Summary
Analysis of mitochondrial respiration function represented by the oxygen consumption rate is necessary for assessing mitochondrial respiration function. This protocol describes steps to evaluate the respiration function of mitochondria in situ in saponin-permeabilized cardiomyocytes. In permeabilized cells, mitochondria are in a relatively integrated cellular system, and mitochondrial respiration is more physiologically relevant than isolated mitochondria.
For complete details on the use and execution of this protocol, please refer to Gong et al. (2015a) and Gong et al. (2015b).
Graphical Abstract

Highlights
-
•
Mitochondria are retained in a relatively integrated cellular system
-
•
Mitochondrial respiration in situ is more physiologically relevant
-
•
Saponin concentration is critical for mitochondrial integrity
Analysis of mitochondrial respiration function represented by the oxygen consumption rate is necessary for assessing mitochondrial respiration function. This protocol describes steps to evaluate the respiration function of mitochondria in situ in saponin-permeabilized cardiomyocytes. In permeabilized cells, mitochondria are in a relatively integrated cellular system, and mitochondrial respiration is more physiologically relevant than isolated mitochondria.
Before You Begin
Timing: 0.2–3 days
-
1.
Prepare freshly isolated or cultured cardiomyocytes according to our step-by-step STAR protocol (Tian et al., 2020) or other protocols before you start the diagnostic study.
Adult cardiomyocytes were cultured in serum-free M199 medium, supplemented with 10 mM glutathione, 26.2 mM sodium bicarbonate, 5 mM creatine, 2 mM L-carnitine, 5 mM taurine, 0.1% insulin-transferrin-selenium-X, 0.02% bovine serum albumin, 50 U/mL penicillin-streptomycin, and 5% fetal bovine serum, for up to 3 days (depend on experiment). Change medium every 2 days.
Note: The protocol is primarily for adult cardiomyocytes. Cardiomyocyte cell lines and other cell types can also be assessed following this protocol, but the concentration of saponin needs to be rigorously screened.
-
2.
Prepare necessary solutions before the respirometric measurements. Refer to Key Resources Table and Materials and Equipment sections for a complete list of materials and equipment.
-
3.
Prepare several microsyringes by cutting off the long needle according to the length of the Transparent polycarbonate plunger (Figure 1).
Note: The length of the needle is 0.5–1 mm longer than the plunger. Too long will reach to the electrode and damage it.
Figure 1.

Prepared Microsyringes
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| M199 | Sigma-Aldrich | Cat# M2520 |
| Glutathione | Sigma-Aldrich | Cat# G6013 |
| NaHCO3 | Sigma-Aldrich | Cat# V900182 |
| Creatine | Sigma-Aldrich | Cat# C3630 |
| L-carnitine | Sigma-Aldrich | Cat# C0158 |
| Insulin-transferrin-selenium-X | Thermo Fisher Scientific | Cat# 51500056 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat# 12483020 |
| Pen/Strep (100×) | Thermo Fisher Scientific | Cat# 10378016 |
| Glutamic acid (Glutamate) | Sigma-Aldrich | Cat# C27647 |
| L-Malic acid (Malate) | Sigma-Aldrich | Cat# M1000 |
| ADP | Sigma-Aldrich | Cat# A5285 |
| TMPD | Sigma-Aldrich | Cat# T3134 |
| L-Ascorbic acid (Ascorbate) | Sigma-Aldrich | Cat# A4034 |
| Oligomycin A | Sigma-Aldrich | Cat# O4876 |
| FCCP | Sigma-Aldrich | Cat# C2920 |
| Cytochrome c | Sigma-Aldrich | Cat# C2506 |
| Pyruvate | Sigma-Aldrich | Cat# P2256 |
| EGTA | Sigma-Aldrich | Cat# E3889 |
| MgCl2.6H2O | Sigma-Aldrich | Cat# V900020 |
| Taurine | Sigma-Aldrich | Cat# T8691 |
| KH2PO4 | Sigma-Aldrich | Cat# V900041 |
| HEPES | Sigma-Aldrich | Cat# V900477 |
| BSA | Sigma-Aldrich | Cat# A6003 |
| Potassium lactobionate | Bio-sugars | Cat# 69313 |
| Mannitol | Sigma-Aldrich | Cat# M9546 |
| Dithiothreitol | Sigma-Aldrich | Cat# V900830 |
| Saponin | Sigma-Aldrich | Cat# 47036 |
| Ethanol Absolute | Sigma-Aldrich | Cat# 51976 |
| Na2SO3 | Sigma-Aldrich | Cat# 71988 |
| KOH | Sigma-Aldrich | Cat# 5958 |
| trypsin-EDTA | Gibico | Cat# 25200056 |
| PBS | ThermoFisher | Cat# 10010001 |
| Experimental Models: Organisms/Strains | ||
| SD rat | Shanghai SLAC | Cat# SlacSD |
| Software and Algorithms | ||
| 782 Oxygen system | Strathkelvin Instruments | https://www.mendeley.com/catalogue/b4bf87c2-4afe-379f-b85d-52b1ca2bc3f4/ |
| Other | ||
| Plastic forceps | zmkam | Cat# 93305 |
| Microsyringe | Sh-Gaoge | Cat# G025 |
| Rinse Bottle | ThermoFisher | Cat# N10809 |
| Bottle Top Filter | ThermoFisher | Cat# 2903345 |
| Cell counter | Countstar | IC1000 |
| Inverted microscope | Leica | DMi8 |
| Refrigerated centrifuge | Hettich | Universal 320R0 |
| Vacuum pump | Qilinbeier | GL-802 |
| pH meter | Mettler Toledo | FE20 plus |
| Mitocell respirometry system | Strathkelvin Instruments | MS200 |
Alternatives: You can use Chlorolab2+ from Hansatech Instruments instead of Mitocell respirometry system.
Materials and Equipment
Solution Preparation
Note: Prepare all solutions using 18.2 Ω MilliQ sterilized H2O or absolute ethanol. Aliquot these solutions and store at −20°C or −70°C.
-
•
Mitochondrial respiration buffer (MRB)
| Reagent | Final Concentration | Amount |
|---|---|---|
| EGTA | 0.5 mM | 0.19 g |
| MgCl2·6H2O | 3 mM | 0.61 g |
| Taurine | 20 mM | 2.502 g |
| KH2PO4 | 10 mM | 1.361 g |
| HEPES | 20 mM | 4.77 g |
| BSA | 0.1% | 1.0 g |
| Potassium lactobionate | 60 mM | 23.93 g |
| Mannitol | 110 mM | 20.04 g |
| Dithiothreitol | 0.3 mM | 0.046 g |
| ddH2O | n/a | ∼1,000 mL |
| Total | n/a | 1,000 mL |
Note: Adjust the pH to 7.4 with 5M KOH, filter with a 0.45 μm bottle top filter, dispense into 50 mL aliquots and store at −20°C.
CRITICAL: Ca2+ overload can cause the dysfunction of mitochondrial. EGTA is used to chelate Ca2+.
-
•
5 M KOH: Dissolve 14.02 g KOH in 50 mL ddH2O. Store at room temperature (20°C–26°C).
-
•
0.05% trypsin-EDTA: Dilute 0.25% trypsin-EDTA with PBS to 0.05%. Store at 4°C.
-
•
0.6 M MgCl2: Dissolve 2.44 g MgCl2·6H2O in 20 mL ddH2O. Store at room temperature (20°C–26°C).
-
•
5 mM Oligmycin A: Dissolve 5 mg Oligmycin A in 1.26 mL absolute ethanol. Dispense into aliquots and store at −20°C. Dilute to 100 μM with ddH2O before use.
-
•
10 mM FCCP: Dissolve 5 mg FCCP in 1.967 mL absolute ethanol. Dispense into aliquots and store at −20°C. Dilute to 20 μM with ddH2O before use.
-
•
400 mM Glutamate: Dissolve 0.748 g glutamate in 10 mL ddH2O. Neutralize with 5 M KOH and check pH. Dispense into aliquots, store at −20°C.
-
•
200 mM Malate: Dissolve 0.268 g malate in 10 mL ddH2O. Neutralize with 5 M KOH and check pH. Dispense into aliquots, store at −20°C.
-
•
80 mM Ascorbate: Dissolve 0.158 g in 10 mL ddH2O.
-
•
1 mg/mL Saponin: Dissolve 10 mg in 10 mL ddH2O.
CRITICAL: Saponin needs to be freshly prepared each day.
-
•
20 mM TMPD solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| TMPD | 20 mM | 4.74 mg |
| 80 mM Ascorbate | 1 mM | 12.5 μL |
| ddH2O | n/a | ∼987.5 μL |
| Total | n/a | 1 mL |
Note: Ascorbate is used to prevent the oxidation of TMPD.
-
•
20 mM ADP solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| ADP | 20 mM | 20.04 mg |
| 0.6 M MgCl2 | 6 mM | 20 μL |
| 5 M KOH | ∼ 1.14 M | ∼18 μL |
| ddH2O | n/a | ∼1,962 μL |
| Total | n/a | 2,000 μL |
Note: ADP is the crucial compound to trigger mitochondria respiration. Use MgCl2 stock solution instead of powder because MgCl2 dissolves in water will release heat, which will decrease the stability of ADP. Aliquot and store at −70°C.
Step-By-Step Method Details
Calibrating the Oxygen Electrodes
Timing: 10 min
-
1.
Turn on the Mitocell respirometry system (Figure 2).
Note: The MT200 mitocell respirometer only needs 100 μL of sample each time, which is far less than the traditional Clark oxygen electrode meter.
-
2.
Turn on the 782 Oxygen system software and check the connection of oxygen meter.
Note: The software cannot connect to the oxygen meter if you turn on the software before step 1.
-
3.
Fill the chamber with MilliQ water using a rinse bottle, and discard the water with a vacuum pump, rinse 3 times (Figures 3A and 3B).
Note: The tip must be inserted against the wall of the oxygen electrode chamber, otherwise it can easily damage the membrane of the electrode.
-
4.
Fill air-saturated water (MilliQ water in contact with air) into the oxygen electrode chamber.
-
5.
Put a micro-magnetic stirrer into the oxygen electrode chamber of MT200 respirometer with a plastic forceps, and start stirring (Figure 3C).
Note: Keep eyes on the tiny magnetic stirrer because it is easily aspirated on the tip. Prepare a big magnetic to find the lost magnetic stirrer.
-
6.
Click on “Calibration high” of calibration panel to start calibrating the high value of the electrode. Please wait until the process is completed.
-
7.
Add a small amount (3–5 mg) of sodium sulfite into the air-saturated water in the oxygen electrode chamber and click on “Calibration zero” to calibrate the zero value of the electrode (Figure 3D).
-
8.
Rinse the chamber 6–10 times to wash out the sodium sulfite. Now, the oxygen meter is ready for use.
Note: Please rinse more times if the sloped basal line occurred.
Pause Point: The calibrated oxygen meter could wait for a break of 2–3 h.
Figure 2.
The Mitocell Respirometry System
The system comprising a 782 oxygen meter and a MT200 respirometer with a 1302 electrode.
Figure 3.
The Process of Calibration
(A) Rinse the oxygen electrode chamber.
(B) The tip site for sucking out the solution in the oxygen electrode chamber.
(C) Put the magnetic stirrer into the oxygen electrode chamber.
(D) Add 3–5 mg sodium sulfite to calibrate the zero of the electrode.
Counting Cardiomyocytes
Timing: 10 min
-
9.
Wash cardiomyocytes twice with 5 mL PBS, detach cells using 1 mL 0.05% trypsin digesting 2–3 min and pipette 5–8 times, and collected cardiomyocytes through 80 g, 2 min centrifugation, and discard the supernatant.
Note: The ability of attachment of adult cardiomyocytes is weaker than the cell lines.
-
10.
Resuspend cells in 1–2 mL mitochondrial respiration medium B (MRB).
-
11.
Pipette the cells well and use one drop to count the number of cells with an automatic cell counter.
-
12.
Dilute the cells to 5 × 105 cells per mL with MRB and keep on ice.
CRITICAL: Appropriate cell number is very important for measurement. Too many cells will cause the quick decline of the basal line without enough time to treat the cells with substrates and inhibitors. Also, few cells will lead to a weak response.
Measuring Mitochondrial Respiration
Timing: 15–20 min
-
13.
Add 100 μL cell suspension (5 × 104 cells) using a pipette to oxygen electrode chambers. Close the oxygen electrode chamber with the Transparent polycarbonate plunger, equilibrate again for 3–5 min.
Note: keep the cardiomyocytes, mitochondrial electron transport chain substrates and inhibitors on the ice during measurement.
-
14.
Start recording oxygen consumption. After 2–3 min of basal line recording, add 5 μL saponin (final concentration is ∼50 μg/mL) using a 25 μL microsyringe for cell membrane permeabilization, and continue recording (Figure 4).
Note: Add the solution of saponin, substrates, or inhibitors slowly; otherwise, it will cause bubbles to disrupt the trace.
CRITICAL: 1–2 min is enough for complete cell membrane permeabilization. Excess of saponin (more than 100 μg/mL) can damage the mitochondrial membrane (Kuznetsov et al., 2008). The permeabilization conditions should be strictly optimized for different cell types to preserve mitochondrial intactness. To check whether the mitochondrial outer membrane is damaged, we can assess the oxygen consumption of TMPD/Ascorbate/ADP and cytochrome c (Please refer to the section of Troubleshooting 2)
-
15.
1–2 min later, add 5 μL Glutamate/Malate solution (final concentration is ∼10 mM glutamate, ∼5 mM malate) to the chamber and record 2 min of resting complex I-supported respiration.
Note: Make a mixed Glutamate/Malate substrate solution by mixing 400 mM Glutamate and 200 mM malate in a 1:1 ratio. Alternatively, you can use 10 mM Pyruvate instead of Glutamate.
-
16.
Add 5 μL 20 mM ADP (final concentration is ∼1 mM) to the chamber, incubate for 3–5 min and record the respiration.
Note: If the slope of trace (state 3) only slightly increases (the ratio of respiration rate of control cells before and after the addition of ADP is less than 2), it indicates that the mitochondria may be damaged by permeabilization. Please perform the oxygen consumption of TMPD/Ascorbate/ADP and cytochrome c (Please refer to the section of Troubleshooting 2)
-
17.
Add 5 μL 100 μM Oligmycin A (final concentration is ∼ 5 μM ) to the chamber, to inhibit ATP synthase and reduce oxygen consumption rate. Incubate for 2–3 min, and record the respiration.
-
18.
Add 5 μL 20 μM FCCP (final concentration is ∼ 1 μM) to the chamber to uncouple oxygen consumption from ATP production, and trigger the maximal oxygen consumption, record the respiration until it reaches zero.
Figure 4.

Add Saponin, Substrates or Inhibitors with a Microsyringe
Cleaning the Oxygen Electrode Chambers
Timing: 35 min
-
19.
Rinse the chamber with MilliQ H2O for several times. Then sterilize with 70% ethanol for 30 min after each experiment.
Expected Outcomes
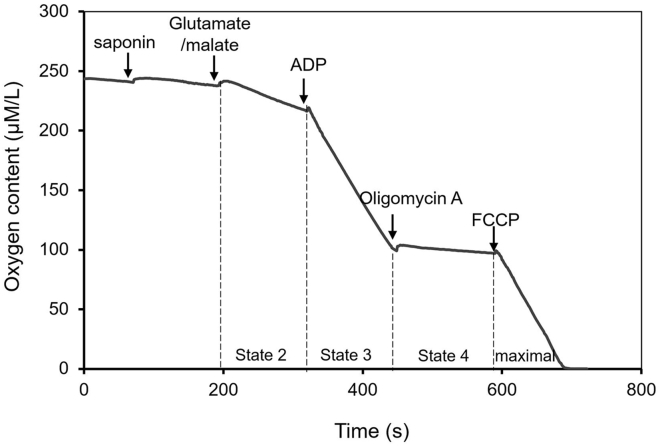
The goal of the method is for analyzing mitochondrial respiration function in permeabilized cardiomyocytes. In our protocol, we evaluate mitochondrial respiration function in permeabilized cardiomyocytes using an MT200 mitocell respirometer. Only 100 μL of the sample (5 × 104 myocytes) is needed each time, which can save precious samples. The mitochondrial respiration is more physiologically relevant than isolated mitochondria due to preserving their essential interactions with other intracellular systems. At the same time, the mitochondria in situ are usually more stable than isolated mitochondria. A typical oxygen concentration traces for successfully permeabilized cardiomyocytes are shown in figure (Figure 5).
Figure 5.
A Representative Trace of Mitochondrial Respiration in Permeabilized Cardiomyocytes
Compounds were added as indicated. The slope of trace represent the different state of mitochondrial respiration. State 2: Mitochondrial respiration stimulated by the substrate in the absence of and added ADP; State 3: Mitochondrial respiration initiated by adding ADP in the presence of substrate; State 4: Mitochondrial respiration in the absence of ATP synthesis, which is uncoupled with the Oligmycin A. Maximal respiration: Mitochondrial respiration triggered by FCCP.
Limitations
The permeabilization method, not specific target to cytoplasm membrane, may also cause damage to the mitochondrial membrane. The permeabilization conditions must be strictly optimized for different cell types to preserve mitochondrial intactness. The cell number is also need be optimized for different cell types. This method is not able to separately investigate distinct mitochondrial subpopulations.
Troubleshooting
Problem 1
The rates of initial respiration before the addition of ADP is high
Potential Solutions
Wash glassware with RMB supplemented with EGTA to remove the calcium. Decrease the cell number.
Problem 2
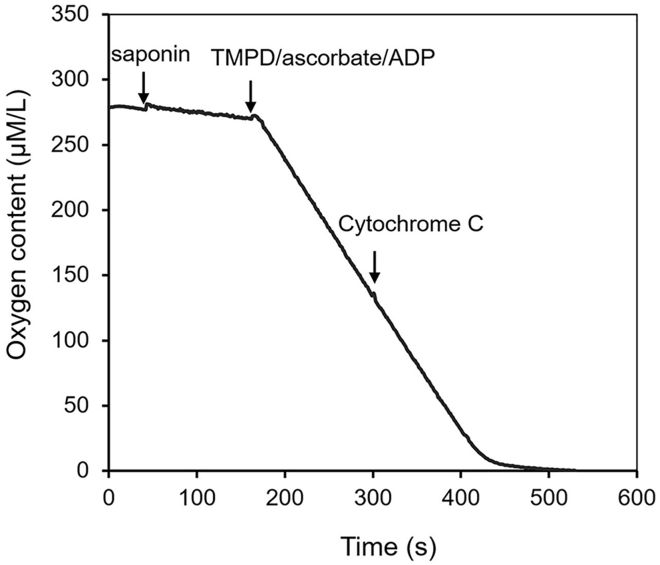
ADP-stimulated respiration is too low.
Potential Solutions
First, perform the oxygen consumption of TMPD/Ascorbate/ADP followed with cytochrome c addition (Figure 6). If cytochrome c markedly increases the oxygen consumption triggered by TMPD/Ascorbate/ADP, please decrease the concentration of saponin to avoid damaging mitochondrial outer membrane. Slightly increase the concentration of saponin if cytochrome c does not affect oxygen consumption.
Figure 6.
A Representative Trace of Mitochondrial Oxygen Consumption Triggered by TMPD/Ascorbate and Cytochrome c in Permeabilized Cardiomyocytes
Compounds were added as indicated.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Guohua Gong (guohgong@tongji.edu.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate any unique datasets or code.
Acknowledgments
This work was supported partly by the National Key Research and Development Program of China (no. 2018YFA0107102 to G.G.), the National Natural Science Foundation of China (no. 31901044, 31771524 to G.G. and no. 81970333 to Y.Q.), and the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (no. TP2017036 to G.G.).
Author Contributions
G.G. conceived, designed, and supervised the project. M.G. and A.L. conducted most of the experiments and performed data analysis. L.-B.L. maintained the Clark electrode. Y.Q. provided valuable suggestions. G.G. and M.G. wrote the manuscript.
Declaration of Interests
The authors declare that they have no competing interests.
Contributor Information
Meng Gao, Email: meng_gao1993@163.com.
Guohua Gong, Email: guohgong@tongji.edu.cn.
References
- Gong G., Song M., Csordas G., Kelly D.P., Matkovich S.J., Dorn G.W., 2nd Parkin-mediated mitophagy directs perinatal cardiac mitochondrial metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G.H., Liu X.Y., Zhang H.L., Sheu S.S., Wang W. Mitochondrial flash as a novel biomarker of mitochondrial respiration in the heart. Am. J. Physiol. Heart Circ. Physiol. 2015;309:H1166–H1177. doi: 10.1152/ajpheart.00462.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.G., Gao M., Li A.Q., Liu B.L., Jiang W.T., Qin Y., Gong G.H. Isolation of viable adult rat cardiomyocytes with high yield. STAR Protoc. 2020;1:100045. doi: 10.1016/j.xpro.2020.100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov A.V., Veksler V., Gellerich F.N., Saks V., Margreiter R., Kunz W.S. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues, and cells. Nat. Protoc. 2008;3:967–976. doi: 10.1038/nprot.2008.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.