Abstract
Background
Numerous investigations have previously evaluated the association of interleukin (IL) 4 gene polymorphisms and the risk of asthma, conferring inconsistent results. To resolve the incongruent outcomes yielded from different single studies, we conducted the most up-to-date meta-analysis of IL4 gene −589C/T (rs2243250) polymorphism and susceptibility to asthma.
Methods
A systematic literature search was performed in ISI web of science, Scopus, Medline/PubMed databases prior to September 2020, and the pooled odds ratio (OR) and their corresponding 95% CI were calculated to determine the association strength.
Results
Literature search led to retrieving of 49 publications (55 case-control studies) containing 9572 cases and 9881 controls. It was revealed that IL4 gene −589C/T polymorphism increased the risk of asthma across all genetic models, including dominant model (OR = 1.22), recessive model (OR = 1.17), allelic model (OR = 1.21), and TT vs. CC model (OR = 1.34), but not the CT vs. TT model. The subgroup analysis by age indicated that IL4 gene -589C/T polymorphism was significantly associated with asthma risk in both pediatrics and adults. Additionally, the subgroup analysis by ethnicity revealed significant association in Asian, American, and Europeans. Finally, subgroup analysis by East Asian and non-East Asian populations indicated significant associations.
Conclusions
The current meta-analysis revealed that IL4 gene -589C/T polymorphism was a susceptibility risk in both pediatrics and adults in the whole and different ethnic groups.
Keywords: Asthma, Interleukin 4, Polymorphism, Meta-analysis, Genetic susceptibility
Background
Asthma is one of the most common atopic disorders of the respiratory tract, which results in wheezing, coughing, breathlessness, and bronchial obstruction [1]. The prevalence and incidence of asthma raised regularly and it estimated more than 300 million persons of the world are affected by this disease [2]. The main causes of asthma are not completely clear. However, is has been postulated that asthma is mediated by interactions between specific external stimulating factors, including pollutants, viral and bacterial infections, allergens, tobacco smokes, etc., and genetics of the patients. Additionally, increasing number of studies recommend that genetic factors play a critical role in the etiology of asthma by their interactions with the environmental elements [3, 4]. The heritability of asthma is estimated to be 35 to 95% [5]. Numerous studies have examined the correlation between genetic variations of pro and anti-inflammatory genes and susceptibility to asthma [6, 7]. In recent decades, single nucleotide polymorphisms (SNP) have become one of the frequently studied models of DNA variation in analyses of the association between genetics and susceptibility to disease [8, 9].
The role of immunological factors especially cytokines in modulating and controlling the inflammatory response of the respiratory tracts is essential in the evolution, progression, and exacerbations of asthma [10]. Interleukin (IL)-4 is a key ingredient of the immune system required in the regulation of response to an allergen through controlling the isotype switching of antibody in B lymphocytes to IgG and IgE class [11]. Elevated serum levels of IgE are suggestive of allergic reactions and resemble a high level of IL-4 mRNA assembly [12]. Moreover, it acts as a growth factor to facilitate the differentiation of T helper (Th) 2 cells and mast cells. These characteristics of IL-4 accentuate on the crucial roles of cytokines in the pathogenesis asthma [13, 14]. Additionally, IL4 gene polymorphisms, like promoter region (C + 33 T) SNP [15], and 3017 G/T SNP in intron 2 [16], have been associated with IgE levels, which might be involved in the pathogenesis of asthma.
The IL4 gene is located on chromosome 5q31 [17]. The -589C/T (rs2243250) polymorphism has been recognized on upstream of the transcription initiation site [18]. It has been demonstrated that the binding of a transcription factor is enhanced by the appearance of the polymorphic T allele that may result in an overexpression of the IL4 gene and, thus, raising the power of any immunological response that dependents on IL-4 [19]. To date, many studies have examined the association between IL4 gene -589 C/T polymorphisms and the risk of asthma, but their outcomes have not been consistent. Therefore, we performed this meta-analysis to analyze the relationship between the -589C/T polymorphisms and susceptibility to asthma.
Methods
This study conducted in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement including; literature review, study selection, inclusion and exclusion criteria, data extraction and quality assessment, and statistical analysis [20]. No ethics committee confirmation was necessary for this meta-analysis, which does not contain any studies with human participants or animals performed by any of the authors.
Literature review
A comprehensive search was performed in the ISI web of science, Scopus, Medline/PubMed databases to retrieve published articles prior to September 2020. The following main key words and Medical Subject Headings (Mesh) were searched: (“asthma” [Mesh] OR “asthmatic”) AND (“interleukin-4” OR “IL-4” OR “rs2243250”) AND (“single nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”). No restrictions were placed on language, sample size, population or publication date.
Study selection
The retrieved publications by primary literature search were imported into Endnote X8 software. The duplicate studies were removed and title and abstract of remain studies were reviewed by two investigators. Full-text verification was performed if we could not categorize studies based on title and abstract. Any disagreements during study selection was discussed and resolved by consensus.
Inclusion and exclusion criteria
The following inclusion criteria were used to distinguish eligible studies: i) studies with distinct case and control group evaluating the association between IL-4 C589T polymorphism and susceptibility to asthma; ii) studies with calculable or extractable data for odds ratio (OR) and 95% confidence intervals (CIs); iii) studies with sufficient data for alleles and genotypes in case and control group. The duplicates, reviews, book chapters, and meta-analysis were excluded. The application of these criteria results in 49 qualified studies for the meta-analysis.
Data extraction and quality assessment
Two of our authors independently and according to an extraction checklist extracted the following data: the first author, journal and year of publication, country of origin, ethnicity, number of subjects in the case and the control groups for each gender, mean or range of age, genotyping method, genotype counts in the case and the control group. The quality of each study was assessed using the Newcastle-Ottawa Scale (NOS) criteria [21]. Studies with scores 0–3, 4–6 or 7–9 were low, moderate or high-quality, respectively.
Statistical analysis
Statistical analyses were carried out using STATA (version 14.0; Stata Corporation, College Station, TX) and SPSS (version 23.0; SPSS, Inc. Chicago, IL). The strength of association between polymorphism and asthma susceptibility was estimated by odd ratios (ORs) and 95% confidence intervals (CIs) for the dominant model, recessive model, allele contrasts, and additive comparison. Heterogeneity among included studies was measured via Q statistics (P value< 0.1 considered statistically significant) and I2-test (I2 values of 25, 50 and 75% were described as low, moderate, and high heterogeneity, respectively). In the presence of heterogeneity random effect model (REM) was used, however fixed effect model (FEM) was applied in homogeneous condition [22, 23]. In order to assessed the predefined sources of heterogeneity among included studies, subgroup analysis and meta-regression analysis based on year of population, the continent of the study population, and genotyping method were performed. The genotypic frequency distribution in the controls was checked for consistency of the Hardy– Weinberg equilibrium (HWE). Furthermore, publication bias was computed by the Begg’s and Egger’s test and visual examination of the funnel plot (P value< 0.05 considered statistically significant) [24, 25]. Additionally, to calculate overall effect size in absence of each study, a sensitivity analysis was conducted.
Results
Search results and characteristics of the selected studies
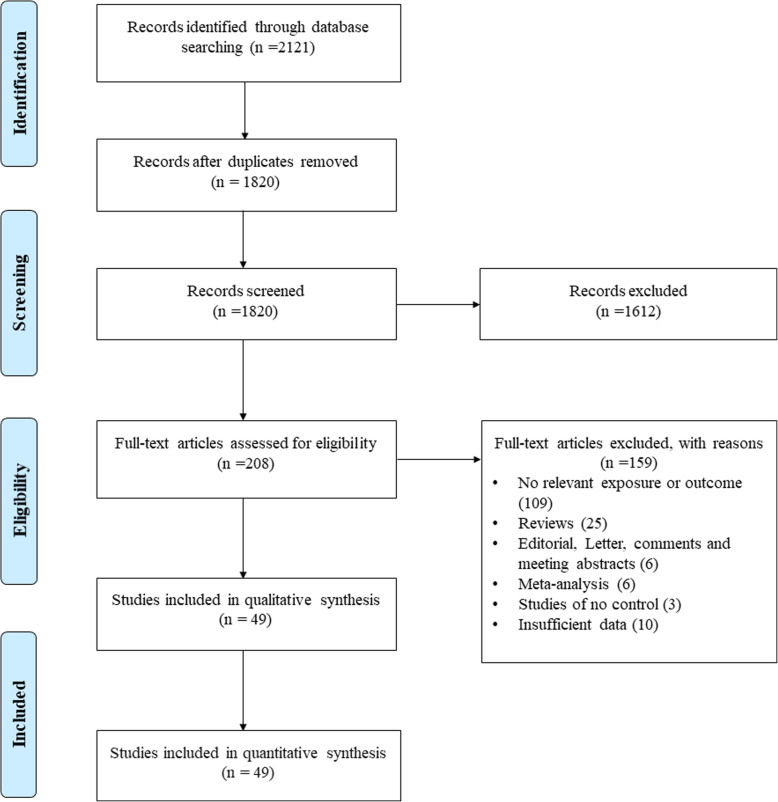
Our primary search retrieved 2121 potential articles. After removing of duplicate articles (n = 301), 1820 articles remain for abstract and full-text screening. Of 1820 articles, 1612 were excluded base on title and abstract and 159 articles based on full-text reading. Ultimately 49 publications with 9579 cases and 9881 controls met the inclusion criteria and their data were extracted for meta-analysis. Among these 49 publications, four of them, including Basehore et al. [16], Donfack et al. [26], Zhang et al. [27], and Baye et al. [28] examined two or three different populations with separate case and control; therefore, we assumed them as 9 case-control studies collectively (55 studies). The detailed information on study selection process is illustrated in Fig. 1, Tables 1, and 2.
Fig. 1.
Flow diagram of study selection process
Table 1.
Characteristics of studies included in meta-analysis of overall asthma
| Study author | Year | Country | Ethnicity 1 (Continent) | Ethnicity 2 | Ethnicity 3 | Age group | Total cases/control | Genotyping method | Quality Score |
|---|---|---|---|---|---|---|---|---|---|
| Walley et al. [29] | 1996 | UK | Europe | non East-Asian | Caucasian | Pediatric | 124 / 59 | PCR-RFLP | 6 |
| Hijazi et al. [30] | 2000 | Kuwait | Asia | non East-Asian | Arab | Mixed | 84 / 100 | PCR-RFLP | 6 |
| Sandford et al. [31] | 2000 | New Zealand | Europe | non East-Asian | Caucasian | Adult | 233 / 143 | PCR-RFLP | 7 |
| Takabayashi et al. [32] | 2000 | Japan | Asia | East-Asian | Caucasian | Pediatric | 100 / 100 | PCR-RFLP | 6 |
| Hakonarson et al. [33] | 2001 | Iceland | Europe | non East-Asian | Caucasian | Mixed | 94 / 94 | PCR | 6 |
| Cui et al. [34] | 2003 | China | Asia | East-Asian | Caucasian | Mixed | 241 / 175 | PCR-RFLP | 7 |
| Basehore et al. (i) [16] | 2004 | USA | America | non East-Asian | African American | Adult | 233 / 245 | PCR | 7 |
| Basehore et al. (ii) [16] | 2004 | USA | America | non East-Asian | African American | Adult | 168 / 269 | PCR | 7 |
| Basehore et al. (iii) [16] | 2004 | USA | America | non East-Asian | African American | Adult | 116 / 130 | PCR | 6 |
| Lee et al. [35] | 2004 | Korea | Asia | East-Asian | Caucasian | Pediatric | 254 / 100 | PCR-RFLP | 6 |
| Park et al. [36] | 2004 | Korea | Asia | East-Asian | Caucasian | Mixed | 532 / 170 | SNaPshot | 8 |
| Wang et al. [37] | 2004 | China | Asia | East-Asian | Caucasian | Adult | 93 / 62 | PCR-RFLP | 6 |
| Adjers et al. [38] | 2004 | Finland | Europe | non East-Asian | Caucasian | Adult | 243 / 401 | PCR-RFLP | 7 |
| Donfack et al. (i) [26] | 2005 | USA | America | non East-Asian | African American | Mixed | 126/ 205 | LAS | 6 |
| Donfack et al. (ii) [26] | 2005 | USA | America | non East-Asian | African American | Mixed | 205 / 183 | LAS | 7 |
| Zhang et al. (i) [27] | 2005 | China | Asia | East-Asian | Caucasian | Adult | 152 / 157 | PCR-RFLP | 6 |
| Zhang et al. (ii) [27] | 2005 | Malaysia | Asia | East-Asian | Caucasian | Adult | 76 / 100 | PCR-RFLP | 6 |
| Zhang et al. (iii) [27] | 2005 | India | Asia | non East-Asian | Caucasian | Adult | 87 / 103 | PCR-RFLP | 6 |
| Gervaziev et al. [39] | 2006 | Russia | Europe | non East-Asian | Caucasian | Adult | 109 / 68 | PCR-RFLP | 6 |
| Schubert et al. [40] | 2006 | Germany | Europe | non East-Asian | Caucasian | Pediatric | 231 / 270 | PCR-RFLP | 7 |
| Kabesch et al. [41] | 2006 | Germany | Europe | non East-Asian | Caucasian | Pediatric | 73 / 773 | PCR-RFLP | 6 |
| Battle et al. [42] | 2007 | USA | America | non East-Asian | African American | Mixed | 255 / 175 | PCR-RFLP | 6 |
| Hosseini-Farahabadi et al. [43] | 2007 | Iran | Asia | non East-Asian | Caucasian | Adult | 30 / 50 | PCR-RFLP | 5 |
| Kamali-Sarvestani et al. [44] | 2007 | Iran | Asia | non East-Asian | Caucasian | Adult | 149 / 112 | PCR-RFLP | 6 |
| Chiang et al. [45] | 2007 | China | Asia | East-Asian | Caucasian | Adult | 167 / 111 | PCR-RFLP | 6 |
| Mak et al. [46] | 2007 | China | Asia | East-Asian | Caucasian | Adult | 289 / 292 | PCR-RFLP | 7 |
| Attab et al. [47] | 2008 | Jordan | Asia | non East-Asian | Arab | Pediatric | 40 / 40 | PCR-RFLP | 5 |
| De Faria et al. [48] | 2008 | Brazil | America | non East-Asian | Caucasian | Pediatric | 88 / 202 | PCR-RFLP | 6 |
| Jiang et al. [49] | 2009 | China | Asia | East-Asian | Caucasian | Adult | 13 / 13 | PCR-RFLP | 5 |
| Amirzargar et al. [50] | 2009 | Iran | Asia | non East-Asian | Caucasian | Mixed | 59 / 139 | PCR-RFLP | 6 |
| Daley et al. [51] | 2009 | Australia | Oceania | non East-Asian | Caucasian | Mixed | 644 / 751 | Illumina Bead array system | 8 |
| Haller et al. [52] | 2009 | USA | America | non East-Asian | African American | Adult | 72 / 70 | PCR-RFLP | 6 |
| Rad et al. [53] | 2010 | Iran | Asia | non East-Asian | Caucasian | Adult | 64 / 65 | PCR-RFLP | 6 |
| Wu et al. [54] | 2010 | China | Asia | East-Asian | Caucasian | Pediatric | 252 / 227 | PCR-RFLP | 7 |
| Beghe et al. [55] | 2010 | UK and Italy | Europe | non East-Asian | Caucasian | Mixed | 299 / 176 | PCR-RFLP | 7 |
| Bijanzadeh et al. [56] | 2010 | India | Asia | non East-Asian | Caucasian | Mixed | 100 / 50 | PCR-RFLP | 6 |
| Fance et al. [57] | 2010 | China | Asia | East-Asian | Caucasian | Adult | 62 / 30 | PCR-RFLP | 6 |
| Baye et al. (i) [28] | 2011 | USA | America | non East-Asian | African American | Pediatric | 413 / 298 | Illumina GoldenGate Assay system | 7 |
| Baye et al. (ii) [28] | 2011 | USA | America | non East-Asian | African American | Pediatric | 315 / 51 | Illumina GoldenGate Assay system | 6 |
| Daneshmandi et al. [58] | 2011 | Iran | Asia | non East-Asian | Caucasian | Adult | 81 / 124 | PCR-RFLP | 7 |
| Huang et al. [59] | 2011 | China | Asia | East-Asian | Caucasian | Pediatric | 100 / 122 | PCR-RFLP | 6 |
| Hwang et al. [60] | 2012 | China | Asia | East-Asian | Caucasian | Pediatric | 188 / 376 | PCR-RFLP | 7 |
| Chiang et al. [61] | 2012 | China | Asia | East-Asian | Caucasian | Adult | 452 / 106 | PCR-RFLP | 6 |
| Micheal et al. [62] | 2013 | Pakistan | Asia | non East-Asian | Caucasian | Mixed | 108 / 120 | PCR-RFLP | 6 |
| Ricciardolo et al. [63] | 2013 | Italy | Europe | non East-Asian | Caucasian | Mixed | 57 / 124 | PCR-SSP | 6 |
| Smolnikova et al. [64] | 2013 | Russia | Europe | non East-Asian | Caucasian | Mixed | 64 / 50 | PCR-RFLP | 6 |
| Li et al. [65] | 2014 | China | Asia | East-Asian | Caucasian | Pediatric | 491 / 503 | PCR-LDR | 7 |
| Wang et al. [66] | 2015 | China | Asia | East-Asian | Caucasian | Mixed | 392 / 849 | Mass array | 7 |
| Dahmani et al. [67] | 2016 | Algeria | Africa | non East-Asian | Arab | Adult | 44 / 19 | PCR-RFLP | 6 |
| Li et al. [68] | 2016 | China | Asia | East-Asian | Caucasian | Pediatric | 317 /351 | PCR and Sequencing | 7 |
| Narozna et al. [69] | 2016 | Poland | Europe | non East-Asian | Caucasian | Mixed | 177 / 189 | Taq Man | 7 |
| Zhang et al. [68] | 2016 | China | Asia | East-Asian | Caucasian | Pediatric | 38 / 35 | PCR and Sequencing | 6 |
| Hussein et al. [70] | 2017 | Iraq | Asia | non East-Asian | Arab | Mixed | 48 / 25 | ARMS-PCR | 6 |
| Abood et al. [71] | 2018 | Iraq | Asia | non East-Asian | Arab | Mixed | 100 / 100 | AS-PCR | 6 |
| Zhang et al. [72] | 2019 | China | Asia | East-Asian | Caucasian | Pediatric | 37 / 29 | PCR and Sequencing | 5 |
Table 2.
Distribution of genotype and allele among asthma patients and controls
| Study author | Asthma cases | Healthy control | P-HWE | MAF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | CC | CT | TT | C | T | |||
| Walley et al. [29] | 56 | 55 | 13 | 167 | 81 | 31 | 23 | 5 | 85 | 33 | 0/8 | 0/72 |
| Hijazi et al. [30] | 5 | 25 | 54 | 35 | 133 | 9 | 31 | 60 | 49 | 151 | 0/1 | 0/245 |
| Sandford et al. [31] | 146 | 78 | 9 | 370 | 96 | 100 | 41 | 2 | 241 | 45 | 0/33 | 0/842 |
| Takabayashi et al. [32] | 6 | 43 | 51 | 55 | 145 | 10 | 39 | 51 | 59 | 141 | 0/53 | 0/295 |
| Hakonarson et al. [33] | 73 | 20 | 1 | 166 | 22 | 67 | 25 | 2 | 159 | 29 | 0/85 | 0/845 |
| Cui et al. [34] | 11 | 89 | 141 | 111 | 371 | 9 | 52 | 114 | 70 | 280 | 0/34 | 0/2 |
| Basehore et al. (i) [16] | 153 | 72 | 8 | 378 | 88 | 181 | 59 | 5 | 421 | 69 | 0/94 | 0/859 |
| Basehore et al. (ii) [16] | 22 | 77 | 69 | 121 | 215 | 29 | 119 | 121 | 177 | 361 | 0/97 | 0/329 |
| Basehore et al. (iii) [16] | 43 | 55 | 18 | 141 | 91 | 55 | 59 | 16 | 169 | 91 | 0/97 | 0/65 |
| Lee et al. [35] | 9 | 77 | 168 | 95 | 413 | 3 | 29 | 68 | 35 | 165 | 0/96 | 0/175 |
| Park et al. [36] | 19 | 164 | 349 | 202 | 862 | 7 | 54 | 109 | 68 | 272 | 0/92 | 0/2 |
| Wang et al. [37] | 29 | 42 | 22 | 100 | 86 | 21 | 26 | 15 | 68 | 56 | 0/22 | 0/548 |
| Adjers et al. [38] | 106 | 103 | 34 | 315 | 171 | 189 | 164 | 48 | 542 | 260 | 0/18 | 0/675 |
| Donfack et al. (i) [26] | 85 | 34 | 7 | 204 | 48 | 144 | 55 | 6 | 343 | 67 | 0/78 | 0/836 |
| Donfack et al. (ii) [26] | 25 | 82 | 98 | 132 | 278 | 24 | 82 | 77 | 130 | 236 | 0/76 | 0/355 |
| Zhang et al. (i) [27] | 4 | 47 | 101 | 55 | 249 | 3 | 45 | 109 | 51 | 263 | 0/5 | 0/162 |
| Zhang et al. (ii) [27] | 11 | 35 | 30 | 57 | 95 | 16 | 43 | 41 | 75 | 125 | 0/4 | 0/375 |
| Zhang et al. (iii) [27] | 50 | 31 | 6 | 131 | 43 | 66 | 30 | 7 | 162 | 44 | 0/17 | 0/786 |
| Gervaziev et al. [39] | 16 | 75 | 18 | 107 | 111 | 18 | 43 | 7 | 79 | 57 | 0/01 | 0/58 |
| Schubert et al. [40] | 143 | 78 | 10 | 364 | 98 | 189 | 74 | 7 | 452 | 88 | 0/93 | 0/837 |
| Kabesch et al. [41] | 42 | 29 | 2 | 113 | 33 | 564 | 188 | 21 | 1316 | 230 | 0/26 | 0/851 |
| Battle et al. [42] | 28 | 113 | 114 | 169 | 341 | 19 | 77 | 79 | 115 | 235 | 0/97 | 0/328 |
| Hosseini-Farahabadi et al. [43] | 17 | 8 | 5 | 42 | 18 | 38 | 12 | 0 | 88 | 12 | 0/33 | 0/88 |
| Kamali-Sarvestani et al. [44] | 139 | 6 | 4 | 284 | 14 | 93 | 18 | 1 | 204 | 20 | 0/9 | 0/91 |
| Chiang et al. [45] | 1 | 19 | 147 | 21 | 313 | 7 | 34 | 70 | 48 | 174 | 0/31 | 0/216 |
| Mak et al. [46] | 15 | 95 | 179 | 125 | 453 | 19 | 87 | 186 | 125 | 459 | 0/05 | 0/214 |
| Attab et al. [47] | 31 | 9 | 0 | 71 | 9 | 33 | 7 | 0 | 73 | 7 | 0/54 | 0/912 |
| De Faria et al. [48] | 38 | 41 | 9 | 117 | 59 | 67 | 108 | 27 | 242 | 162 | 0/1 | 0/599 |
| Jiang et al. [49] | 0 | 8 | 5 | 8 | 18 | 1 | 9 | 3 | 11 | 15 | 0/13 | 0/423 |
| Amirzargar et al. [50] | 0 | 59 | 0 | 59 | 59 | 10 | 129 | 0 | 149 | 129 | < 0.001 | 0/535 |
| Daley et al. [51] | 476 | 155 | 13 | 1107 | 181 | 549 | 186 | 16 | 1284 | 218 | 0/95 | 0/854 |
| Haller et al. [52] | 6 | 30 | 36 | 42 | 102 | 7 | 31 | 32 | 45 | 95 | 0/89 | 0/321 |
| Rad et al. [53] | 46 | 18 | 0 | 110 | 18 | 42 | 23 | 0 | 107 | 23 | 0/08 | 0/823 |
| Wu et al. [54] | 6 | 83 | 163 | 95 | 409 | 11 | 84 | 132 | 106 | 348 | 0/61 | 0/233 |
| Beghe et al. [55] | 232 | 63 | 4 | 527 | 71 | 136 | 37 | 3 | 309 | 43 | 0/79 | 0/877 |
| Bijanzadeh et al. [56] | 92 | 4 | 4 | 188 | 12 | 48 | 1 | 1 | 97 | 3 | < 0.001 | 0/97 |
| Fance et al. [57] | 38 | 13 | 11 | 89 | 35 | 27 | 1 | 2 | 55 | 5 | < 0.001 | 0/916 |
| Baye et al. (i) [28] | 267 | 130 | 16 | 664 | 162 | 233 | 61 | 4 | 527 | 69 | 0/99 | 0/884 |
| Baye et al. (ii) [28] | 35 | 140 | 140 | 210 | 420 | 12 | 25 | 14 | 49 | 53 | 0/89 | 0/48 |
| Daneshmandi et al. [58] | 63 | 15 | 3 | 141 | 21 | 94 | 26 | 4 | 214 | 34 | 0/2 | 0/862 |
| Huang et al. [59] | 1 | 19 | 80 | 21 | 179 | 4 | 43 | 75 | 51 | 193 | 0/46 | 0/209 |
| Hwang et al. [60] | 1 | 51 | 136 | 53 | 323 | 12 | 89 | 275 | 113 | 639 | 0/15 | 0/15 |
| Chiang et al. [61] | 13 | 110 | 329 | 136 | 768 | 7 | 34 | 65 | 48 | 164 | 0/38 | 0/226 |
| Micheal et al. [62] | 26 | 63 | 19 | 115 | 101 | 31 | 84 | 5 | 146 | 94 | < 0.001 | 0/608 |
| Ricciardolo et al. [63] | 35 | 19 | 3 | 89 | 25 | 109 | 12 | 3 | 230 | 18 | < 0.001 | 0/927 |
| Smolnikova et al. [64] | 36 | 28 | 0 | 100 | 28 | 39 | 11 | 0 | 89 | 11 | 0/38 | 0/89 |
| Li et al. [65] | 17 | 150 | 324 | 184 | 798 | 21 | 144 | 338 | 186 | 820 | 0/26 | 0/184 |
| Wang et al. [66] | 50 | 177 | 165 | 277 | 507 | 104 | 412 | 333 | 620 | 1078 | 0/17 | 0/365 |
| Dahmani et al. [67] | 13 | 19 | 12 | 45 | 43 | 6 | 11 | 2 | 23 | 15 | 0/35 | 0/605 |
| Li et al. [68] | 112 | 0 | 205 | 224 | 410 | 138 | 0 | 213 | 276 | 426 | < 0.001 | 0/393 |
| Narozna et al. [69] | 117 | 55 | 5 | 289 | 65 | 133 | 53 | 3 | 319 | 59 | 0/37 | 0/843 |
| Zhang et al. [68] | 8 | 11 | 19 | 27 | 49 | 17 | 13 | 5 | 47 | 23 | 0/34 | 0/671 |
| Hussein et al. [70] | 42 | 5 | 1 | 89 | 7 | 8 | 13 | 4 | 29 | 21 | 0/73 | 0/58 |
| Abood et al. [71] | 66 | 17 | 17 | 149 | 51 | 7 | 90 | 3 | 104 | 96 | < 0.001 | 0/52 |
| Zhang et al. [72] | 7 | 13 | 17 | 27 | 47 | 11 | 15 | 3 | 37 | 21 | 0/51 | 0/637 |
P-HWE p-value for Hardy–Weinberg equilibrium, MAF minor allele frequency of control group
Meta-analysis of IL-4 SNP (C-589 T) and the risk of asthma
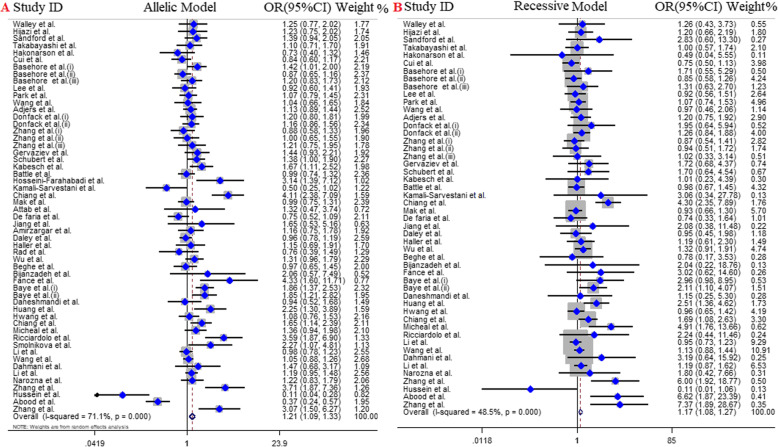
Overall, 55 studies with 9572 cases and 9881 controls included in quantitative analysis of the association between IL-4 gene -589C/T polymorphism and the risk of asthma. Of those, 11 articles were conducted in European countries [29, 31, 33, 38–41, 55, 63, 64, 69], 32 articles were in Asian countries [27, 30, 32, 34–37, 43–46, 49, 50, 53, 54, 56–62, 65, 66, 68, 70–73], 10 articles in American countries [16, 26, 28, 42, 48, 52] and one article in each Algeria [67] and Australia country [51]. The analysis of overall population revealed the significant positive association between IL4 gene -589C/T polymorphism and the risk of asthma across all genetic models; including dominant model (OR = 1.22, 95% CI = 1.04–1.44, P = 0.01, REM), recessive model (OR = 1.17, 95% CI = 1.08–1.27, P < 0.001, FEM), allelic model (OR = 1.21, 95% CI = 1.09–1.33, P < 0.001, REM), and TT vs. CC model (OR = 1.34, 95% CI = 1.18–1.52, P < 0.001, FEM), except CT vs. TT model (OR = 1.13, 95% CI = 0.95–1.34, P = 0.17, REM) (Fig. 2). Additionally, along with subgroup analysis based on age we stratified the analysis by ethnicity in three conditions.
Fig. 2.
Pooled OR and 95% CI of individual studies and pooled data for the association between Il-4 C589T polymorphism and asthma risk in; a allelic model, b recessive Model
Subgroup analysis by age
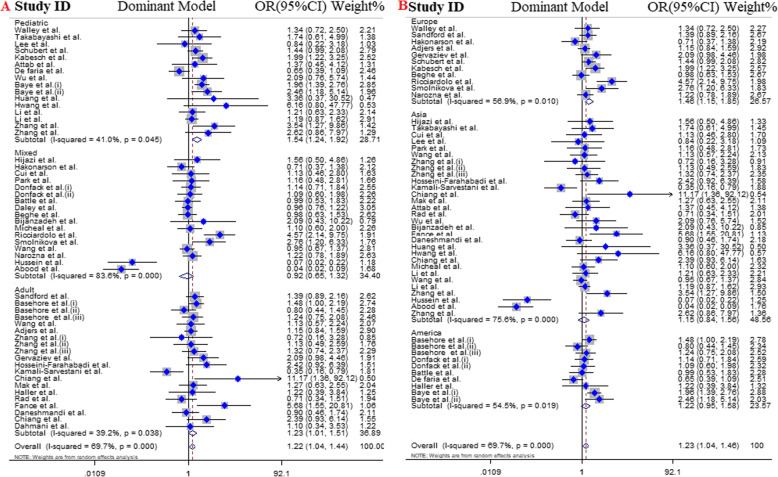
We stratified eligible articles into three groups including: pediatrics (16 articles), adults (21 articles) and mixed (cover both range;18 articles). The results highlighted a predisposing role of IL4 gene -589C/T polymorphism for the asthma risk in pediatrics and adults under all genotype models. However, no significant association was detected in mixed group (Table 3, Fig. 3).
Table 3.
Main results of pooled ORs in meta-analysis of IL-4 gene polymorphisms in asthmatic patients
| Subgroup | Sample size | Test of association | Test of heterogeneity | Test of publication bias (Begg’s test) | Test of publication bias (Egger’s test) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genetic model | Case/Control | OR | 95% CI (p-value) | I2 (%) | P | z | P | t | P | |
| Overall | Dominant model | 9579 / 9881 | 1.22 | 1.04–1.44 (0.01) | 69.7 | < 0.001 | - 1.33 | 0.24 | - 1.17 | 0.39 |
| Recessive model | 9579 / 9881 | 1.17 | 1.08–1.27 (< 0.001) | 48.5 | < 0.001 | −1.38 | 0.16 | −0.60 | 0.55 | |
| Allelic model | 9579 / 9881 | 1.21 | 1.09–1.33 (< 0.001) | 71.1 | < 0.001 | − 1.05 | 0.41 | −1.82 | 0.07 | |
| TT vs. CC | 9579 / 9881 | 1.34 | 1.18–1.52 (< 0.001) | 30.5 | 0.02 | −1.25 | 0.24 | −1.90 | 0.65 | |
| CT vs. CC | 9579 / 9881 | 1.13 | 0.95–1.34 (0.17) | 68.7 | < 0.001 | −2.06 | 0.33 | −1.73 | 0.09 | |
| Age groups | ||||||||||
| Pediatrics | Dominant model | 3061 / 3536 | 1.54 | 1.24–1.92 (< 0.001) | 41 | 0.04 | − 1.93 | 0.05 | −1.63 | 0.23 |
| Recessive model | 3061 / 3536 | 1.20 | 1.05–1.37 (< 0.001) | 58.3 | < 0.001 | −0.36 | 0.71 | −1.14 | 0.27 | |
| Allelic model | 3061 / 3536 | 1.37 | 1.16–1.63 (< 0.001) | 68 | < 0.001 | −1.53 | 0.12 | − 1.99 | 0.06 | |
| TT vs. CC | 3061 / 3536 | 1.51 | 1.22–1.87 (< 0.001) | 51.6 | 0.01 | −1.44 | 0.15 | −1.47 | 0.24 | |
| CT vs. CC | 3061 / 3536 | 1.49 | 1.23–1.81 (< 0.001) | 10.6 | 0.33 | −1.92 | 0.05 | −1.22 | 0.42 | |
| Adults | Dominant model | 2933 / 2670 | 1.23 | 1.01–1.51 (0.04) | 35.2 | 0.066 | −2.10 | 0.03 | −1.86 | 0.08 |
| Recessive model | 2933 / 2670 | 1.21 | 1.04–1.40 (0.01) | 46 | 0.01 | −0.91 | 0.36 | −0.71 | 0.48 | |
| Allelic model | 2933 / 2670 | 1.24 | 1.05–1.47 (< 0.001) | 63.8 | < 0.001 | −0.97 | 0.33 | −1.45 | 0.16 | |
| TT vs. CC | 2933 / 2670 | 1.37 | 1.09–1.72 (< 0.001) | 5 | 0.39 | −1.01 | 0.47 | −1.77 | 0.19 | |
| CT vs. CC | 2933 / 2670 | 1.15 | 0.96–1.39 (0.13) | 23 | 0.17 | −2.13 | 0.03 | −1.56 | 0.13 | |
| Mixed | Dominant model | 3585 / 3675 | 0.92 | 0.65–1.32 (0.65) | 83.6 | < 0.001 | −0.09 | 0.92 | − 1.05 | 0.31 |
| Recessive model | 3585 / 3675 | 1.12 | 0.97–1.28 (0.11) | 45.4 | 0.02 | −0.41 | 0.68 | 0.39 | 0.70 | |
| Allelic model | 3585 / 3675 | 1.03 | 0.85–1.24 (0.78) | 76.3 | < 0.001 | −0.72 | 0.47 | 0.02 | 0.98 | |
| TT vs. CC | 3585 / 3675 | 1.14 | 0.91–1.42 (0.24) | 20.8 | 0.21 | −0.18 | 0.85 | −0.28 | 0.87 | |
| CT vs. CC | 3585 / 3675 | 0.87 | 0.59–1.28 (0.48) | 84.9 | < 0.001 | 0 | 1 | −1.11 | 0.28 | |
| Ethnicity-1 (Continent) | ||||||||||
| Asia | Dominant model | 5196 / 4936 | 1.15 | 0.84–1.56 (0.39) | 75.6 | < 0.001 | −1.86 | 0.06 | −1.44 | 0.20 |
| Recessive model | 5196 / 4936 | 1.16 | 1.06–1.28 (< 0.001) | 65 | < 0.001 | −1.62 | 0.10 | −0.60 | 0.55 | |
| Allelic model | 5196 / 4936 | 1.17 | 1–1.37 (0.04) | 76.7 | < 0.001 | −1.72 | 0.08 | −1.04 | 0.30 | |
| TT vs. CC | 5196 / 4936 | 1.34 | 1.13–1.58 (< 0.001) | 42.7 | 0.01 | −1.48 | 0.13 | −1.15 | 0.40 | |
| CT vs. CC | 5196 / 4936 | 1 | 0.70–1.42 (0.97) | 75.1 | < 0.001 | −2 | 0.04 | −1.42 | 0.20 | |
| Europe | Dominant model | 1704 / 2347 | 1.46 | 1.15–1.85 (< 0.001) | 56.9 | 0.01 | 0 | 1 | −0.70 | 0.49 |
| Recessive model | 1704 / 2347 | 1.35 | 0.98–1.86 (0.06) | 0 | 0.94 | − 1.58 | 0.11 | −1.91 | 0.08 | |
| Allelic model | 1704 / 2347 | 1.34 | 1.12–1.61 (< 0.001) | 51 | 0.02 | −1.03 | 0.30 | −1.50 | 0.16 | |
| TT vs. CC | 1704 / 2347 | 1.53 | 1.10–2.14 (0.01) | 0 | 0.80 | 0.16 | 0.87 | −0.87 | 0.40 | |
| CT vs. CC | 1704 / 2347 | 1.44 | 1.13–1.83 (< 0.001) | 55.6 | 0.01 | 0.78 | 0.43 | 0.33 | 0.74 | |
| America | Dominant model | 1991 / 1828 | 1.22 | 0.95–1.58 (0.11) | 54.5 | 0.01 | −1.33 | 0.27 | −2.05 | 0.07 |
| Recessive model | 1991 / 1828 | 1.15 | 0.96–1.39 (0.12) | 24.3 | 0.22 | −1.34 | 0.18 | 0.99 | 0.35 | |
| Allelic model | 1991 / 1828 | 1.19 | 0.99–1.44 (0.06) | 64.8 | < 0.001 | − 0.98 | 0.32 | −0.48 | 0.64 | |
| TT vs. CC | 1991 / 1828 | 1.27 | 0.98–1.64 (0.07) | 43.7 | 0.06 | − 1.52 | 0.12 | −1.91 | 0.09 | |
| CT vs. CC | 1991 / 1828 | 1.18 | 0.94–1.48 (0.15) | 39.3 | 0.09 | −1.52 | 0.12 | −1.94 | 0.08 | |
| Ethnicity-2 | ||||||||||
| East-Asian | Dominant model | 4246 / 3908 | 1.43 | 1.14–1.79 (< 0.001) | 26.3 | 0.14 | −1.08 | 0.28 | 1.53 | 0.29 |
| Recessive model | 4246 / 3908 | 1.14 | 1.03–1.26 (< 0.001) | 66.6 | < 0.001 | −1.02 | 0.27 | −1.51 | 0.36 | |
| Allelic model | 4246 / 3908 | 1.29 | 1.10–1.52 (< 0.001) | 72 | < 0.001 | −1.79 | 0. 58 | −3.10 | 0.06 | |
| TT vs. CC | 4246 / 3908 | 1.33 | 1.11–1.59 (< 0.001) | 41.8 | 0.02 | −1.27 | 0.29 | −1.39 | 0.31 | |
| CT vs. CC | 4246 / 3908 | 1.24 | 1.00–1.53 (0.04) | 0 | 0.74 | −1.89 | 0.68 | −1.71 | 0.10 | |
| Non-East-Asian | Dominant model | 5333 / 5973 | 1.10 | 0.90–1.36 (0.35) | 77.4 | < 0.001 | −0.80 | 0.42 | −1.18 | 0.35 |
| Recessive model | 5333 / 5973 | 1.25 | 1.08–1.45 (< 0.001) | 21.9 | 0.14 | 0.59 | 0.55 | 0.73 | 0.47 | |
| Allelic model | 5333 / 5973 | 1.15 | 1–1.32 (0.04) | 71.5 | < 0.001 | −1.05 | 0.48 | −1.82 | 0.07 | |
| TT vs. CC | 5333 / 5973 | 1.34 | 1.12–1.61 (< 0.001) | 24 | 0.11 | −0.37 | 0.70 | −1.04 | 0.30 | |
| CT vs. CC | 5333 / 5973 | 1.03 | 0.83–1.28 (0.78) | 77.9 | < 0.001 | −1.16 | 0.24 | −1.93 | 0.06 | |
| Ethnicity 3 | ||||||||||
| Caucasian | Dominant model | 7360 / 7971 | 1.30 | 1.12–1.51 (< 0.001) | 49.2 | < 0.001 | −1.04 | 0.48 | −1.51 | 0.18 |
| Recessive model | 7360 / 7971 | 1.16 | 1.06–1.27 (< 0.001) | 49.7 | < 0.001 | −1.31 | 0.24 | −2.77 | 0.09 | |
| Allelic model | 7360 / 7971 | 1.25 | 1.12–1.39 (< 0.001) | 65 | < 0.001 | 1.40 | 0.17 | −1.12 | 0.38 | |
| TT vs. CC | 7360 / 7971 | 1.34 | 1.16–1.56 (< 0.001) | 24.9 | 0.09 | −1.52 | 0.16 | −1.34 | 0.29 | |
| CT vs. CC | 7360 / 7971 | 1.22 | 1.05–1.42 (< 0.001) | 39.6 | < 0.001 | −1.54 | 0.12 | −1.80 | 0.08 | |
| Arab | Dominant model | 316 / 284 | 0.36 | 0.07–1.88 (0.22) | 91.5 | < 0.001 | 0.68 | 0.49 | −0.17 | 0.83 |
| Recessive model | 316 / 284 | 1.53 | 0.27–1.48 (0.09) | 87.4 | < 0.001 | 0 | 1 | −1.67 | 0.19 | |
| Allelic model | 316 / 284 | 0.63 | 0.67–3.68 (0.29) | 85.4 | < 0.001 | 0.49 | 0.62 | −0.11 | 0.92 | |
| TT vs. CC | 316 / 284 | 0.93 | 0.43–1.99 (0.85) | 66.6 | 0.02 | 0.68 | 0.49 | 1.25 | 0.33 | |
| CT vs. CC | 316 / 284 | 0.29 | 0.05–1.84 (0.19) | 92.3 | < 0.001 | 0 | 1 | −0.71 | 0.55 | |
| African-American | Dominant model | 1903 / 1626 | 1.34 | 1.07–1.67 (0.01) | 35.3 | 0.13 | −1.67 | 0.09 | 1.97 | 0.27 |
| Recessive model | 1903 / 1626 | 1.18 | 0.98–1.43 (0.07) | 24.7 | 0.22 | 0.63 | 0.53 | 1.11 | 0.30 | |
| Allelic model | 1903 / 1626 | 1.25 | 1.04–1.50 (0.01) | 58.9 | 0.01 | −1.46 | 0.14 | −0.81 | 0.44 | |
| TT vs. CC | 1903 / 1626 | 1.37 | 1.04–1.80 (0.02) | 36.2 | 0.12 | −1.67 | 0.09 | −1.44 | 0.40 | |
| CT vs. CC | 1903 / 1626 | 1.30 | 1.06–1.58 (0.01) | 13.9 | 0.31 | −1.67 | 0.09 | −1.46 | 0.41 | |
Fig. 3.
Pooled odds ratio and 95% confidence interval of individual studies and pooled data for the association between IL-4 C589T polymorphism and asthma risk in different subgroups for; a dominant model [age subgroup], b dominant model [continent]
Subgroup analysis by ethnicity 1 (continent)
In this subgroup we categorized studies by their continent: including Asia (32 articles), Europe (11 articles), America (10 articles), Africa (1 article), and Oceania (1 article). Since there was only one study for each one of the African and Australian population, these studies were excluded from the analysis. The results indicated that presence of IL4 gene -589C/T SNP in Asian population increased susceptibility of asthma across all genotype models except dominant model (OR = 1.15, 95% CI = 0.84–1.56, P = 0. 39, REM) and CT vs. CC model (OR = 1, 95% CI = 0.70–1.42, P = 0. 97, REM). Moreover, in contrast with effect of IL4 gene -589C/T SNP on the risk of asthma in American populations, a significant positive association was detected in European population thorough dominant model (OR = 1.46, 95% CI = 1.15–1.85, P < 0.001, REM), allelic model (OR = 1.34, 95% CI = 1.12–1.61, P < 0.001, REM), TT vs. CC model (OR = 1.53, 95% CI = 1.10–2.14, P = 0.01, FEM), and CT vs.CC model (OR = 1.44, 95% CI = 1.13–1.83, P < 0.001, REM) (Table 3, Fig. 3).
Subgroup analysis by ethnicity 2 (east and non-east Asian)
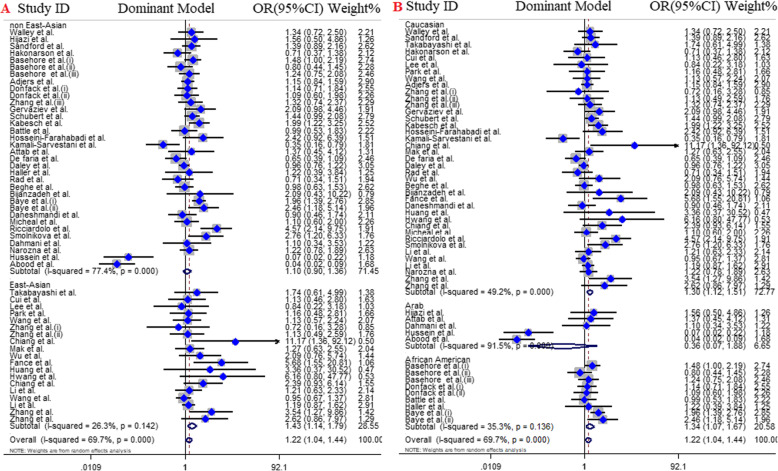
The subgroup analysis according to East Asian (20 articles) and non-East Asian (35 articles) title revealed the significant association between IL4 gene -589C/T polymorphism and the risk of asthma across in all genotype models of East Asians and three genotype models of non-East Asian including; recessive model (OR = 1.25, 95% CI = 1.08–1.45, P < 0.001, FEM), allelic model (OR = 1.15, 95% CI = 1–1.32, P = 0.04, REM), TT vs. CC model (OR = 1.34, 95% CI = 1.12–1.61, P < 0.001, FEM) (Table 3, Fig. 4).
Fig. 4.
Pooled odds ratio and 95% confidence interval of individual studies and pooled data for the association between IL-4 C589T polymorphism and asthma risk in different subgroups for; a dominant model [East and non-East Asian], b dominant model [ethnicity]
Subgroup analysis by ethnicity 3
Finally, subgroup analysis of eligible articles according ethnicity including Caucasians (41 articles), African-Americans (9 articles), and Arabs (5 articles) showed that there was no significant association between IL4 gene -589C/T SNP and asthma risk in Arab population. Also, except recessive model (OR = 1.18, 95% CI = 0.98–1.43, P = 0.07, FEM) other genotype models in African-American population were significant including dominant model (OR = 1.34, 95% CI = 1.07–1.67, P = 0.01, FEM), allelic model (OR = 1.25, 95% CI = 1.04–1.50, P = 0.01, REM), TT vs. CC model (OR = 1.37, 95% CI = 1.04–1.80, P = 0.02, FEM), and CT vs. CC model (OR = 1.30, 95% CI = 1.06–1.58, P = 0.01, FEM). Conversely, all genotype models were significant in Caucasians and presence of IL4 gene -589C/T SNP increase risk of asthma (Table 3, Fig. 4).
Meta-regression analyses
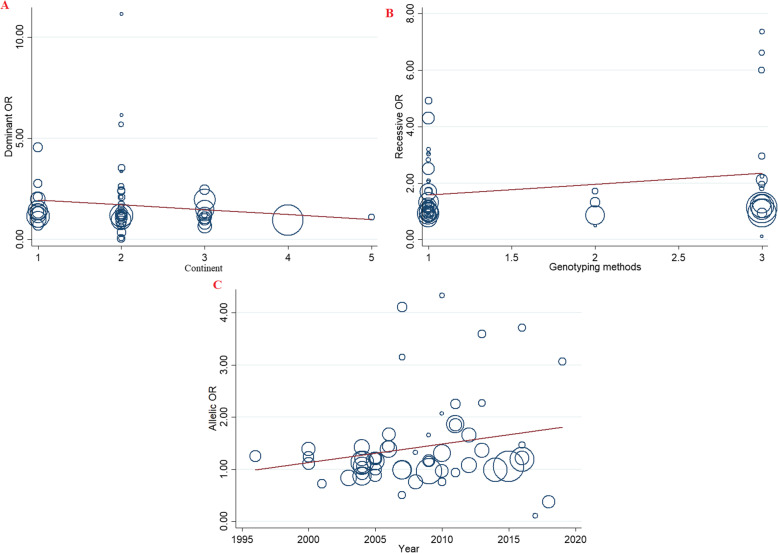
Meta-regression analyses were performed to explore potential sources of heterogeneity among included studies (Table 4). The findings indicated that none of the expected heterogeneity parameter were the source of heterogeneity (Fig. 5).
Table 4.
Meta-regression analyses of potential source of heterogeneity
| Heterogeneity Factors | Coefficient | SE | T | P-value | 95% CI | ||
|---|---|---|---|---|---|---|---|
| UL | LL | ||||||
| Publication Year | Dominant model | 0.035 | 0.041 | 0.85 | 0.40 | −0.048 | 1.119 |
| Recessive model | 0.140 | 0.036 | 3.81 | 0.07 | −0.066 | 0.213 | |
| Allelic model | 0.035 | 0.022 | 1.58 | 0.11 | −0.009 | 0.080 | |
| TT vs. CC | 0.123 | 0.064 | 1.91 | 0.06 | −0.006 | 0.254 | |
| CT vs. CC | 0.020 | 0.035 | 0.58 | 0.56 | −0.050 | 0.090 | |
| continent | Dominant model | −0.238 | 0.265 | −0.90 | 0.37 | −0.772 | 0.294 |
| Recessive model | 0.022 | 0.274 | 0.08 | 0.93 | −0.530 | 0.574 | |
| Allelic model | −0.116 | 0.146 | −0.79 | 0.43 | −0.410 | 0.177 | |
| AA vs. CC | −0.096 | 0.435 | −0.22 | 0.82 | −0.973 | 0.780 | |
| CA vs. CC | −0.265 | 0.209 | −1.27 | 0.21 | −0.685 | 0.154 | |
| Genotyping methods | Dominant model | −0.137 | 0.241 | −0.57 | 0.57 | −0.621 | 0.346 |
| Recessive model | 0.382 | 0.232 | 1.65 | 0.10 | −0.084 | 0.849 | |
| Allelic model | 0.039 | 0.130 | 0.30 | 0.76 | −0.221 | 0.300 | |
| TT vs. CC | 0.056 | 0.388 | 0.14 | 0.88 | −0.726 | 0.838 | |
| CT vs. CC | −0.114 | 0.199 | −0.57 | 0.57 | −0.515 | 0.287 | |
Fig. 5.
Meta-regression plots of the association between IL-4 C589T polymorphism and risk of asthma based on; a Continent (dominant), b Genotyping methods (recessive), c Publication year (Allelic)
Publication bias
To check existence of publication, Egger’s linear regression and Begg’s funnel plot test were used. The shape of funnel plot did not disclose obvious asymmetry under all genotype model of the IL4 gene -589C/T polymorphism (Fig. 6).
Fig. 6.
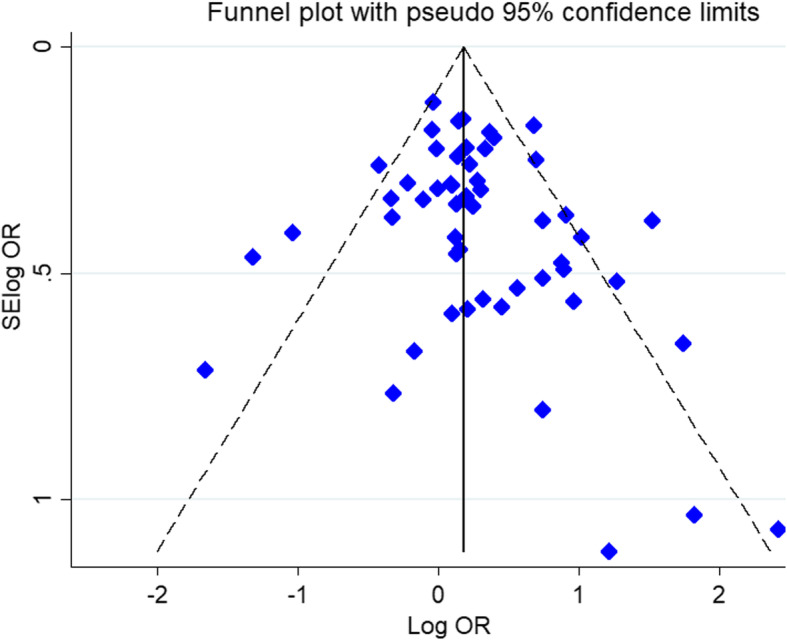
Begg’s funnel plot for publication bias test. Dominant model C598T. Each point represents a separate study for the indicated association
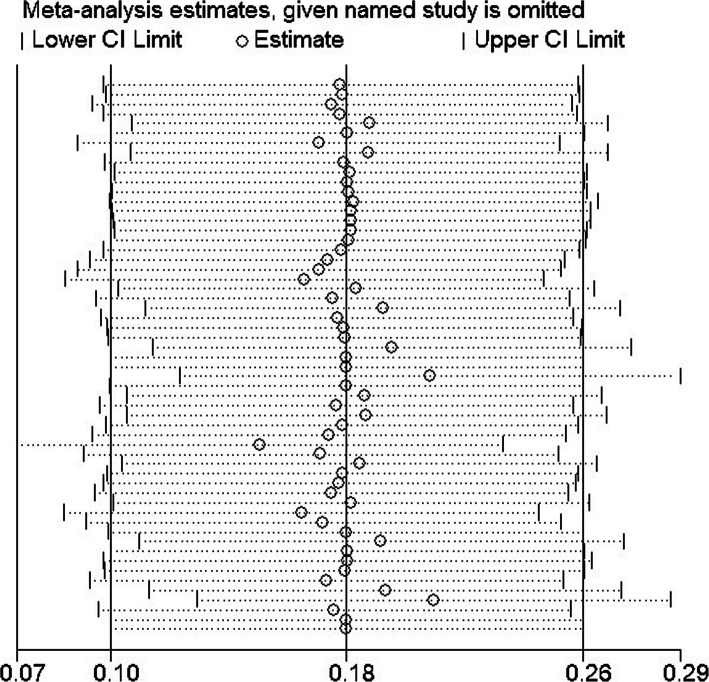
Sensitivity analysis
The impact of individual study on pooled OR was evaluated by sequential omission of each studies. The result showed that no individual study significantly affected the pooled ORs under all genotype models of the IL4 gene -589C/T polymorphism (Fig. 7).
Fig. 7.
Sensitivity analysis in present meta-analysis investigates the single nucleotide polymorphisms of IL-4 C589T contribute to risk for asthma
Discussion
To date, several individual case-control replication studies have attempted to divulge the association of IL4 gene -589C/T polymorphism and risk of asthma. Due to some differences, however, these disperse investigation demonstrated incongruous reports. The differences in the race of study subjects, diversity in the diagnostic criteria of the patients, limited sample sizes may be the cause of such inconsistent results [74]. On the other hand, meta-analysis is a tool that has the potential to solve the problem of inconsistency by removing the confining issues of insufficient statistical power in the individual studies. Therefore, to resolve the mentioned confining factors about the IL4 gene -589C/T polymorphism, the present most up-to-date meta-analysis was conducted to determine a bona fide estimation of the association between IL4 gene -589C/T polymorphism and susceptibility to asthma. Our analysis indicated that this SNP was associated with increased risk of asthma in the overall population as well as during subgroup analysis by age groups and ethnicity/continent.
Asthma is a complicated pulmonary disease, characterized by airway hyperresponsiveness, airway inflammation, and airway remodeling [75, 76]. During asthma, there is a hyperactivity of Th2 responses, in which the cytokines of the type 2 immunity, such as IL-4, IL-5, and IL-13 promote the harmful inflammatory events in the airways. Studies have reported that local administration of IL-4 gene plasmids prior to antigen challenge could stimulate the airway hyperresponsiveness and accumulation of eosinophils in mice [77]. This phenotype of asthma is commonly referred to “eosinophilic” asthma. On the other side, “noneosinophilic” asthma is characterized by low frequency of eosinophils in the involved sites, but other inflammatory cells are dominant in the effector phase, such as neutrophils, mixed granulocyte inflammatory cells, or even little number of inflammatory cells, called paucigranulocytic inflammation. Th17 mediated IL-17 axis and lack of significant Th2/Th17 inflammation have been attributed to the noneosinophilic asthma [78]. Among the SNPs in the IL4 gene, the -589C/T (rs2243250) polymorphism has been widely investigated in susceptibility to asthma. It has been shown that the T allele of this SNP leads to increased affinity of the binding of transcription factors in comparison to the C allele, leading to overexpression of IL4 mRNA [79, 80]. As a consequence, it is a biological justification that IL4 gene −589C/T SNP impresses the IL-4 expression and, hence, could affect the asthma susceptibility.
Previously, three meta-analysis studies have attempted to disclose the association of IL4 gene −589C/T SNP with the risk of asthma. Wang et al. in 2012 indicated that the T allele of IL4 gene −589C/T SNP increased the risk of asthma (OR = 1.12). Basically, individuals carrying the T allele had a 24% increased risk of asthma in comparison to the CC homozygote model. Subgroup analysis revealed the association of this polymorphism in the Caucasians [81]. In addition, Nie et al. in 2013 included 40 studies involving 7345 cases and 7819 controls in their meta-analysis [18]. This meta-analysis indicated that TT vs. CC (OR = 1.40) and CT vs. CC (OR = 1.22) models were significantly associated with increased risk of asthma. In the subgroup analysis by ethnicity, significant associations were found among Asians and Caucasians, but not in the African-Americans. In addition, the subgroup analysis by atopic status revealed no significant association among atopic asthma patients and non-atopic asthma patients. On the other side, Zhang et al. [75] by evaluating pediatric asthma risk by evolving 17 case-control studies (15 publications) containing 3427 cases and 4247 controls revealed that IL4 -589C/T polymorphism was associated with increased risk of asthma in pediatrics. Furthermore, the subgroup analyses by ethnicity, indicated significant association in Caucasians and Asians.
Our analysis was performed on 55 case-control studies containing 9572 cases and 9881 controls. It was observed that IL4 gene -589C/T polymorphism increased the risk of asthma across all genetic models, including dominant model (OR = 1.22), recessive model (OR = 1.17), allelic model (OR = 1.21), and TT vs. CC model (OR = 1.34), but not the CT vs. TT model. Furthermore, subgroup analysis by age indicated that IL4 gene -589C/T polymorphism was significantly associated with asthma risk in both pediatrics and adults. The subgroup analysis by ethnicity revealed significant association in Asian, American, and Europeans. Finally, subgroup analysis by East Asian and non-East Asian populations indicated significant associations.
This meta-analysis bears some limitations and caveats. First, the analysis was according to crude estimation of IL4 gene -589C/T polymorphism association with asthma susceptibility, regardless of the effect of confounding factors, like age, sex, environmental factors, and contribution of other genes in LD with IL4 gene. Second, we did not analyze other genes that could be contributing in understanding of cytokine involvement in the susceptibility to asthma.
Conclusion
All in all, here we carried out the most up-to-date analysis of the IL4 gene 589C/T polymorphism and asthma risk prior to September 2020. Our meta-analysis further confirmed some results of the previously performed meta-analysis, while rejected some of them. In a whole, IL4 gene -589C/T polymorphism increased the risk of asthma across all genetic models. Moreover, the subgroup analysis by age indicated that IL4 gene -589C/T polymorphism was significantly associated with asthma risk in both pediatrics and adults. Also, the subgroup analysis by ethnicity revealed significant association in Asian, American, and Europeans. Ultimately, subgroup analysis by East Asian and non-East Asian populations indicated significant associations.
Acknowledgements
Not applicable.
Abbreviations
- IL
Interleukin
- Th
T helper
- CI
Confidence interval
- OR
Odds ratio
- SNP
Single-nucleotide polymorphism
- PRISMA
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- NOS
Newcastle–Ottawa scale
- HWE
Hardy–Weinberg equilibrium
Authors’ contributions
BR and DI originated the study, acquired data. AK, AMG, and AMF analyzed and interpreted the data. MH, MA, and DI prepared the original draft. BR, DI, and MJM critically revised the paper. SA and HM supervised the project. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data that support the conclusions of this manuscript are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mojgan Hosseini, Email: mojgan-hosseini@iiau.ac.ir.
Haleh Mikaeili, Email: mikaeilihale@gmail.com.
References
- 1.Makoui MH, Imani D, Motallebnezhad M, Azimi M, Razi B. Vitamin D receptor gene polymorphism and susceptibility to asthma: meta-analysis based on 17 case-control studies. Ann Allergy Asthma Immunol. 2020;124(1):57–69. doi: 10.1016/j.anai.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Peters SP, Ferguson G, Deniz Y, Reisner C. Uncontrolled asthma: a review of the prevalence, disease burden and options for treatment. Respir Med. 2006;100(7):1139–1151. doi: 10.1016/j.rmed.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Subbarao P, Mandhane PJ, Sears MR. Asthma: epidemiology, etiology and risk factors. Cmaj. 2009;181(9):E181–E190. doi: 10.1503/cmaj.080612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sykes A, Johnston SL. Etiology of asthma exacerbations. J Allergy Clin Immunol. 2008;122(4):685–688. doi: 10.1016/j.jaci.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242(1):10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malerba G, Pignatti PF. A review of asthma genetics: gene expression studies and recent candidates. J Appl Genet. 2005;46(1):93–104. [PubMed] [Google Scholar]
- 7.Noutsios GT, Floros J. Childhood asthma: causes, risks, and protective factors; a role of innate immunity. Swiss Med Wkly. 2014;144(5152). [DOI] [PubMed]
- 8.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Kendler KS, Chen X, Dick D, Maes H, Gillespie N, Neale MC, Riley B. Recent advances in the genetic epidemiology and molecular genetics of substance use disorders. Nat Neurosci. 2012;15(2):181. doi: 10.1038/nn.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai D, Brightling C. Cytokine and anti-cytokine therapy in asthma: ready for the clinic? Clin Exp Immunol. 2009;158(1):10–19. doi: 10.1111/j.1365-2249.2009.03998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulsen LK, Hummelshoj L. Triggers of IgE class switching and allergy development. Ann Med. 2007;39(6):440–456. doi: 10.1080/07853890701449354. [DOI] [PubMed] [Google Scholar]
- 12.Bao K, Reinhardt RL. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine. 2015;75(1):25–37. doi: 10.1016/j.cyto.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rengarajan J, Szabo SJ, Glimcher LH. Transcriptional regulation of Th1/Th2 polarization. Immunol Today. 2000;21(10):479–483. doi: 10.1016/S0167-5699(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 14.Weiss DL, Brown MA. Regulation of IL-4 production in mast cells: a paradigm for cell-type-specific gene expression. Immunol Rev. 2001;179(1):35–47. doi: 10.1034/j.1600-065X.2001.790104.x. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki I, Hizawa N, Yamaguchi E, Kawakami Y. Association between a C+ 33T polymorphism in the IL-4 promoter region and total serum IgE levels. Clin Exp Allergy. 2000;30(12):1746–1749. doi: 10.1046/j.1365-2222.2000.00983.x. [DOI] [PubMed] [Google Scholar]
- 16.Basehore MJ, Howard TD, Lange LA, Moore WC, Hawkins GA, Marshik PL, Harkins MS, Meyers DA, Bleecker ER. A comprehensive evaluation of IL4 variants in ethnically diverse populations: association of total serum IgE levels and asthma in white subjects. J Allergy Clin Immunol. 2004;114(1):80–87. doi: 10.1016/j.jaci.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 17.Hegab AE, Sakamoto T, Saitoh W, Massoud HH, Massoud HM, Hassanein KM, Sekizawa K. Polymorphisms of IL4, IL13, and ADRB2 genes in COPD. Chest. 2004;126(6):1832–1839. doi: 10.1378/chest.126.6.1832. [DOI] [PubMed] [Google Scholar]
- 18.Nie W, Zhu Z, Pan X, Xiu Q. The interleukin-4− 589C/T polymorphism and the risk of asthma: a meta-analysis including 7345 cases and 7819 controls. Gene. 2013;520(1):22–29. doi: 10.1016/j.gene.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 19.Rosenwasser L, Borish L. Promoter polymorphisms predisposing to the development of asthma and atopy. Clin Exp Allergy. 1998;28:13–15. doi: 10.1046/j.1365-2222.1998.028s5013.x. [DOI] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 21.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 22.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994:1088–101. [PubMed]
- 25.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donfack J, Schneider DH, Tan Z, Kurz T, Dubchak I, Frazer KA, Ober C. Variation in conserved non-coding sequences on chromosome 5q and susceptibility to asthma and atopy. Respir Res. 2005;6(1):145. doi: 10.1186/1465-9921-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Zhang X, Qiu D. Association of interleukin-4 and interleukin-4 receptor gene polymorphism and serum IgE levels in Chinese, Malayan and Hindoo. Zhonghua Jie He He Hu Xi Za Zhi. 2005;28:489–490. [Google Scholar]
- 28.Baye TM, Kovacic MB, Myers JMB, Martin LJ, Lindsey M, Patterson TL, He H, Ericksen MB, Gupta J, Tsoras AM. Differences in candidate gene association between European ancestry and African American asthmatic children. PLoS One. 2011;6(2). [DOI] [PMC free article] [PubMed]
- 29.Walley A, Cookson W. Investigation of an interleukin-4 promoter polymorphism for associations with asthma and atopy. J Med Genet. 1996;33(8):689–692. doi: 10.1136/jmg.33.8.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hijazi Z, Haider M. Interleukin-4 gene promoter polymorphism [C590T] and asthma in Kuwaiti Arabs. Int Arch Allergy Immunol. 2000;122(3):190–194. doi: 10.1159/000024396. [DOI] [PubMed] [Google Scholar]
- 31.Sandford AJ, Chagani T, Zhu S, Weir TD, Bai TR, Spinelli JJ, FitzGerald JM, Behbehani NA, Tan WC, Paré PD. Polymorphisms in the IL4, IL4RA, and FCERIB genes and asthma severity. J Allergy Clin Immunol. 2000;106(1):135–140. doi: 10.1067/mai.2000.107926. [DOI] [PubMed] [Google Scholar]
- 32.Takabayashi A, Ihara K, Sasaki Y, Suzuki Y, Nishima S, Izuhara K, Hamasaki N, Hara T. Childhood atopic asthma: positive association with a polymorphism of IL-4 receptor α gene but not with that of IL-4 promoter or fc ε receptor I β gene. Exp Clin Immunogenet. 2000;17(2):63–70. doi: 10.1159/000019125. [DOI] [PubMed] [Google Scholar]
- 33.Hákonarson H, Bjornsdottir US, Ostermann E, Arnason T, Adalsteinsdottir AE, Halapi E, Shkolny D, Kristjansson K, Gudnadottir SA, Frigge ML. Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med. 2001;164(11):2036–2044. doi: 10.1164/ajrccm.164.11.2101086. [DOI] [PubMed] [Google Scholar]
- 34.Cui T, Wu J, Pan S, Xie J. Polymorphisms in the IL-4 and IL-4R [α] genes and allergic asthma. Clin Chem Lab Med. 2003;41(7):888–892. doi: 10.1515/CCLM.2003.134. [DOI] [PubMed] [Google Scholar]
- 35.Lee SG, Kim BS, Kim JH, Lee SY, Choi SO, Shim JY, Hong TJ, Hong SJ. Gene–gene interaction between interleukin-4 and interleukin-4 receptor α in Korean children with asthma. Clin Exp Allergy. 2004;34(8):1202–1208. doi: 10.1111/j.1365-2222.2004.02015.x. [DOI] [PubMed] [Google Scholar]
- 36.Park BL, Kim LH, Choi YH, Lee J-H, Rhim T, Lee YM, Uh S-T, Park H-S, Choi BW, Hong S-J. Interleukin 3 (IL3) polymorphisms associated with decreased risk of asthma and atopy. J Hum Genet. 2004;49(10):517–527. doi: 10.1007/s10038-004-0184-x. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Halmurat W, Yilihamu S, Xiang Y, Ablikemu A. A study on the relationship between interleukin-4 promoter polymorphism and asthma in a Xinjiang Uyger population. Zhonghua Jie He He Hu Xi Za Zhi. 2004;27(7):460–464. [PubMed] [Google Scholar]
- 38.Ådjers K, Karjalainen J, Pessi T, Eklund C, Hurme M. Epistatic effect of TLR4 and IL4 genes on the risk of asthma in females. Int Arch Allergy Immunol. 2005;138(3):251–256. doi: 10.1159/000088726. [DOI] [PubMed] [Google Scholar]
- 39.Gervaziev YV, Kaznacheev V, Gervazieva V. Allelic polymorphisms in the interleukin-4 promoter regions and their association with bronchial asthma among the Russian population. Int Arch Allergy Immunol. 2006;141(3):257–264. doi: 10.1159/000095295. [DOI] [PubMed] [Google Scholar]
- 40.Schubert K, Von Bonnsdorf H, Burke M, Ahlert I, Braun S, Berner R, Deichmann K, Heinzmann A. A comprehensive candidate gene study on bronchial asthma and juvenile idiopathic arthritis. Dis Markers. 2006;22(3):127–132. doi: 10.1155/2006/373620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, von Mutius E. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117(2):269–274. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 42.Battle NC, Choudhry S, Tsai H-J, Eng C, Kumar G, Beckman KB, Naqvi M, Meade K, Watson HG, LeNoir M. Ethnicity-specific gene–gene interaction between IL-13 and IL-4Rα among African Americans with asthma. Am J Respir Crit Care Med. 2007;175(9):881–887. doi: 10.1164/rccm.200607-992OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosseini-Farahabadi S, Tavakkol-Afshari J, Rafatpanah H, Farid-Hosseini R, Daluei MK. Association between the polymorphisms of IL-4 gene promoter (−590C> T), IL-13 coding region (R130Q) and IL-16 gene promoter (−295T> C) and allergic asthma. Iran J Allergy Asthma Immunol. 2007;6:9–14. [PubMed] [Google Scholar]
- 44.Kamali-Sarvestani E, Ghayomi M, Nekoee A. Association of TNF-alpha-308 G/a and IL-4-589 C/T gene promoter polymorphisms with asthma susceptibility in the south of Iran. J Investig Allergol Clin Immunol. 2007;17(6):361. [PubMed] [Google Scholar]
- 45.Chiang CH, Tang YC, Lin MW, Chung MY. Association between the IL-4 promoter polymorphisms and asthma or severity of hyperresponsiveness in Taiwanese. Respirology. 2007;12(1):42–48. doi: 10.1111/j.1440-1843.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- 46.Mak JC, Ko FW, Chu CM, Leung HC, Chan HW, Cheung AH, Ip MS, Chan-Yeung M. Polymorphisms in the IL-4, IL-4 receptor α chain, TNF-α, and lymphotoxin-α genes and risk of asthma in Hong Kong Chinese adults. Int Arch Allergy Immunol. 2007;144(2):114–122. doi: 10.1159/000103222. [DOI] [PubMed] [Google Scholar]
- 47.Attab KA, Al-Qaoud KM, Al-Bataieneh K, Ajlouni MJ. Association of SNP in the IL-4, IL-18 and eotaxin genes with asthma in a Jordanian population. Int J Integrative Biol. 2008;4(2):86. [Google Scholar]
- 48.De Faria IC, De Faria EJ, Toro AA, Ribeiro JD, Bertuzzo CS. Association of TGF-β1, CD14, IL-4, IL-4R and ADAM33 gene polymorphisms with asthma severity in children and adolescents. J Pediatr. 2008;84:203-10. [DOI] [PubMed]
- 49.Jiang P, Liu J, Xue-Bo Y, Rong-Yu L. Several interleukin-4 and interleukin-13 gene single nucleotide polymorphisms among Chinese asthmatic patients. In: Allergy and asthma proceedings: OceanSide Publications; 2009. p. 413. [DOI] [PubMed]
- 50.Amirzargar A, Movahedi M, Rezaei N, Moradi B, Dorkhosh S, Mahloji M, Mandaviani S. 2 polymorphisms in IL4 and IL4RA confer susceptibility to asthma. J Investig Allergol Clin Immunol. 2009;19(6):433. [PubMed] [Google Scholar]
- 51.Daley D, Lemire M, Akhabir L, Chan-Yeung M, He JQ, McDonald T, Sandford A, Stefanowicz D, Tripp B, Zamar D. Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet. 2009;125(4):445–459. doi: 10.1007/s00439-009-0643-8. [DOI] [PubMed] [Google Scholar]
- 52.Haller G, Torgerson DG, Ober C, Thompson EE. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J Allergy Clin Immunol. 2009;124(6):1204–1209.e1209. doi: 10.1016/j.jaci.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdi RI, Bagheri M, Rahimirad MH, Moradi Z. IFN-γ+ 874 and IL-4-590 polymorphisms and asthma susceptibility in north west of Iran. 2010. [Google Scholar]
- 54.Wu X, Li Y, Chen Q, Chen F, Cai P, Wang L, Hu L. Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle China. J Asthma. 2010;47(3):238–244. doi: 10.3109/02770900903509099. [DOI] [PubMed] [Google Scholar]
- 55.Beghé B, Hall I, Parker S, Moffatt M, Wardlaw A, Connolly M, Fabbri L, Ruse C, Sayers I. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy. 2010;65(4):474–481. doi: 10.1111/j.1398-9995.2009.02167.x. [DOI] [PubMed] [Google Scholar]
- 56.Bijanzadeh M, Ramachandra NB, Mahesh P, Mysore RS, Kumar P, Manjunath B, Jayaraj B. Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung. 2010;188(5):415–422. doi: 10.1007/s00408-010-9247-2. [DOI] [PubMed] [Google Scholar]
- 57.Fan C, Liu Y, Ma Y, Zhang W. Susceptibility gene polymorphism and bronchial asthma. Prog Modn Biomed. 2010;10(17):3264–3267. [Google Scholar]
- 58.Daneshmandi S, Pourfathollah AA, Pourpak Z, Heidarnazhad H, Kalvanagh PA. Cytokine gene polymorphism and asthma susceptibility, progress and control level. Mol Biol Rep. 2012;39(2):1845–1853. doi: 10.1007/s11033-011-0927-7. [DOI] [PubMed] [Google Scholar]
- 59.Huang H-R, Zhong Y-Q, Wu J-F. The association between IFN-γ and IL-4 genetic polymorphisms and childhood susceptibility to bronchial asthma. Gene. 2012;494(1):96–101. doi: 10.1016/j.gene.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 60.Hwang B-F, Liu I-P, Huang T-P. Gene–environment interaction between interleukin-4 promoter and molds in childhood asthma. Ann Epidemiol. 2012;22(4):250–256. doi: 10.1016/j.annepidem.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 61.Chiang C-H, Lin M-W, Chung M-Y, Yang U-C. The association between the IL-4, ADRβ2 and ADAM 33 gene polymorphisms and asthma in the Taiwanese population. J Chin Med Assoc. 2012;75(12):635–643. doi: 10.1016/j.jcma.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 62.Micheal S, Minhas K, Ishaque M, Ahmed F, Ahmed A. IL4 gene polymorphisms and their association with atopic asthma and allergic rhinitis in Pakistani patients. 2013. [PubMed] [Google Scholar]
- 63.Ricciardolo FLM, Sorbello V, Silvestri M, Giacomelli M, Debenedetti V, Malerba M, Ciprandi G, Rossi G, Rossi A, Bontempelli M. TNF-α, IL-4Rα and IL-4 polymorphisms in mild to severe asthma from Italian Caucasians. Int J Immunopathol Pharmacol. 2013;26(1):75–84. doi: 10.1177/039463201302600107. [DOI] [PubMed] [Google Scholar]
- 64.Smolnikova MV, Smirnova SV, Freidin MB, Tyutina OS. Immunological parameters and gene polymorphisms (C-590T IL4, C-597A IL10) in severe bronchial asthma in children from the Krasnoyarsk region, West Siberia. Int J Circumpolar Health. 2013;72(1):21159. doi: 10.3402/ijch.v72i0.21159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, Lin L-H, Wang J, Peng X, Dai H-R, Xiao H, Li F, Wang Y-P, Yang Z-J, Li L. Interleukin-4 and interleukin-13 pathway genetics affect disease susceptibility, serum immunoglobulin E levels, and gene expression in asthma. Ann Allergy Asthma Immunol. 2014;113(2):173–179.e171. doi: 10.1016/j.anai.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 66.Wang R-S, Jin H-X, Shang S-Q, Liu X-Y, Chen S-J, Jin Z-B. Relación entre la expresión de IL-2 e IL-4 y sus polimorfismos y los riesgos de padecer infección por Mycoplasma pneumoniae y asma en niños. Arch Bronconeumol. 2015;51(11):571–578. doi: 10.1016/j.arbres.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 67.Dahmani DI, Sifi K, Salem I, Chakir J, Hanachi S, Bachtarzi MZ, Rouabah L, Abadi N, Bougrida M, Rouabhia M. The C-589T IL-4 single nucleotide polymorphism as a genetic factor for atopic asthma, eczema and allergic rhinitis in an eastern Algerian population. Int J Pharm Sci Rev Res. 2016;37(1):213–223. [Google Scholar]
- 68.Li L, Li Y, Zeng X, Li J, Du X. Role of interleukin-4 genetic polymorphisms and environmental factors in the risk of asthma in children. Genet Mol Res. 2016;15(4):534–543. doi: 10.4238/gmr15048873. [DOI] [PubMed] [Google Scholar]
- 69.Narożna B, Hoffmann A, Sobkowiak P, Schoneich N, Bręborowicz A, Szczepankiewicz A. Polymorphisms in the interleukin 4, interleukin 4 receptor and interleukin 13 genes and allergic phenotype: a case control study. Adv Med Sci. 2016;61(1):40–45. doi: 10.1016/j.advms.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Hussein IA, Jaber SH. Genotyping of IL-4− 590 (C> T) gene in Iraqi asthma patients. Dis Markers. 2017;2017. [DOI] [PMC free article] [PubMed]
- 71.Abood SH, Mohanad A-E. Study of the correlation between total immunoglobulin-E levels and inter-leukin-4 polymorphism in asthmatic children. Int J Res Pharm Sci. 2018;9(4):1515–1523. [Google Scholar]
- 72.Zhang J-H, Zhang M, Wang Y-N, Zhang X-Y. Correlation between IL-4 and IL-13 gene polymorphisms and asthma in Uygur children in Xinjiang. Exp Ther Med. 2019;17(2):1374–1382. doi: 10.3892/etm.2018.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, Zhou G, Wei T, Chang Z. Association between the interleukin 4 gene-590C> T promoter polymorphism and asthma in Xinjiang Uighur children. Genet Mol Res. 2016;15(3). [DOI] [PubMed]
- 74.Lilly CM. Diversity of asthma: evolving concepts of pathophysiology and lessons from genetics. J Allergy Clin Immunol. 2005;115(4):S526–S531. doi: 10.1016/j.jaci.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 75.Zhang S, Li Y, Liu Y. Interleukin-4-589C/T polymorphism is associated with increased pediatric asthma risk: a meta-analysis. Inflammation. 2015;38(3):1207–1212. doi: 10.1007/s10753-014-0086-9. [DOI] [PubMed] [Google Scholar]
- 76.Carpaij OA, Burgess JK, Kerstjens HA, Nawijn MC, van den Berge M. A review on the pathophysiology of asthma remission. Pharmacol Ther. 2019;201:8–24. doi: 10.1016/j.pharmthera.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Fu C-L, Ye Y-L, Lee Y-L, Chiang B-L. Effects of overexpression of IL-10, IL-12, TGF-β and IL-4 on allergen induced change in bronchial responsiveness. Respir Res. 2006;7(1):72. doi: 10.1186/1465-9921-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carr TF, Zeki AA, Kraft M. Eosinophilic and Noneosinophilic asthma. Am J Respir Crit Care Med. 2018;197(1):22–37. doi: 10.1164/rccm.201611-2232PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akkad D, Arning L, Ibrahim S, Epplen J. Sex specifically associated promoter polymorphism in multiple sclerosis affects interleukin 4 expression levels. Genes Immun. 2007;8(8):703–706. doi: 10.1038/sj.gene.6364429. [DOI] [PubMed] [Google Scholar]
- 80.Rosenwasser LJ, Borish L. Genetics of atopy and asthma: the rationale behind promoter-based candidate gene studies (IL-4 and IL-10) Am J Respir Crit Care Med. 1997;156(4):S152–S155. doi: 10.1164/ajrccm.156.4.12tac-14. [DOI] [PubMed] [Google Scholar]
- 81.Wang Z-d, Lian D, Shen J-L, Sun R, Xu W, Xin Z, Lei L, Jin L-H. Association between the interleukin-4, interleukin-13 polymorphisms and asthma: a meta-analysis. Mol Biol Rep. 2013;40(2):1365–1376. doi: 10.1007/s11033-012-2180-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that support the conclusions of this manuscript are included within the article.