Summary
Previously developed senescent primary fibroblast models have limited relevance to study age-related changes in metabolically active tissues such as the liver. Here, we describe a protocol to generate senescent cells from the mouse hepatic cell line, AML12. These senescent cells exhibit molecular and metabolic signatures that are similar to those observed in livers from aged mice. These senescent AML12 cells should be a useful in vitro model to study the metabolic effects of aging in the liver.
For complete details on the use and execution of this protocol, please refer to Singh et al. (2020).
Graphical Abstract
Highlights
-
•
We have developed an in vitro hepatic aging model as senescent AML12 cells
-
•
This in vitro model exhibited hepatic age-related molecular and metabolic signatures
-
•
This method will contribute to the study of in vitro effects of hepatic aging
Previously developed senescent primary fibroblast models have limited relevance to study age-related changes in metabolically active tissues such as the liver. Here, we describe a protocol to generate senescent cells from the mouse hepatic cell line, AML12. These senescent cells exhibit molecular and metabolic signatures that are similar to those observed in livers from aged mice. These senescent AML12 cells should be a useful in vitro model to study the metabolic effects of aging in the liver.
Before You Begin
-
1.
Prepare culture medium:
Timing: approximately 30 min
-
a.
Complete growth medium (Table 1): Add 50 mL FBS (final 10%) + 5 mL penicillin/streptomycin 100× (final concentration 100 units/mL and 1,000 μg/mL respectively) + 5 mL ITS 100× (final 1× that correspond to 0.01 g/L insulin, 0.0055 g/L transferrin, and 0.0000067 g/L sodium selenite) + Dexamethasone (final 100 nM) in 440 mL DMEM:F12 medium for AML12 maintenance.
Table 1.
Solutions/Media Preparation
| Stock Concentration | Final Concentration | |
|---|---|---|
| Solution: Complete Growth Medium for Maintenance | ||
| DMEM:F12 medium | NA | NA |
| FBS | NA | 10% (v/v) |
| Penicillin-Streptomycin | 100× | 100 units/mL and 1,000 μg/mL |
| ITS | 100× | 0.01 g/L insulin, 0.0055 g/L transferrin, and 0.0000067 g/L sodium selenite |
| Dexamethasone | NA | 100 nM |
| Solution: Basal Medium for Treatment | ||
| DMEM:F12 medium | NA | NA |
| Penicillin-Streptomycin | 100× | 100 units/mL and 1,000 μg/mL |
-
b.
Basal DMEM:F12 medium (Table 1) with only penicillin/streptomycin (without FBS, ITS, and Dexamethasone) for H2O2 treatment.
-
2.
Thaw and culture AML12 cells (ATCC® CRL-2254™) in 5 mL complete medium in a T25 flask.
Timing: 15 min
-
3.
Incubate the cells in an incubator with 5% CO2/Air environment, 95% humidity and 37°C for 3–5 days so that the cells are approximately 90% confluent.
Timing: 2–5 days
-
4.
Once cells are 90% confluent, subculture AML12 cells in one T75 or two T25 flasks.
Timing: 30 min
-
5.
Prepare and autoclave 1× phosphate buffered saline (PBS).
Timing: 1.5–2 h
-
6.
Aliquot 30% H2O2 solution (9.7 M) in small vials (preferably 15–20 μL in 100–200 μL centrifuge tubes) and store at −20°C. Aliquoting in small volume tubes decreases the amount of oxidizing air volume.
Timing: 30 min
-
7.
Remember to pre-warm the media, reagents, buffer to 37°C before using them in this protocol
CRITICAL: H2O2 vapors are corrosive and irritating to the respiratory tract so handle H2O2 solution carefully in the hood. In the event of accidental skin contact, flush skin immediately with copious amounts of water for at least 15 min and also remove any contaminated clothing and shoes.
Note: Standard protocol for AML12 thawing and sub-culturing can be found at https://www.atcc.org/Products/All/CRL-2254.aspx.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM:F12 medium | Gibco/ThermoFisher | Cat# 11330032 |
| FBS | Sigma-Aldrich | Cat# F4135 |
| ITS; Insulin-Transferrin-Selenium (100×) | Gibco/ThermoFisher | Cat# 41400045 |
| Penicillin-Streptomycin (10,000 U/mL) (100×) | Gibco/ThermoFisher | Cat# 15140163 |
| Dexamethasone-Water Soluble | Sigma-Aldrich | Cat# D2915 |
| Hydrogen peroxide 30% stabilized, AnalaR NORMAPUR® | VWR chemicals Singapore | Cat# VWRC23619.297 |
| Phosphate buffered saline (10×) | Sigma-Aldrich | Cat# P5493 |
| TrypLE™ Express Enzyme (1×), phenol red | Gibco/ThermoFisher | Cat# 12605028 |
| Hoechst 33342 | Abcam | Cat# ab228551 |
| Alexa Fluor 488 antibody | Molecular Probes, ThermoFisher | Cat# A-11094 |
| γH2A.X antibody | Abcam /ThermoFisher | Cat# ab81299 |
| Critical Commercial Assays | ||
| Cellular Senescence Assay kit | Merck Millipore | Cat# KAA002 |
| Experimental Models: Cell Lines | ||
| AML12 cells | ATCC | ATCC® CRL-2254™ |
| Software and Algorithms | ||
| PRISM 8.0 | Graphpad | Version 8.0 |
| Other | ||
| Biological Safety Cabinet | Esco Micro Pte. Ltd. | Model abculture® Class II, Type A2 (E-Series) |
| Incubator | NuAire | Model No NU-5500E |
| Water bath | Julabo | Model No TW12 |
| Inverted microscope | Nikon Eclipse TS100 | N/A |
| Benchtop Refrigerated Centrifuge | Eppendorf/VWR Singapore Pte Ltd | Cat# EPPE5811000.320 Model 5810 R |
| Countess II automated Cell counter | ThermoFisher | Cat# AMQAX1000 |
| Nunc™ Cell Culture Treated Flasks with Filter Caps (T175 and T25) | ThermoFisher | Cat# 178885 (T175) Cat# 136196 (T25) |
| Falcon™ 15 mL Conical Centrifuge Tubes | Fisher Scientific | Cat# 14-959-49B |
Step-By-Step Method Details
Seeding Cells (Day 0)
Timing: approximately 30 min
This step describes how to seed AML12 cells
-
1.
Remove and discard culture medium from the precultured AML12 flask.
-
2.
Briefly rinse the cell layer with prewarmed 1× autoclaved PBS to remove all traces of serum.
-
3.
Add 2.0 mL of TrypLE™ to T25 flask, incubate at 37°C for 3–5 min and observe cells under an inverted microscope. Extend incubation time (if required) so that the cell monolayer is dispersed (usually within 5–10 min).
Note: Do not agitate the cells by hitting or shaking the flask to avoid clumping.
As TrypLE™ is a proprietary product and concentration is not disclosed, it is used as 1× final concentration. Alternatively, 0.05% Trypsin-EDTA solution can be used for 5–15 min.
-
4.
Add 8.0 mL of complete growth medium and aspirate cells by gently pipetting.
-
5.
Transfer cell suspension to a 15 mL conical centrifuge tube and centrifuge at 250 x g for 5 min.
Note: Do not use higher x g for centrifugation to avoid clumping.
-
6.
Suspend cell pellet in 1.0 mL complete medium and check cell number using cell counter.
-
7.
Seed 4 × 106 cell in 20 mL complete medium into one T175 flask.
CRITICAL: Do not use >4 × 106 cells while seeding to avoid >50% confluency next day (day 1).
Senescence Induction (Day 1)
Timing: 1–1.5 h
This step describes how using high dose H2O2 treatment induces senescence.
-
8.
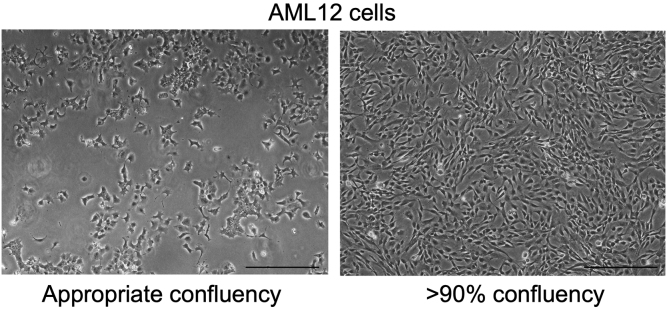
Check T175 flask containing AML12 under inverted microscope to make sure they have reached approximately 50% confluency as shown in Figure 1
Figure 1.
Confluency of AML 12 Cells before H2O2 Treatment
Left panel represents an appropriate confluency (approximately 50%) to start H2O2 treatment for senescence induction. Right panel represents that cells are fully confluent and cannot be used for senescence induction. Scale bars as 100 μm.
-
9.
If cells are appropriately confluent, change complete growth media from the flask to prewarmed basal media containing 1.0 mM H2O2.
-
10.
Incubate the cells for 1 h in cell culture incubator at 37°C.
-
11.
After 1 h, change the basal media to complete growth media for recovery and incubate cells in cell culture incubator at 37°C for next 23 h.
Note: If cells are less confluent and look sparse, wait until cells reach approximately 50% confluency. If cells are, or almost are confluent (>75%), split and reseed cells as described in day 0.
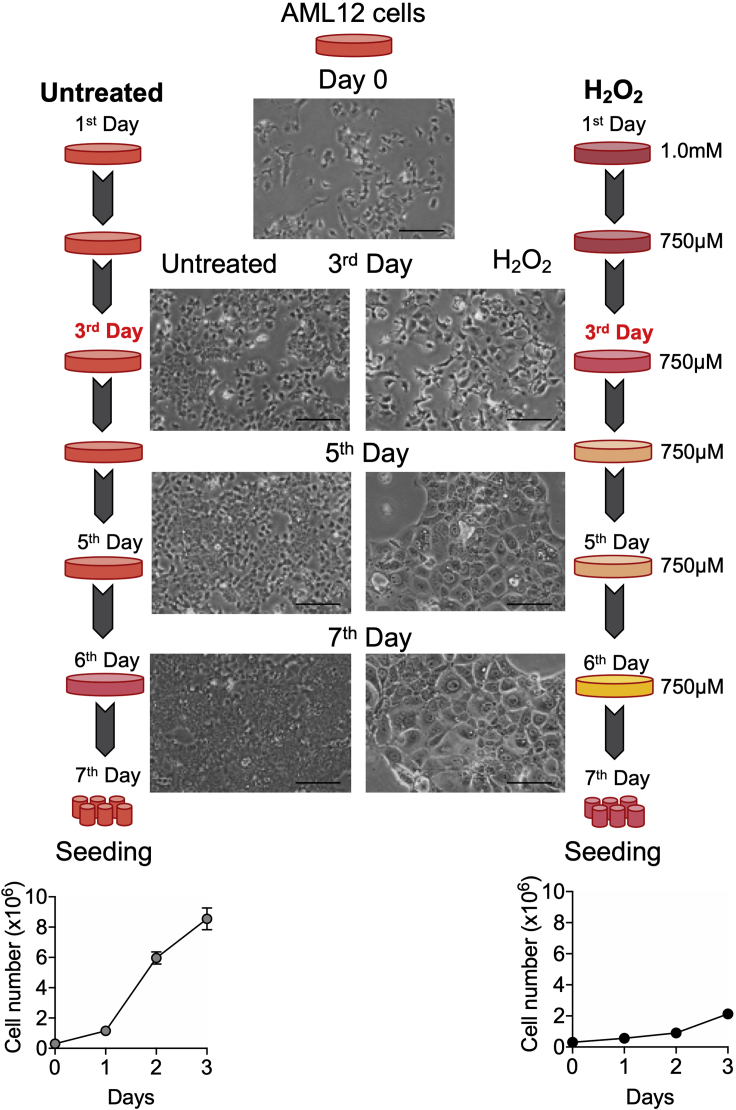
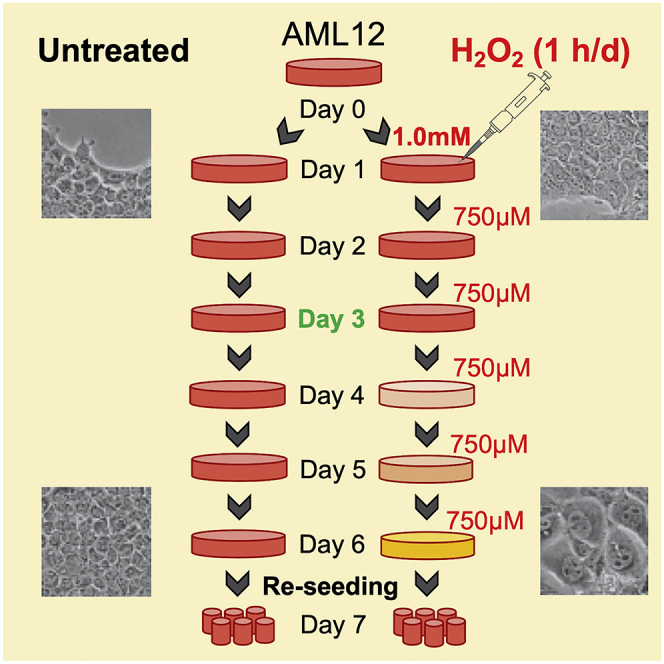
CRITICAL: Appropriate confluency (about 50%–60%) is important to achieve senescence-induced morphological changes in cells such as cellular hypertrophy shown in Figure 2.
Figure 2.
Schematic Representation of H2O2 Treatment for Senescence Induction in AML12 Cells
Shown are morphological changes during senescence induction, and the effect on cell number for 3 days after the last re-seeding of senescent AML12 cells. Scale bars as 100 μm.
Senescence Induction (Days 2–6)
Timing: 1–1.5 h each day
Periodic treatment of low dose H2O2 from day 2 to day 6 for senescence induction
-
12.
Check flask containing AML12 under inverted microscope for cell health.
-
13.
Change complete growth media from the flask to prewarmed basal media containing 750 μM H2O2.
-
14.
Incubate the cell for 1 h in cell culture incubator at 37°C.
-
15.
After 1 h, change the basal media complete growth media for recovery, and incubate cells in cell culture incubator at 37°C for the next 23 h.
-
16.
Repeat the steps 12–15 4 times until day 6.
CRITICAL: If cells become confluent at day 3 or 4, they need to be sub-cultured at 1:3 (if it is day 2) or 1:2 (if it is day 4) ratio as described in the day 0 section, steps 1–5 followed by suspending the cell pellet in 60 or 40 mL of complete growth media and seeding 20 mL in each T175 flask. This plating density will allow the cells to achieve senescence-induced hypertrophy.
Note: The time of the H2O2 treatment should be kept the same each day to allow sufficient recovery time (22–24 h) to induce senescence successfully.
Note: A change in the media color (from reddish/orange to more orange/yellow) can be visualized when senescence progresses each day of senescence induction (as shown in Figure 2). Senescent cells are metabolically more active for anaerobic glycolysis and generate lactic acid. Therefore, the media becomes more acidic as senescence progresses.
Note: For ease, notes can be written on the flask as shown on Figure 3.
Figure 3.
Marking the Date and Time of H2O2 Treatments on the Flask
Splitting Senescent Cells (Day 7)
Timing: ∼1 h
Sub-culturing/seeding of senescent cells for experiments
-
17.
Check flask containing senescent AML12 under inverted microscope for cell health and morphological changes as shown in Figure 2.
-
18.
Remove and discard culture medium from the flask.
-
19.
Briefly rinse the cell layer with prewarmed 1× autoclaved PBS to remove all traces of serum.
-
20.
Add 2.0 to 3.0 mL of TrypLE™ to a flask, incubate at 37°C for 3–5 min and observe cells under an inverted microscope. Extend incubation time (if required) so that the cell monolayer is dispersed (usually within 5–10 min).
Note: Do not agitate the cells by hitting or shaking the flask to avoid clumping.
-
21.
Add 6.0 to 8.0 mL of complete growth medium and aspirate cells by gently pipetting.
-
22.
Transfer cell suspension to a 15 mL conical centrifuge tube and centrifuge at 250 x g for 5 min at 4°C.
Note: Do not centrifuge at higher than 250 x g to avoid clumping.
-
23.
Suspend the cell pellet in 1.0 mL complete medium and check cell number using a cell counter.
-
24.
Seed the cells in appropriate cultureware (6-well plate, chambered slides, culture dishes, etc.) according to experimental requirements.
CRITICAL: Senescent cells exhibit cellular hypertrophy as a morphological change. Thus, a smaller number of senescent cells than control cells (non-senescent cells) are required for seeding if the experiment will be performed the next day. However, if the experiment needs several days to be completed, more senescent cells than controls cells will need to be seeded, since senescent cells have a lower population doubling time compared to control non-senescent AML12 cells (shown in Figure 2). Final results should be normalized appropriately to either protein content or cell number.
Pause Point: These senescent cells can be cryopreserved for future experiments at this point. Details for cryopreservation can be found https://www.atcc.org/Products/All/CRL-2254.aspx.
Expected Outcomes
Previous oxidative stress-induced senescence protocols have used one to two brief H2O2 exposures or prolonged exposure on hepatoma cells (Aravinthan et al., 2014; Chen et al., 2007; Duan et al., 2005; Irvine et al., 2014; Wang et al., 2013). However, these cells are likely to have limited relevance to aging in the liver due to differences in tissue origin or malignancy.
Here, we modified and optimized oxidative stress-induced premature senescence protocol using mouse hepatic AML12 cells which are derived from a transgenic mouse over-expressing human TGFα (Wu et al., 1994). The resulting senescent AML12 cells are morphologically distinct from control non-senescent cells (as shown in the Figure 2) from day 3 onwards and can be visualized under an inverted microscope.
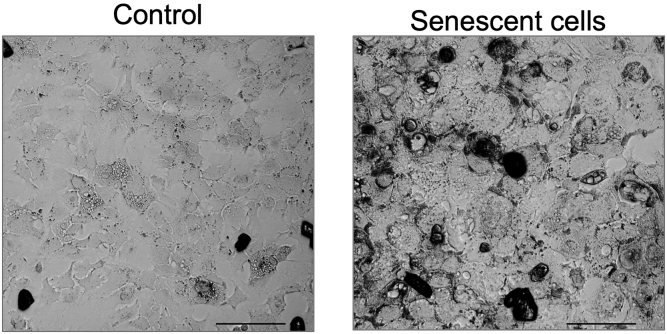
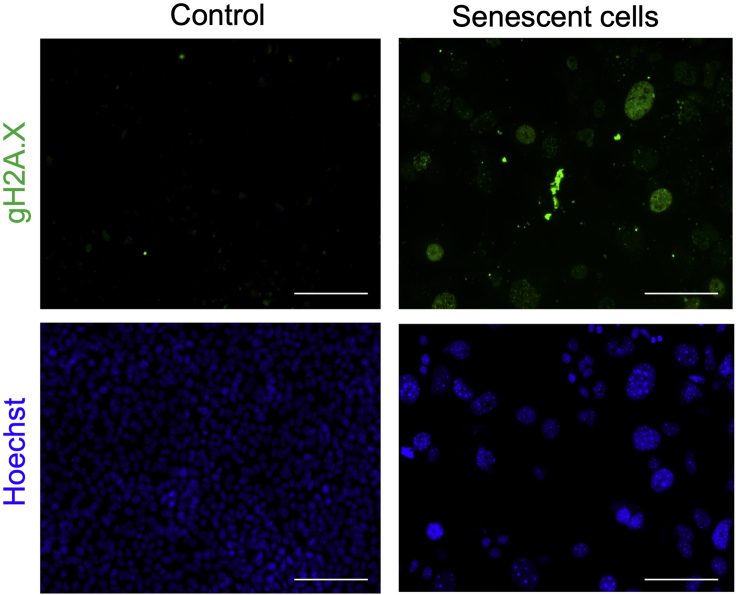
To validate senescence, we performed the following assays: senescence-associated β-galactosidase staining, γH2A.X staining, and qPCR to monitor expression of senescence genes (e.g., P16, P21, P53) (Chen et al., 2007; Dimri et al., 1995; Mah et al., 2010; Wang et al., 2013).
For senescence-associated β-galactosidase staining, we recommend using the Cellular Senescence Assay kit (Merck Millipore: KAA002) according to the kit protocol. It takes 2 days to complete the staining experiment (Figure 4).
Figure 4.
Senescence-Associated β-Galactosidase Staining in AML12 Cells Using Cellular Senescence Assay kit (Merck Millipore: KAA002)
Scale bars as 50 μm.
For immunofluorescence staining for γH2A.X, we suggest using Cell Signaling Technology’s standard protocol available https://www.cellsignal.com/contents/resources-protocols/immunofluorescence-general-protocol/if and the primary γH2A.X antibody (Abcam ab81299) in 1:100 dilution overnight at 4°C and secondary 1:200 Alexa Fluor 488 antibody (Molecular Probes, ThermoFisher). Cells were also counter-stained with 5 μM Hoechst 33342 (Abcam: ab228551) for 5 min to delineate the nucleus (Figure 5).
Figure 5.
Immunofluorescence Staining for γH2A.X and Nuclei (Hoechst 33342) in AML12 Cells
Scale bars as 100 μm.
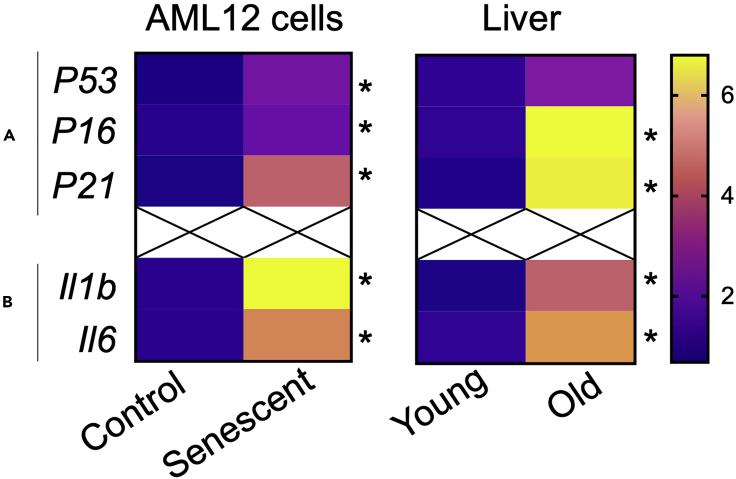
Gene expression analysis can be done using reverse transcriptase qPCR (RT-qPCR) using gene-specific SYBR green optimized primers (KiCqStart ® SYBR ® Green Primers, Sigma-Aldrich) and default qPCR cycle program as shown in Figure 6. Gene expression of senescence and pro-inflammatory genes were significantly increased in these senescent cells and were similar to changes found in the livers of aged mice.
Figure 6.
Gene Expression of Senescence and Inflammation Markers
Senescence genes (A) and pro-inflammatory gene (B) analysis in senescent AML12 as well as in livers from young (12–20 weeks) and old (108–128 weeks) mice. Statistical significance was calculated as ∗p < 0.05 using Graphpad PRISM 8.0.
Limitations
Replicative senescence during aging is believed to be because of telomere shortening, including other features such as increased expression of cell cycle arrest genes and inefficient repair of DNA double strand breaks (DDSB) (Bernadotte et al., 2016). It has been shown that acute H2O2 treatment is unable to produce DDSB whereas prolonged H2O2 treatment causes significant telomere shortening in human fibroblast cells (Duan et al., 2005; Wang et al., 2013). The senescent AML12 cells developed by this protocol did not show telomere shortening (data not shown). However, immunofluorescence staining for γH2A.X in the senescent AML 12 cells produced from our protocol show that these cells have DDSB. These studies suggesting that H2O2 treatment for 6 days may induce telomere-independent senescence in AML12. Thus, we believe that our protocol is an improvement upon earlier protocols (Aravinthan et al., 2014; Chen et al., 2007; Duan et al., 2005; Irvine et al., 2014; Wang et al., 2013). Furthermore, we have not tested this protocol in other cell lines. Hence, researchers may want to optimize H2O2 treatment dosage to achieve cellular senescence in different cells by using various dose and for different time duration.
Troubleshooting
Problem 1
If there is no cell death at day 2 of H2O2 treatment.
Potential Solution
It is normal to see some cell death (usually 3%–5% floating cells) on day 2 after the initial 1 mM H2O2 treatment. However, if there is no cell death observed after the day 1 H2O2 treatment, we recommend checking the H2O2 stock solution concentration. The easiest way to determine H2O2 solution concentration is to measure the absorbance at 240 nm and use a molar extinction coefficient of 43.6 M−1 cm−1 (Noble and Gibson, 1970). A standard 30% H2O2 solution is 9.77 M and a 1:1,000 dilution should have an A240 of 0.388. After measurement, adjust the final concentration of H2O2 treatment to generate a 1 mM solution for day 1 and 0.75 mM for days 2–6.
Problem 2
If cells are confluent during the course of H2O2 treatments.
Potential Solution
It was observed that if the cells are confluent (>90%) at day 2–3, induction of senescence is affected and not complete. So, one has to split the cells back to 50%–60% confluency as described in the steps 12–15, so the induction of senescence is efficient.
Resource Availability
Lead Contact
Brijesh Kumar Singh; singhbrijeshk@duke-nus.edu.sg.
Materials Availability
We did not generate any new materials.
Data and Code Availability
We did not generate a dataset or code.
Acknowledgments
This research was funded by the Ministry of Health (MOH) and National Medical Research Council (NMRC), Singapore, grant numbers NMRC/OFYIRG/0002/2016 and MOH-000319 (MOH-OFIRG19may-0002) to B.K.S.; NMRC/OFYIRG/077/2018 to M.T.; and CSAI19may-0002 to P.M.Y.
Author Contributions
M.T. and B.K.S. optimized the protocol. M.T., B.K.S., and P.M.Y. arranged funding support for the study. M.T., B.K.S., and P.M.Y. wrote the protocol.
Declaration of Interests
The authors declare no competing interests.
References
- Aravinthan A., Shannon N., Heaney J., Hoare M., Marshall A., Alexander G.J. The senescent hepatocyte gene signature in chronic liver disease. Exp. Gerontol. 2014;60:37–45. doi: 10.1016/j.exger.2014.09.011. [DOI] [PubMed] [Google Scholar]
- Bernadotte A., Mikhelson V.M., Spivak I.M. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging (Albany NY) 2016;8:3–11. doi: 10.18632/aging.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Ozanne S.E., Hales C.N. Methods of cellular senescence induction using oxidative stress. Methods Mol. Biol. 2007;371:179–189. doi: 10.1007/978-1-59745-361-5_14. [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Duan J., Zhang Z., Tong T. Irreversible cellular senescence induced by prolonged exposure to H2O2 involves DNA-damage-and-repair genes and telomere shortening. Int. J. Biochem. Cell Biol. 2005;37:1407–1420. doi: 10.1016/j.biocel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Irvine K.M., Skoien R., Bokil N.J., Melino M., Thomas G.P., Loo D., Gabrielli B., Hill M.M., Sweet M.J., Clouston A.D. Senescent human hepatocytes express a unique secretory phenotype and promote macrophage migration. World J. Gastroenterol. 2014;20:17851–17862. doi: 10.3748/wjg.v20.i47.17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah L.J., El-Osta A., Karagiannis T.C. GammaH2AX as a molecular marker of aging and disease. Epigenetics. 2010;5:129–136. doi: 10.4161/epi.5.2.11080. [DOI] [PubMed] [Google Scholar]
- Noble R.W., Gibson Q.H. The reaction of ferrous horseradish peroxidase with hydrogen peroxide. J. Biol. Chem. 1970;245:2409–2413. [PubMed] [Google Scholar]
- Singh B.K., Tripathi M., Sandireddy R., Tikno K., Zhou J., Yen P.M. Development of an in vitro senescent hepatic cell model for metabolic studies in aging. bioRxiv. 2020 doi: 10.18632/aging.103740. 2020.03.31.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wei D., Xiao H. Methods of cellular senescence induction using oxidative stress. Methods Mol. Biol. 2013;1048:135–144. doi: 10.1007/978-1-62703-556-9_11. [DOI] [PubMed] [Google Scholar]
- Wu J.C., Merlino G., Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. U S A. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We did not generate a dataset or code.