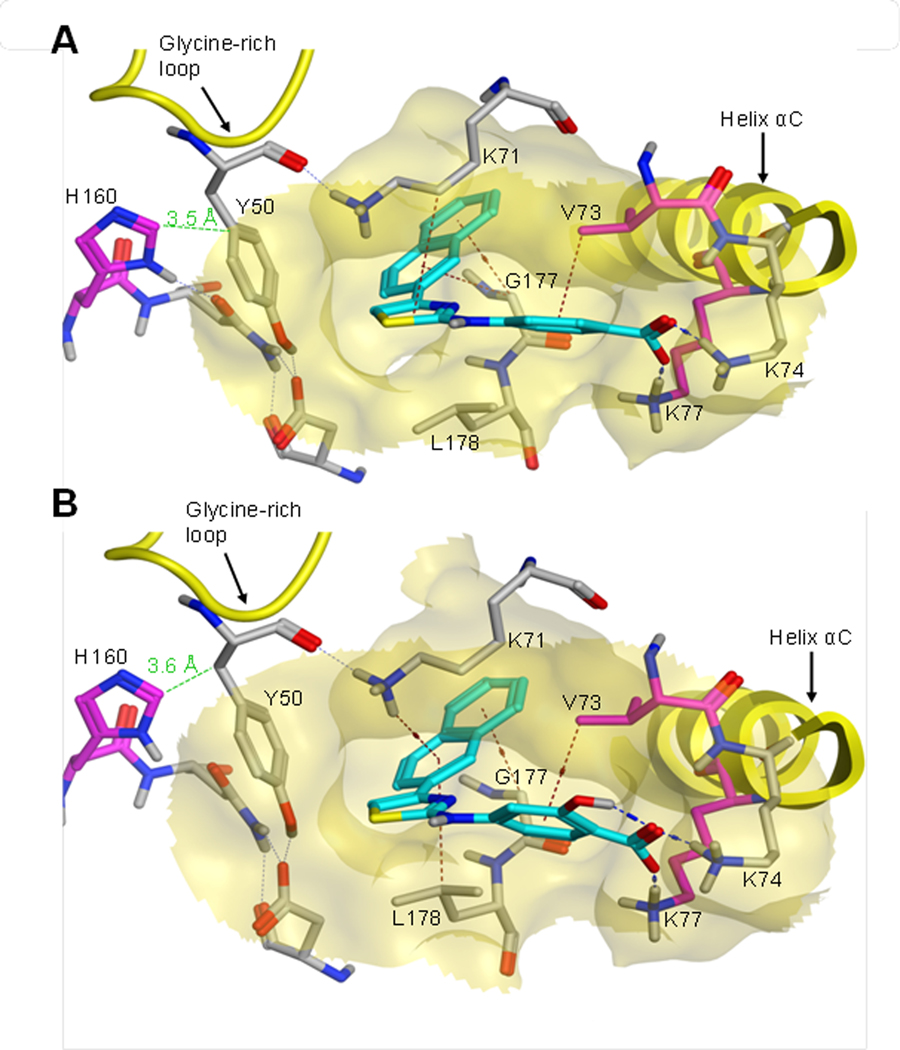
Predicted binding mode of the naphthalen-2-yl-thiazolamine derivatives in the allosteric pocket. Compounds 19 (A) and 27 (B) (cyan) were docked into the hydrophobic pocket between the glycine–rich loop and the αC-helix in the crystallized inactive conformation of CK2α (PDB entry 3FWQ) using MOE. Residues that are potentially relevant to the binding are labeled. Residues that caused a drop of 27 inhibitory potency when mutated to alanine are colored in magenta. The carboxylate forms salt bridges with Lys74 and Lys77, while the naphthalene ring is buried in the mostly hydrophobic cavity, additionally anchored by cation–π (with Lys71) and/ or CH–π interactions (with Gly177/Leu178). A further CH–π interaction was consistently observed between Val73 and the benzoic acid ring. Interactions are indicated by dashed lines (H-bonds/salt bridges: blue; CH–π interactions: brown). The green dashed line denotes the shortest distance between His160 and Tyr50.
The predicted tighter fitting of the naphthalene moiety imposed a higher constraint on the orientation of the residual molecule than it was the case with compounds 1–3, potentially shifting the relative position of the carboxylate. Given that an optimal ionic interaction was crucial for the overall binding affinity, it seemed worthwhile to revisit the carboxylic acid substitution on the 4-naphthalen-2-yl-thiazol-2-yl-arylamines. Several derivatives 23–27 were synthesized (Table 3) and evaluated for their CK2α inhibitory properties. Compounds 23 without acid function and 24 with a 4-cyano substitution were lacking CK2α inhibition, thus confirming the necessity of an acidic function also in the naphthalenyl series. However, in contrast to the situation with the 3-nitrophenyl series (Table 1), where the p-carboxyl derivative was substantially more potent than the meta analog (cf. compounds 1 and 2), the carboxylic acid function was now even slightly preferred in the meta position (Table 3, comparison between 25 and 19). Because our docking simulation had predicted a CH–π interaction of the benzoic acid moiety with Val73, we aimed at increasing the π-electron density using a methoxy subsituent. Compared with the parent compound 25, the 6-methoxy-3-benzoic acid analog 26 exhibited a reduced potency on CK2α, which might be explained either by a steric clash of the methoxy group with the binding pocket, unfavorable conformational effects or the decreased acidity of the essential carboxyl function. To avoid these potential problems, while yet increasing both electron density and acidity of the benzoic acid moiety, we synthesized and tested the salicylic acid analog. Salicylic acid is known for its more than 16 fold stronger dissociation in water to the anion compared with benzoic acid,31 which should lead to an enhancement of the ionic interactions. In agreement with these considerations, the salicylic acid derivative 27 exhibited a 5-fold higher potency than the corresponding benzoic acid analog 19, reaching a submicromolar IC50 value (IC50 = 0.6 μM, Table 3). As expected, docking simulations predicted a binding mode for 27 similar to that of 19, with a slightly shifted orientation of the salicylic acid head (Figure 5B). Also considering the higher ionization rate, the binding pose obtained for 27 was in line with the observed increase in potency.
Table 3.
Optimization of the 4-naphthalen-2-yl-thiazol-2-yl-arylamine derivatives.
IC50 values are the mean of ≥ two experiments with S.D. ≤ 20%.
Kd values determined by at least three runs, S.D.<20%; n.d., not determined.
Carboxyl group replacement and prodrug synthesis.
Since our goal was to develop inhibitors suitable for cellular studies, we further considered bioisosteric replacements of the carboxylic acid as well as a prodrug approach, in order to circumvent problems associated with poor cell permeabilities of carboxylic acid derivatives.32, 33 A set of appropriate derivatives of 2 and 19 were synthesized and tested for their cellular efficacy using the clear cell renal cell carcinoma cell line 786-O (Table 4). This cell line was chosen because it displays a strong decrease of cell viability following to inhibition of the intracellular CK2.5, 34 The acylsulfonamides 32 and 33, which were supposed to preserve the acidic properties (pKa ≈ 4–5), did not reach the cell-free potency of the parent compound 19; however, the tosyl derivative 33 was more efficient than 19 in decreasing the cell viability at 25 μM (87% vs. 43% reduction) and as efficient as the best compound of this series, 27. Altogether, this might indicate a better cellular permeability of the methylbenzenesulfonamide 33 compared with the carboxylic acid derivatives, as could be expected based on previous experience.35 On the other hand, it could not be ruled out that off-target interactions in the cells in addition to CK2α inhibition could be responsible for the effects on cell viability. Therefore, in light of the markedly higher potency of 27 toward purified CK2α, we concluded that any effects exerted by this compound in cells will more likely be mediated through CK2 inhibition.
Table 4.
Cell-free and cellular effects of selected carboxylic acid bioisosteres and prodrugs.
 | ||||
|---|---|---|---|---|
| Cpd | R1 | R2 | % inhibition of CK2α at 10 μMa (IC50b) | % inhibition of 786-O cell growth at 25 μMc |
| 19 |  |
 |
67 (3) | 43 |
| 27 |  |
 |
96 (0.6) | 87 |
| 32 |  |
 |
41 | 23 |
| 33 |  |
 |
54 | 87 |
| 34 |  |
 |
7 | 42 |
| 35 |  |
 |
37 | 48 |
| 36 |  |
 |
41 | 45 |
Percent inhibition of recombinant CK2α, S.D. ≤ 10%.
IC50 values are the mean of ≥ two experiments with S.D. ≤ 20%.
Cells were incubated for 24 h24h with 25 μM of respective compound or DMSO as a control (set to 100 % growth); S.D. <10%.
The acetoxymethyl ester prodrugs26 34–36 were expectedly less potent than the parent compounds under cell-free conditions, however, none of them led to an improvement in cellular efficacy compared with 19 (Table 4). Further experiments evaluating the stability of these prodrugs have to be carried out before envisaging new derivatives.
Investigation of the mode of action.
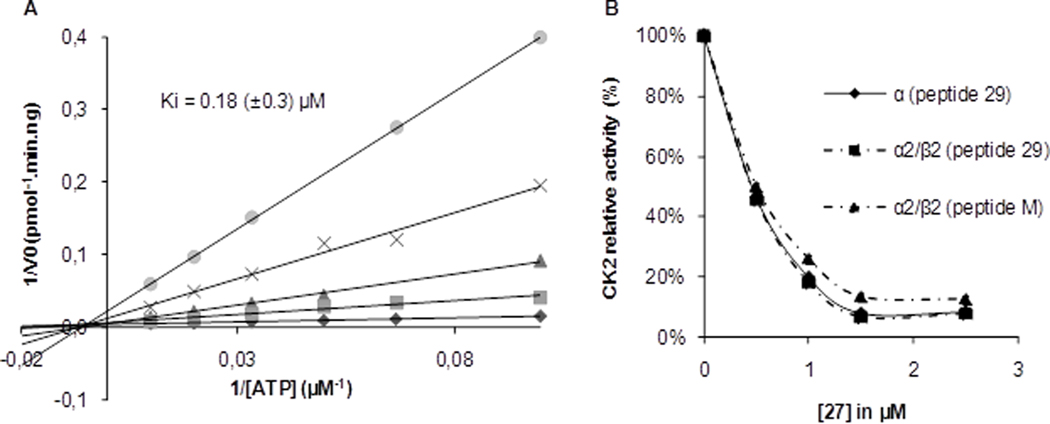
Before continuing cellular studies, it was important to verify that our optimized lead compound 27 still exhibits non-ATP competitive behavior in the CK2α inhibition assay, as demonstrated earlier for the starting compounds 2 and 4.20 The Lineweaver-Burk analysis for 27 is depicted in Figure 2A, confirming a non-ATP competitive mechanism of CK2 inhibition, with a Ki value of 0.18 μM.
Figure 2.
Enzymatic characterization of compound 27. (A) Lineweaver-Burk inhibition plot of human recombinant CK2α by compound 27 at various concentrations: 0, 0.5, 1, 1.5, 2.5 μM. Ki was determined by plotting the slopes at varying inhibitor concentration from three independent experiments. (B) CK2α and CK2α2β2 activity with two peptide substrates in the presence of increasing concentrations of compound 27.
It was also of interest to investigate a potential preference of the inhibitor for either the catalytic CK2α subunit or the holoenzyme CK2α2β2. In addition, two substrate peptides, representative of differentially recognized CK2 substrate proteins, were also tested: a CK2β–independent peptide substrate (peptide 29) and a CK2β–dependent peptide substrate (peptide M, derived from eIF2β). As shown in Figure 2B, the resulting three experimental settings gave similar results, indicating that i) 27 is able to inhibit CK2α and the holoenzyme complex with the same potency, thereby ruling out any binding to the CK2α–β subunit interface pocket,36 and ii) the inhibition mode is not sensitive against extended substrate interactions as exerted by the eIF2β–derived peptide M.37
Binding characterization using Surface Plasmon Resonance (SPR) and isothermal titration calorimetry (ITC) analysis.
To determine the affinity constants of our allosteric inhibitors, we performed SPR analysis after coupling CK2α to the biochip surface. Intriguingly, the analysis revealed that the Kd values fully correlated with the IC50s of the respective compounds, and moreover, that both parameters even showed mostly similar absolute values, as indicated by the slope of 1.1 in the IC50 vs. Kd plot (r2 = 0.91, Figure S1, Supporting Information). Moreover, these findings corroborated that compound binding in the enzymatic inhibition assay was not subject to competition neither with ATP nor substrate peptide, which would have raised the IC50 over the Kd. In addition, the close correlation between these parameters indicated that the binding of the allosteric ligands consistently leads to full inhibition, in line with the stabilization of an inactive conformation of CK2α.
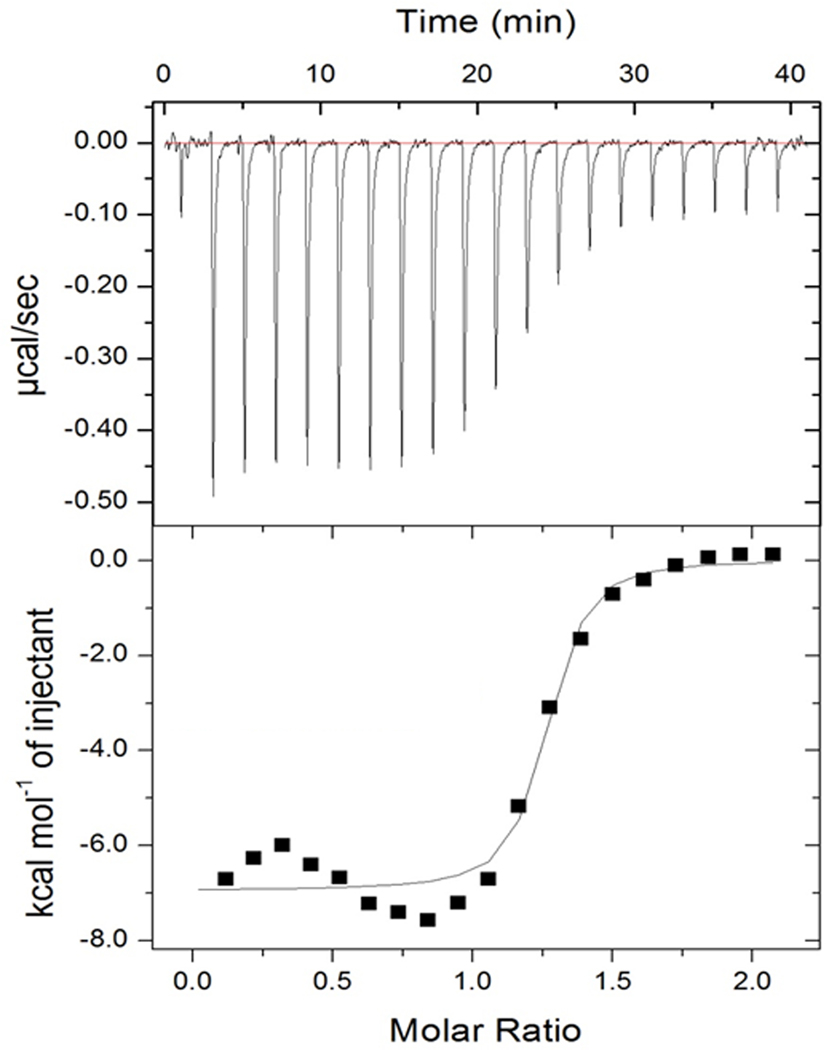
To complement the SPR analysis data, the thermodynamic characteristics of binding were determined for 27 using ITC (Figure 3). Under these conditions, we obtained a Kd value of 0.11 μM (±15 % S.D.) for 27, which was somewhat lower than that found by SPR analysis (0.3 μM), possibly reflecting that the allosteric binding site was better accessible when the protein was in solution than when it was immobilized on the SPR sensor chip. The thermodynamic parameters calculated for ΔH (−6.9 ± 0.17 kcal/mol) and TΔS (2.53 ± 0.29 kcal/mol) indicated a mainly enthalpy–driven binding, in agreement with the essential contribution of the ionic interaction to the binding affinity. As described above, the replacement of the carboxylate in 2 by the methyl ester function in 31 had abolished the inhibitory potency (Table 1), consistent with a surface localization of the ester function, where water molecules strongly compete with the potential H-bond interactions of the ligand.
Figure 3.
Thermodynamic characterization of binding of the allosteric inhibitor 27 to CK2α by ITC. The top panel shows the raw heat signal for successive injections of dissolved compound 27 into a CK2α solution at 21 °C. The bottom panel shows the integrated heats of injections corrected for heats of dilution, with the solid line corresponding to the best fit of the data to a bimolecular binding model. Shown is one experiment out of two that gave similar values.
A comparison of the thermodynamic binding signature of 27 with that of the previously analyzed compound 4-(4-(pyridin-2-yl)thiazol-2-ylamino)benzoic acid (compound 9 in ref. 20) (ΔH = −13.3 kcal/mol; TΔS = - 6.6 kcal/mol; Kd(ITC) = 14.4 μM; Figure S2) revealed that the increase in binding affinity achieved with 27 was solely attributable to the enhancement of hydrophobic interaction. The exchange of the too polar pyridin-2-yl moiety by naphthalenyl in 27 turned the negative entropic contribution to a positive one, however, it can also be seen that this was accompanied by a decrease in binding enthalpy. This phenomenon, known as enthalpy–entropy compensation, is not uncommon in lead optimization programmes; based on previous experience, the thermodynamic profile of 27, with its still predominant enthalpy term, is in an ideal range for a lead compound, predicting that the binding affinity can be further enhanced by increasing ΔS, which is usually more easily achieved than the subsequent optimization of enthalpy.38
Mapping of the allosteric binding site targeted by 27.
To this end, compound 27 was screened against a panel of 13 GST-CK2α mutants, possessing point mutations mainly located in the area of the binding site as described previously for compound 4 (Figure 4, Table S1).20 Three CK2α mutants were significantly less sensitive toward inhibition by 27 than the other mutants or the wild type enzyme: Val73Ala, Lys77Ala and His160Ala (Figure 4) and therefore selected for dose–response experiments. Compared with the wild type GST-CK2α (IC50 = 0.6 μM), the IC50s of compound 27 were significantly elevated with the following three mutants: a 2.8-fold increase was noted for CK2α-Val73Ala (IC50 = 1.7 μM), a 4.3-fold increase for CK2α-Lys77Ala (IC50 = 2.6 μM), and a 1.7-fold increase for CK2α-His160Ala (IC50 = 1 μM).
The loss of potency toward the Val73Ala and Lys77Ala mutants can be explained on the basis of the predicted direct interaction of the respective side chains with 27, thereby corroborating these docking results (Figure 5B). The loss of the Lys74 side chain in the silent Lys74Ala mutant might be compensated by Lys77 and Arg191, which could rotate to the former positions of Lys74 and Lys77, respectively. Surprisingly, the exchange of Lys71 by Ala also did not influence the affinity of 27 to the binding pocket. It could be speculated that this exchange leads to a conformational rearrangement of side chains close to the binding pocket, e.g. involving Ser51 and Leu70, which could then replace the interactions formerly predicted for Lys71 by OH–π and/or CH–π contacts. It is noteworthy that a reduced activity against the Lys77Ala and His160Ala mutants was observed with both 27 and the previously described analogue 4-[4-(3-Bromo-phenyl)-thiazol-2-ylamino]-benzoic acid (compound 7 in ref. 20), corroborating that the compounds were targeting the same binding site. In addition, the repeated identification of His160 in our alanine scan strengthens the previous hypothesis that this residue, being too far away to contact the inhibitor itself, may play a role in the formation and/or stabilization of the allosteric pocket in the inactive conformation. 20 In the original coordinates of the PDB entry 3FWQ, His160 is relatively close to Tyr50 (~5.6 Å), and a further convergence of the two residues was observed during the docking simulations (Figure 5). Thus, at least in some conformations, His160 may have the function to fix the position of Tyr50, which forms one of the pocket walls, thus stabilizing the hydrophobic part of the allosteric binding site. Altogether, the CK2α mutant screening results supported the predicted binding model shown in Figure 5.
Compound 27 is a selective inhibitor of CK2.
Selectivity is an important feature for kinase inhibitors to avoid off-target related side effects.39, 40 Allosteric kinase inhibitors of the so-called type III target binding sites that are less conserved among the protein kinase families than the ATP binding pocket, usually providing high selectivity for the kinase of interest.39 Therefore, a pronounced initial selectivity should be observed with an inhibitor targeting an allosteric site, even before optimization of this parameter. For our study, compound 27 was evaluated against a selected panel of 54 kinases (Supplementary Table S2), including closely related kinases from the CMGC family as well as kinases from other branches of the kinome. Only four other kinases besides CK2α were inhibited by more than 50% in the presence of 5 μM 27 (8.3 times IC50 against CK2α): ErbB2 (70%), Aurora-B (62%), Pim-1 (60%) and PI3K (53%). It was noteworthy that the main off-targets of CX-4945, Clk2 and Dyrk1A, were not or only weakly inhibited by 27, respectively. The overall selectivity of 27, as expressed by a Gini coefficient41 of 0.78, was higher than that of CX-4945 (Gini coefficient = 0.67) and of other CK2 inhibitors described in the literature.34, 42, 43 The high selectivity of 27 is consistent with binding to an allosteric site that is not strongly conserved in the human kinome.
The selectivity of the most potent compound from our first study, 4-[4-(3-Bromo-phenyl)-thiazol-2-ylamino]-benzoic acid (compound 7 in ref. 20), was only evaluated in a smaller screening panel (32 kinases); however, comparing the data from the 20 kinases common to both screenings, it can be concluded that our optimization efforts led to an improved selectivity: Under comparable screening conditions, the most strongly affected off-targets from the first study, GSK3β, CK1γ1, ACVR1 (ALK) and Clk4, which were all inhibited from 60 to 70 %,20 displayed only 41, 32, 0 and 23 % inhibition, respectively, in the presence of 27. Further kinases, including Clk2, Abl, CHK2, CDK1/cyclinB and Clk1, which had shown some weak inhibition (from 20 to 30 %) with the previous analogue20, were not affected by 27. Only Pim-1 and EGFR were inhibited slightly stronger by 27 (60 and 21 %, respectively) than by the previous compound20 (43 and 0 % inhibition, respectively).
Inhibition of CK2 cellular activity and induction of apoptosis by 27.
CK2 inhibitors are known to decrease the viability of cancer cells.5, 34 Consequently, to assess the cellular potency of 27, the compound was screened against several cell lines of diverse tissue origins (Table 5), and the half maximal growth-inhibitory concentration (GI50) was determined. Among the tested cancer cell lines, 27 exhibited GI50 values ranging from 5 μM in the clear cell renal cell carcinoma cell line 786-O to 20 μM in prostate cancer cells (DU145). Importantly, compound 27 was clearly less toxic against the non-cancerous (benign) kidney cells RPTEC, displaying a 5-fold higher GI50 (27 μM) than against the kidney cancer cell line 786-O. This result is consistent with the dependency of cancer cells on a high level of CK2α expression for promoting cell growth and survival, as reported earlier.4 Of note, the ATP-competitive inhibitor CX-4945, which is currently evaluated in clinical trials, was similarly effective against 786-O cancer cells (GI50 = 6 μM, Supplementary Figure S3), but showed an overall less pronounced tumor cell selectivity than 27 in our assays; for instance, CX-4945 was acting weaker against the tumor cell lines MCF7, DU145 and U138 than against RPTEC cells (Supplementary Figure S4). Furthermore, while 27 showed no difference in potency between the non-tumor fibrocystic mammary epithelial cell line MCF10A and the mammary tumor cell line MCF7, CX-4945 inhibited the cell growth of MCF10A cells even 5 times stronger than that of the MCF7 tumor cells (Figure S4). Overall, the in vitro tumor cell selectivity of 27 was higher than that of CX-4945.
Table 5.
Effect of compound 27 on various cell lines.
| Tissue of origin | Cell name | Cpd 27 GI50 in μM |
|---|---|---|
| Kidney | 786-O | 5 (± 2) |
| RPTEC | 27 (± 5) | |
| Breast | MCF7 | 10 (± 2) |
| MCF10A | 10 (± 3) | |
| Lung | H1299 | 9 (± 4) |
| A549 | 12 (± 3) | |
| Prostate | PC3 | 7 (± 2) |
| DU145 | 20 (± 6) | |
| Glioblastoma | U138 | 13 (± 3) |
Cells were grown as described in the experimental part, and GI50 values were determined by linear interpolation after transformation to log[c] scale. Standard deviations from a minimum of two independent experiments are shown between brackets.
786-O cells are known to be resistant to classical chemotherapy and are frequently used as a model for clear cell renal cell carcinoma, as they share many characteristics with the clinical tumor cells.44, 45 Moreover, it was already demonstrated that the inhibition of CK2α efficiently blocks the growth of these renal cancer cells.46 Therefore, the 786-O cell line was selected as a model for the in-depth characterization of the effects of 27 in cancer cells.
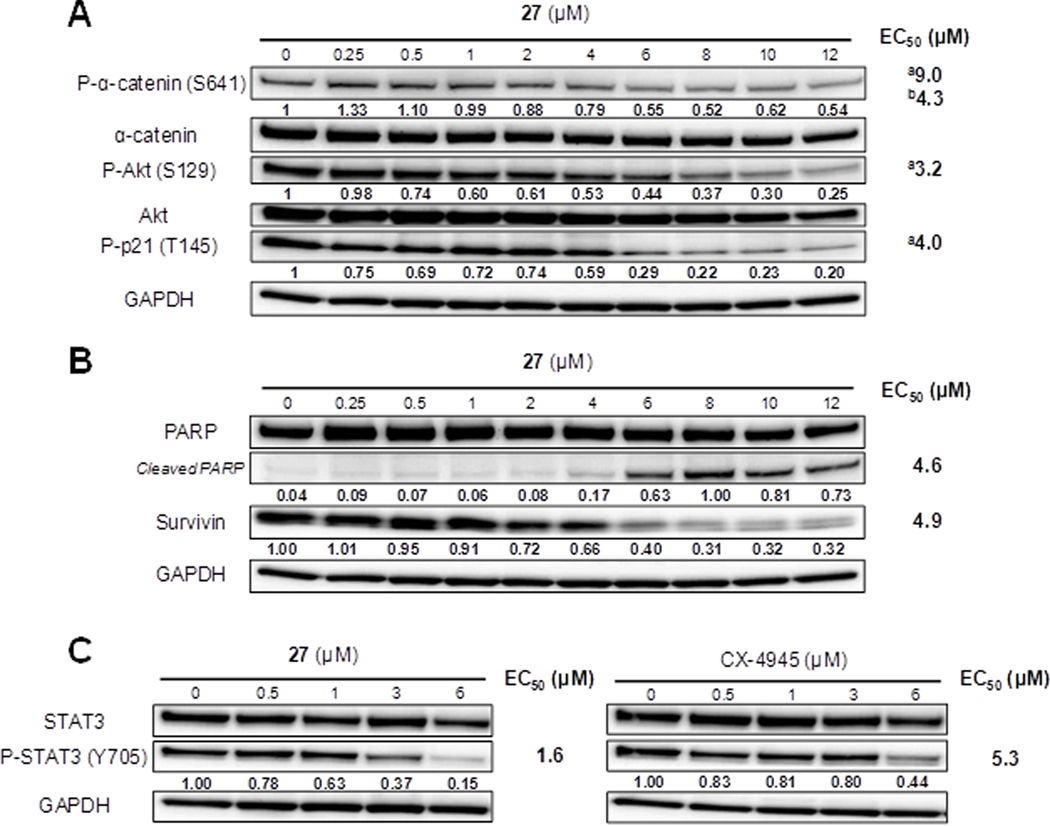
First, the potency of 27 to inhibit the CK2 activity in intact cells was evaluated. Two protein substrates, Akt1 and α-catenin, known to be phosphorylated by CK2 on residues Ser12947 and Ser64148, respectively, were used as a reporter of CK2 cellular activity. A concentration–dependent decrease in phosphorylation of Akt1 (Ser129) and α-catenin (Ser641) was observed after a 24 h treatment with compound 27 (Figure 6A). In addition, numerous dead cells were observed microscopically during the cell viability experiments (data not shown). Since the CK2-catalyzed phosphorylation of Akt1 on Ser129 was associated with a hyperactivation of the latter kinase,47 we analyzed whether Akt downstream signaling was also affected by our CK2 inhibitor. Indeed, the phosphorylation of the Akt1 phosphorylation site (Thr145) on p21/Waf1 was reduced in the presence of 27 in a concentration-dependent manner (EC50 = 4.0 μM, Figure 6A). Altogether, we can conclude that compound 27 inhibited CK2 cellular activity, blocked Akt1 downstream signaling and decreased cell viability.
Figure 6.
Compound 27 inhibits CK2–dependent substrate phosphorylations in cells and induces apoptosis. (A) Cellular inhibition of CK2 by compound 27. 786-O cells were plated and incubated for 24 h in the presence of various concentrations of 27. The phosphorylation status of the CK2 protein substrates were measured by Western Blot analysis of cell extracts. Band intensities were quantified using ImageJ and normalized using anti-GAPDH antibody signals as loading control. aEC50 calculated assuming that phosphorylation levels will approach zero with increasing concentrations of 27; bEC50 calculated assuming a basal level of non-responsive P-α-catenin of 50% of the DMSO control. (B) Apoptosis induction upon compound 27 treatment. Appearance of cleaved PARP and decrease of survivin levels were observed by Western Blot analysis of cell extracts. (C) 27 shows a higher potency to inhibit the STAT3 activation in 786-O cells than CX-4945.
In light of the inhibitory effects of 27 on the Akt1 activity, we surmised that the observed cell death is due to the induction of apoptosis. Two apoptosis markers, the appearence of cleaved PARP and the decrease in survivin protein levels, were analyzed by Western Blotting. Both hallmarks of apoptosis induction were observed simultaneously after a 24 h treatment by compound 27 (Figure 6B). The concentration of 27 needed for apoptosis induction (about 4.6–4.9 μM) was consistent with the GI50 (about 5 μM) and the cellular inhibition of CK2 activity (about 3 μM as monitored with P-Akt1(Ser129)), suggesting that CK2 inhibition was responsible for the induction of apoptosis and the subsequent cell death. Notably, the EC50 of 27 for the inhibition of cellular CK2, at least according to the Phospho-Akt1-Ser129 reporter signal (EC50 = 3.2 μM), was only about 5 times higher than the IC50 toward purified CK2α (0.6 μM) (Figure 6A). In contrast, under the same experimental conditions, we observed a 70-fold drop of potency with CX-4945 in living cells (IC50 cellular CK2 ≈ 1 μM / IC50 cell-free CK2 = 14.7 nM) (Figure 7C), which is in the expected range for ATP-competitive inhibitors of kinases with Km(ATP) values in the low μM range49 (Km(ATP) for CK2 ≈ 10 μM50). Assuming that the cellular uptake of both compounds was comparable, the distinct behavior of 27 added to the evidence that it was acting in a non-ATP competitive manner in living cells. A similar behavior had been observed with CI-1040, a highly selective and non-ATP competitive inhibitor of MEK1/2, that showed a ratio of about 5 to 6-fold between enzymatic potency (IC50 = 17 nM) and cellular efficacy (EC50 = 100 nM).51, 52
Figure 7.
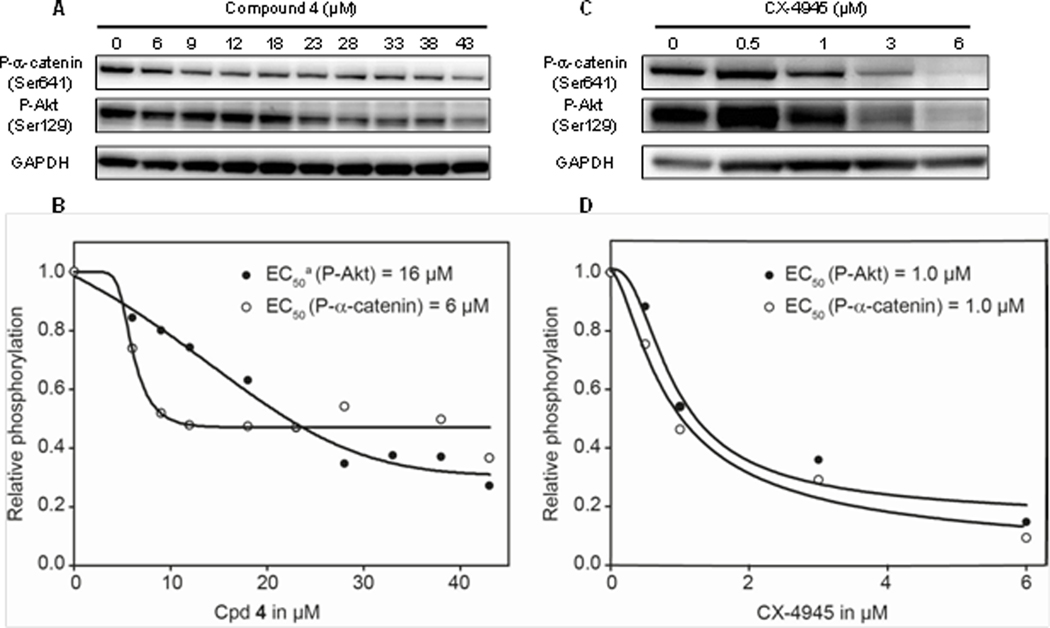
The potency of 4 but not of CX-4945 to inhibit CK2–catalyzed phosphorylations in cells depends on the substrate. 786-O cells were incubated with various concentrations of 4 (A, B) or CX-4945 (C, D) for 24 h, and the phosphorylation status of two protein substrates of CK2 was measured by Western Blot analysis of the cell extracts. GAPDH was probed as loading control. (B, D) Western Blot quantification was performed using ImageJ and fitted to a sigmoid equation using SigmaPlot to determine the EC50s. aHalf-maximum decrease relating to the basal level of non-responsive α-catenin phosphorylation (ca. 50% of the control).
The inhibitory potency of the allosteric ligands is substrate-dependent.
Given that more than 300 proteins were identified as potential CK2 substrates, it is fair to ask how the catalytic activity towards these numerous substrates is regulated.1 Although CK2β was found to modulate substrate specificity of the catalytic subunit upon complex formation,7, 8 it is unlikely that this is the only physiological mechanism of controlling the CK2 activity.53 On the basis of our data and previous speculations about the potential role of Tyr50 in the transition between CK2 conformational states29, 50 it might be hypothesized that adoption of an inactive conformation is not only artificially induced by our ligands but also has a physiological, regulatory function. Postulating that either active or inactive states of CK2 could be stabilized in vivo by interaction with proteins or other effectors, the binding affinities of different CK2 conformations toward our allosteric inhibitors are expected to vary. In support of this, we obtained first hints from the finding that the phosphorylation of α-catenin-Ser641 in cells was less efficiently suppressed by 27 than the phosphorylation of Akt1-Ser129 (Figure 6A), whereas both CK2 target sites were invariantly affected by the bona fide ATP-competitive inhibitor CX-4945 (Figure 7C and D). To rule out that this result was due to a particularity of 27, we repeated the same experiment with the previous, structurally somewhat different compound 4,20 that did not contain the naphthalene ring and carried a 3-carboxyphenyl instead of the salicylic acid (Figure 1). With 4, the differential response of the substrate phosphorylations toward the inhibitor became even more evident (Figure 7A and B): While the Phospho-α-catenin(Ser641) signal showed a steep initial decrease (EC50 for the partial inhibition = 6 μM), about half of the phosphorylations could not be inhibited further by 4, or this required much higher concentrations (Figure 7B), basically confirming the tendency already seen with 27 (cf. Figure 6A). In contrast, the CK2–catalyzed Ser129-phosphorylation on Akt1 could be suppressed to a greater extent by 4, exhibiting an overall EC50 of 16 μM. The latter correlated well with the EC50 = 25 μM for the growth inhibition of the 786-O cells by this compound (data not shown).
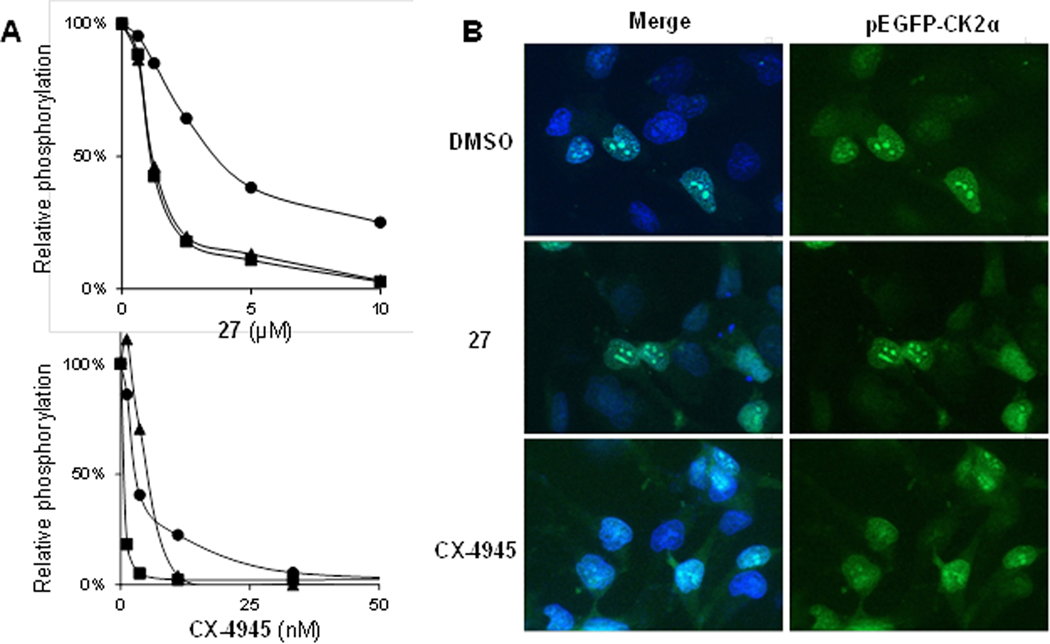
Next, we aimed at corroborating the above observations by analyzing the phosphorylation of further known CK2 protein substrates (nucleolin, Six1 and the CK2β subunit), using defined cell-free conditions (Figure 8A). Compound 27 inhibited the Six1 and the CK2β phosphorylation with about the same efficacy (IC50 ≈ 1 μM) as the canonical peptide substrate used in the previous dose–response assay (IC50 = 0.6 μM), whereas a clear drop of potency was observed in the case of nucleolin (IC50 ≈ 3.8 μM) (Figure 8A). Comparable potency differences against Six1 and CK2β phosphorylations on the one hand (IC50s ≈ 7–8 μM) and nucleolin phosphorylation on the other hand (IC50 ≈ 42 μM) were also noted for compound 4, just shifted to higher concentration ranges (Figure S5A). Under the same conditions, CX-4945 inhibited the phosphorylation of nucleolin, Six1 and the CK2β subunit with IC50s between 2 and 6 nM, in the range of that found with the peptide substrate (IC50 = 5 nM).
Figure 8.
Differential effects of the allosteric inhibitor 27 and the ATP-site directed inhibitor CX-4945 on the CK2-catalyzed phosphorylation of nucleolin and cellular consequences. (A) Phophosphorylation of substrate proteins (● nucleolin, ■ GST-Six1, ▲ CK2β auto-phosphorylation) by CK2α2β2 in the presence of various concentrations of compound 27 (upper panel) or the ATP-competitive inhibitor CX-4945 (lower panel). Graphs represent the average of two measurements. (B) HeLa pEGFP-CK2α cell imaging showing the subcellular localization of CK2α after 12 h treatment with DMSO, compound 27 (15 μM) or CX-4945 (8 μM). Nuclei were stained with Hoechst-33342 and merged images as well as GFP single channel were depicted for clarity.
The above data were in agreement with the expectation that CX-4945 can enter and block the ATP-binding site in the fully active state of CK2 as well as in less active conformational states. On the other hand, our allosteric inhibitor might bind with reduced affinity when the fully active conformation of the kinase is stabilized through interaction with a particular substrate or other effectors, as it could be the case with nucleolin. Similarly, it can be speculated that a certain part of the cellular α-catenin pool might be associated with CK2 in a tight complex, thereby stabilizing the active conformation of the CK2α subunit and rendering it less sensitive towards allosteric inhibition. It should be noted that an interaction with substrates was proposed earlier by Lolli and Battistutta as a possible mechanism to activate the autoinhibited CK2α2/β2 oligomers found in cells.54
Another major survival pathway which is constitutively upregulated in cancer with the participation of CK2 is the STAT3 pathway. Because of its importance as a target for potential anticancer therapies, we wondered whether the CK2 recruited to this pathway would be sensitive toward our allosteric inhibitors or show only a low response like in the case of nucleolin. STAT3-Tyr705 phosphorylation is the critical step in triggering the transcription factor activity of STAT3, while Ser727 phosphorylation is involved in the regulation of the phospho-Tyr705 levels.55 Inhibition or overexpression of CK2 was shown to influence the Ser727 phosphorylation levels, but, dependent on the cell type, either a positive56, 57 or negative55 correlation with the Tyr705 phosphorylation status was observed. Because STAT3-Tyr705 phosphorylation had been consistently diminished in all cell types after treatment by CK2 inhibitors, we used an anti-phospho-STAT3-Tyr705 antibody to analyze the effects of 27 and CX-4945 in 786-O cells. Interestingly, 27 had a somewhat stronger impact on the STAT3-Tyr705 phosphorylation levels than CX-4945 (Figure 6C), suggesting that the cellular CK2 pool engaged in the STAT3 activation pathway is – at least in the 786-O renal cancer cells – more amenable to allosteric inhibition than to ATP binding site-directed inhibition.
In summary, our results indicated that the sensitivity of cellular CK2 toward allosteric inibition varied to a considerable extent, depending on the protein substrate and/or the signalling complex environment.
Different subcellular localization of CK2 after treatment of cells by 27 vs. CX-4945.
Next we analyzed whether the differing sensitivity of the CK2–mediated nucleolin phosphorylation toward allosteric vs. ATP-competitive inhibition also translates into a different localization of the nucleolin/CK2 complex in cells after treatment with the two inhibitor types. Nucleolin is an abundant nucleolar protein known to bind to CK2α with very high affinity.58 Due to this complex formation, a significant portion of CK2α in growing cells is usually concentrated in the nucleoli.59 CK2 phosphorylates the bipartite nuclear localization signal near the N-terminus of nucleolin, thus promoting the translocation of the nucleolin–CK2 complex to the nucleolus.60 In consequence, the phosphorylation status of the CK2 phospho-acceptor site in nucleolin is expected to determine the subcellular localization of the high-affinity complex, which can be monitored by a fluorescent label. Using an EGFP-CK2α construct, we analyzed whether compound 27 and CX-4945 differently influence the subcellular localization of CK2α in HeLa cells. Indeed, we found that CX-4945 at 8 μM induced a significant translocation of EGFP-CK2α from the nucleoli to the nuclear matrix and the cytoplasm compartments (Figure 8B), whereas in the presence of compound 27, the nucleolar localization pattern of EGFP-CK2α was mostly retained at a concentration of 15 μM. A comparable difference was also found with 4, at a higher concentration (40 μM, Figure S5B). These results were consistent with the observed potency differences between compound CX-4945 and 4 or 27, respectively, with respect to inhibition of the nucleolin phosphorylation in the cell free assay. Thus, the alternative mode of action of our compounds, that rendered its potency substrate–dependent, further translated into a different impact on the subcellular localization pattern of a high affinity CK2α/substrate complex when compared with the ATP-competitive inhibitor CX-4945.
CONCLUSIONS
Supported by a binding model (Figure 5A), the previous hit compounds20 were systematically optimized to achieve a submicromolar potency with compound 27. Hence, we were able to demonstrate that the allosteric site targeted by our CK2 modulators is druggable and holds the potential for the development of high affinity ligands without having to critically increase the molecular weight. This view is particularly supported by the high cellular efficacy of 27: Although the IC50 of 0.6 μM was still about 40 times higher than that of the CK2 inhibitor CX-4945, which is currently in clinical phase II trials, the cellular potency of 27 was even superior in one of the analyzed CK2–dependent pathways, the STAT3 activation (EC50 = 1.6 μM). The comparably high cellular activity of the allosteric inhibitor 27 can probably be attributed to the absence of binding competition with the high intracellular ATP levels (~2–5 mM), which severly limited the cellular efficacy of CX-4945 on the other hand. As expected for an inhibitor targeting a non-ATP binding site, the kinase selectivity profile was also superior to that of CX-4945. Although the allosteric inhibitors were modulators of CK2 (also see below) with a varying impact on specific pathways rather than being total blockers of activity, the most important anticancer effects of CK2 inhibitors, i. e. induction of apoptosis and cell growth inhibition, were fully retained. Hence, there is an excellent perspective to develop highly efficient and selective allosteric CK2 inhibitors by optimizing further the binding affinity of the current lead compound 27.
Most notably, we could demonstrate that the potencies of 27 and 4 to inhibit CK2 depend on the individual substrate; particular pools of cellular CK2 that are engaged by different signaling pathways varied in their sensitivity towards allosteric but not ATP binding site–directed inhibition. In view of this particular outcome, the 2-aminothiazolyl benzoic acid derivatives will have a pharmacological profile distinct from any ATP-competitive CK2 inhibitor, which is worth being investigated further in in vivo models. It will also be interesting to examine whether fewer side effects can be expected for the novel compounds due to their less complete CK2 inhibition.
For most kinases, the equilibrium between the active and inactive state of the catalytic domain is regulated by phosphorylation of the activation loop. Nevertheless, it is increasingly recognized that even kinases that were a priori activated through phosphorylation exist in a dynamic equilibrium between multiple conformations in solution.61–64 Therefore, it can be assumed that CK2, which is not regulated at all by phosphorylation of a typical regulatory element on the catalytic subunit, also exists in a dynamic equilibrium between different conformations. Other kinases, for which the activation loop phosphorylation is not sufficient or not the only way to fully activate the catalytic domain, include PDK165 and Aurora-A66. Notably, the active conformation in these cases is stabilized by transient interactions with substrate proteins (PDK165) or with a complex binding partner, which is the spindle-associated protein Tpx2 in the case of Aurora-A.67 Analogously, our data provided first hints that the interactions of CK2 with its substrates might influence the conformational state of the catalytic domain; e.g., nucleolin, one of the substrates analyzed in the present study, might a priori stabilize the active conformation of CK2α, as it was found to counteract the allosteric inhibition by our compounds (Figures 8 and S5). On the other hand, a substrate–induced conformational stabilization is not necessarily sufficient to ensure steady phosphorylation of the substrate by CK2: in the case of nucleolin, it was reported that an additional interaction between the growth factor FGF-2 and CK2β in the CK2–nucleolin complex was required to trigger the phosphorylation.68 Nevertheless, our novel ligands might be useful as tools to probe the conformational state of different CK2 populations bound in complexes with individual substrates, in order to elucidate the relevance of CK2 conformational stabilization for the regulation of its activity.
EXPERIMENTAL SECTION
Chemistry.
3-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoic acid (1), 4-((4-(3-Nitrophenyl)thiazol-2-yl)amino)benzoic acid (2) and 3-((4-(pyridin-2-yl)thiazol-2-yl)amino)benzoic acid (17) were purchased from Sigma-Aldrich. All commercially available chemicals, 2-bromo-1-(3-nitrophenyl)ethanone (6c), 2-bromo-1-(2,4-dimethoxyphenyl)ethanone (6d), 2-bromo-1-(2,4-dihydroxyphenyl)ethanone (6e), 2-bromo-1-(4-bromo-2-hydroxyphenyl)ethanone (6f), 1-bromo-3,3-dimethylbutan-2-one (6g), 2-bromo-1-(thiophen-3-yl)ethanone (6h), 2-bromo-1-(naphthalen-2-yl)ethanone (6i), 3-(2-bromoacetyl)-2H-chromen-2-one (6j), 1-(benzofuran-2-yl)-2-bromoethanone (6k), 1-(benzo[b]thiophen-5-yl)-2-bromoethanone (6l), other starting materials and solvents were purchased from Sigma-Aldrich, Acros Organics, Fischer scientific or Alfa Aesar and were used without further purification. Reactions were monitored by Thin Layer Chromatography (TLC Silica gel 60 F254) purchased from Merck and observed under UV light (254 nm). Purification by semi-preparative HPLC was carried out on an Agilent 1200 series HPLC system (Agilent Technologies) using an Agilent C18 column (30 × 100 mm/10 μm) as stationary phase and a gradient of water and acetonitrile as eluent. 1H and 13C NMR spectra were obtained in dimethylsulphoxide-d6, acetone-d6 or CDCl3 on a Bruker DRX-500 instrument, operating at 500 MHz for the 1H and at 125 MHz for the 13C at 300K, using residual signal of deuterated NMR solvent as internal reference.69 Chemical shifts are reported in parts per million (ppm), multiplicity of the signals are indicated by lower-case letters (singlet s, doublet d, triplet t, quadruplet q, multiplet m, broad singlet br s, or combination of letters). DEPT 135 was used to determine carbon multiplicity. Analytical HPLC was performed using a SpectraSYSTEM™ (ThermoFisher) with a Macherey-Nagel C18 column (3 ˣ 125 mm/5 μm). The purity of the final compounds was determined using HPLC–coupled ElectroSpray Ionisation (ESI) mass spectrometry (Finnigan Surveyor MSQ Plus mass spectrometer, ThermoFisher). The UV traces at 254 nm thus obtained were used to calculate the purity of the final compounds, which was at least 95 % in all cases. Melting points (mp) of solids were determined by SMP3 melting point apparatus from Bibby Sterling. High-resolution mass spectra (HRMS) were recorded on a Bruker MicroTOF Q mass spectrometer.
General procedure 1 for the preparation of 2’-bromo-acetophenone analogs from acetophenones.23
Bromine (1 eq) was added dropwise over 5 min to a stirred and warmed (40°C) solution of acetophenone derivative (1 eq) in CHCl3. At the end of the addition, the mixture was cooled down at RT; Et2O was then added, followed by a saturated aqueous solution of NaHCO3 and the resulting mixture was then stirred for 30 min. The organic layer was separated, washed with a saturated aqueous solution of NaHCO3, dried with Na2SO4 and filtered. The volatiles were removed under reduced pressure to afford the product as a solid, which was used in the next step without further purification.
2’-Bromo-3-methoxy-acetophenone (6a).
The reaction was carried out according to general procedure 1, scale: 3-methoxyacetophenone (3.33 mmol, 459 μL), bromine (3.33 mmol, 172 μL), CHCl3 (10 mL), Et2O (100 mL), to afford the title product as a yellow solid (3.33 mmol, 762 mg). Yield = quantitative. LC-UV purity = 75%. The compound was not ionized by ESI+. 1H-NMR (500 MHz, CDCl3) δ = 3.70 (s, 3H, CH3-O), 4.3 (s, 2H, CH2-Br), 6.99 (ddd, J = 1.0; 2.5; 8.2 Hz, 1H), 7.24 (t, J = 8.2 Hz, 1H), 7.34 (dd, J = 1.6, 2.5 Hz, 1H), 7.39 (ddd, J = 1.0, 1.6, 7.6 Hz, 1H); spectral data are in accordance with literature data.70
2’-Bromo-4-methoxy-acetophenone (6b).
The reaction was carried out according to general procedure 1, scale: 4-methoxyacetophenone (3.33 mmol, 500 mg), bromine (3.33 mmol, 172 μL), CHCl3 (10 mL), Et2O (100 mL), to afford the title product as a grey solid (3.33 mmol, 765 mg). Yield = quantitative. LC-UV purity = 78%. LC-MS (ESI+) m/z = 229.31 for [M+H]+. 1H-NMR (500 MHz, CDCl3) δ = 3.89 (s, 3H, CH3-O), 4.40 (s, 2H, CH2-Br), 6.95 – 6.97 (m, 2H), 7.96 – 7.98 (m, 2H); spectral data are in accordance with literature data.71, 72
General procedure 2 for the preparation of thiourea analogs from amines.
Carbon disulfide (3 eq) was slowly added to a mixture of aminobenzoic acid derivative (1 eq) in a 1/1 THF/H2O (0.7 mol.L−1) solvent mixture and Et3N (2.5 eq). The resulting mixture was stirred at RT for 24h. Then iodine (1.05 eq) in THF (5 mL) was added dropwise at 0 °C and the mixture was stirred for 3 h. A 1M HCl aqueous solution (7.5 mL) and Na2SO3 (0.2 eq) were added and the stirring was continued for 15 min. The aqueous layer was extracted with EtOAc (3 × 50 mL). The combined organic layers were washed twice with brine (25 mL) and dried on Na2SO4. After filtration, the volatiles were removed under reduced pressure to give the isothiocyanate intermediate as a slightly yellow solid, which was converted to the thiourea derivative by stirring at RT in NH4OH (100 mL) for 6 h. NH4OH refers to a 35% w/w ammonia solution in water. After removal of the solids by filtration on a sintered-glass funnel, the filtrate was concentrated to afford the thiourea product, which was used in the next step without further purification.
Phenylthiourea (8a).
Phenylisothiocyanate (8.37 mmol, 1 mL) was added to a solution of NH4OH(aq) (40 mL) and stirred at RT for 12 h. After removal of volatile in vacuo, the white residue was triturated in water and filtered to obtain the title product as a white solid (8.15 mmol, 1.24 g). Yield = 97%. LC-UV purity > 99%. LC-MS (ESI+) m/z = 153.1 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.12 (t, J = 7.3 Hz, 1H), 7.32 (t, J = 7.3 Hz, 2H), 7.40 (d, J = 7.6 Hz, 2H), 9.70 (s, 1H); spectral data are in accordance with literature data.73
2-(4-Thioureidophenyl)acetic acid (8b).
The reaction was carried out according to the general procedure 2, scale: 2-(4-aminophenyl)acetic acid (3.31 mmol, 500 mg), to afford the title product as an off-white solid (3.1 mmol, 651 mg). Yield = 93%. LC-UV purity = 97%. LC-MS (ESI+) m/z = 211.05 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 3.34 (s, 2H), 5.00 (br s, 3H), 7.15 (d, J = 8.5 Hz, 2H), 7.30 (d, J = 8.2 Hz, 2H), 7.58 (br s, 1H), 10.2 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 43.0, 122.7, 129.2, 133.4, 137.2, 173.9, 181.0. mp = 139–141 °C.
4-Thioureidobenzoic acid (8c).
The reaction was carried out according to the general procedure 2, scale: 4-aminobenzoic acid (7.3 mmol, 1 g), to afford the title product as an off-white solid (7.3 mmol, 1.43 g). Yield = quantitative. LC-UV purity = 80%. The compound was not ionized by ESI+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.56 – 7.58 (m, 2H), 7.80 – 7.82 (m, 2H); spectral data are in accordance with literature data.74
1-(4-Cyanophenyl)thiourea (8d).
A solution of 4-cyanophenyl isothiocyanate (6.24 mmol, 1 g) in NH4OH(aq) (80 mL) was stirred at RT for 12 h. After cooling at 0 °C, the mixture was filtrated to afford the title product as an off-white solid (4.85 mmol, 860 mg). Yield = 78%. LC-UV purity > 99%. The compound was not ionized by ESI+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.75 (m, 4H), 10.07 (s, 1H); spectral data are in accordance with literature data.75
3-Thioureidobenzoic acid (8e).
The reaction was carried out according to the general procedure 2, scale: 3-aminobenzoic acid (7.3 mmol, 1 g), to afford the title product as an off-white solid (7.3 mmol, 1.43 g). Yield = quantitative. LC-UV purity = 96%. The compound was not ionized by ESI+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.28 (t, J = 7.8 Hz, 1H), 7.57 – 7.59 (m, 2H), 8.04 (d, J = 6.5 Hz, 1H), 8.92 (br s, 1H), 10.96 (br s, 1H). mp = 183–185 °C.
4-Methoxy-3-thioureidobenzoic acid (8f).
The reaction was carried out according to the general procedure 2, scale: 3-amino-4-methoxybenzoic acid (7.3 mmol, 1.22 g) to afford the title product as an off-white solid (7.3 mmol, 1.65 g). Yield = quant. LC-UV purity = 97%. LC-MS (ESI+) m/z = 226.79 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 3.80 (s, 3H), 5.33 (br s, labile H), 6.97 (d, J = 8.8 Hz, 1H), 7.52 (br s, 1H), 7.70 (dd, J = 1.9, 8.5 Hz, 1H), 8.10 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 55.7, 110.4, 126.5, 127.5, 127.7, 129.6, 154.2, 169.3, 181.6. mp = 241–243 °C.
2-Hydroxy-4-thioureidobenzoic acid (8g).
The reaction was carried out according to the general procedure 2, scale: 4-aminosalicylic acid (7.3 mmol, 1.12 g) to afford the title product as an off-white solid (7.3 mmol, 1.55 g). Yield = quantitative. LC-UV purity = 70%. The compound was not ionized by ESI+. 1H-NMR (500 MHz, DMSO-d6): δ = 6.73 (dd, J = 2.2, 8.2 Hz, 1H), 6.96 (s, 1H), 7.30 – 7.50 (br s, labile H), 7.60 (d, J = 8.2 Hz, 1H), 7.56 – 7.70 (br s, labile H), 10.00 (s, 1H). mp = 185–187 °C, melting point is in accordance with lit. data (179–180 °C).76
General procedure 3 for the preparation of 2-aminothiazole analogs.
An ethanolic solution of 2’-bromo-acetophenone derivative and aryl thiourea derivative at eqimolar ratio, corrected with respect to LC-UV purity, was heated at 80 °C for 3–12 h (reaction monitoring by TLC). The mixture was then cooled down to RT and water (twice more than EtOH v/v) was added. The mixture was stirred for 30 min and then filtrated. The obtained solid was washed twice with water to afford the product as a solid. When necessary, purification was performed (i) by recrystallization or washing with boiling MeCN (ii) by recrystallization or washing in H2O/MeOH (iii) by HPLC preparative to obtain the product with LC-UV purity higher than 95%.
4-(3-Nitrophenyl)-N-phenylthiazol-2-amine (9).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-3’-nitro-acetophenone (0.66 mmol, 161 mg), phenylthiourea (0.66 mmol, 100 mg), absolute EtOH (5 mL). After removal of the solvents under reduced pressure, water (10 mL) and MeOH (2 mL) were added. The pH of the solution was adjusted to 9 with solid Na2CO3 and then stirred for 1h. After filtration and washing of the residual solid with water (3 × 5 mL), the product was obtained as a yellow solid (0.66 mmol, 196 mg). Yield = quantitative. LC-UV purity = 99%. LC-MS (ESI+) m/z = 298.06 for [M+H]+.1H-NMR (500 MHz, DMSO-d6): δ = 6.99 (t, J = 6.9 Hz, 1H), 7.36 (t, J = 6.6 Hz, 2H), 7.65 – 7.71 (m, 4H), 8.14 (d, J = 7.3 Hz, 1H), 8.35 (d, J = 6.9 Hz, 1H), 8.69 (s, 1H), 10.39 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 105.6 (CH), 116.9 (2 x CH), 119.8 (CH), 121.4 (CH), 121.9 (CH), 129.0 (2 x CH), 130.1 (CH), 131.7 (CH), 135.9 (C), 140.9 (C), 147.6 (C), 148.2 (C), 163.5 (C). mp = 136–138 °C. HRMS (ESI+): m/z calcd for C15H12N3O2S [M+H]+: 298.0645; found: 298.0633.
2-(4-(4-(3-Nitrophenyl)thiazol-2-ylamino)phenyl)acetic acid (10).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-3’-nitro-acetophenone (0.48 mmol, 117 mg), 2-(4-thioureidophenyl)acetic acid (0.48 mmol, 100 mg), EtOH (5 mL) to afford ethyl 2-(4-((4-(3-nitrophenyl)thiazol-2-yl)amino)phenyl) (0.41 mmol, 158 mg). Then, a mixture of ethyl 2-(4-((4-(3-nitrophenyl)thiazol-2-yl)amino)phenyl)acetate in THF (5 mL) and NaOH 0.5 M (1.3 mmol, 2.6 mL) was heated at 80 °C for 4 h. After removal of THF under reduced pressure, 1M HCl(aq) was added until pH = 1. The product was obtained after filtration as a black solid (0.22 mmol, 78 mg). Yield = 73%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 355.74 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 3.53 (s, 2H), 7.24 (d, J = 8.2 Hz, 2H), 7.64 (d, J = 8.2 Hz, 2H), 7.69 (s, 1H), 7.73 (t, J = 7.9 Hz, 1H), 8.16 (d, J = 7.9 Hz, 1H), 8.37 (d, J = 7.9 Hz, 1H), 8.70 (s, 1H), 10.43 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 39.7 (CH2), 105.5 (CH), 116.9 (CH), 119.8 (CH), 122.0 (CH), 128.0 (C), 129.0 (CH), 130.2 (CH), 131.7 (CH), 135.9 (C), 139.5 (C), 147.6 (C), 148.2 (C), 163.6 (C), 172.8 (C). mp = 205–207 °C. HRMS (ESI+): m/z calcd for C17H14N3O4S [M+H]+: 356.0700; found: 356.0691.
4-((4-(3-Methoxyphenyl)thiazol-2-yl)amino)benzoic acid (11).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-3’-methoxy-acetophenone (0.66 mmol, 152 mg), 4-thioureidobenzoic acid (0.51 mmol, 100 mg), EtOH (10 mL). Purification was performed by washing the residue with boiling MeCN (5 mL) and hot filtration to give a crude solid which was then purified by flash chromatography (cyclohexane/Acetone 50/50) to afford the title product as a yellow solid (0.09 mmol, 29 mg). Yield = 17%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 327.10 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 3.83 (s, 3H), 6.91 (dd, J = 8.2, 2.2 Hz, 1H), 7.36 (t, J = 8.2 Hz, 1H), 7.48 (s, 2H), 7.53 (d, J = 7.9 Hz, 1H), 7.81 (d, J = 8.8 Hz, 2H), 7.94 (d, J = 8.8 Hz, 2H), 10.69 (s, 1H), 12.56 (br s, 1H).13C-NMR (125.7 MHz, DMSO-d6): δ = 55.0 (CH3), 104.5 (CH), 111.1 (CH), 113.2 (CH), 115.8 (CH), 118.1 (CH), 122.8 (C), 129.7 (CH), 130.8 (CH), 135.6 (C), 144.8 (C), 150.0 (C), 159.5 (C), 162.1 (C), 167.0 (C). mp = 246–248 °C. HRMS (ESI+): m/z calcd for C17H15N2O3S [M+H]+: 327.0798; found: 327.0786.
4-((4-(4-Methoxyphenyl)thiazol-2-yl)amino)benzoic acid (12).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-4’-methoxy-acetophenone (0.71 mmol, 163 mg), 4-thioureidobenzoic acid (0.51 mmol, 100 mg), absolute EtOH (10 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford the title product as a grey solid (0.15 mmol, 49 mg). Yield = 29%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 327.09 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 3.80 (s, 3H), 7.00 (d, J = 7.3 Hz, 2H), 7.29 (s, 1H), 7.81 (d, J = 7.9 Hz, 2H), 7.87 (d, J = 7.9 Hz, 2H), 7.93 (d, J = 7.9 Hz, 2H), 10.65 (s, 1H), 12.54 (br s, 1H, O-H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 55.1 (CH3), 101.9 (CH), 114.0 (2 x CH), 115.9 (2 x CH), 122.7 (C), 127.1 (2 x CH), 127.2 (C), 130.8 (2 x CH), 145.0 (C), 150.1 (C), 158.9 (C), 162.1 (C), 167.0 (C). mp = 266–268 °C. HRMS (ESI+): m/z calcd for C17H15N2O3S [M+H]+: 327.0798; found: 327.0789.
4-((4-(2,4-Dimethoxyphenyl)thiazol-2-yl)amino)benzoic acid (13).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(2,4-dimethoxyphenyl)ethanone (0.38 mmol, 100 mg), 4-thioureidobenzoic acid (0.58 mmol, 114 mg), EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford an off-white solid (0.25 mmol, 89 mg). Yield = 66%. LC-UV purity = 97%. LC-MS (ESI+) m/z = 356.78 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6) δ = 3.81 (s, 3H), 3.91 (s, 3H), 6.65 – 6.67 (m, 2H), 7.31 (s, 1H), 7.81 (td, J = 2.2, 9.1 Hz, 2H), 7.93 (td, J = 2.2 Hz, 8.83 Hz, 2H), 8.08 – 8.10 (m, 1H), 10.58 (s, 1H), 12.56 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 55.2 (CH3), 55.5 (CH3), 98.5 (CH), 105.2 (CH), 105.5 (CH), 115.7 (CH), 115.8 (C), 122.5 (C), 130.2 (CH), 130.7 (CH), 145.0 (C), 146.2 (C), 157.7(C), 159.8 (C), 160.3 (C), 167.0 (C). mp = 271–273 °C. HRMS (ESI+): m/z calcd for C18H17N2O4S [M+H]+: 357.0904; found: 357.0902.
4-((4-(2,4-Dihydroxyphenyl)thiazol-2-yl)amino)benzoic acid (14).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(2,4-dihydroxyphenyl)ethanone (0.43 mmol, 100 mg), 4-thioureidobenzoic acid (0.65 mmol, 127 mg), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford the title product as yellow solid (0.38 mmol, 125 mg). Yield = 89%. LC-UV purity = 95% LC-MS (ESI+) m/z = 328.84 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 6.34 (dd, J = 8.4, 2.4 Hz, 1H), 6.36 (d, J = 2.3 Hz, 1H), 7.26 (s, 1H), 7.67 (d, J = 8.8 Hz, 2H), 7.76 (d, J = 8.2 Hz, 1H), 7.94 (d, J = 8.8 Hz, 2H), 9.48 (br s, 1H), 10.67 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 102.2 (CH), 102.8 (CH), 107.1 (CH), 111.6 (C), 116.0 (CH), 122.9 (C), 129.0 (CH), 130.8 (CH), 144.7 (C), 147.6 (C), 156.2(C), 158.1 (C), 161.2 (C), 166.9 (C). mp = 367–369 °C. HRMS (ESI+): m/z calcd for C16H13N2O4S [M+H]+: 329.0591; found: 329.0581.
4-((4-(5-Bromo-2-hydroxyphenyl)thiazol-2-yl)amino)benzoic acid (15).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(5-bromo-2-hydroxyphenyl)ethanone (0.34 mmol, 100 mg), 4-thioureidobenzoic acid (0.51 mmol, 100 mg), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford a yellow solid (0.31 mmol, 120 mg). Yield = 90%. LC-UV purity = 98%. LC-MS (ESI+) m/z = 390.65/392.62 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6) δ = 6.91 (d, J = 8.5 Hz, 1H), 7.31 (dd, J = 2.5, 8.5 Hz, 1H), 7.64 (s, 1H), 7.68 (d, J = 8.8 Hz, 2H), 7.94 (d, J = 9.1 Hz, 2H), 8.12 (d, J = 2.5 Hz, 1H), 10.74 (s, 1H), 10.84 (s, 1H), 12.60 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 107.7 (CH), 110.3 (C), 116.0 (CH), 118.4 (CH), 122.1 (C), 123.1 (C), 130.3 (CH), 130.8 (CH), 130.9 (CH), 144.6 (C), 145.4 (C), 154.2 (C), 161.4 (C), 166.9 (C). mp = 312–314 °C. HRMS (ESI+): m/z calcd for C16H12BrN2O3S [M+H]+: 390.9747; found: 390.9742.
4-((4-(tert-Butyl)thiazol-2-yl)amino)benzoic acid (16).
The reaction was carried out according to the general procedure 3, scale: 1-bromo-3,3-dimethylbutan-2-one (0.45 mmol, 60 μL), 4-thioureidobenzoic acid (0.41 mmol, 80 mg), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford the title product as a white solid (0.10 mmol, 28 mg). Yield = 25%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 277.04 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 1.28 (s, 9H), 6.53 (s, 1H), 7.70 (d, J = 8.5 Hz, 2H), 7.88 (d, J = 8.5 Hz, 2H), 10.48 (s, 1H), 12.49 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 29.5 (CH3), 34.3 (C), 100.5 (CH), 115.5 (CH), 122.3 (C), 130.6 (CH), 145.1 (C), 161.5 (C), 161.6 (C), 166.9 (C). mp = 280–282 °C. HRMS (ESI+): m/z calcd for C14H17N2O2S [M+H]+: 277.1005; found: 277.0995.
4-((4-(Thiophen-3-yl)thiazol-2-yl)amino)benzoic acid (18).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(thiophen-3-yl)ethanone (0.41 mmol, 84 mg), 4-thioureidobenzoic acid (0.41 mmol, 80 mg), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration by hot filtration to afford the title product as a brown solid (0.20 mmol, 59 mg). Yield = 48%. LC-UV purity = 96%.LC-MS (ESI+) m/z = 302.86 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.27 (s, 1H), 7.59 (m, 2H), 7.82 (m, 2H), 7.91 (m, 3H), 10.65 (s, 1H), 12.56 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 103.4 (CH), 115.9 (2 x CH), 121.8 (CH), 122.7 (C), 125.9 (CH), 126.8 (CH), 130.7 (2 x CH), 136.6 (C), 144.8 (C), 146.6 (C), 162.1 (C), 166.9 (C). mp = 278–280 °C. HRMS (ESI+): m/z calcd for C14H11N2O2S2 [M+H]+: 303.0256; found: 303.0245.
4-((4-(Naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (19).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(naphthalen-2-yl)ethanone (0.41 mmol, 102 mg), 4-thioureidobenzoic acid (0.41 mmol, 80 mg), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford the title product as a brown solid (0.17 mmol, 59 mg). Yield = 42%. LC-UV purity = 95%. LC-MS (ESI+) m/z = 346.57 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.53 (m, 2H), 7.60 (s, 1H), 7.89 (d, J = 8.8 Hz, 2H), 7.93 (d, J = 7.9 Hz, 2H), 7.98 (m, 3H), 8.04 (d, J = 8.5 Hz, 1H), 8.08 (dd, J = 8.5, 1.9 Hz, 1H), 8.50 (s, 1H), 10.73 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 104.8 (CH), 116.0 (2 x CH), 122.8 (C), 123.9 (CH), 124.3 (CH), 126.0 (CH), 126.3 (CH), 127.5 (CH), 128.1 (CH),128.2 (CH), 130.8 (2 x CH), 131.7 (C), 132.4 (C), 133.1 (C), 144.8 (C), 150.1 (C), 162.3 (C), 167.0 (C). mp = 294–296 °C. HRMS (ESI+): m/z calcd for C20H15N2O2S [M+H]+: 347.0849; found: 347.0838.
4-((4-(2-Oxo-2H-chromen-3-yl)thiazol-2-yl)amino)benzoic acid (20).
The reaction was carried out according to the general procedure 3, scale: 3-(bromoacetyl)coumarin (0.45 mmol, 120 mg), 4-thioureidobenzoic acid (0.54 mmol, 106 mg), EtOH absolute (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford the title product as a yellow solid (0.33 mmol, 120 mg). Yield = 73%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 364.86 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.42 (t, J = 8.2 Hz, 1H), 7.46 (d, J = 8.5 Hz, 2H), 7.64 (t, J = 8.2 Hz, 1H), 7.88 (m, 3H), 7.99 (d, J = 8.8 Hz, 2H), 8.02 (d, J = 7.9 Hz, 1H), 8.75 (s, 1H), 10.75 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 110.8 (CH), 115.8 (CH), 116.2 (CH), 119.2 (C), 120.1 (C), 122.9 (C), 124.6 (CH), 129.0 (CH), 130.9 (CH), 131.7 (CH), 138.9 (CH), 143.6 (C), 144.6 (C), 152.3 (C), 158.7 (C), 161.7 (C), 167.0 (C). mp = 343–345 °C. HRMS (ESI+): m/z calcd for C19H13N2O4S [M+H]+: 365.0591; found: 365.0589.
4-((4-(Benzofuran-2-yl)thiazol-2-yl)amino)benzoic acid (21).
The reaction was carried out according to the general procedure 3, scale: 1-(benzofuran-2-yl)-2-bromoethanone (0.42 mmol, 100 mg), 4-thioureidobenzoic acid (0.50 mmol, 99 mg), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford a brown solid which was further purified by semi-preparative HPLC (H2O/MeCN + 0.1% TFA: from 70/30 to 0/100 in 45 min) to afford the title product as a yellow solid (0.19 mmol, 63 mg). Yield = 45%. LC-UV purity = 99%. LC-MS (ESI+) m/z = 336.96 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.28 (m, 2H), 7.34 (dd, J = 8.2, 1.3 Hz, 1H), 7.45 (s, 1H), 7.62 (d, J = 7.6 Hz, 1H), 7.70 (d, J = 7.3, 1H), 7.84 (d, J = 8.8 Hz, 2H), 7.95 (d, J = 8.8 Hz, 2H), 10.83 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 103.0 (CH), 106.4 (CH), 110.9 (CH), 116.1 (CH), 121.4 (CH), 123.0 (CH), 123.2 (C), 124.7 (CH), 128.4 (C), 130.7 (CH), 141.4 (C), 144.6 (C), 151.6 (C), 154.1 (C), 163.0 (C), 166.9 (C). mp = 296–298 °C. HRMS (ESI+): m/z calcd for C18H13N2O3S [M+H]+: 337.0641; found: 337.0630.
4-((4-(Benzo[b]thiophen-5-yl)thiazol-2-yl)amino)benzoic acid (22).
The reaction was carried out according to the general procedure 3, scale: 1-(1-benzothiophen-5-yl)-2-bromo-1-ethanone (0.39 mmol, 100 mg), 4-thioureidobenzoic acid (0.59 mmol, 115 mg), absolute EtOH (5 mL). The obtained solid was stirred in boiling MeCN (20 mL) and filtrated. The filtrate layer was concentrated and the remaining solid was then dissolved in hot MeOH (10 mL). After cooling at RT, water (10 mL) was added and the obtained solid was filtrated and washed with water (2 × 5 mL) to obtain the title product as a yellow solid (0.15 mmol, 51 mg). Yield = 38%. LC-UV purity = 95%. LC-MS (ESI+) m/z = 352.76 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.48 (s, 1H), 7.56 (d, J = 5.4 Hz, 1H), 7.79 (d, J = 5.4 Hz, 1H), 7.88 (d, J = 8.8 Hz, 2H), 7.94 – 7.99 (m, 3H), 8.04 (d, J = 8.5 Hz, 1H), 8.48 (d, J = 1.6 Hz, 1H), 10.70 (s, 1H), 12.52 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 103.7 (CH), 115.9 (2 x CH), 120.7 (CH), 122.3 (CH), 122.7 (CH), 122.8 (C), 124.3 (CH), 128.0 (CH), 130.8 (3 x CH), 138.4 (C), 139.9 (C), 144.9 (C), 150.3 (C), 162.2 (C), 167.0 (C). mp = 353–355 °C. HRMS (ESI+): m/z calcd for C18H13N2O2S2 [M+H]+: 353.0413; found: 353.0400.
4-(Naphthalen-2-yl)-N-phenylthiazol-2-amine (23).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(naphthalen-2-yl)ethanone (0.66 mmol, 164 mg), phenylthiourea (0.66 mmol, 100 mg), absolute EtOH (15 mL). After water (25 mL) addition, the pH was adjusted to 9 with aqueous Na2CO3 (1M). The resulting mixture was cooled at 0 °C and the precipitate was filtrated to afford the title product as a white solid (0.65 mmol, 195 mg). Yield = 98%. LC-UV purity> 99%.LC-MS (ESI+) m/z = 302.85 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.00 (t, J = 7.3 Hz, 1H), 7.39 (t, J = 8.2 Hz, 2H), 7.48 (s, 1H), 7.52 (qi, J = 8.2 Hz, 2H), 7.79 (d, J = 7.6 Hz, 2H), 7.91 (d, J = 7.9 Hz, 1H), 7.95 (d, J = 8.8 Hz, 1H), 8.00 (d, J = 7.9 Hz, 1H), 8.07 (dd, J = 8.5, 1.5 Hz, 1H), 10.32 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 103.6 (CH), 116.8 (CH), 121.2 (CH), 124.0 (CH), 124.1 (CH), 125.9 (CH), 126.3 (CH), 127.5 (CH), 128.0 (CH), 128.1 (CH), 129.0 (CH), 131.9 (C), 132.4 (C), 133.1 (C), 141.1 (C), 150.0 (C), 163.1 (C). mp = 148–150 °C. HRMS (ESI+): m/z calcd for C19H15N2S [M+H]+: 303.0950; found: 303.0939.
4-((4-(Naphthalen-2-yl)thiazol-2-yl)amino)benzonitrile (24).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-2’acetonaphtone (1.41 mmol, 351 mg), 1-(4-cyanophenyl)thiourea (1.41 mmol, 250 mg), EtOH absolute (5 mL). The resulting mixture was cooled at RT and aqueous Na2CO3 (1 M) was added dropwise until pH = 8. After cooling at 0 °C,the precipitate was filtrated and washed with H2O (2 × 5 mL) to afford the title product as a slightly yellow solid (462 mg, 1.41 mmol). Yield = quantitative. LC-UV purity = 99%. LC-MS (ESI+) m/z = 327.94 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.51 (dt, J = 6.9, 1.3 Hz, 1H), 7.55 (dt, J = 6.9, 1.6 Hz, 1H), 7.61 (s, 1H), 7.81 (d, J = 8.8 Hz, 2H), 7.92 (d, J = 8.2 Hz, 1H), 7.96 (m, 3H), 8.01 (d, J = 7.9 Hz, 1H), 8.08 (dd, J = 8.5, 1.3 Hz, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 101.8 (C), 105.2 (CH), 116.8 (CH), 119.6 (C), 123.9 (CH), 124.3 (CH), 126.0 (CH), 126.4 (CH), 127.5 (CH), 128.1 (CH), 131.6 (C), 132.4 (C), 133.1 (C), 133.5 (2 x CH), 145.2 (C), 150.2 (C), 162.4 (C). mp = 228–230 °C. HRMS (ESI+): m/z calcd for C20H14N3S [M+H]+: 328.0903; found: 328.0894.
3-((4-(Naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (25).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(naphthalen-2-yl)ethanone (0.40 mmol, 100 mg), 3-thioureidobenzoic acid (0.52 mmol, 102 mg), EtOH absolute (5 mL). The title product was obtained pure after filtration as a yellow solid (0.34 mmol, 118 mg). Yield = 86%. LC-UV purity = 98%. LC-MS (ESI+) m/z = 346.80 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.49 – 7.56 (m, 4H), 7.59 (d, J = 7.6 Hz, 1H), 7.90 – 7.98 (m, 4H), 8.08 (dd, J = 1.9, 8.5 Hz, 1H), 8.55 (s, 1H), 8.77 (s, 1H), 10.56 (s, 1H), 13.01 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 104.1 (CH), 117.7 (CH), 120.8 (CH), 121.9 (CH), 123.8 (CH), 124.5 (CH), 126.0 (CH), 126.4 (CH), 127.5 (CH), 128.0 (CH), 128.1 (CH), 129.2 (CH), 131.5 (C), 131.8 (C), 132.4 (C), 133.2 (C), 141.3 (C), 149.8 (C), 162.7 (C), 167.4 (C). mp = 275–277 °C. HRMS (ESI+): m/z calcd for C20H15N2O2S [M+H]+: 347.0849; found: 347.0843.
4-Methoxy-3-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (26).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(naphthalen-2-yl)ethanone (1 mmol, 250 mg), 4-methoxy-3-thioureidobenzoic acid (1.3 mmol, 294 mg), EtOH absolute (25 mL). Product was obtained pure after filtration as a yellow solid (0.94 mmol, 353 mg). Yield = 94%. LC-UV purity = 97%. LC-MS (ESI+) m/z = 376.82 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6) δ = 3.97 (s, 3H), 7.14 (d, J = 8.5 Hz, 1H), 7.49 – 7.55 (m, 3H), 7.64 (dd, J = 2.2, 8.2 Hz, 1H), 7.91 – 7.96 (m, 3H), 8.07 (dd, J = 1.9, 8.5 Hz, 1H), 8.60 (d, J = 0.6 Hz, 1H), 9.70 (d, J = 2.2 Hz, 1H), 9.90 (s, 1H), 12.71 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 55.9 (CH3), 104.7 (CH), 110.0 (CH), 118.8 (CH), 122.9 (C), 123.5 (CH), 123.6 (CH), 124.5 (CH), 125.9 (CH), 126.3 (CH), 127.5 (CH), 127.9 (CH), 128.0 (CH), 129.9 (C), 131.8 (C), 132.3 (C), 133.2 (C), 149.1 (C), 150.8 (C), 162.7 (C), 167.5 (C). mp = 256–258 °C. HRMS (ESI+): m/z calcd for C21H17N2O3S [M+H]+: 377.0954; found: 377.0937.
2-Hydroxy-4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (27).
The reaction was carried out according to the general procedure 3, scale: 2-bromo-1-(naphthalen-2-yl)ethanone (0.50 mmol, 124 mg), 2-hydroxy-4-thioureidobenzoic acid (0.70 mmol, 149 mg), EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford a yellow solid (0.37 mmol, 133 mg). Yield = 73%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 362.84 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.19 (dd, J = 1.9, 8.8 Hz, 1H), 7.49 – 7.57 (m, 3H), 7.61 (s, 1H), 7.80 (d, J = 8.5 Hz, 1H), 7.92 (d, J = 7.9 Hz, 1H), 7.98 (d, J = 8.2 Hz, 2H), 8.07 (d, J = 8.5 Hz, 1H), 8.46 (s, 1H), 10.75 (s, 1H), 11.8 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 103.0 (CH), 105.2 (CH), 105.6 (C), 108.4 (CH), 124.0 (CH), 124.2 (CH), 126.0 (CH), 126.5 (CH), 127.6 (CH), 128.1 (CH), 128.2 (CH), 131.3 (CH), 131.7 (C), 132.5 (C), 133.1 (C), 147.0 (C), 150.2 (C), 162.1 (C), 162.6 (C), 171.7 (C). mp = 221–223 °C. HRMS (ESI+): m/z calcd for C20H15N2O3S [M+H]+: 363.0798; found: 363.0786.
2-((4-(3-Nitrophenyl)thiazol-2-yl)amino)benzoic acid (30).
A solution of HCl 37% (1 mL) and 1,4-dioxane (5 mL) containing 2-bromo-4-phenylthiazole (2.08 mmol, 500 mg) and 2-aminobenzoic acid (2.08 mmol, 286 mg) was heated at reflux for 60h. After solvents removal, the residual solid was washed with water (5 mL) and then with cold acetone (5 mL) to obtain a white solid which was further purified by flash chromatography (cyclohexane/EtOAc, from 7/3 to 0/100) to obtain a second white solid containing 75% of product (by HPLC). After a second purification by preparative HPLC (H2O/MeCN - 1% formic acid, from 70/30 to 0/100), the pure title product was obtain as a white solid (0.04 mmol, 12 mg). Yield = 2%. LC-UV purity = 97%. LC-MS (ESI+) m/z = 297.08 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 7.07 (t, J = 7.9 Hz, 1H), 7.34 (t, J = 7.6 Hz, 1H), 7.45 (t, J = 7.6 Hz, 2H), 7.53 (s, 1H), 7.69 (td, J = 7.6, 1.6 Hz, 1H), 7.94 (d, J = 6.9 Hz, 2H), 8.02 (dd, J = 7.9, 1.6 Hz, 1H), 8.62 (d, J = 8.5 Hz, 1H), 11.48 (s, 1H).13C-NMR (125.7 MHz, DMSO-d6): δ = 104.6 (CH), 113.7 (C), 116.9 (CH), 120.4 (CH), 125.7 (CH), 127.8 (CH), 128.6 (CH), 131.4 (CH), 134.1, 134.7 (CH), 142.9 (C), 150.3 (C), 161.8 (C), 170.0 (C). mp = 95–97 °C. HRMS (ESI+): m/z calcd for C16H12N3O4S [M+H]+: 342.0543; found: 342.0538.
Methyl 4-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoate (31).
Three drops of 95% H2SO4 were added to a mixture of 4-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoic acid (2) (0.14 mmol, 46 mg) in MeOH (5 mL) and the mixture was then heated at reflux for 2 h. Na2CO3 1 M was then added until pH = 10–11 and MeOH was removed under vacuo. The obtained solid was filtrated and washed with water (2 × 5 mL) to afford the title product as a yellow solid (0.14 mmol, 49 mg). Yield = quantitative. LC-UV purity = 97%. LC-MS (ESI+) m/z = 356.01 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 3.82 (s, 3H), 7.74 – 7.84 (m, 4H), 7.96 (s, 2H), 8.17 (s, 1H), 8.40 (s, 1H), 8.71 (s, 1H), 10.83 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ = 51.7 (CH3), 107.0 (CH), 116.1 (2 x CH), 119.9 (CH), 121.8 (C), 122.1 (CH), 130.3 (CH), 130.6 (2 x CH), 131.9 (CH), 135.7 (C), 144.9 (C), 147.8 (C), 148.3 (C), 162.6 (C), 165.8 (C). mp = 242–244 °C. HRMS (ESI+): m/z calcd for C17H14N3O4S [M+H]+: 356.0700; found: 356.0695.
N-(Methylsulfonyl)-4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzamide (32).
A solution of 4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (19) (0.23 mmol, 80 mg), HATU (0.28 mmol, 106 mg) and DIPEA (0.69 mmol, 120 μL) in dry DMF (2 mL) was stirred under N2 for 30 min. And then added to a solution of NaH (0.92 mmol, 22 mg) and MeSO2NH2 (1.15 mmol, 110 mg) in dry THF (2 mL) beforehand stirred at RT under N2 for 30 min. The resulting mixture was stirred at RT for 4 h. After removal of solvents under vacuo, residue was purified by semi-preparative HPLC (H2O/MeCN + 0.1% TFA: from 40/60 to 0/100 in 50 min) followed by a second purification by semi-preparative HPLC (H2O/MeCN + 0.1% TFA: from 25/75 to 20/80 in 45 min) to obtain the title product as a white solid (0.026 mmol, 11 mg). Yield = 11%. LC-UV purity = 99%. LC-MS (ESI+) m/z = 423.73 for [M+H]+. 1H-NMR (500 MHz, acetone-d6) δ = 3.40 (s, 3H), 7.48 (s, 1H), 7.49 – 7.56 (m, 2H), 7.92 (d, J = 7.9 Hz, 1H), 7.96 (d, J = 8.6 Hz, 1H), 7.99 (d, J = 8.0 Hz, 1H), 8.01 – 8.04 (m, 2H), 8.08 – 8.12 (m, 3H), 8.56 (s, 1H), 9.89 (brs, 1H), 10.49 (brs, 1H). 13C-NMR (125.7 MHz, acetone-d6) δ = 42.8 (CH3), 106.2 (CH), 118.3 (CH), 126.0 (CH), 126.2 (C), 126.8 (CH), 128.0 (CH), 128.3 (CH), 129.5 (CH), 130.1 (CH), 130.2 (CH), 131.8 (3 x CH), 134.0 (C), 135.1 (C), 135.7 (C), 153.1 (C), 167.1 (C). mp = 240–242 °C. HRMS (ESI+): m/z calcd for C21H18N3O3S2 [M+H]+: 424.0784; found: 424.0784.
4-((4-(Naphthalen-2-yl)thiazol-2-yl)amino)-N-tosylbenzamide (33).
4-methylbenzenesulfonamide (0.23 mmol, 39.5 mg), N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (0.46 mmol, 88 mg) and 4-dimethylaminopyridine (0.46 mmol, 56 mg) were added to a solution of 4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (19) (0.23 mmol, 80 mg) in CH2Cl2 (10 mL). The resulting mixture was stirred at RT under N2 atmosphere for 36 h. NH4Cl(aq) (10 mL) was added, and then H2O (10 mL). The aqueous layer was extracted with CH2Cl2 (2 × 20 mL). The combined organic layers were dried with Na2SO4, filtrated and concentrated to give a brown solid which was further purified by semi-preparative HPLC (H2O/MeCN + 0.1% TFA: from 30/70 to 0/100 in 45 min) to obtain the title product as a white solid (0.026 mmol, 13 mg). Yield = 11%. LC-UV purity = 97%. LC-MS (ESI+) m/z = 499.79 for [M+H]+. 1H-NMR (500 MHz, aceton-d6) δ = 2.43 (s, 3H), 7.43 (dt, J = 7.9, 0.6 Hz, 2H), 7.52 (m, 2H), 7.91 (dd, J = 7.9, 0.6 Hz, 1H), 7.94 (d, J = 9.1 Hz, 1H), 7.99 (m, 8H), 8.09 (dd, J = 8.5, 1.9 Hz, 1H), 8.54 (s, 1H). 13C-NMR (125.7 MHz, Acetone-d6) δ = 22.5 (CH3), 118.2 (CH), 125.9 (CH), 126.1 (C), 126.8 (CH), 127.9 (CH), 128.3 (CH), 129.5 (CH), 130.1 (CH), 130.2 (CH), 130.2 (CH), 131.2 (CH), 131.7 (CH), 133.9 (C), 135.0 (C), 135.6 (C), 139.2 (C), 146.3 (C), 147.3 (C), 152.8 (C), 164.2 (C), 165.9 (C). mp = 223–225 °C. HRMS (ESI+): m/z calcd for C27H22N3O3S2 [M+H]+: 500.1044; found: 500.1046.
General procedure 4 for the preparation of acetoxymethyl prodrug.
To a solution of the acid derivative (1 eq) in dry DMF (5 mL) under N2 flow was added triethylamine (5 eq) and bromo-methylacetate (3 eq). After stirring for 12 h, water (10 mL) was added and a precipitate occurred. The solid obtained after filtration was washed three times with water (5 mL) and further purified by semi-preparative HPLC (H2O/MeCN: from 40/60 to 0/100 in 50 min) to obtain the desired product.
Acetoxymethyl 4-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoate (34).
The reaction was carried out according to the general procedure 4, scale: 4-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoic acid (2) (0.26 mmol, 90 mg), Et3N (1.3 mmol, 183 μL), bromo-methylacetate (0.79 mmol, 78 μL), DMF (5 mL), then H2O (10 mL). After purification by semi-preparative HPLC, the product was obtained as an orange solid (0.11 mmol, 46 mg). Yield = 43%. LC-UV purity > 99%. LC-MS (ESI+) m/z = 413.60 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ = 2.11 (s, 3H), 5.91 (s, 2H), 7.73 (t, J = 8.2 Hz, 1H), 7.80 (s, 1H), 7.86 (d, J = 8.8 Hz, 2H), 7.97 (d, J = 8.8 Hz, 2H), 8.16 (ddd, J = 8.2, 2.2, 1.0 Hz, 1H), 8.40 (dt, J = 8.2, 1.0 Hz, 1H), 8.70 (t, J = 1.9 Hz, 1H), 10.94 (s, 1H).13C-NMR (125.7 MHz, DMSO-d6): δ = 20.5 (CH3), 79.4 (CH2), 107.2 (CH), 116.2 (CH), 119.9 (CH), 120.4 (C), 122.1 (CH), 130.3 (CH), 131.1 (CH), 131.9 (CH), 135.7 (C), 145.6 (C), 147.8 (C), 148.3 (C), 162.4 (C), 164.0 (C), 169.4 (C). mp = 169–171 °C. HRMS (ESI+): m/z calcd for C19H16N3O6S [M+H]+: 414.0754; found: 414.0738.
Acetoxymethyl 4-((4-(thiophen-3-yl)thiazol-2-yl)amino)benzoate (35).
The reaction was carried out according to the general procedure 4, scale: 4-((4-(thiophen-3-yl)thiazol-2-yl)amino)benzoic acid (18) (0.25 mmol, 77 mg), Et3N (1.25 mmol, 176 μL), bromo-methylacetate (0.76 mmol, 75 μL), DMF (5 mL), then H2O (10 mL). After purification by semi-preparative HPLC, the product was obtained as an off-white solid (0.04 mmol, 14 mg). Yield = 16%. LC-UV purity = 96%. LC-MS (ESI+) m/z = 374.66 for [M+H]+. 1H-NMR (500 MHz, aceton-d6) δ = 2.10 (s, 3H), 5.96 (s, 2H), 7.15 (s, 1H), 7.51 (dd, J = 5.0, 3.2 Hz, 1H), 7.60 (dd, J = 5.0, 1.3 Hz, 1H), 7.93 (dd, J = 3.2, 1.3 Hz, 1H), 7.98 (d, J = 9.0 Hz, 2H), 8.03 (d, J = 9.0 Hz, 2H), , 9.83 (s, 1H). 13C-NMR (125.7 MHz, acetone-d6) δ = 21.6 (CH3), 81.2 (CH2), 105.0 (CH), 118.3 (CH), 123.2 (C), 123.8 (CH), 127.7 (CH), 128.1 (CH), 133.1 (CH), 139.0 (C), 148.0 (C), 149.5 (C), 164.2 (C), 166.2 (C), 171.7 (C). mp = 136–138 °C. HRMS (ESI+): m/z calcd for C17H15N2O4S2 [M+H]+: 375.0387; found: 375.0388.
Acetoxymethyl 4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoate (36).
The reaction was carried out according to the general procedure 4, scale: 4-((4-(naphthalen-2-yl)thiazol-2-yl)amino)benzoic acid (19) (0.14 mmol, 50 mg), Et3N (0.7 mmol, 98 μL), bromo-methylacetate (0.43 mmol, 42 μL), DMF (5 mL), then H2O (10 mL). After purification by semi-preparative HPLC, the product was obtained as an off-white solid (0.098 mmol, 41 mg). Yield = 70%. LC-UV purity = 95%. LC-MS (ESI+) m/z = 418.61 for [M+H]+. 1H-NMR (500 MHz,aceton-d6) δ = 2.11 (s, 3H), 5.98 (s, 2H), 7.47 (s, 1H), 7.49 – 7.55 (m, 2H), 7.91 (d, J = 8.2 Hz, 1H), 7.96 (d, J = 8.8 Hz, 1H), 8.03 – 8.06 (m, 5H), 8.12 (dd, J = 8.5, 1.9 Hz, 1H), 8.59 (s, 1H), 9.90 (s, 1H). 13C-NMR (125.7 MHz, aceton-d6) δ = 21.6 (CH3), 81.2 (CH2), 106.2 (CH), 118.3 (CH), 123.3 (C), 125.9 (CH), 126.8 (CH), 127.9 (CH), 128.2 (CH), 129.5 (CH), 130.1 (CH), 130.3 (CH), 133.2 (CH), 134.04 (C), 135.1 (C), 135.7 (C), 148.0 (C), 153.2 (C), 164.3 (C), 166.2 (C), 171.1 (C). mp = 148–150 °C. HRMS (ESI+): m/z calcd for C23H19N2O4S [M+H]+: 419.0992; found: 419.0990.
Biology and modeling
Radiometric kinase assays.
CK2 radiometric kinase assay was performed as previously described in Prudent et al.77 Production and purification of GST-rhCK2α mutants were performed as described in the paper of Moucadel et al.78 and inhibitors were tested following the CK2 radiometric kinase assay procedure. ATP concentrations were 100 μM if not stated otherwise. Substrate dependent phosphorylation assays on nucleolin, GST-Six1 and CK2β auto-phosphorylation were performed following the CK2 radiometric kinase assay with CK2α2β2, the reaction was quenched by Laemmli buffer and samples were submitted to electrophoresis in NuPAGE buffer (150 V for 75 min) using pre-cast 4–12% gradient gel (Bio-Rad). Proteins were stained using Coomassie blue (InstantBlue™, Expedeon, Cambridgeshire, UK) and phosphorylation status measured using a phosphorimager PMI (BIO-RAD) and quantified with ImageJ. Nucleolin protein was purified as described previously.68 The GST C-terminal HSIX1 fusion protein was induced and purified on glutathione beads as described.79
Surface Plasmon Resonance analysis.
SPR binding studies were performed using a Reichert SR7000DC instrument optical biosensor (Reichert Technologies) equipped with a CMD500m sensor chip obtained from XanTec Bioanalytics. CK2α was immobilized by a capturing approach using a monoclonal anti-GST antibody (27 kDa, Clone GST-R 6G9 produced in rat, SAB4200055 Sigma Aldrich). Prior to use, the anti-GST antibodies were purified twice by micro dialysis at 4°C (Membrane: dialysis tubing benzoylated, 9 mm, D2272–5FT) in 10 mM sodium acetate, pH = 5. Anti-GST antibodies were immobilized using amine-coupling chemistry at 12 °C. The surfaces of all two flow cells were activated for 7 min with a 1:1 mixture of 0.1 M NHS (N-hydroxysuccinimide) and 0.1 M EDC (3-(N,N-dimethylamino) propyl-N-ethylcarbodiimide) at a flow rate of 10 μL/min. The antibody at a concentration of 30 μg/ml in 10 mM sodium acetate, pH 5.0, was immobilized at a density of 2000 RU on flow cell 2; flow cell 1 was left blank to serve as a reference surface. All the surfaces were blocked with a 3 min injection at 10 μL/min of 1 M ethanolamine, pH 8.0. GST tagged CK2α at a concentration of 50 μg/mL in 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) polysorbate 20, pH = 7.4 was captured at a density of 900 RU on flow cell 2.
Kd determination by surface plasmon resonance (SPR) analysis.
To collect kinetic binding data, analytes in 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) polysorbate 20, 1% DMSO (v/v), pH = 7.4 were injected over the two flow cells at concentrations of 3, 1.5, 0.75, 0.375, 0.187, 0.094 μM at a flow rate of 25 μl/min and at a temperature of 25 °C. The complex was allowed to associate and dissociate for 120 s. Duplicate injections (in random order) of each sample and a buffer blank were flowed over the two surfaces. Affinities were obtained after treatments (DMSO calibration, blank and references subtractions) using the software Scrubber 2.0c.
Isothermal Titration Calorimetry (iTC).
ITC experiments were performed on an Auto-iTC200 instrument (Malvern Instruments, UK) at 25 ºC. 30 μM of human CK2α1−335 in 50 mM Tris-HCl pH 8.5, 500 mM NaCl was placed into the cell. Compound 27 was used at a concentration of 300 μM and solubilized in the same buffer as CK2α1−335, with the ligand solution placed in the syringe. Each titration experiment consisted of 20 injections of 2 μL every 120 s, with stirring at 1,000 rpm. A tiny first injection of 0.2 μL injection was performed during each titration and the data omitted from analysis. DMSO was used at a final concentration of 5% (v/v). Data were fitted to a one-site model and analyzed using the MicroCal Origin software.
Cell culture.
The human kidney carcinoma cell line (786-O) was cultured in RPMI 1640 medium (Gibco) supplemented with 10% (v/v) fetal calf serum, penicillin (100 U/mL) and streptomycin (100 μg/mL) at 37 °C in 5% CO2 atmosphere. Experiments were performed in RPMI 1640 (GIBCO) supplemented with 0.1% or 0.5% (v/v) fetal serum, penicillin (100 U/mL) and streptomycin (100 μg/mL) 37 °C in 5% CO2 atmosphere.
Cell viability assay.
Cells were plated into 96-well plates at 1 × 104 cells/well. The following day, the culture medium was replaced with fresh medium containing the inhibitors at various concentrations. DMSO (0.25%) and cell free wells were used as control. After 24 h, 50 μL of (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT) solution in PBS (5 mg/mL) were added in each well and the plate incubated for 2 h at 37 °C in 5 % CO2 atmosphere. Then, 80 μL of SDS (10%) containing 0.01 M HCl were added in each well and gently stirred at RT for 4 h. Absorbance was measured at 570 nm with a microplate reader (Infinite 200 Pro, Tecan). Results are the average of a triplicate experiment with a standard deviation lower than 10%. EC50 was determined by linear interpolation after transformation to log[c] scale.
CK2 inhibition in cells.
Cells were plated into 6-well plates at 3 ˣ 105 cells/well. The following day, the culture medium was replaced with fresh medium containing the inhibitors at various concentrations or DMSO (0.5%) as reference. After incubation for 24 h, medium was removed, cells were washed with cold PBS and frozen at −80 °C. The phosphorylation status of two protein substrates of CK2 (α-catenin and Akt1) was measured by Western Blot analysis of cell extracts.
Western Blot analysis.
Primary antibodies were α-catenin (phospho-Ser641) antibody from SAB Signalway Antibody, Akt1-(phosphor-Ser129) antibody from Abgent, anti-GAPDH mAb from Ambion, Akt1, α-catenin, PARP, STAT3 and STAT3-(phosphor-Tyr705) antibodies from Cell Signaling, survivin antibody from Novus biological, p21-(phosphor-Thr145) antibody from Abcam. Secondary antibodies were peroxidase-conjugated affinity pure Goat anti-rabbit IgG (#111035003) and peroxidase-conjugated affinity pure Goat anti-mouse IgG (#115035003) from Jackson Immuno Research. Cells were lysed in RIPA buffer (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% DOC and 1 mM EDTA) containing both protease- and phosphatase-inhibitor cocktails (Sigma-Aldrich; P8340, P2850, P5726). Cell homogenates were quantified using BCA protein Assay kit (Thermo Scientific). SDS-PAGE was performed using pre-cast 4–12% gradient gel (Bio-Rad) and submitted to electrophoresis in NuPAGE buffer (150 V for 75 min). Separated proteins at 20 μg/lane were transferred to PVDF membranes (100 V for 60 min). Blotted membranes were blocked during 1 h at room temperature with saturation buffer (1% BSA in Tris Buffer Saline 10 mM, Tween 0.1% (TBST)), and then incubated with primary antibody diluted in saturation buffer, for 2 h or overnight. After 3 washes with TBST, secondary antibodies were added for 1 h. Luminata Forte Western HRP substrate (Millipore) was added and membranes were read with Fusion Fx7 (PerkinElmer). Anti-GAPDH was used as loading control. Images were analyzed and band intensities were quantified using ImageJ software.
Molecular docking procedure.
In silico docking simulations were performed using MOE2010 as previously described.80 Side chains in the putative allosteric binding pocket in PDB entry 3FWQ (chain B) were defined as flexible within a radius of 8 Å. The initial binding poses were evaluated for compatibility with the experimental data reported previously.20 Suitable poses were used to define a pharmacophore comprising the position of the naphthalenyl ring and of the benzoic acid ring within a tolerance radius of 1.3 Å, and refined through targeted re-docking using the pharmacophore definition. The final docking pose with the highest score was optimized using energy minimization of the protein–ligand complex followed by the LigX routine implemented in MOE, using the OPLS-AA forcefield.
Cell transfection and imaging.
The cDNA encoding GFP-CK2α was subcloned from a pEGFP-CK2α vector into the pMSCV-puro plasmid.81 Viral particles were produced and stable expression of EGFP-CK2α was accomplished by transfection of HeLa cells followed by 1 μg/ml puromycin–mediated selection. HeLa pEGFP-CK2α cells were seeded on Lab-Tek™ slides. After treatment with DMSO, compound 4 (40 μM), compound 27 (15 μM) or CX-4945 (8 μM) for 12 h, cells were washed twice with PBS and fixed with 4 % paraformaldehyde for 10 min at 20 °C. Nuclei were stained with Hoechst-33342 (Sigma-Aldrich) for 30 min and cells were washed with PBS (3 × 15 min). Images were acquired with a Zeiss ApoTome microscope and analyzed using Zen software. Fluorescence intensity, exposure times and post-acquisition modifications were kept identical for all the samples.
Supplementary Material
ACKNOWLEDGEMENTS
M.E. received funding from the Deutsche Forschungsgemeinschaft (DFG) (EN381/2–3). B.B., T.L. and M.L.B., received financial support from “Région Rhône-Alpes” (grant “ARC 1 Santé” 12–008707-01). B.B. received funding from the “Cancéropôle Lyon Rhône-Alpes Auvergne (CLARA)” for a fellowship “Mobilité Jeunes Chercheurs” (INCA Grant N°2011–097). The authors would like to acknowledge Heide Ford for the GST-Six1 plasmid.
ABBREVIATIONS
- AMPPNP
Adenosine-5’-[(β, γ)-imido]triphosphate
- Akt
Ak mouse thymoma
- CK2
Casein Kinase 2
- Clk
Cdc2-like kinase
- Cpd
compound
- DIPEA
diisopropylethylamine
- Dyrk
dual specificity tyrosine–regulated kinase
- EDC
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride
- EGFP
enhanced green fluorescent protein
- eIF2β
β subunit of eukaryotic translation initiation factor 2
- ErbB2
erythroblastic leukemia viral oncogene homolog 2
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GI50
half maximal growth-inhibitory concentration
- HATU
1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- ITC
isothermal titration calorimetry
- MEK1/2
mitogen-activated protein kinase/extracellular signal-regulated protein kinase 1 and 2
- p21/Waf1
21 kDa protein wild-type activating fragment-1, PARP, poly(ADP-ribose)-polymerase
- PBS
phosphate–buffered saline
- PDK1
3-phosphoinositide–dependent protein kinase-1
- Pim-1
proviral integration site for moloney murine leukemia virus 1
- Six1
sine oculis–related homeobox 1
- SPR
surface plasmon resonance
- SR
serine/arginine–rich
- STAT3
signal transducer and activator of transcription 3
- STD
saturation-transfer difference
- TPX2
targeting protein for Xklp2
- Wnt
wingless–related integration site
Footnotes
The authors declare no competing financial interest.
ANCILLARY INFORMATION
ASSOCIATED CONTENT
Suporting Information. The Supporting Information is available free of charge on the ACS Publications Website at DOI: (to be inserted by publisher)
Inhibition of GST-CK2α wild type vs. single alanine mutants by compound 27, Residual kinase activity in the presence of compound 27 (5 μM), Correlation between biochemical inhibition (IC50) and binding affinity constants, Isothermal titration calorimetry of CK2α with compound 9, Viability of the kidney cancer cell line 786-O in the presence of CX-4945, Cell growth inhibition of various cell lines by 27 vs. CX-4945, Differential effects of the allosteric inhibitor 4 and the ATP-site directed inhibitor CX-4945 on the CK2–catalyzed phosphorylation of nucleolin and cellular consequences, 1H and 13C NMR spectra for compounds 8b; 8f; 9–16; 18–27; 30–36, HPLC chromatograms and mass spectra of key compounds 12, 15, 18, 19, 22, 25 and 27, 3D coordinates from docking models, Molecular formula strings (CSV).
REFERENCES
- (1).Meggio F; Pinna LA, One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [DOI] [PubMed] [Google Scholar]
- (2).Litchfield DW, Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003, 369, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chua MM; Ortega CE; Sheikh A; Lee M; Abdul-Rassoul H; Hartshorn KL; Dominguez I, CK2 in cancer: cellular and biochemical mechanisms and potential therapeutic target. Pharmaceuticals (Basel) 2017, 10, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ruzzene M; Pinna LA, Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim. Biophys. Acta 2010, 1804, 499–504. [DOI] [PubMed] [Google Scholar]
- (5).Siddiqui-Jain A; Drygin D; Streiner N; Chua P; Pierre F; O’Brien SE; Bliesath J; Omori M; Huser N; Ho C; Proffitt C; Schwaebe MK; Ryckman DM; Rice WG; Anderes K, CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [DOI] [PubMed] [Google Scholar]
- (6).Niefind K; Guerra B; Ermakowa I; Issinger OG, Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 2001, 20, 5320–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bibby AC; Litchfield DW, The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2β. Int. J. Biol. Sci. 2005, 1, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Martel V; Filhol O; Nueda A; Cochet C, Dynamic localization/association of protein kinase CK2 subunits in living cells: a role in its cellular regulation? Ann. N. Y. Acad. Sci. 2002, 973, 272–277. [DOI] [PubMed] [Google Scholar]
- (9).Guerra B; Issinger OG, Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 1999, 20, 391–408. [DOI] [PubMed] [Google Scholar]
- (10).Cozza G; Meggio F; Moro S, The dark side of protein kinase CK2 inhibition. Curr. Med. Chem. 2011, 18, 2867–2884. [DOI] [PubMed] [Google Scholar]
- (11).Chua P; Pierre F; Whitten J, Serine-Threonine Protein Kinase and Parp Modulators [Internet] - WO/2008/028168. http://patentscope.wipo.int/search/en/WO2008028168, 2008.
- (12).Ferguson AD; Sheth PR; Basso AD; Paliwal S; Gray K; Fischmann TO; Le HV, Structural basis of CX-4945 binding to human protein kinase CK2. FEBS Lett. 2011, 585, 104–110. [DOI] [PubMed] [Google Scholar]
- (13).Kim H; Choi K; Kang H; Lee SY; Chi SW; Lee MS; Song J; Im D; Choi Y; Cho S, Identification of a novel function of CX-4945 as a splicing regulator. PLoS One 2014, 9, e94978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kim H; Lee KS; Kim AK; Choi M; Choi K; Kang M; Chi SW; Lee MS; Lee JS; Lee SY; Song WJ; Yu K; Cho S, A chemical with proven clinical safety rescues Down-syndrome-related phenotypes in through DYRK1A inhibition. Dis. Model. Mech. 2016, 9, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Srivastava A; Hirota T; Irle S; Tama F, Conformational dynamics of human protein kinase CK2α and its effect on function and inhibition. Proteins 2018, 86, 344–353. [DOI] [PubMed] [Google Scholar]
- (16).Niefind K; Issinger OG, Conformational plasticity of the catalytic subunit of protein kinase CK2 and its consequences for regulation and drug design. Biochim. Biophys. Acta 2010, 1804, 484–492. [DOI] [PubMed] [Google Scholar]
- (17).Raaf J; Brunstein E; Issinger OG; Niefind K, The CK2 alpha/CK2 beta interface of human protein kinase CK2 harbors a binding pocket for small molecules. Chem. Biol. 2008, 15, 111–117. [DOI] [PubMed] [Google Scholar]
- (18).De Fusco C; Brear P; Iegre J; Georgiou KH; Sore HF; Hyvönen M; Spring DR, A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066. Bioorg. Med. Chem. 2017, 25, 3471–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Iegre J; Brear P; De Fusco C; Yoshida M; Mitchell SL; Rossmann M; Carro L; Sore HF; Hyvönen M; Spring DR, Second-generation CK2α inhibitors targeting the αD pocket. Chem. Sci. 2018, 9, 3041–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bestgen B; Krimm I; Kufareva I; Kamal AAM; Seetoh W-G; Abell C; Hartmann RW; Abagyan R; Cochet C; Le Borgne M; Engel M; Lomberget T, 2-Aminothiazole derivatives as selective allosteric modulators of the protein kinase CK2. Part 1: Identification of an allosteric binding site. J. Med. Chem. 2018, submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lindsley JE; Rutter J, Whence cometh the allosterome? Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 10533–10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Barril X, Druggability predictions: methods, limitations, and applications. WIREs Comput. Mol. Sci. 2013, 3. [Google Scholar]
- (23).Clive DLJ; Hisaindee S; Coltart DM Derivatized amino acids relevant to native peptide synthesis by chemical ligation and acyl transfer. J. Org. Chem. 2003, 68, 9247–9254. [DOI] [PubMed] [Google Scholar]
- (24).Marzabadi MR; Wong WC; Noble SA; Desai MN Selective NPY (Y5) antagonists. US6989379B1, 2006. [Google Scholar]
- (25).Nakano S; Saito D Method for Producing Isothiocyanate Compound Having Carboxyl Group. US8367857B2, 2013. [Google Scholar]
- (26).Wilhelm A; Lopez-Garcia LA; Busschots K; Fröhner W; Maurer F; Boettcher S; Zhang H; Schulze JO; Biondi RM; Engel M 2-(3-Oxo-1,3-diphenylpropyl)malonic acids as potent allosteric ligands of the PIF pocket of phosphoinositide-dependent kinase-1: development and prodrug concept. J. Med. Chem. 2012, 55, 9817–9830. [DOI] [PubMed] [Google Scholar]
- (27).Guerra B; Bischoff N; Bdzhola VG; Yarmoluk SM; Issinger OG; Golub AG; Niefind K A Note of caution on the role of halogen bonds for protein kinase/inhibitor recognition suggested by high- and low-salt CK2α complex structures. ACS Chem. Biol. 2015, 10, 1654–1660. [DOI] [PubMed] [Google Scholar]
- (28).Lolli G; Ranchio A; Battistutta R Active form of the protein kinase CK2 α2β2 holoenzyme is a strong complex with symmetric architecture. ACS Chem. Biol. 2014, 9, 366–371. [DOI] [PubMed] [Google Scholar]
- (29).Raaf J; Issinger OG; Niefind K First inactive conformation of CK2 alpha, the catalytic subunit of protein kinase CK2. J. Mol. Biol. 2009, 386, 1212–1221. [DOI] [PubMed] [Google Scholar]
- (30).Lolli G; Pinna LA; Battistutta R Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization. ACS Chem. Biol. 2012, 7, 1158–1163. [DOI] [PubMed] [Google Scholar]
- (31).Dunn GE; Kung FL Effect of intramolecular hydrogen bonding on ionization constants of substituted salicylic acids. Can. J. Chem. 1966, 44, 1261–1269. [Google Scholar]
- (32).Li C; Benet LZ; Grillo MP Studies on the chemical reactivity of 2-phenylpropionic acid 1-O-acyl glucuronide and S-acyl-CoA thioester metabolites. Chem. Res. Toxicol. 2002, 15, 1309–1317. [DOI] [PubMed] [Google Scholar]
- (33).Regan SL; Maggs JL; Hammond TG; Lambert C; Williams DP; Park BK Acyl glucuronides: the good, the bad and the ugly. Biopharm. Drug Dispos. 2010, 31, 367–395. [DOI] [PubMed] [Google Scholar]
- (34).Cozza G; Mazzorana M; Papinutto E; Bain J; Elliott M; di Maira G; Gianoncelli A; Pagano MA; Sarno S; Ruzzene M; Battistutta R; Meggio F; Moro S; Zagotto G; Pinna LA Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem. J. 2009, 421, 387–395. [DOI] [PubMed] [Google Scholar]
- (35).Meanwell NA Synopsis of some recent tactical application of bioisosteres in drug design. J. Med. Chem. 2011, 54, 2529–2591. [DOI] [PubMed] [Google Scholar]
- (36).Bestgen B; Belaid-Choucair Z; Lomberget T; Le Borgne M; Filhol O; Cochet C In search of small molecule inhibitors targeting the flexible CK2 subunit interface. Pharmaceuticals (Basel) 2017, 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Poletto G; Vilardell J; Marin O; Pagano MA; Cozza G; Sarno S; Falqués A; Itarte E; Pinna LA; Meggio F The regulatory beta subunit of protein kinase CK2 contributes to the recognition of the substrate consensus sequence. A study with an eIF2 beta-derived peptide. Biochemistry 2008, 47, 8317–8325. [DOI] [PubMed] [Google Scholar]
- (38).Ladbury JE; Klebe G; Freire E Adding calorimetric data to decision making in lead discovery: a hot tip. Nat. Rev. Drug Discov. 2010, 9, 23–27. [DOI] [PubMed] [Google Scholar]
- (39).Davis MI; Hunt JP; Herrgard S; Ciceri P; Wodicka LM; Pallares G; Hocker M; Treiber DK; Zarrinkar PP Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1046–1051. [DOI] [PubMed] [Google Scholar]
- (40).Anastassiadis T; Deacon SW; Devarajan K; Ma H; Peterson JR Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Graczyk PP Gini Coefficient: A new way to express selectivity of kinase inhibitors against a family of kinases. J. Med. Chem. 2007, 50, 5773–5779. [DOI] [PubMed] [Google Scholar]
- (42).Battistutta R; Cozza G; Pierre F; Papinutto E; Lolli G; Sarno S; O’Brien SE; Siddiqui-Jain A; Haddach M; Anderes K; Ryckman DM; Meggio F; Pinna LA Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 2011, 50, 8478–8488. [DOI] [PubMed] [Google Scholar]
- (43).Pagano Mario A.; Bain J; Kazimierczuk Z; Sarno S; Ruzzene M; Di Maira G.; Elliott M; Orzeszko A; Cozza G; Meggio F; Pinna Lorenzo A. The selectivity of inhibitors of protein kinase CK2: an update. Biochem. J. 2008, 415, 353–365. [DOI] [PubMed] [Google Scholar]
- (44).Brodaczewska KK; Szczylik C; Fiedorowicz M; Porta C; Czarnecka AM Choosing the right cell line for renal cell cancer research. Mol. Cancer 2016, 15, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Singer EA; Gupta GN; Marchalik D; Srinivasan R Evolving therapeutic targets in renal cell carcinoma. Curr. Opin. Oncol. 2013, 25, 273–280. [DOI] [PubMed] [Google Scholar]
- (46).Roelants C; Giacosa S; Duchemin-Pelletier E; McLeer-Florin A; Tisseyre C; Aubert C; Champelovier P; Boutonnat J; Descotes JL; Rambeaud J-J; Arnoux V; Long J-A; Pasquier D; Laramas M; Kassem M; David-Boudet L; Schoutteten L; Bestgen B; Pillet C; Cochet C; Filhol O Dysregulated Expression of Protein Kinase CK2 in Renal Cancer In Protein Kinase CK2 Cellular Function in Normal and Disease States, Ahmed K; Issinger O-G; Szyszka R, Eds. Springer International Publishing: Cham, 2015; pp 241–257. [Google Scholar]
- (47).Di Maira G; Salvi M; Arrigoni G; Marin O; Sarno S; Brustolon F; Pinna LA; Ruzzene M Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005, 12, 668–677. [DOI] [PubMed] [Google Scholar]
- (48).Ji H; Wang J; Nika H; Hawke D; Keezer S; Ge Q; Fang B; Fang X; Fang D; Litchfield DW; Aldape K; Lu Z EGF-induced ERK activation promotes CK2-mediated disassociation of α-catenin from β-catenin and transactivation of β-catenin. Mol. Cell 2009, 36, 547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Knight ZA; Shokat KM Features of selective kinase inhibitors. Chem. Biol. 2005, 12, 621–637. [DOI] [PubMed] [Google Scholar]
- (50).Allende JE; Allende CC Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995, 9, 313–323. [DOI] [PubMed] [Google Scholar]
- (51).Sebolt-Leopold JS; Dudley DT; Herrera R; Van Becelaere K; Wiland A; Gowan RC; Tecle H; Barrett SD; Bridges A; Przybranowski S; Leopold WR; Saltiel AR Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999, 5, 810–816. [DOI] [PubMed] [Google Scholar]
- (52).Solit DB; Garraway LA; Pratilas CA; Sawai A; Getz G; Basso A; Ye Q; Lobo JM; She Y; Osman I; Golub TR; Sebolt-Leopold J; Sellers WR; Rosen N BRAF mutation predicts sensitivity to MEK inhibition. Nature 2006, 439, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Filhol O; Cochet C, Protein kinase CK2 in health and disease: Cellular functions of protein kinase CK2: a dynamic affair. Cell. Mol. Life Sci. 2009, 66, 1830–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lolli G; Battistutta R, Structural Basis of CK2 Regulation by Autoinhibitory Oligomerization In Protein Kinase CK2 Cellular Function in Normal and Disease States., Ahmed K; Issinger O; Szyszka R, Eds. Springer: Cham, 2015; Vol. 12, pp 35–47. [Google Scholar]
- (55).Mandal T; Bhowmik A; Chatterjee A; Chatterjee U; Chatterjee S; Ghosh MK Reduced phosphorylation of Stat3 at Ser-727 mediated by casein kinase 2 - protein phosphatase 2A enhances Stat3 Tyr-705 induced tumorigenic potential of glioma cells. Cell. Signal. 2014, 26, 1725–1734. [DOI] [PubMed] [Google Scholar]
- (56).Manni S; Brancalion A; Mandato E; Tubi LQ; Colpo A; Pizzi M; Cappellesso R; Zaffino F; Di Maggio SA; Cabrelle A; Marino F; Zambello R; Trentin L; Adami F; Gurrieri C; Semenzato G; Piazza F Protein kinase CK2 inhibition down modulates the NF-κB and STAT3 survival pathways, enhances the cellular proteotoxic stress and synergistically boosts the cytotoxic effect of bortezomib on multiple myeloma and mantle cell lymphoma cells. PLoS One 2013, 8, e75280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Piazza FA; Ruzzene M; Gurrieri C; Montini B; Bonanni L; Chioetto G; Di Maira G; Barbon F; Cabrelle A; Zambello R; Adami F; Trentin L; Pinna LA; Semenzato G Multiple myeloma cell survival relies on high activity of protein kinase CK2. Blood 2006, 108, 1698–1707. [DOI] [PubMed] [Google Scholar]
- (58).Li D; Dobrowolska G; Krebs EG The physical association of casein kinase 2 with nucleolin. J. Biol. Chem. 1996, 271, 15662–15668. [DOI] [PubMed] [Google Scholar]
- (59).Pfaff M; Anderer FA Casein kinase II accumulation in the nucleolus and its role in nucleolar phosphorylation. Biochim. Biophys. Acta 1988, 969, 100–109. [DOI] [PubMed] [Google Scholar]
- (60).Schwab MS; Dreyer C Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. Eur. J. Cell Biol. 1997, 73, 287–297. [PubMed] [Google Scholar]
- (61).Engen JR; Wales TE; Hochrein JM; Meyn MA; Banu Ozkan S; Bahar I; Smithgall TE Structure and dynamic regulation of Src-family kinases. Cell. Mol. Life Sci. 2008, 65, 3058–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Shan Y; Seeliger MA; Eastwood MP; Frank F; Xu H; Jensen M; Dror RO; Kuriyan J; Shaw DE A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Marino KA; Sutto L; Gervasio FL The effect of a widespread cancer-causing mutation on the inactive to active dynamics of the B-Raf kinase. J. Am. Chem. Soc. 2015, 137, 5280–5283. [DOI] [PubMed] [Google Scholar]
- (64).Lovera S; Morando M; Pucheta-Martinez E; Martinez-Torrecuadrada JL; Saladino G; Gervasio FL Towards a molecular understanding of the link between imatinib resistance and kinase conformational dynamics. PLoS Comput. Biol. 2015, 11, e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Emmanouilidi A; Falasca M 3-Phosphoinositide-Dependent Kinase 1 (PDK1) In Encyclopedia of Signaling Molecules, Choi S, Ed. Springer New York: New York, NY, 2017; pp 1–4. [Google Scholar]
- (66).Cyphers S; Ruff EF; Behr JM; Chodera JD; Levinson NM A water-mediated allosteric network governs activation of Aurora kinase A. Nat. Chem. Biol. 2017, 13, 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Bayliss R; Sardon T; Vernos I; Conti E Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 2003, 12, 851–862. [DOI] [PubMed] [Google Scholar]
- (68).Bonnet H; Filhol O; Truchet I; Brethenou P; Cochet C; Amalric F; Bouche G Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J. Biol. Chem. 1996, 271, 24781–24787. [DOI] [PubMed] [Google Scholar]
- (69).Gottlieb HE; Kotlyar V; Nudelman A NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 1997, 62, 7512–7515. [DOI] [PubMed] [Google Scholar]
- (70).Pravst I; Zupan M; Stavber S Halogenation of ketones with N-halosuccinimides under solvent-free reaction conditions. Tetrahedron 2008, 64, 5191–5199. [Google Scholar]
- (71).Kumar RS; Kulangiappar K; Kulandainathan MA Convenient electrochemical method for the synthesis of α-bromo alkyl aryl ketones. Synth. Comm. 2010, 40, 1736–1742. [Google Scholar]
- (72).Borowitz IJ; Parnes H Kinetics and mechanism of the reaction of triphenylphosphine with alpha-haloacetophenones. J. Org. Chem. 1967, 32, 3560–3564. [Google Scholar]
- (73).Katritzky AR; Kirichenko N; Rogovoy BV; Kister J; Tao H Synthesis of mono- and N,N-disubstituted thioureas and N-acylthioureas. Synthesis 2004, 11, 1799–1805. [Google Scholar]
- (74).Guha-Sircar S; Patnaik; K. K Preparation of antibacterials from organomercurials. J. Indian Chem. Soc. 1950, 27, 535–538. [Google Scholar]
- (75).Hay MP; Turcotte S; Flanagan JU; Bonnet M; Chan DA; Sutphin PD; Nguyen P; Giaccia AJ; Denny WA 4-Pyridylanilinothiazoles that selectively target von Hippel−Lindau deficient renal cell carcinoma cells by inducing autophagic cell death. J. Med. Chem. 2010, 53, 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Seligman RB; Bost RW; McKee RL Some derivatives of p-aminosalicylic acid. J. Am. Chem. Soc. 1953, 75, 6334–6335. [Google Scholar]
- (77).Prudent R; Moucadel V; Laudet B; Barette C; Lafanechère L; Hasenknopf B; Li J; Bareyt S; Lacôte E; Thorimbert S; Malacria M; Gouzerh P; Cochet C Identification of polyoxometalates as nanomolar noncompetitive inhibitors of protein kinase CK2. Chem. Biol. 2008, 15, 683–692. [DOI] [PubMed] [Google Scholar]
- (78).Moucadel V; Prudent R; Sautel CF; Teillet F; Barette C; Lafanechere L; Receveur-Brechot V; Cochet C Antitumoral activity of allosteric inhibitors of protein kinase CK2. Oncotarget 2011, 2, 997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Ford HL; Landesman-Bollag E; Dacwag CS; Stukenberg PT; Pardee AB; Seldin DC Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 2000, 275, 22245–22254. [DOI] [PubMed] [Google Scholar]
- (80).ElHady AK; Abdel-Halim M; Abadi AH; Engel M Development of selective Clk1 and −4 inhibitors for cellular depletion of cancer-relevant proteins. J. Med. Chem. 2017, 60, 5377–5391. [DOI] [PubMed] [Google Scholar]
- (81).Filhol O; Nueda A; Martel V; Gerber-Scokaert D; Benitez MJ; Souchier C; Saoudi Y; Cochet C Live-cell fluorescence imaging reveals the dynamics of protein kinase CK2 individual subunits. Mol. Cell. Biol. 2003, 23, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.