Summary
Myeloid cells, including dendritic cells (DCs), granulocytes, monocytes, monocyte-derived cells and macrophages, are important players in the immune response, but their identification is not as clear as lymphocytes, especially in tissues. This protocol details the step-by-step procedure for the analysis of myeloid populations in various mouse tissues by flow cytometry.
For complete details on the use and execution of this protocol, please refer to Liu et al. (2019).
Graphical Abstract
Highlights
-
•
Preparation of single-cell suspension from mouse tissues
-
•
Analysis of tissue-resident macrophages in mouse tissues
-
•
Gating strategy for myeloid cells in mouse organs
Myeloid cells, including dendritic cells (DCs), granulocytes, monocytes, monocyte-derived cells and macrophages, are important players in the immune response, but their identification is not as clear as lymphocytes, especially in tissues. This protocol details the step-by-step procedure for the analysis of myeloid populations in various mouse tissues by flow cytometry.
BEFORE YOU BEGIN
Antibody Mix Preparation
Timing: 1 h
Prepare the antibody mix for each sample before starting the experiment.
-
1.
Make antibody mix for mouse peripheral blood as the flow panel below. The volume of the antibody mix for 1 blood sample is 7 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| FITC | NK1.1 | eBioscience | 11-5941-82 | PK136 | 1/100 | 1 μL |
| PerCP-eFluor710 | CD172a | eBioscience | 46-1721-82 | P84 | 1/200 | 0.5 μL |
| Alexa Fluor 647 | Siglec F | BD | 562680 | E50-2440 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| APC-eFluor780 | CD3e | eBioscience | 47-0031-82 | 145-2C11 | 1/200 | 0.5 μL |
| eFluor450 | CD19 | eBioscience | 48-0193-82 | eBio1D3 (1D3) | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV605 | CD115 | Biolegend | 135517 | AFS98 | 1/200 | 0.5 μL |
| BV650 | CD11b | Biolegend | 101259 | M1/70 | 1/200 | 0.5 μL |
| BV711 | Ly6G | Biolegend | 127643 | 1A8 | 1/200 | 0.5 μL |
| BV785 | Ly6C | Biolegend | 128041 | HK1.4 | 1/200 | 0.5 μL |
| PE-Cy7 | FceR1 | eBioscience | 25-5898-80 | MAR-1 | 1/200 | 0.5 μL |
| BUV395 | CD45 | BD | 565967 | 30-F11 | 1/200 | 0.5 μL |
-
2.
Make antibody mix for mouse spleen as the panel below. The volume of the antibody mix for 1 spleen sample is 7 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| FITC | CD11b | Biolegend | 101206 | M1/70 | 1/200 | 0.5 μL |
| PerCP-eFluor710 | CD172a | ebioscience | 46-1721-82 | P84 | 1/200 | 0.5 μL |
| APC | Lin CD3e | Biolegend | 100312 | 145-2C11 | 1/200 | 0.5 μL |
| APC | Lin CD19 | eBioscience | 17-0193-82 | eBio1D3 | 1/200 | 0.5 μL |
| APC | Lin CD49b | Biolegend | 108910 | DX5 | 1/200 | 0.5 μL |
| APC | Lin Ly6G | Biolegend | 127614 | 1A8 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| eFluor450 | B220 | eBioscience | 48-0452-82 | RA3-6B2 | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV605 | CD115 | Biolegend | 135517 | AFS98 | 1/200 | 0.5 μL |
| BV650 | XCR1 | Biolegend | 148220 | ZET | 1/200 | 0.5 μL |
| BV785 | Ly6C | Biolegend | 128041 | HK1.4 | 1/200 | 0.5 μL |
| PE-Cy7 | F4/80 | Biolegend | 123113 | BM8 | 1/200 | 0.5 μL |
| BUV395 | CD45 | BD | 565967 | 30-F11 | 1/200 | 0.5 μL |
-
3.
Make antibody mix for mouse lung and brain as the panel below. Use biotin labelled F4/80 as primary antibody and use PE-Cy7 conjugated streptavidin (Cat# 25-4317-82, eBioscience) to detect biotin. The volume of the antibody mix for 1 sample is 4.25 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| Alexa Fluor 488 | Ly6C | Biolegend | 128022 | HK1.4 | 1/400 | 0.25 μL |
| APC | CD64 | Biolegend | 139306 | X54-5/7.1 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| APC-Cy7 | CD45 | BD | 557659 | 30-F11 | 1/200 | 0.5 μL |
| BV421 | Siglec F | BD | 562681 | E50-2440 | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV650 | CD11b | Biolegend | 101259 | M1/70 | 1/200 | 0.5 μL |
| BV711 | Ly6G | Biolegend | 127643 | 1A8 | 1/200 | 0.5 μL |
| Biotin | F4/80 | eBioscience | 13-4801-81 | BM8 | 1/200 | 0.5 μL |
-
4.
Make antibody mix for mouse kidney, epidermis and gut as the panel below. The volume of the antibody mix for 1 sample is 4.25 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| Alexa Fluor 488 | Ly6C | Biolegend | 128022 | HK1.4 | 1/400 | 0.25 μL |
| APC | CD64 | Biolegend | 139306 | X54-5/7.1 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| APC-Cy7 | CD45 | BD | 557659 | 30-F11 | 1/200 | 0.5 μL |
| BV421 | Siglec F | BD | 562681 | E50-2440 | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV650 | CD11b | Biolegend | 101259 | M1/70 | 1/200 | 0.5 μL |
| BV711 | Ly6G | Biolegend | 127643 | 1A8 | 1/200 | 0.5 μL |
| PE-Cy7 | F4/80 | Biolegend | 123113 | BM8 | 1/200 | 0.5 μL |
-
5.
Make antibody mix for mouse peritoneal lavage as the panel below. The volume of the antibody mix for 1 sample is 4.75 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| Alexa Fluor 488 | Ly6C | Biolegend | 128022 | HK1.4 | 1/400 | 0.25 μL |
| Alexa Fluor 647 | Tim-4 | Biolegend | 130008 | RMT4-54 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| APC-Cy7 | CD45 | BD | 557659 | 30-F11 | 1/200 | 0.5 μL |
| BV421 | Siglec F | BD | 562681 | E50-2440 | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV605 | CD115 | Biolegend | 135517 | AFS98 | 1/200 | 0.5 μL |
| BV650 | CD11b | Biolegend | 101259 | M1/70 | 1/200 | 0.5 μL |
| BV711 | Ly6G | Biolegend | 127643 | 1A8 | 1/200 | 0.5 μL |
| PE-Cy7 | F4/80 | Biolegend | 123113 | BM8 | 1/200 | 0.5 μL |
-
6.
Make antibody mix for mouse dermis as the panel below. The volume of the antibody mix for 1 sample is 4.75 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| FITC | CD24 | eBioscience | 11-0242-81 | M1/69 | 1/400 | 0.25 μL |
| APC | CD64 | Biolegend | 139306 | X54-5/7.1 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| APC-Cy7 | CD45 | BD | 557659 | 30-F11 | 1/200 | 0.5 μL |
| BV421 | Siglec F | BD | 562681 | E50-2440 | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV650 | CD11b | Biolegend | 101259 | M1/70 | 1/200 | 0.5 μL |
| BV711 | Ly6G | Biolegend | 127643 | 1A8 | 1/200 | 0.5 μL |
| BV785 | Ly6C | Biolegned | 128041 | HK1.4 | 1/200 | 0.5 μL |
| PE-Cy7 | F4/80 | Biolegend | 123113 | BM8 | 1/200 | 0.5 μL |
-
7.
Make antibody mix for mouse liver as the panel below. The volume of the antibody mix for 1 sample is 5.75 μL.
| Fluorophore | Marker | Company | Cat# | Clone | Final Dilution | Volume |
|---|---|---|---|---|---|---|
| Alexa Fluor 488 | Ly6C | Biolegend | 128022 | HK1.4 | 1/400 | 0.25 μL |
| PerCP-eFluor710 | Tim-4 | eBioscience | 46-5866-80 | RMT4-54 | 1/200 | 0.5 μL |
| APC | Lin CD3e | Biolegend | 100312 | 145-2C11 | 1/200 | 0.5 μL |
| APC | Lin CD19 | eBioscience | 17-0193-82 | eBio1D3 | 1/200 | 0.5 μL |
| APC | Lin CD49b | Biolegend | 108910 | DX5 | 1/200 | 0.5 μL |
| APC | Lin Ly6G | Biolegend | 127614 | 1A8 | 1/200 | 0.5 μL |
| Alexa Fluor 700 | MHCII | eBioscience | 56-5321-82 | M5/114.15.2 | 1/200 | 0.5 μL |
| APC-Cy7 | CD45 | BD | 557659 | 30-F11 | 1/200 | 0.5 μL |
| BV421 | CD64 | Biolegend | 139309 | X54-5/7.1 | 1/200 | 0.5 μL |
| BV510 | CD11c | Biolegend | 117353 | N418 | 1/200 | 0.5 μL |
| BV650 | CD11b | Biolegend | 101259 | M1/70 | 1/200 | 0.5 μL |
| PE-Cy7 | F4/80 | Biolegend | 123113 | BM8 | 1/200 | 0.5 μL |
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse B220 eFluor 450 (RA3-6B2) | eBioscience | Cat# 48-0452-82; RRID: AB_1548761 |
| Anti-mouse CD3e APC (145-2C11 ) | Biolegend | Cat# 100312; RRID: AB_312677 |
| Anti-mouse CD3e APC-eFluor780 (145-2C11) | eBioscience | Cat# 47-0031-82; RRID: AB_11149861 |
| Anti-mouse CD11b BV650 (M1/70) | Biolegend | Cat# 101259; RRID: AB_2566568 |
| Anti-mouse CD11b FITC (M1/70) | Biolegend | Cat# 101206; RRID: AB_312789 |
| Anti-mouse CD11c BV510 (N418) | Biolegend | Cat# 117353; RRID: AB_2686978 |
| Anti-mouse CD16/32 purified (2.4G2) | BD | Cat# 553142; RRID: AB_394657 |
| Anti-mouse CD19 APC (eBio1D3) | eBioscience | Cat# 17-0193-82; RRID: AB_1659676 |
| Anti-mouse CD19 eFluor450 (eBio1D3) | eBioscience | Cat# 48-0193-82; RRID: AB_2043815 |
| Anti-mouse CD24 FITC (M1/69) | eBioscience | Cat# 11-0242-81; RRID: AB_464987 |
| Anti-mouse CD45 APC-Cy7 (30-F11) | BD | Cat# 557659; RRID: AB_396774 |
| Anti-mouse CD45 BUV395 (30-F11) | BD | Cat# 565967; RRID: AB_2739420 |
| Anti-mouse CD49b APC (DX5) | Biolegend | Cat# 108910; RRID: AB_313417 |
| Anti-mouse CD64 APC (X54-5/7.1) | biolegend | Cat# 139306; RRID: AB_11219391 |
| Anti-mouse CD64 BV421 (X54-5/7.1) | Biolegend | Cat# 139309; RRID: AB_2562694 |
| Anti-mouse CD115 BV605 (AFS98) | Biolegend | Cat# 135517; RRID: AB_2562760 |
| Anti-mouse CD172a PerCP-eFluor® 710 (P84) | eBioscience | Cat# 46-1721-82; RRID: AB_10804639 |
| Anti-mouse F4/80 Biotin (BM8) | eBioscience | Cat# 13-4801-81; RRID: AB_466656 |
| Anti-mouse F4/80 PE-Cy7 (BM8) | Biolegend | Cat# 123113; RRID: AB_893490 |
| Anti-mouse FceRI PE-Cy7 (MAR-1) | eBioscience | Cat# 25-5898-80; RRID: AB_2573492 |
| Anti-mouse Ly6C Alexa Fluor 488 (HK1.4) | Biolegend | Cat# 128022; RRID: AB_10639728 |
| Anti-mouse Ly6C BV785 (HK1.4) | Biolegend | Cat# 128041; RRID: AB_2565852 |
| Anti-mouse Ly6G APC (1A8) | Biolegend | Cat# 127614; RRID: AB_2227348 |
| Anti-mouse Ly6G BV711 (1A8) | Biolegend | Cat# 127643; RRID: AB_2565971 |
| Anti-mouse MHCII (I-A/I-E) Alexa Fluor 700 (M5/114.15.2) | eBioscience | Cat# 56-5321-82; RRID: AB_494009 |
| Anti-mouse NK1.1 FITC (PK136) | eBioscience | Cat# 11-5941-82; RRID: AB_465318 |
| Anti-mouse Siglec F Alexa Fluor 647 (E50-2440) | BD | Cat# 562680; RRID: AB_2687570 |
| Anti-mouse Siglec F BV421 (E50-2440) | BD | Cat# 562681; RRID: AB_2722581 |
| Anti-mouse TIM-4 Alexa Fluor 647 (RMT4-54) | Biolegend | Cat# 130008; RRID: AB_2271648 |
| Anti-mouse TIM-4 PerCP-eFluor® 710 (RMT4-54) | eBioscience | Cat# 46-5866-80; RRID: AB_2573780 |
| Anti-mouse XCR1 BV650 (ZET) | Biolegend | Cat# 148220; RRID: AB_2566410 |
| Streptavidin PE-Cy7 | eBioscience | Cat# 25-4317-82; RRID: AB_10116480 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase type IV | Sigma | Cat# C5138 |
| DNase I | Roche | Cat# 1 0104159 001 |
| Dispase | Gibco | Cat# 17105-041 |
| DAPI | ThermoFisher | Cat# D1306 |
| Software and Algorithms | ||
| FlowJo V10 | FlowJo LLC | https://www.flowjo.com |
| Other | ||
| Centrifuge | Eppendorf | 5810 R |
| Cell culture CO2 incubator | Thermofisher | N/A |
| Countess II | Thermofisher | Cat# AMQAX1000 |
| Symphony flow cytometer | BD | Symphony |
MATERIALS AND EQUIPMENT
Collagenase IV Stock
For 10 mg/mL Collagenase IV stock, dissolve 100 mg Collagenase IV in 10 mL RPMI1640. Store at −20°C in 200 μL aliquots for up to 6 months.
DNase I
For 10 mg/mL DNase I stock, dissolve 100 mg DNase I in 5 mL DNase storage buffer (20 mM Tris, 1 mM MgCl2) and 5 mL glycerol. Store at −20°C in 500 μL aliquots for up to 6 months.
Digest Solution
Prepare 40 mL of digest solution for one mouse by adding 800 μL Collagenase IV stock (final concentration 0.2 mg/mL) and 200 μL DNase I stock (final concentration 0.05 mg/mL) into 39 mL RPMI1640 supplemented with 10% fetal bovine serum (FBS). Always prepare fresh.
Dispase
For 100 U/mL Dispase stock solution, dissolve 1.2 g Dispase in 20 mL distilled H2O (do not use PBS as it will result in sedimentation). Store at −20°C in 500 μL aliquots for up to 6 months. Dilute the stock 50 times with Hanks' blanced salt solution (HBSS) just before use.
Ammonium-Chloride-Potassium (ACK) Lysis Buffer
For 1 L of 10× ACK Lysis Buffer, dissolve 82.9 g NH4Cl (final concentration 1.55 M), 10 g KHCO3 (final concentration 100 mM), 0.372 g Na2-EDTA (or 2 mL 0.5 M EDTA solution) in 800 mL autoclaved distilled H2O, adjust the pH to 7.2 with 1 M NaOH and the volume to 1000 mL with distilled H2O and pass through a 0.22 μm filter. Can be stored at 4°C for several months. Dilute 10 times with ddH2O before use.
FACs Buffer
To make 1 L of FACs buffer, dissolve 5 g BSA (final concentration 0.5%) in 900 mL sterile PBS and 4 mL of 0.5 M EDTA (final concentration 2 mM), and adjust the volume to 1000 mL with PBS. Store at 4°C for up to 3 months.
4′,6-Diamidino-2-phenylindole (DAPI) Solution
To make 5 mg/mL DAPI stock, dissolve 10 mg (1 bottle) in 2 mL ddH2O, store at −20°C in 200 μL aliquots for up to 3 years. For 1 μg/mL working solution, add 10 μL DAPI stock into 50 mL PBS, store at 4°C in the dark by covering with aluminium foil.
40% and 80% Percoll
To make 100% isotonic Percoll, mix 1 mL of 10× PBS with 9 mL of Percoll; make 80% Percoll by mixing 10 mL of 100% Percoll with 2.5 mL of RPMI 1640 medium; make 40% Percoll by mixing 5 mL of 80% with 5 mL of RPMI 1640 medium. Always prepare fresh.
Fc Blocking Buffer
To make Fc blocking buffer for 10 samples, add 5 μL of anti-CD16/32 antibody (Key Resources Table) into 700 μL FACs buffer. Always prepare fresh.
STEP-BY-STEP METHOD DETAILS
Sampling
Timing: 30 min
This step details how to anaesthetize and perfuse the mice, and take samples.
-
1.
Terminally anaesthetize mice by intraperitoneal (i.p.) injection of 250 μL of 0.9% sodium pentobarbital solution.
-
2.
Collect blood by cardiac puncture, and transfer to an anticoagulant blood collection tube.
-
a.
Add 10 mL of 1× ACK lysis buffer into a new 50-mL tube, transfer the blood into the ACK lysis buffer, incubate at 20°C–26°C for 5 min.
-
b.
Add 20 mL of PBS to stop the lysis, and centrifuge at 1350 rpm (375 × g) for 5 min at 4°C.
-
c.
Discard the supernatant and add 5 mL of 1× ACK lysis buffer into the tube to resuspend the pellet. Incubate at 20°C–26°C for 5 min.
-
d.
Add 20 mL of PBS to stop the lysis, and centrifuge at 1350 rpm (375 × g) for 5 min at 4°C.
-
e.
Discard the supernatant and resuspend with 2 mL of FACs buffer, keep on ice until staining.
-
3.
Euthanize the terminally anaesthetized mice by cervical dislocation.
-
4.
Collect the peritoneal lavage.
-
a.
Inject 5 mL of PBS containing 2 mM EDTA into the peritoneal cavity with a 5-mL syringe, gently massage the abdomen for 2 min.
-
b.
Open the skin to expose the abdomen wall, retrieve the maximal amount of peritoneal lavage with a syringe, transfer into a 15-mL tube.
-
c.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant and resuspend with 2 mL of FACs buffer, keep on ice until staining.
-
5.
Make a cut on the right atria, then perfuse the mice with 10 mL of PBS using a 10-mL syringe via the left ventricle.
CRITICAL: It is critical to perfuse all the blood out, otherwise there will be circulating blood cells contamination to the tissue myeloid cells. Perfuse with more PBS until the wash out is clear and free of red blood cells.
-
6.
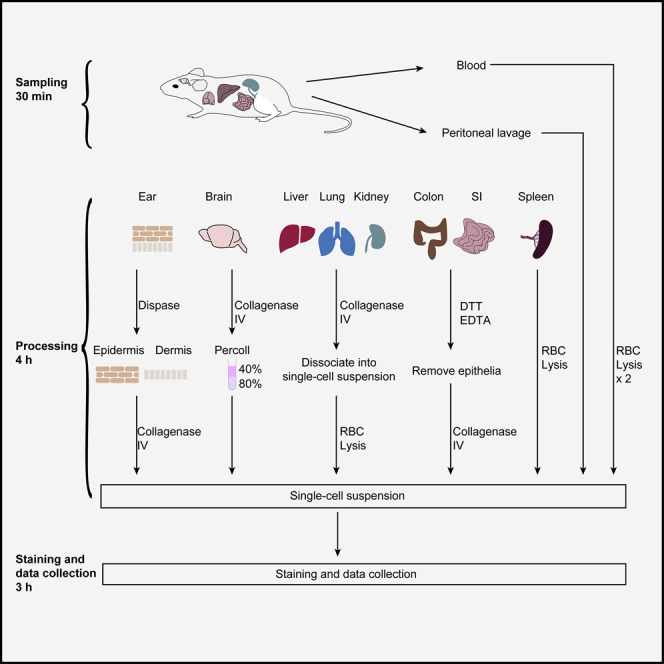
Take the lung, spleen, liver (one lobe), kidney (one side), colon and small intestine, two ears and half of the brain. Place them into a petri dish with PBS, and keep on ice until processing.
Processing the Organs
Timing: 4 h
This step details how to digest the tissues into single-cell suspensions.
-
7.
Skin (ear) preparation (Tamoutounour et al., 2013).
-
a.
Split the ears between the dorsal and ventral sections (Figure 1A), and digest with 1 mL of Dispase solution in a 2-mL tube at 37°C for 90 min (process other organs during this time).
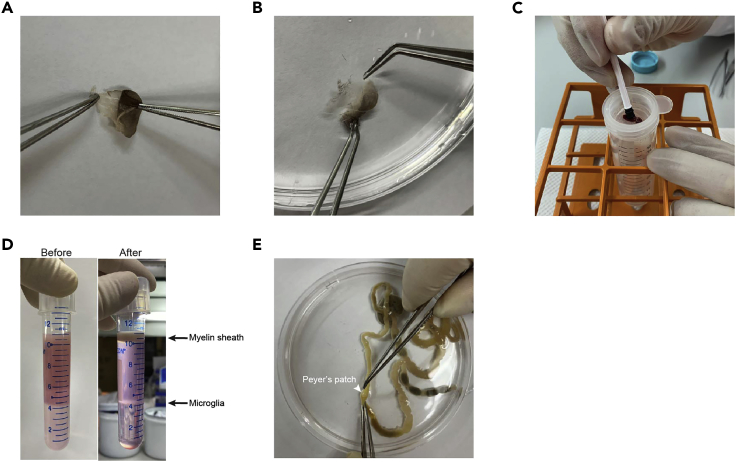
Figure 1.
Processing the Tissues
(A) Separation of the dorsal and ventral sections of the ear.
(B) Separation of the dermis and epidermis after digestion with Dispase solution.
(C) Grinding the spleen on the 70 μm cell strainer with a syringe plunger.
(D) Image of brain suspension before and after gradient centrifugation.
(E) Removing Peyer’s patches from the intestine.
-
b.
After Dispase disgestion, put the ear pieces into a petri dish with 10 mL of PBS and separate the epidermis from dermis. Process epidermis and dermis separately (Figure 1B).
-
c.
Cut the tissues into small pieces with scissors in 2-mL round-bottom tubes containing 1 mL of digest solution.
-
d.
Pour the sample into one well of a 6-well plate and add 2 mL of digest solution (3 mL in total), incubate at 37°C for 60 min.
-
e.
Use a syringe with a 1.2 mm inner diameter needle to dissociate the digested tissue until no obvious debris are present.
-
f.
Pass the suspension through a 70 μm cell strainer into a new 50-mL tube and use 10 mL of PBS to pass through the strainer to wash it.
-
g.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant and resuspend with 2 mL of FACs buffer. Keep on ice until staining.
-
8.
Spleen preparation.
-
a.
Place a 70 μm cell strainer in a 50-mL tube, and rinse with 2 mL of PBS.
-
b.
Put the spleen in the cell strainer and grind with a syringe plunger, use 10 mL of PBS to pass through the strainer to wash it (Figure 1C).
-
c.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant.
-
d.
Add 10mL of 1× ACK lysis buffer to resuspend the pellet and incubate for 5 min at 20°C–26°C.
-
e.
Add 20 mL of PBS to stop the lysis, centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant.
-
f.
Resuspend the pellet with 2 mL of FACs buffer. Keep on ice until staining.
-
9.
Microglia preparation (Ginhoux et al., 2010, Thion et al., 2018).
-
a.
Cut the brain into small pieces with scissors in a 2-mL round-bottom tube containing 1 mL of digest solution.
-
b.
Pour the sample into one well of a 6-well plate and add 2 mL of digest solution into the well (3 mL in total), incubate at 37°C for 60 min.
-
c.
Use a syringe with a 1.2 mm inner diameter needle to dissociate the digested tissue until no obvious debris are present.
-
d.
Pass the suspension through a 70 μm cell strainer into a new 50-mL tube and use 10 mL of PBS to pass through the strainer to wash it.
-
e.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant and resuspend the pellet with 8 mL of 40% Percoll.
-
f.
Transfer 5 mL of 80% Percoll into a Polypropylene Round-Bottom Tube (Falcon 352059).
-
g.
Layer 8 mL of brain cell suspension onto the 80% Percoll with a pasteur pipette.
-
h.
Centrifugate at 2800 rpm (1,578 × g) for 20 min at 20°C–26°C with low acceleration (acceleration 1) and no brake.
-
i.
Collect the middle interface layer (Figure 1D) and transfer into a new 15-mL tube containing 10 mL of PBS.
-
j.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant and resuspend with 2 mL of FACs buffer. Keep on ice until staining.
-
10.
Lung, liver and kidney preparation. These three organs share the same digest protocol, process them separately in parallel.
-
a.
Cut the organs into small pieces with scissors in 2-mL round-bottom tubes containing 1 mL of digest solution.
-
b.
Pour the sample into one well of a 6-well plate and add 2 mL of digest solution into the well (3 mL in total), incubate at 37°C for 60 min.
-
c.
Use a syringe with a 1.2 mm inner diameter needle to dissociate the digested tissue until no obvious debris are present.
-
d.
Pass the suspension through a 70 μm cell strainer into 50-mL tubes and use 10 mL of PBS to pass through the strainer to wash it.
-
e.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant.
-
f.
Add 5 mL of 1× ACK lysis buffer to resuspend the pellet and incubate for 5 min at 20°C–26°C.
-
g.
Add 20 mL of PBS to stop the lysing, centrifuge at 1,350 rpm (375 × g) for 5 min at 4°C, discard the supernatant.
-
h.
Resuspend with 2 mL of FACs buffer. Keep on ice until staining.
-
11.
Gut preparation (Bain et al., 2014).
-
a.
Remove all fat tissue and Peyer’s patches from the intestine in a 10-cm dish (Figure 1E).
-
b.
Process the colon and small intestine separately.
-
c.
Open the intestines longitudinally, cut into 0.5 cm long sections and wash 4 times with 20 mL of PBS in 50-mL tubes.
-
d.
After washing, add 12.5 mL of fresh calcium/magnesium-free PBS containing 5 mM EDTA and 2 mM DTT to the 50-mL tubes and incubate at 37°C with agitation for 20 min to detach the epithelial cells.
-
e.
Remove the epithelial sheet by vigorously shaking up-and-down for 15 s by hand.
-
f.
Pour the suspension into a 10-cm dish and pick out the tissue and put into a new 50-mL tube,
-
g.
Add 20 mL of PBS, wash the remaining tissue by vigorously shaking for 15 s by hand.
-
h.
Wash the tissue again by repeating steps f-g.
-
i.
Pour the suspension into a 10-cm dish and pick out the remaining tissue and put into 2-mL round-bottom tubes containing 1 mL of digest solution.
-
j.
Cut the tissue into small pieces with scissors.
-
k.
Pour the sample into one well of a 6-well plate and add 2 mL of digest solution into the well (3 mL in total), incubate at 37°C for 60 min.
-
l.
Use a syringe with a 1.2 mm inner diameter needle to dissociate the digested tissue until no obvious debris are present.
-
m.
Pass the suspension through a 70 μm cell strainer into 50-mL tubes and use 10 mL of PBS to pass through the strainer to wash it.
-
n.
Centrifuge at 1350 rpm (375 × g) for 5 min at 4°C, discard the supernatant and resuspend with 2 mL of FACs buffer. Keep on ice until staining.
Pause Point: The cell suspensions can be stored at 4°C and stained the next day but will lead to increased cell death and should be done only as the last option.
-
12.
Perform cell counting using Countess II.
Staining
Timing: 1.5 h
This step details how to stain tissue cells with indicated antibody panels for myeloid cell analysis.
-
13.
Label FACs tubes for each sample, and add 1 mL of FACs buffer into each tube.
-
14.
Transfer the tissue cell suspension containing ~2 million cells into FACs tubes, vortex briefly and centrifuge at 1350 rpm (375 × g) for 5 min at 4°C.
-
15.
Discard the supernatant and resuspend the cells with 70 μL Fc blocking buffer (see Reagent setup), incubate at 4°C for 15 min.
-
16.
Add indicated volume of the antibody mix (see Antibody mix preparation) into the suspension and vortex briefly, incubate at 4°C for 30 min in the dark by covering with aluminum foil.
-
17.
Wash with 1 mL of FACs buffer, vortex briefly and centrifuge at 1350 rpm (375 × g) for 5 min at 4°C.
-
18.
For samples other than lung and brain, discard the supernatant and resuspend with 120 μL FACs buffer and 40 μL DAPI solution (final concentration 250 ng/mL).
-
19.
For lung and brain sample, discard the supernatant and resuspend with 70 μL FACS buffer containing 0.25 μL PE-Cy7 conjugated streptavidin and vortex briefly, incubate at 4°C for 30 min in the dark by covering with aluminum foil.
-
20.
Wash with 1 mL of FACs buffer, vortex briefly and centrifuge at 1350 rpm (375 × g) for 5 min at 4°C.
-
21.
Discard the supernatant and resuspend with 120 μL FACs buffer and 40 μL DAPI solution (final concentration 250 ng/mL).
Data Collection
Timing: 1.5 h
This step details how to collect flow data on a BD symphony cytometer
-
22.
Collect data on a BD Symphony flow cytometer. Unstained samples and single-stained samples are used to set appropriate PMT voltages that have clear positive and negative populations. Compensation beads (Anti-Rat and Anti-Hamster Ig κ /Negative Control Compensation Particles Set, Cat# 552845, BD) were used to do auto-compensation. Refer to Cossarizza et al. for the guidelines for the use of flow cytometry in immunological studies (Cossarizza et al., 2019).
EXPECTED OUTCOMES
Following this protocol, we developed gating strategies for myeloid cells in different organs of the mouse. The principle of the gating strategy is to use negative markers to exclude unwanted cells, while using positive markers to define the wanted populations.
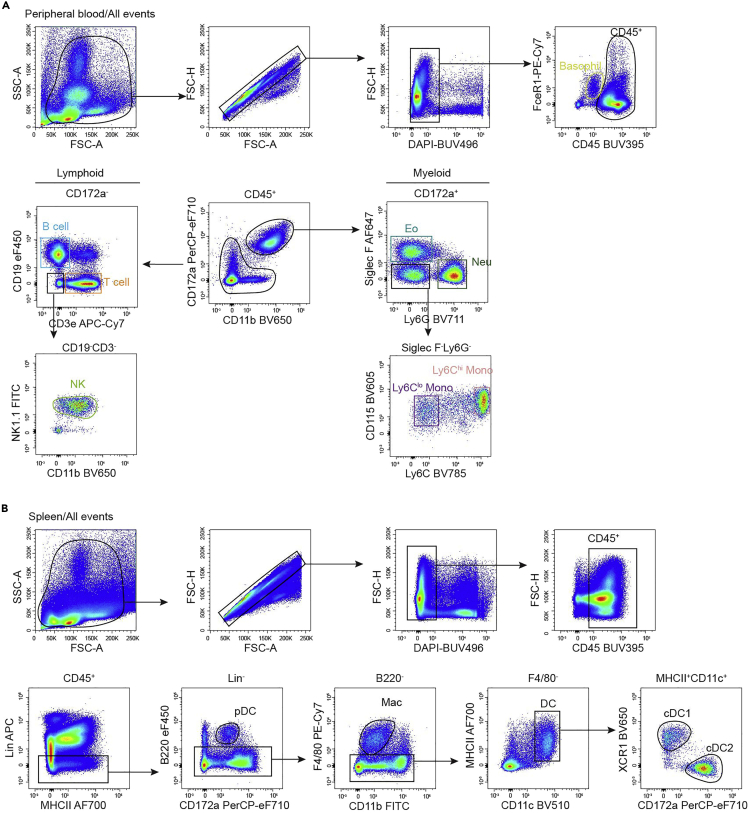
In the peripheral blood (Figure 2A), we firstly gated basophils since they are CD45int that different from other CD45+ cells. The CD45+ cells were divided into myeloid cells and lymphoid cells based on the expression of CD172a and CD11b. In the myeloid gate (CD11b+CD172a+), neutrophils are Ly6G+, eosinophils are Siglec F+, monocytes are Siglec F−Ly6G−CD115+ and form a continuum from Ly6Chi to Ly6Clo. In the lymphoid gate (CD172a−CD11blo-neg), B cells are CD19+MHCII+, T cells are CD19−CD3e+, NK cells are CD19−CD3e−NK1.1+.
Figure 2.
Gating Strategy for Myeloid Cells in Peripheral Blood and Spleen
(A) The gating strategy of lymphoid and myeloid lineages in the peripheral blood of 8-week-old mice.
(B) The gating strategy of pDCs (Lin−B220+CD172a+), cDCs (Lin−B220−F4/80−CD11c+MHCII+) and macrophages (Lin−B220−F4/80+CD11b+) in the spleen of 8-week-old mice. Lineage markers include CD3e, CD19, CD49b and Ly6G.
Spleen contains large amounts of lymphocytes that will affect the flow cytometry analysis of myeloid cells, so we put antibodies for T cells (anti-CD3e), B cells (anti-CD19), NK cells (anti-CD49b) and neutrophils (anti-Ly6G) in the lineage to exclude lymphocytes and neutrophils. In the lineage negative fraction, pDCs are CD172a+B220+, macrophages are B220−F4/80+, cDCs are B220−F4/80−CD11c+MHCII+ and contain XCR1+ cDC1 and CD172a+ cDC2 (Figure 2B).
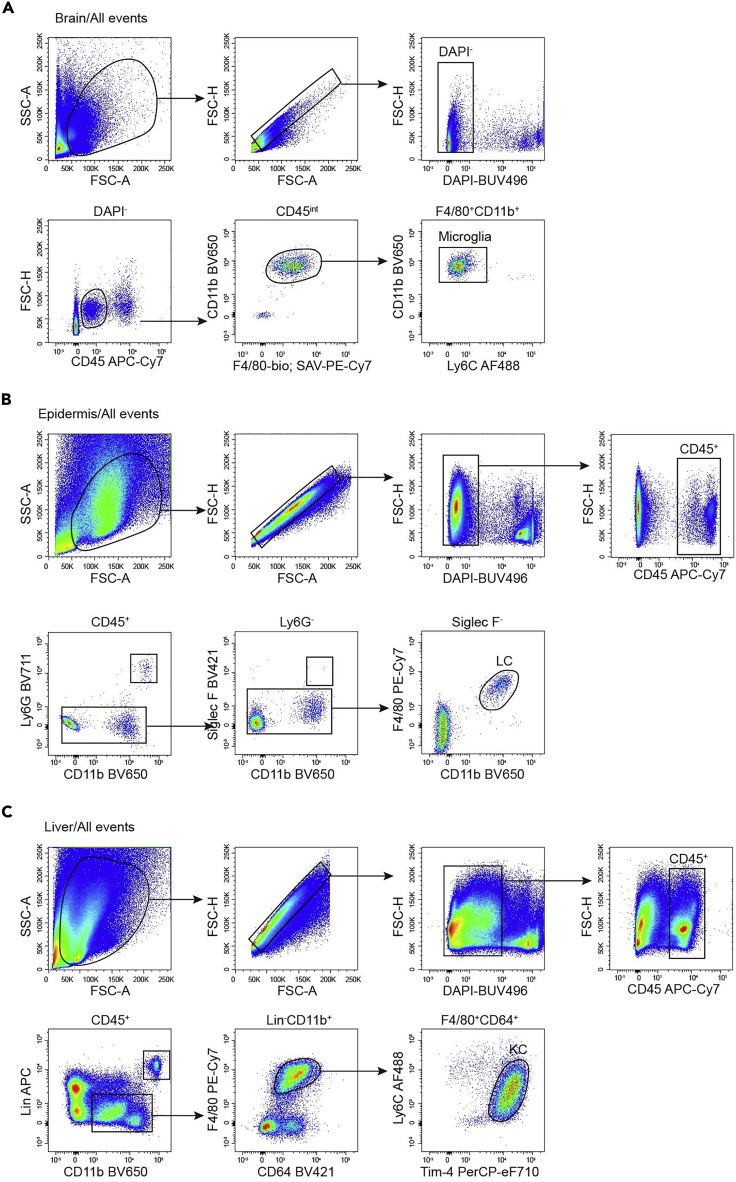
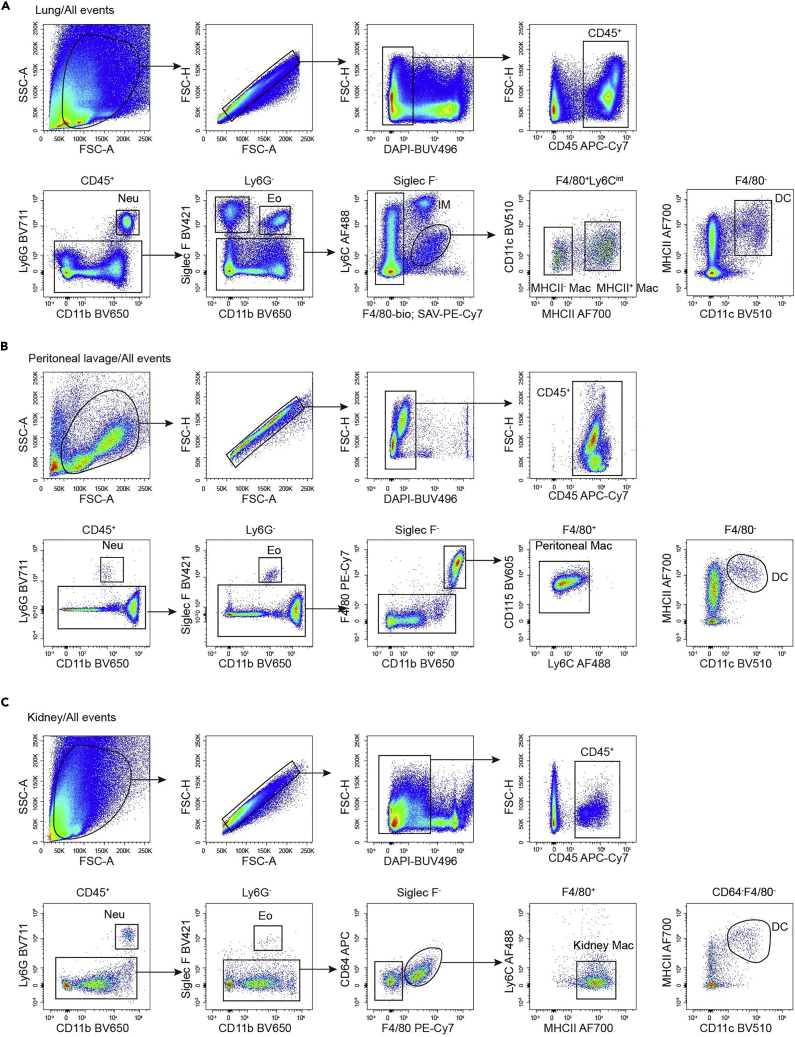
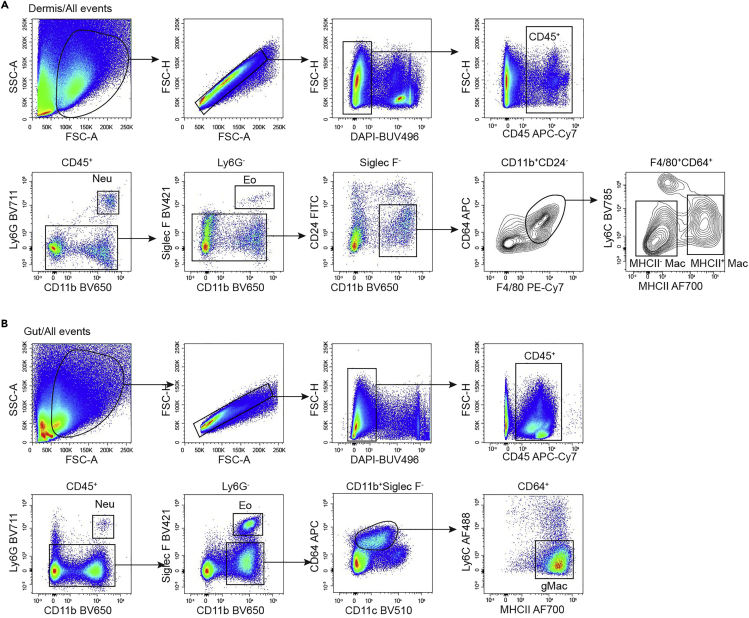
Microglia are the macrophages in the brain and are easily identified as CD45intCD11b+F4/80+Ly6C− (Figure 3A). In the epidermis, there are two main populations of immune cells, Langerhans cells (LCs) and dendritic epidermal T cells (DETCs). LCs are CD11b+F4/80+ and also express CD11c and MHCII (Figure 3B). Kupffer cells (KCs) are the macrophages in the liver and are Lin−CD11b+F4/80+Tim-4+ (Figure 3C). In the lung, alveolar macrophages (AMs) are CD45+Siglec F+CD11b− and are also positive for CD11c. Interstitial macrophages in the lung are F4/80+Ly6C−, and can be further divided into MHCII+ and MHCII− populations. DCs in the lung are F4/80−CD11c+MHCII+ (Figure 4A). In the peritoneal lavage, the most abundant cells are B cells and macrophages. Peritoneal macrophages are CD11bhiF4/80+CD115+Ly6C− (Figure 4B). Kidney macrophages are gated as F4/80+CD11b+MHCII+Ly6C−, DCs are F4/80−MHCII+CD11c+ (Figure 4C). In the dermis, macrophages are CD11b+CD24−CD64+F4/80+, and can be further divided into MHCII+ and MHCII− populations (Figure 5A). Gut macrophages are CD11b+CD64+MHCII+Ly6C− (Figure 5B).
Figure 3.
Gating Strategy for Myeloid Cells in Brain, Epidermis and Liver
(A) Gating strategy of brain microglia (CD45intCD11b+F4/80+Ly6C−) of 8-week-old mice.
(B) Gating strategy of epidermal Langerhans cells (LCs) (CD45+Ly6G−Siglec F−CD11b+F4/80+) of 8-week-old mice.
(C) Gating strategy of liver Kupffer cells (KCs) (CD45+Lin−CD11b+F4/80+Tim-4+) of 8-week-old mice. Lineage markers include CD3e, CD19, CD49b and Ly6G.
Figure 4.
Gating Strategy for Myeloid Cells in Lung, Peritoneal Lavage and Kidney
(A) Gating strategy of lung alveolar macrophages (AMs) (CD45+Ly6G−Siglec F+), interstitial macrophages (IM) ( CD45+Ly6G−Siglec F−F4/80+Ly6Clo) and DCs (CD45+Ly6G−Siglec F−F4/80−CD11c−MHCII−) of 8-week-old mice.
(B) Gating strategy of peritoneal macrophages (CD45+Ly6G−Siglec F−CD11bhiF4/80hiCD115+Ly6C−) and DCs (CD45+Ly6G−Siglec F−F4/80−CD11c+MHCII+) of 8-week-old mice.
(C) Gating strategy of kidney macrophages (CD45+Ly6G−Siglec F−CD11b+F4/80+MHCII+Ly6C−) and DCs (CD45+Ly6G−Siglec F−F4/80−CD11c+MHCII+) of 8-week-old mice.
Figure 5.
Gating Strategy for Myeloid Cells in Dermis and Gut
(A) Gating strategy of dermal MHCII+ macrophages (CD45+Ly6G−Siglec F−CD11b+CD24−F4/80+CD64+MHCII+) and MHCII− macrophages (CD45+Ly6G−Siglec F−CD11b+CD24−F4/80+CD64+MHCII−) of 8-week-old mice.
(B) Gating strategy of gut macrophages (CD45+Ly6G−Siglec F−CD11b+CD64+CD11cintMHCII+Ly6C−) of 8-week-old mice.
LIMITATIONS
This protocol meets the basic needs for analyzing myeloid cells in most organs, but was not optimized for all organs. Since this experiment is labor intensive, to keep the experiment simple, we used Collagenase IV for the digestion of all organs. Although it works efficiently for most of the organs, it may not be the most suitable enzyme for digesting certain organs, such as the lung. Although the panels we use are able to distinguish most of the myeloid cell types in tissue, recent researches have reported new subsets of macrophages with new markers, including three macrophage subsets in the gut which can be identified with CD4 and Tim-4 (Shaw et al., 2018), and two newly defined interstitial macrophages in most tissues (Chakarov et al., 2019). More markers are needed to analyze these new subsets. In all the panels, we leave PE channel empty, so that the panels can be easily expended, new markers and fluorescent proteins (mCherry and tdTomato) can be added to or detected with PE channel.
In this protocol, we use DAPI to counter stain the dead cells, thus we are unable to fix the samples for later analysis. This limitation can be overcome by using a fixable live/dead dye, for example LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit from Invitrogen (L23105), instead of DAPI.
TROUBLESHOOTING
Problem 1
No positive staining of some markers.
Potential Solution
Several reasons may result in this problem. 1) use of a different clone, or forgetting to add the antibody; 2) Inappropriate PMT voltages. To overcome this problem, use single stains to find an appropriate PMT voltage that gives a clear positive staining and separation; 3) for lung and brain, the staining of F4/80 is not bright. To overcome this problem, use biotin-labelled F4/80 and PE-Cy7 conjugated streptavidin to amplify the signal.
Problem 2
Red blood cell residue in the blood samples.
Potential Solution
The potential reason for this problem might be that the ACK lysis buffer has expired. ACK lysis buffer is unstable. Use fresh 1× ACK lysis buffer every time to circumvent this problem. Do not keep the 10× ACK lysis buffer stock at 20°C–26°C for an extended time.
Problem 3
Low cellular viability.
Potential Solution
This may happen with gut samples as there are plenty of epithelial cells in the gut and they are very vulnerable. To avoid this situation, we recommend washing the tissue intensively and keep it on ice during the whole procedure except for at steps 11d and 11k.
Problem 4
Inadequate cell number.
Potential Solution
Depending on the volume of blood collected, the cell number of blood samples may vary. If there are less than 2 million cells, use all the cells for staining. For peritoneal lavage and brain, we can normally get 3 million cells. For other organs, there will be far more than 2 million cells.
ACKNOWLEDGMENTS
F.G. is an EMBO YIP awardee and is supported by Singapore Immunology Network (SIgN) core funding as well as a Singapore National Research Foundation Senior Investigatorship (NRFI) NRF2016NRF-NRFI001-02. This work was supported by the National Natural Science Foundation of China (31900630 to Z.L.).
AUTHOR CONTRIBUTIONS
Z.L., Y.G., A.S., and S.Z. conducted the experiments; Z.L. and F.G. analyzed the data and wrote the paper; F.G. supervised the project and conceptualized the study.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Contributor Information
Zhaoyuan Liu, Email: zhaoyuan_liu@sjtu.edu.cn.
Florent Ginhoux, Email: florent_ginhoux@immunol.a-star.edu.sg.
References
- Bain C.C., Bravo-Blas A., Scott C.L., Gomez Perdiguero E., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., Mowat A.M. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarov S., Lim H.Y., Tan L., Lim S.Y., See P., Lum J., Zhang X.M., Foo S., Nakamizo S., Duan K. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019;363 doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- Cossarizza A., Chang H.D., Radbruch A., Acs A., Adam D., Adam-Klages S., Agace W.W., Aghaeepour N., Akdis M., Allez M. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition) Eur. J. Immunol. 2019;49:1457–1973. doi: 10.1002/eji.201970107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gu Y., Chakarov S., Bleriot C., Kwok I., Chen X., Shin A., Huang W., Dress R.J., Dutertre C.A. Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell. 2019;178:1509–1525.e.19. doi: 10.1016/j.cell.2019.08.009. [DOI] [PubMed] [Google Scholar]
- Shaw T.N., Houston S.A., Wemyss K., Bridgeman H.M., Barbera T.A., Zangerle-Murray T., Strangward P., Ridley A.J.L., Wang P., Tamoutounour S. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med. 2018;215:1507–1518. doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour S., Guilliams M., Montanana Sanchis F., Liu H., Terhorst D., Malosse C., Pollet E., Ardouin L., Luche H., Sanchez C. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Thion M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., Blecher R., Ulas T., Squarzoni P., Hoeffel G. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 2018;172:500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]