Abstract
Background
Genomic studies of high-grade/progressive meningiomas have reported a heterogeneous mutation spectrum, identifying few recurrently mutated genes. Most studies have been underpowered to detect genomic subclasses of aggressive meningiomas due to relatively small number of available samples. Here, we present a genomic survey of one of the largest multi-institutional cohorts of high-grade/progressive meningiomas to date.
Methods
850 high-grade/progressive meningiomas, including 441 WHO grade 2 and 176 WHO grade 3 meningiomas and 220 progressive WHO grade 1 meningiomas, were tested as part of a clinical testing program by hybridization capture of 406 cancer-related genes to detect base substitutions, indels, amplifications, deletions, and rearrangements. Information from pathology reports, histopathology review, and patient clinical data was assessed.
Results
Genomic analyses converged to identify at least three distinct patterns of biologically-aggressive meningiomas. The first and most common contained NF2-mutant tumors (n = 426, 50%), was associated with male sex (64.4% %, p = 0.0001) and often harbored additional mutations in CDKN2A/B (24%), and the chromatin regulators ARID1A (9%), and KDM6A (6%). A second group (NF2-agnostic) featured TERT promoter (TERTp; n = 56) or TP53 mutations (n = 25) and were either NF2-mutant or wild-type, and displayed no association with either sex (p = 0.39). The remaining group generally lacked NF2 mutations, and accounted for 40% of the cases—with three subgroups. One consistent primarily of grade 3 lesions harboring alterations in chromatin regulators BAP1 (n = 22) or PBRM1 (n = 16). A second subgroup contained AKT1 (n = 26), PIK3CA (n = 14) and SMO (n = 7) mutant skull-based meningiomas, and a third mixed subgroup included 237 meningiomas with a heterogeneous spectrum of low frequency and non-recurrent alterations.
Conclusions
Our findings indicate that the patterns of genomic alterations in high-grade/progressive meningiomas commonly group into three different categories. The most common NF2-associated canonical group frequently harbored CDKN2A/B alterations, which is potentially amenable to targeted therapies. An NF2-agnostic group harbored frequent TERTp and TP53 mutations. The final subclass, distinct from the canonical NF2 mutant associated pathway, was partly characterized by BAP1/PBRM1 alterations (rhabdoid/papillary histology) or skull-base disease. Overall, these data increase our understanding of the pathobiology of high-grade/progressive meningiomas and can guide the design of clinical trials.
IRB approval status
Reviewed and approved by Western IRB; Protocol No. 20152817.
Electronic supplementary material
The online version of this article (10.1186/s40478-020-01040-2) contains supplementary material, which is available to authorized users.
Keywords: Meningioma, 1p loss, Progressive, NF2, TERT, PBRM1, CDKN2A, KDM6A, Metastatic
Introduction
Our understanding of the molecular biology of meningiomas has significantly improved in the last decade. This new era began with the publication of two landmark genetic sequencing studies in 2013, in which AKT1, SMO, TRAF7 and KLF4 were identified as frequently mutated genes in skull base, WHO grade 1 meningiomas [5, 6]. Since that time, much work has been carried out to detect additional recurrently mutated genes involved in the molecular pathogenesis of these tumors [1, 7, 37].
WHO grade 2 and 3 meningiomas have high-grade histologic features and represent up to 25% of all meningiomas. Affected patients frequently experience disease progression despite treatment, with aggressive regrowth resulting in high morbidity and mortality. There is substantial heterogeneity in clinical presentation amongst these tumors, as some initially present as benign grade 1 disease, and only manifest biologically-aggressive progression after many years (either as recurrent grade 1, or transformed higher-grade disease), while other tumors are higher grade (2 or 3) at initial presentation. Investigators have long sought effective predictors of meningioma recurrence and malignant transformation, to guide the administration of intensified treatments such as radiation or radical surgery, or to identify genomic subsets that are amenable to targeted therapies.
The genetic profiles of high-grade meningiomas are dominated by inactivation of the NF2 tumor suppressor gene. However, more data have accumulated regarding alterations in genes not previously implicated in meningiomas that appear to delineate subsets of progressive meningiomas with unfavorable clinical prognoses. These aberrations include promoter mutations and rearrangements of TERT, and inactivating mutations in BAP1, PBRM1, ARID1A, and SMARCE1 as well as focal deletions of the dystrophin gene (DMD) [2, 3, 14, 19–21, 28, 33, 41]. Mutations in some of these genes are enriched in tumors with distinct histologic features such as SMARCE1 in clear cell meningioma, and BAP1 and PBRM1 in rhabdoid and papillary meningiomas, respectively [33, 36, 41].
Despite this progress in identifying recurrently mutated genes in aggressive meningiomas, these genes are altered in fewer than 35% of high-grade/progressive meningiomas, suggesting the existence of additional aberrations in unidentified or underrepresented cancer genes. Notably, most genomic studies of high-grade meningiomas have been underpowered to detect recurrent clinically actionable mutations and distinct high-grade meningioma molecular subclasses. Consequently, most studies have reported a heterogeneous spectrum of mutations with few recurrently mutated genes [27, 28, 32] and have not provided detailed information regarding how often these mutations co-occur with one another.
In this study, we performed comprehensive genomic profiling of one of the largest sets of WHO grades 2 and 3 meningiomas analyzed to date. We also included analysis of clinically-progressive grade 1 meningiomas. We aimed to assess clinically actionable mutations to facilitate identifying patients amenable to targeted treatment trials. In addition, we sought to improve our understanding of genomic events underlying high-grade/progressive meningiomas.
Methods
Tumor samples and molecular genetic analysis
Samples from 850 meningiomas were analyzed as part of a clinical care for patients using comprehensive genomic profiling (CGP) in a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited laboratory (Foundation Medicine, Cambridge, MA). Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817). The pathologic diagnosis of each case was first made in the referring center and was then confirmed in our facility (Foundation Medicine, Cambridge, MA) on routine hematoxylin and eosin stained slides. All samples that contained a minimum of 20% tumor nuclear nuclei were forwarded for DNA and/or RNA extraction. WHO grade was not available for a small subset of samples (n = 13).
Technical descriptions and validation of the genomic profiling assays used to analyze these samples in the course of clinical care have been published previously [12, 16]. In brief, ≥ 50 ng DNA was extracted from 40 µm scrolls from formalin-fixed, paraffin-embedded tissue blocks of tumor. The samples were assayed by adaptor ligation hybrid capture, performed for all coding exons of 236 (v1), 315 (v2), or 405 (v3) cancer-related genes plus select introns from 19 (v1), 28 (v2), or 31 (v3) genes frequently rearranged in cancer (Additional file 1: Table S1) [12, 16]. For those samples for which RNA was available, targeted RNA-seq. was performed for rearrangement analysis in 265 genes [20]. RNA sequences were analyzed for the presence of rearrangements only. Sequencing of captured libraries was performed using an Illumina technology to a mean exon coverage depth of 593 ×, and resultant sequences were analyzed for base substitutions, insertions, deletions, copy number alterations (focal amplifications and homozygous deletions), and select gene fusions, as previously described [12, 16]. Clinically relevant genomic alterations (CRGA) were defined as alterations that are targetable by anticancer drugs currently available on the market or in registered clinical trials. Germline variants documented in the dbSNP database (dbSNP142; http://www.ncbi.nlm.nih.gov/SNP/), with two or more counts in the ExAC database (http://exac.broadinstitute.org/), or recurrent variants of unknown significance that were predicted by an internally developed algorithm to be germline were removed, with the exception of known driver germline events (e.g., documented hereditary BRCA1/2 and deleterious TP53 mutations). Known confirmed somatic alterations deposited in the Catalog of Somatic Mutations in Cancer were highlighted as biologically significant [11]. All inactivating events (i.e., truncations and deletions) in known tumor suppressor genes were also called as significant. To maximize mutation-detection accuracy (sensitivity and specificity) in impure clinical specimens, the test was previously optimized and validated to detect base substitutions at a ≥ 5% mutant allele frequency (MAF), indels with a ≥ 10% MAF with ≥ 99% accuracy, and fusions occurring within baited introns/exons with > 99% sensitivity [12].
Molecular assignment of subclasses
Based upon mutational phenotype in each sample, we assigned the tumors to at least three subclasses. The used molecular assignment relies on the growing body of literature of molecular patterns that have been recently described in meningiomas [25, 32, 42, 43].
Statistical analysis
The statistical association of detected somatic alterations with other factors, including age, sex, and tumor location were analyzed using the Fisher exact and Mann–Whitney-U tests. Cases with unavailable molecular or histology data were excluded from the final correlation analysis. A two-tailed p value of < 0.05 was considered to be statistically significant. Furthermore, R statistics system version 3.6.1 (https://www.r-project.org/) together with the UpSetR library [8] was used to construct the diagrams of set intersections used in this paper.
Results
Patient demographics
We identified 850 biologically-aggressive meningiomas (defined further below) with available molecular data that were profiled in the comprehensive genomic profiling (CGP) program at Foundation Medicine between 2013 and 2019. These tumors were resected from different patients; 466 females and 384 males (ratio 1.2:1). The median patient age was 57 years (range 0–89 + years). While the cohort included meningiomas of all WHO grades (1, 2 and 3), it predominantly consisted of high-grade meningiomas (441 WHO grade 2 and 176 WHO grade 3), in addition to 220 “progressive” WHO grade 1 meningioma defined as relapsed or “under-treatment” progressive tumors. Tumor locations included the skull base (n = 243), supratentorial area (non-skull base, n = 430), non-CNS sites including the skin of the head and distant metastases (n = 32), and spine (n = 30; three of these spine meningiomas were clinically considered as likely drop metastases). The data about tumor location was not available for 114 cases. (Additional file 1: Table S1).
Genomic alterations
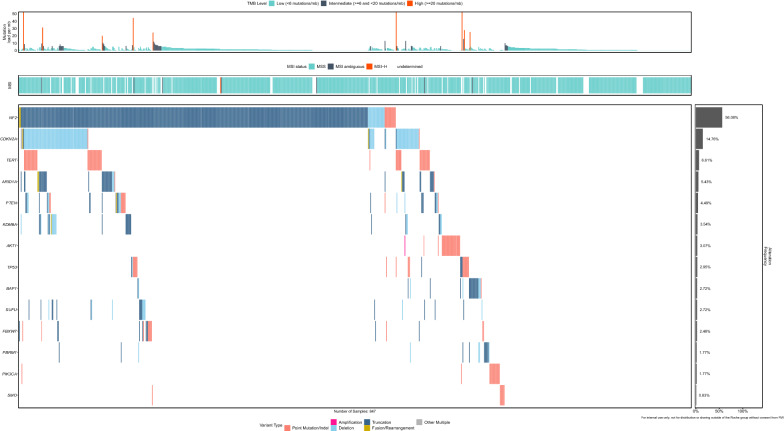
Loss of chromosome 22q (84.1% of eligible cases) and loss of chromosome 1p (68.8%) were the most common copy number alterations in our cohort (Additional file 2: Figure S1). NF2 mutations represented the most frequent gene mutations (n = 474/850, 55.7%). While NF2 mutations and 22q loss events were correlated, there were several cases with 22q loss without detectable NF2 mutation, raising the possibility of cryptic NF2 inactivation. The frequency of NF2 mutations increased significantly with the WHO grade: 81/220 of grade 1 (36.8%), 265/441 of grade 2 (60.1%, p = 0.0001) and 122/176 of grade 3 (69.3%, p = 0.0001) meningiomas harbored NF2 mutations. Moreover, male patients (n = 245/384, 64%) were significantly more affected by NF2-mutant meningiomas than female patients (n = 227/466, 48.7%, p = 0.0001). We detected frequent genomic alterations of genes encoding regulators of the cyclin-dependent kinase inhibitor pathway, including CDKN2A/B deletions (n = 123/850, 14.4%), CDKN2C mutations (n = 16/850, 1.9%), and CDK4 amplifications (17/850 cases with high level amplifications ≥ 4 gene copies, 2%). Additional frequently altered genes included: TERT (7.1%, 56 promoter hotspot mutations of 789 sequenced cases), ARID1A (5.4%, 46 cases, including one gene structural translocation), PTEN (4.3%, 27 mutations, 10 focal gene deletions and one PTEN_TMEM38A_rearrangement), KDM6A (3.5%; 30 cases, including focal exon deletions), SUFU (2.7%, 17 mutations, 5 focal exon deletions and one gene rearrangement), TP53 (2.9%, 23 mutations, 1 focal deletion and 1 gene translocation), BAP1 (2.7%, 18 mutations and 5 exon deletions) and PBRM1 (1.8%, 10 mutations and 5 focal exon deletions). This comprehensive genomic characterization suggested that high-grade/progressive meningiomas could be separate into largely distinct classes; for further analysis, we divided the tumors into three overall subclasses (Figs. 1, 2). All genomic data are shown in Additional file 1: Table S1.
Fig. 1.
Tile plot to summarize molecular alterations in progressive/high-grade meningiomas
Fig. 2.
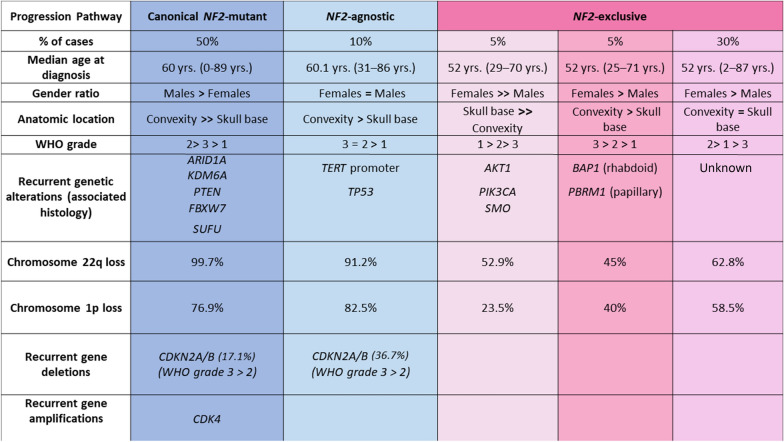
Comparison of three distinct patterns of biologically-aggressive meningiomas. The first and most common subclass (50%) contained NF2-mutant tumors, was associated with male sex and harbored additional alterations in CDKN2A/B, ARID1A, PTEN, and KDM6A. A second group featured TERTp or TP53 mutations and were either NF2-mutant or wild-type and with no association with either sex. The remaining group which accounted for 40% of the cases, generally lacked NF2 mutations and harbored alterations in BAP1/PBRM1, with a further subgroup containing AKT1, PIK3CA and SMO mutant meningiomas and a third with a heterogeneous spectrum of low frequency and non-recurrent alterations
The NF2-associated pathway of meningiomas (the canonical pathway)
This group included 426 meningiomas (50.1%), and represented the largest group of tumors. The NF2 alterations involved 393 NF2 missense mutations (93%), 21 two copy deletions (4.5%) and 12 NF2 structural rearrangements (2.5%). The vast majority of tumors (99.7%) showed synchronous chromosome 22q LOH and 76.9% showed a 1p chromosome loss. The median age of patients was 59 years (range 0–89 + years). We also observed an enrichment in alterations in CDKN2A/B, KDM6A, ARID1A, PTEN, FBXW7, and SUFU in comparison with NF2-wt meningiomas (Additional file 3: Figure S2).
Bi-allelic CDKN2A/B deletions occurred in 93/472 of all NF2-mutant (19.7%) versus 30/377 in NF2-wt meningiomas (8.0%) (p = 0.0001). In the canonical NF2-mutant group, 17.1% (n = 72/426) harbored a CDKN2A/B deletion (Fig. 2). Interestingly, meningiomas harboring CDKN2A/B alterations were significantly more common in males (n = 77/123, 62.6%) than in females (p = 0.0001). CDKN2A/B alterations were significantly enriched in WHO grade 3 meningiomas (n = 75/176, 42.6%) in comparison to WHO grade 2 (n = 47/441, 10.6%, p = 0.0001). Only one WHO grade 1 meningioma had a CDKN2A/B alteration (n = 1/220, 0.4%, p = 0.0001).
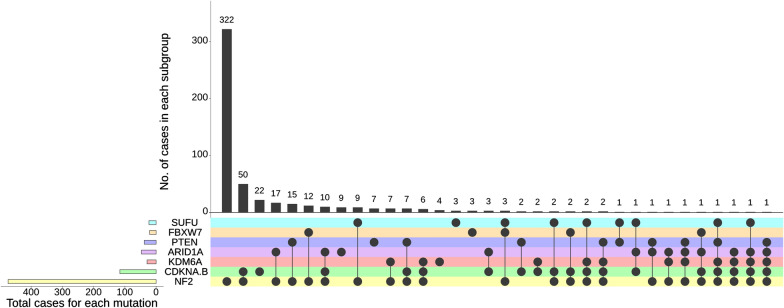
Furthermore, were observed a significant association of alterations in the chromatin regulator ARID1A in NF2-mutant meningiomas (n = 34/472, 7%) versus NF2-wt meningiomas (n = 12/377, 3.2%, p = 0.0138). ARID1A alterations were significantly more common in WHO grade 3 meningiomas (n = 26/176, 14.8%) than in WHO grade 2 (n = 18/441, 4.1%, p = 0.0001) and WHO grade 1 meningiomas (n = 1/220, 0.4%, p = 0.0001). ARID1A alterations were significantly more frequently seen in male patients (29/46, 63%, p = 0.0146). Moreover, a mutual co-occurrence of ARID1A mutations was detected in a subset of meningiomas that harbored aberrations of CDKN2A/B (n = 18), KDM6A (n = 16), PTEN (n = 13), FBXW7 (n = 3) and SUFU (n = 7), as shown in Fig. 3.
Fig. 3.
Diagram of set intersection between the most frequently detected genomic alterations in the NF2-associated subclass in meningiomas
PTEN mutations were identified in 28/472 of all NF2-mutant (5.9%) and in 10/376 of NF2-wt meningiomas (2.6%) (p = 0.0287). Additionally, PTEN mutations were found to be more frequently associated with a WHO grade 3 (n = 15/176, 8.5%) than grade 2 (n = 17/441, 3.8%, p = 0.025) or grade 1 (5/220, 2.3%, p = 0.0094) meningioma in our cohort. Notably, PTEN mutations occurred mutually concurrent with CDKN2A/B (n = 13), KDM6A (n = 5) and SUFU aberrations (n = 2), as shown in Fig. 3.
Importantly, we detected KDM6A alterations in 23/472 of all NF2-mutant (4.9%) versus 7/377 in NF2-wt meningiomas (1.9%) (p = 0.0234). Similarly, KDM6A alterations occurred significantly more often in WHO grade 3 meningiomas (n = 15/176, 9%) versus 13/441 (2.9%, p = 0.0046) and 2/220 (0.9%, p = 0.0001) in WHO grade 2 and 1 meningiomas, respectively.
In addition, the meningiomas of 23 patients showed SUFU mutations, 18 in NF2-mutant (3.7%) and 5 in NF2-wt meningiomas (1.3%, p = 0.033). SUFU mutations were found more frequently in WHO grade 3 (n = 11/176, 6.3%) than grade 2 (n = 11/441, 2.5%, p = 0.030) or grade 1 meningiomas (2/220, 0.9%, p = 0.0036) in our cohort.
FBXW7 mutations were present in 23/472 of all NF2-mutant (4.9%) and in 3/377 of NF2-wt meningiomas (0.8%) (p = 0.0004). No significant difference was identified between grades. Interestingly, we observed that FBXW7 mutations were mutually exclusive to PTEN or KDM6A mutations (Fig. 3).
Finally, in contrast to NF2, CDKN2A/B, and ARID1A alterations, male sex was not significantly associated with gene alterations in KDM6A, PTEN, FBXW7 or SUFU.
The NF2-agnostic group of meningiomas
This group (n = 79, 9.3%) was characterized by TERTp and TP53 mutations. The median age of patients was 60.1 years (range 31–86 years). TERTp mutations occurred in 35/441 (8.1%) of NF2-mutant meningiomas and in 21/348 of NF2-wt patients (5.7%, p = 0.21). WHO grade 3 meningiomas harbored the highest percentage of TERTp mutations (23/159, 14.5%), followed by WHO grade 2 (n = 28/421, 6.7%, p = 0.0048) and WHO grade 1 meningiomas (n = 5/199, 2.5%, p = 0.0001, Additional file 3: Figure S2).
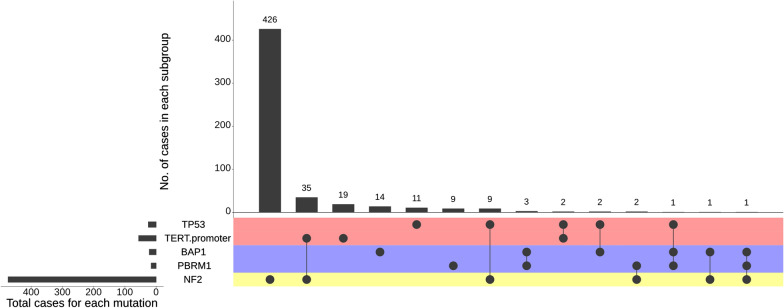
Likewise, TP53 hotspot mutations were detected in 9/472 NF2-mutant meningiomas (1.9%), compared with 16/373 NF2-wt meningiomas (4.2%, p = 0.063). TP53 mutations occurred mainly in WHO grade 3 meningiomas (n = 12/176, 6.8%) followed by WHO grade 2 (n = 10/441, 2.2%, p = 0.0135) and a significantly lower percentage in grade 1 (n = 2/220, 0.9%, p = 0.0018) meningiomas. Notably, two meningiomas harbored simultaneous TERTp and TP53 mutations (Fig. 4). Neither TERTp (25 females, 31 males) nor TP53 (13 females, 12 males) mutations were significantly associated with patient sex. Further frequent genomic alterations in this group were CDKN2A/B deletions (n = 29/79, 36.7%), chromosome 22q loss (91.2%) and chromosome 1p loss (82.5%), Fig. 2).
Fig. 4.
Diagram of set intersection between the most frequently detected genomic alterations in the NF2-agnostic subclass in meningiomas
The NF2 exclusive group of meningiomas contains three subclasses
There were several mutations identified in this third group of high-grade/progressive meningiomas which we further split into three largely distinct subclasses. In one of these subclasses, we found frequent BAP1 (n = 22) and PBRM1 (n = 16) alterations—including 5 cases with BAP1 and PBRM1 aberrations in the same tumor (total n = 33 tumors, Fig. 4). In addition, 81% of BAP1 and 87.5% PBRM1 mutant meningiomas were classified as WHO grades 2 and 3 meningiomas. Simultaneous NF2 mutations were very infrequently detected (n = 4/33 cases) in this subgroup. Likewise, only 45% and 40% of the meningiomas in this group harbored chromosome 22q and 1p loss, respectively. The median age of patients was 52 years (range 25–76 years). The hallmark of this subgroup is the substantial enrichment for rhabdoid and papillary histology and the not infrequent co-occurrence of these histologic features in the same tumor resection. As previously reported, 11 of 16 PBRM1 mutations (68.7%) occurred in meningioma with papillary histologic features [41].
A second subclass within this group contained AKT1 (n = 26), PIK3CA (n = 14) and SMO (n = 6) mutant meningiomas (total n = 46, Additional file 2: Figure S1). We observed a marked predominance of female patients in this subgroup (37 females vs. 9 males). In addition, the majority of these NF2-wt tumors were classified as WHO grade 1 meningioma (n = 31, 70.4%) and were located almost exclusively in the skull base (Fig. 2). Notwithstanding, this subclass also contained 14 WHO grade 2 meningiomas (8 AKT1, 5 PIK3CA, and 1 SMO case), and a subset of these higher grade cases harbored additional genomic alterations such as TERTp mutation (one AKT1-mutant meningioma), TP53 mutation (one PIK3CA-mutant case), PTEN alterations (2 meningiomas with an AKT1 mutation), and CDKN2A 2 copy loss (1 AKT1 case; 1 PIK3CA case). This finding suggests, due to anatomic location, a more biological aggressive course can occur during progression in these meningiomas that are largely regarded as “benign” and that the presence of these mutations is not invariably indicative of a WHO grade 1 designation.
The last subclass in this group contained a mix of 237 meningiomas with a heterogeneous spectrum of mutations outside of the genes mutated in other subclasses such as NF2, TERTp, TP53, BAP1, PBRM1, AKT1, SMO and PIK3CA. The majority of tumors in this subgroup occurred in women (n = 140, 59%). Chromosome 22q monosomy was detected in 112 cases (47.2%). The WHO grading was 88, 120 and 24 of grade 1, 2 and 3, respectively. This group will require further genomic characterization; some of the tumors in this class may ultimately be assigned to one of the other classes (for instance, 22q loss cases resolving to the NF2-associated canonical pathway above), whereas others may contain genomic or non-genomic drivers that are not yet associated with meningioma pathogenesis.
Disseminated meningiomas
Our cohort includes genomic data from 35 (4%) metastatic meningiomas (21 females and 14 males) with 3 occurring as metastases in the spine and 32 occurring outside of the CNS. Extracranial systemic metastases (e.g., in lung, kidney, liver) were reported in 17 cases (49%), whereas the remaining cases disseminated to skin and spine. The majority of disseminated meningiomas were WHO grade 2 (n = 16) and grade 3 (n = 11). Metastatic meningiomas were observed across all three molecular subclasses. Alterations in NF2 (n = 19, 54.3%), CDKN2A (n = 8, 22.9%), BAP1 (n = 4, 11.4%), ARID1A (n = 4, 11.4%), TERTp (n = 3, 9.4%), and TP53 (n = 2, 5.7%) were the most frequent relevant alterations detected in these cases. Interestingly, we did not observe significant association between any of these alterations with dissemination.
Finally, we did not detect recurrent clinically targetable gene rearrangements in our cohort (Additional file 1: Table S1).
Discussion
This series of 850 meningiomas represents the largest collection of high-grade/progressive meningiomas with comprehensive genomic profiles presented to date. The samples originating from over 35 institutions were analyzed as part of a clinical care at Foundation Medicine. With the advantage of broad coverage of greater than 300 genes on our next-generation sequencing panel, the cohort is powered to examine clinical associations and gene–gene pathway interactions for a survey of the dominant pathways that contribute to the development of biologically-aggressive meningiomas. Taken together, we have identified at least three distinct molecular subclasses that define specific genomic tumor subgroups. Assigning meningiomas into distinct molecular subclasses will allow for a more detailed understanding of tumor-specific molecular features, for dissection of biological pathways, and for guiding approaches for diagnosis, treatment decisions and clinical trial design [17].
The first group, which we termed “canonical”, was the most commonly represented, and was associated with NF2-mutation and male-sex, and also harbored additional, previously described mutations in genes including CDKN2A/B, KDM6A, ARID1A, PTEN, FBXW7, and SUFU [2, 4, 20, 22, 30, 40]. Notably from a clinical standpoint, alterations of genes encoding regulators of the cyclin-dependent kinase (CDK) inhibitor pathway occurred in ~ 20% of NF2-mutant high-grade/progressive meningiomas. This frequent genomic finding is relevant because it indicates an opportunity for testing CDK4/CDK6 inhibitors in genomically-guided clinical trials (e.g., Alliance A071401 trial, NCT02523014). Furthermore, we found that the tumor suppressor gene and chromatin regulator KDM6A, which has been identified at low frequency in prior studies with smaller cohorts of meningioma, is more frequently altered in NF2-mutant high-grade meningiomas than previously noted [5, 20]. Indeed, previous work has observed that increased H3K27me3 levels and a hyper-methylated phenotype occupying the polycomb repressive complex (PRC2), is one of the hallmarks of NF2-mutant high-grade meningiomas [15, 22]. Taking into account that KDM6A, which is located on chromosome X and escapes X-inactivation, acts antagonistically to PRC2 and promotes H3K27 demethylation, our findings may partly explain the upregulated PRC2 activity in the NF2-mutant group of meningiomas after inactivation of KDM6A [10, 24, 38, 39].
The second group we identified contains both NF2 mutant and wild-type tumors, and was not associated with either sex. We termed these poor prognosis meningiomas as “NF2-agnostic,” reflecting lack of associations with the presence or absence of NF2 alterations in this cohort. TERTp mutations or TP53 mutations typified this group, with recurrent mutations in the TERTp found in a substantial fraction of these meningiomas. In our large cohort, we estimate the frequency of TERTp mutations ranging from 2.5 to 14% in WHO grade 1 and WHO grade 3 meningiomas, respectively. These estimates must be tempered as the frequency of TERTp mutations might be underestimated in high-grade meningiomas due to intratumoral heterogeneity, the late emergence during tumor evolution, or the occurrence of other TERT alterations, such as gene rearrangements [15, 20, 21]. Some investigators have proposed a designation of “grade 4” meningioma for these meningiomas [25]. Interestingly, in our cohort, this group includes a relatively large subset of high-grade meningiomas that harbored TP53 mutations, a gene alteration that has been previously seldom described in meningiomas or during meningioma progression [18, 20, 40]. This poor prognosis group may prove difficult to treat with targeted agents.
The final group is essentially exclusive of NF2 mutations, containing at least three subgroups. These tumors are less frequent than canonical pathway lesions. The first subgroup is characterized by mutations in BAP1 and/or PBRM1. Although NF2-wt cases have been less frequently linked with increased risk of progression, this NF2-wt subset is uniquely aggressive, which is captured within the current WHO classification that grades the majority of tumors in this subset as grade 3. Indeed, inactivation of the tumor suppressor gene BAP1 has been linked to aggressive meningiomas with rhabdoid histo-morphology and was associated with a shorter progression-free survival [33, 34]. In addition, we have recently reported frequent PBRM1 mutations in meningiomas with papillary histologic features [41]. BAP1 and PBRM1 inactivation can co-occur, and these tumors can have complex mixed histology patterns [33, 41]. Importantly, the frequent alterations of genes that encode epigenetic factors, such as the SWI/SNF complex genes PBRM1 and ARID1A, in addition to KDM6A, BAP1 and FBXW7 indicate that dysregulation of chromatin remodeling is a common feature across all higher-grade meningiomas, regardless of the molecular subgroup [2, 20, 24, 33, 34, 39].
An additional subgroup of the NF2-exclusive category consists of the previously described subset of skull base meningiomas with AKT1, SMO or PIK3CA mutations and predominantly WHO 1 grading [1, 5, 6, 31]. It is important to note that tumors with these mutations are generally grade 1 but their presence in higher grade and progressive meningiomas suggests that they are not exclusively indicators of non-aggressive behavior. This may, in part, be due to the confined anatomic location of these tumors.
In contrast to this subgroup, the last subgroup in the NF2-exclusive category included meningiomas with unknown drivers and with heterogeneous molecular mutations that lacked NF2, TERTp, TP53, BAP1, PBRM1, AKT1, SMO and PIK3CA mutations. In half of the cases in this subgroup, chromosome 22 monosomy was detected and was associated with a higher WHO grade. It is possible that some of these lesions may contain cryptic NF2 inactivation that was not detected with our sequencing methods. Nevertheless, further genomic large-scale analyses are needed to collect more genomic information and accurately subdivide the latter subgroup.
Interestingly, we did not detect frequent clinically targetable gene rearrangements in any of the subclasses in our study, indicating that progressive/high-grade meningiomas are seldom driven by gene rearrangements that are characterized in our genomic assay and are ineligible for targeted therapeutic agents against fusion oncogenes (e.g., NTRK, ALK, etc.). Moreover, none of the cases in our series harbored hotspot IDH1 or IDH2 mutations, which contrasts with recent reports [13, 29].
Finally, we identified and genotyped 35 disseminated meningiomas, with half of the cases with metastases outside of the CNS. Our study adds to the existing literature by providing genomic data on metastatic meningiomas which are quite rare (2%) [9]. We show that metastatic meningiomas are derived from all three subclasses in our study, with half of cases harboring NF2-mutations. Interestingly, loss of merlin, the protein encoded by NF2, has been linked to higher cell motility and tumor invasion [23, 35]. However, further functional studies are needed to explore the role of NF2 loss in tumor dissemination. One limitation of our study is the lack of treatment and survival data, which prevents comparison of the outcomes between the different molecular groups. In addition, although our NGS panel enables a broad survey of known cancer genes, it does not screen for relevant genomic alterations such as DMD deletions or TERT gene translocations both of which have been shown to be associated with a poor outcome in progressive/high-grade meningiomas [20]. Therefore, further studies with whole exome sequencing are warranted to confirm and extend our findings. Future studies will additionally need to address intra-tumor heterogeneity, since our sub-classification of meningiomas does not capture the evolving landscape of intra-tumor heterogeneity and evolutionary progression in high-grade/progressive meningiomas. In addition, comparison to newly established DNA methylation-based classifications and to studies of genome-wide copy number alterations has the potential to provide further important insights into biologically-aggressive meningiomas. Previous studies have developed and validated models using DNA methylation-based arrays to provide important prognostic information to guide therapeutic interventions in meningioma patients [26, 32].
In summary, our analysis identified at least three distinct genomic groupings within high-grade/progressive meningiomas, findings that can outline a genotype-driven sub-typing of these tumors. As with medulloblastomas, gliomas and other CNS tumors that are now defined by molecular parameters in addition to histology, we envision that such findings will aid in the transition towards a similarly integrated diagnostic approach for the molecular subclassification of meningiomas. In addition, we anticipate that molecular subclasses of aggressive meningiomas will have significant implications for the design of therapeutic trials in this patient population.
Supplementary information
Additional file 1: Supp. Table 1: Genomic data with all detected single nucleotide variants, small insertions/deletions, and chromosome arm data. The samples were assayed by adaptor ligation hybrid capture, performed for all coding exons of 236 (v1), 315 (v2), or 405 (v3) cancer-related genes plus select introns from 19 (v1), 28 (v2), or 31 (v3) genes frequently rearranged in cancer.
Additional file 2: Figure S1 shows the genome-wide copy-number alteration data of eligible cases. Loss of chromosome 22q (84.1%) and loss of chromosome 1p (68.8%) were the most common copy number alterations.
Additional file 3: Figure S2: a stacked bar graph demonstrates the most frequently detected mutations in meningiomas, based on their WHO grading. In 13 cases, WHO grading was not available. Those cases included: four meningiomas with NF2 mutations and alterations in PTEN (n= 1), ARID1A (n= 1), BAP1 (n= 1) and PTEN (n= 1).
Authors’ contributions
E.A.W. and T.A.J.: Conception/design. E.W.A., R.S., A.R., P.S., B.M.A. and J.S.R.: Collection and/or assembly of data. E.A.W., S.S., D.P.C., J.S.R. and T.A.J.: Data analysis and interpretation. E.A.W., S.S., D.P.C. and T.A.J.: Manuscript writing. All authors: Final approval of manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. None.
Competing interests
EAW, RS, AR, JMV, BMA, JSR, and SHR are employees or consultants of Foundation Medicine, Inc., a wholly owned subsidiary of Roche Holdings, Inc. and Roche Finance Ltd, and these employees have equity interest in an affiliate of these Roche entities. SS is a consultant for RareCyte. PKB reports honoraria for consulting from Tesaro, Lilly, Angiochem, and Genentech-Roche; speaker’s honoraria from Genentech-Roche and Merck; and research funding (to Massachusetts General Hospital) from BMS, Pfizer, Lilly and Merck. DPC reports receiving honoraria from Merck, Lilly and Boston Pharmaceuticals outside the submitted work. TAJ received honoraria from CSL Behring and Roche Pharma AG unrelated to the submitted work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daniel P. Cahill, Shakti H. Ramkissoon, and Tareq A. Juratli contributed equally to this work as co-senior authors
Contributor Information
Erik A. Williams, Email: erwilliams@foundationmedicine.com
Tareq A. Juratli, Email: tareq.juratli@uniklinikum-dresden.de
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40478-020-01040-2.
References
- 1.Abedalthagafi M, Bi WL, Aizer AA, Merrill PH, Brewster R, Agarwalla PK, Listewnik ML, Dias-Santagata D, Thorner AR, Van Hummelen P, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18:649–655. doi: 10.1093/neuonc/nov316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abedalthagafi MS, Bi WL, Merrill PH, Gibson WJ, Rose MF, Du Z, Francis JM, Du R, Dunn IF, Ligon AH, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015;208:345–350. doi: 10.1016/j.cancergen.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi WL, Greenwald NF, Abedalthagafi M, Wala J, Gibson WJ, Agarwalla PK, Horowitz P, Schumacher SE, Esaulova E, Mei Y, et al. Genomic landscape of high-grade meningiomas. NPJ Genom Med. 2017 doi: 10.1038/s41525-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boström J, Meyer-Puttlitz B, Wolter M, Blaschke B, Weber RG, Lichter P, Ichimura K, Collins VP, Reifenberger G. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159:661–669. doi: 10.1016/S0002-9440(10)61737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, Avşar T, Li J, Murray PB, Henegariu O, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark VE, Harmanci AS, Bai H, Youngblood MW, Lee TI, Baranoski JF, Ercan-Sencicek AG, Abraham BJ, Weintraub AS, Hnisz D, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48:1253–1259. doi: 10.1038/ng.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalle Ore CL, Magill ST, Yen AJ, Shahin MN, Lee DS, Lucas CG, Chen WC, Viner JA, Aghi MK, Theodosopoulos PV, et al. Meningioma metastases: incidence and proposed screening paradigm. J Neurosurg. 2019 doi: 10.3171/2019.1.JNS181771. [DOI] [PubMed] [Google Scholar]
- 10.Ezponda T, Licht JD. Molecular pathways: deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin Cancer Res. 2014;20:5001–5008. doi: 10.1158/1078-0432.CCR-13-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia MM, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill CM, Loewenstern J, Rutland JW, Arib H, Francoeur N, Wang YC, Fishman N, Pain M, Umphlett M, Kinoshita Y, et al. Recurrent IDH mutations in high-grade meningioma. Neuro Oncol. 2020 doi: 10.1093/neuonc/noaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014;24:184–189. doi: 10.1111/bpa.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmanci AS, Youngblood MW, Clark VE, Coskun S, Henegariu O, Duran D, Erson-Omay EZ, Kaulen LD, Lee TI, Abraham BJ, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017 doi: 10.1038/ncomms14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, Patel J, Krivstov A, Frampton GM, Young LE, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127:3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horbinski C, Ligon KL, Brastianos P, Huse JT, Venere M, Chang S, Buckner J, Cloughesy T, Jenkins RB, Giannini C, et al. The medical necessity of advanced molecular testing in the diagnosis and treatment of brain tumor patients. Neuro Oncol. 2019;21:1498–1508. doi: 10.1093/neuonc/noz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joachim T, Ram Z, Rappaport ZH, Simon M, Schramm J, Wiestler OD, von Deimling A. Comparative analysis of the NF2, TP53, PTEN, KRAS, NRAS and HRAS genes in sporadic and radiation-induced human meningiomas. Int J Cancer. 2001;94:218–221. doi: 10.1002/ijc.1467. [DOI] [PubMed] [Google Scholar]
- 19.Juratli TA, Brastianos PK, Cahill DP. TERT alterations in progressive treatment-resistant meningiomas. Neurosurgery. 2018;65:66–68. doi: 10.1093/neuros/nyy154. [DOI] [PubMed] [Google Scholar]
- 20.Juratli TA, McCabe D, Nayyar N, Williams EA, Silverman IM, Tummala SS, Fink AL, Baig A, Martinez-Lage M, Selig MK, et al. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol. 2018;136:779–792. doi: 10.1007/s00401-018-1899-7. [DOI] [PubMed] [Google Scholar]
- 21.Juratli TA, Thiede C, Koerner MVA, Tummala SS, Daubner D, Shankar GM, Williams EA, Martinez-Lage M, Soucek S, Robel K, et al. Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget. 2017;8:109228–109237. doi: 10.18632/oncotarget.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz LM, Hielscher T, Liechty B, Silverman J, Zagzag D, Sen R, Wu P, Golfinos JG, Reuss D, Neidert MC, et al. Loss of histone H3K27me3 identifies a subset of meningiomas with increased risk of recurrence. Acta Neuropathol. 2018 doi: 10.1007/s00401-018-1844-9. [DOI] [PubMed] [Google Scholar]
- 23.Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17:1090–1100. doi: 10.1101/gad.1054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao L, Testa JR, Yang H. The roles of chromatin-remodelers and epigenetic modifiers in kidney cancer. Cancer Genet. 2015;208:206–214. doi: 10.1016/j.cancergen.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirian C, Duun-Henriksen AK, Juratli T, Sahm F, Spiegl-Kreinecker S, Peyre M, Biczok A, Tonn JC, Goutagny S, Bertero L, et al. Poor prognosis associated with TERT gene alterations in meningioma is independent of the WHO classification: an individual patient data meta-analysis. J Neurol Neurosurg Psychiatry. 2020;91:378–387. doi: 10.1136/jnnp-2019-322257. [DOI] [PubMed] [Google Scholar]
- 26.Nassiri F, Mamatjan Y, Suppiah S, Badhiwala JH, Mansouri S, Karimi S, Saarela O, Poisson L, Gepfner-Tuma I, Schittenhelm J, et al. DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro Oncol. 2019;21:901–910. doi: 10.1093/neuonc/noz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassiri F, Mamatjan Y, Suppiah S, Badhiwala JH, Mansouri S, Karimi S, Saarela O, Poisson L, Gepfner-Tuma I, Schittenhelm J, et al. DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro-Oncol. 2019;21:901–910. doi: 10.1093/neuonc/noz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paramasivam N, Huebschmann D, Toprak UH, Ishaque N, Neidert M, Schrimpf D, Stichel D, Reuss D, Sievers P, Reinhardt A, et al. Mutational patterns and regulatory networks in epigenetic subgroups of meningioma. Acta Neuropathol. 2019;138:295–308. doi: 10.1007/s00401-019-02008-w. [DOI] [PubMed] [Google Scholar]
- 29.Pepe F, Pisapia P, Basso Del, de Caro ML, Conticelli F, Malapelle U, Troncone G, Martinez JC. Next generation sequencing identifies novel potential actionable mutations for grade I meningioma treatment. Histol Histopathol. 2019 doi: 10.14670/HH-18-195. [DOI] [PubMed] [Google Scholar]
- 30.Peters N, Wellenreuther R, Rollbrocker B, Hayashi Y, Meyer-Puttlitz B, Duerr EM, Lenartz D, Marsh DJ, Schramm J, Wiestler OD, et al. Analysis of the PTEN gene in human meningiomas. Neuropathol Appl Neurobiol. 1998;24:3–8. doi: 10.1046/j.1365-2990.1998.00093.x. [DOI] [PubMed] [Google Scholar]
- 31.Peyre M, Gaillard S, de Marcellus C, Giry M, Bielle F, Villa C, Boch AL, Loiseau H, Baussart B, Cazabat L, et al. Progestin-associated shift of meningioma mutational landscape. Ann Oncol. 2018;29:681–686. doi: 10.1093/annonc/mdx763. [DOI] [PubMed] [Google Scholar]
- 32.Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S, Okonechnikov K, Koelsche C, Reuss DE, Capper D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 33.Shankar GM, Abedalthagafi M, Vaubel RA, Merrill PH, Nayyar N, Gill CM, Brewster R, Bi WL, Agarwalla PK, Thorner AR, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19:535–545. doi: 10.1093/neuonc/now235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shankar GM, Santagata S. BAP1 mutations in high-grade meningioma: implications for patient care. Neuro Oncol. 2017;19:1447–1456. doi: 10.1093/neuonc/nox094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw RJ, McClatchey AI, Jacks T. Regulation of the neurofibromatosis type 2 tumor suppressor protein, merlin, by adhesion and growth arrest stimuli. J Biol Chem. 1998;273:7757–7764. doi: 10.1074/jbc.273.13.7757. [DOI] [PubMed] [Google Scholar]
- 36.Smith MJ, O’Sullivan J, Bhaskar SS, Hadfield KD, Poke G, Caird J, Sharif S, Eccles D, Fitzpatrick D, Rawluk D, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45:295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- 37.Strickland MR, Gill CM, Nayyar N, D’Andrea MR, Thiede C, Juratli TA, Schackert G, Borger DR, Santagata S, Frosch MP, et al. Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J Neurosurg. 2017;127:438–444. doi: 10.3171/2016.8.JNS161076. [DOI] [PubMed] [Google Scholar]
- 38.Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber RG, Boström J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA. 1997;94:14719–14724. doi: 10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams EA, Wakimoto H, Shankar GM, Barker FG, 2nd, Brastianos PK, Santagata S, Sokol ES, Pavlick DC, Shah N, Reddy A, et al. Frequent inactivating mutations of the PBAF complex gene PBRM1 in meningioma with papillary features. Acta Neuropathol. 2020 doi: 10.1007/s00401-020-02161-7. [DOI] [PubMed] [Google Scholar]
- 42.Youngblood MW, Clark V, Harmanci AS, Bai HW, Mora DD, Montejo J, Li C, Zhu HD, Erson-Omay EZ, Bilguvar K, et al. Clinical and molecular features of genomic subgroups in meningioma. J Neurosurg. 2017;126:A1424. [Google Scholar]
- 43.Youngblood MW, Duran D, Montejo JD, Li C, Omay SB, Ozduman K, Sheth AH, Zhao AY, Tyrtova E, Miyagishima DF, et al. Correlations between genomic subgroup and clinical features in a cohort of more than 3000 meningiomas. J Neurosurg. 2019 doi: 10.3171/2019.8.JNS191266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supp. Table 1: Genomic data with all detected single nucleotide variants, small insertions/deletions, and chromosome arm data. The samples were assayed by adaptor ligation hybrid capture, performed for all coding exons of 236 (v1), 315 (v2), or 405 (v3) cancer-related genes plus select introns from 19 (v1), 28 (v2), or 31 (v3) genes frequently rearranged in cancer.
Additional file 2: Figure S1 shows the genome-wide copy-number alteration data of eligible cases. Loss of chromosome 22q (84.1%) and loss of chromosome 1p (68.8%) were the most common copy number alterations.
Additional file 3: Figure S2: a stacked bar graph demonstrates the most frequently detected mutations in meningiomas, based on their WHO grading. In 13 cases, WHO grading was not available. Those cases included: four meningiomas with NF2 mutations and alterations in PTEN (n= 1), ARID1A (n= 1), BAP1 (n= 1) and PTEN (n= 1).