Summary
This protocol describes the isolation, handling, culture of, and experiments with human colon stem cell organoids in the context of cystic fibrosis (CF). In human colon organoids, the function of cystic fibrosis transmembrane conductance regulator (CFTR) protein and its rescue by CFTR modulators can be quantified using the forskolin-induced swelling assay. Implementation procedures and validation experiments are described for six CF human colon organoid lines, and representative CFTR genotypes are tested for basal CFTR function and response to CFTR-modulating drugs. For complete details on the use and execution of this protocol, please refer to Dekkers et al (2016) and Berkers and van Mourik (2019).
Graphical Abstract
Highlights
-
•
Rectal biopsies are used to efficiently establish human colon organoid cultures
-
•
Human colon organoids can be cultured and biobanked for prolonged periods
-
•
Human colon organoids can be used to measure function of the CFTR protein using FIS
-
•
Reference CF organoid lines are available for validation of the FIS assay
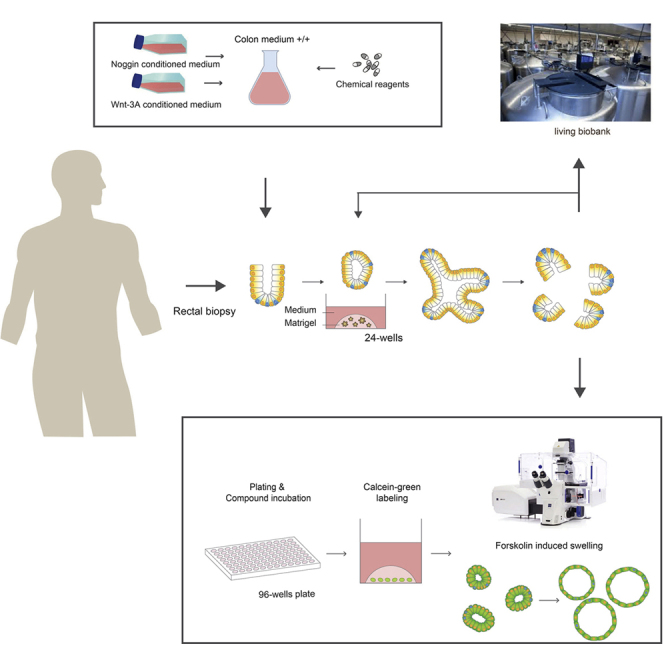
Human colon organoids are used to measure the function of the cystic fibrosis transmembrane conductance regulator (CFTR) protein and its rescue by CFTR modulators through the forskolin-induced swelling assay. This protocol describes the isolation, culture of, and experiments with human colon stem cell organoids for studying cystic fibrosis.
Before You Begin
CRITICAL: All laboratory procedures related to colon organoid or cell cultures should be performed in a laminar flow to avoid contaminations and to protect the operator from biological risks.
Preparations
-
1.
Prepare Dulbecco’s Modified Eagles Medium with additional 1% (v/v) pen- strep (10,000 U/mL) and 10% (v/v) FBS (DMEM +/+) and pre=warm to 37° C
Wnt-3A Conditioned Medium (WCM) Production
Note: it is strongly advised to produce and mix multiple batches (2–3) of WCM to compensate for batch-batch variation during WCM production. Each round can produce around 1,100 mL of WCM leading to a WCM pool of 2,200 (mix of 2 batches)– 3,300 mL (mix of 3 batches) of which the total production time is between 20 (two batches) and 24 (3 batches) days.
-
1.
Thaw a cryovial with L-Wnt-3A cells by putting 13 mL of pre-warmed DMEM +/+ medium in a 15 mL tube (when thawing multiple cryovials, use a 50 mL tube with 30 mL DMEM +/+).
-
2.
Transfer the cells from the cryovial to the 15 mL tube by resuspending the cells in DMEM +/+.
-
3.
Centrifuge the 15 mL tube at 450 g for 5 min. Remove the supernatant and resuspend the cell pellet in 25 mL pre-warmed DMEM +/+ with additional 31.25 μL zeocin (final concentration 125 μg/mL).
-
4.
Make sure that a homogenous cell suspension is obtained and transfer the cell suspension to a T175 flask.
-
5.
Incubate the T175 flask for 3–4 days at 37 °C, 5% CO2.
-
6.
Split the cells when 90%–100% confluency is achieved into 6 x T175 flasks:
-
7.
Remove the DMEM +/+ from the T175 flask.
-
8.
Gently wash the cells once with 5 mL PBS0 and discard the PBS0.
-
9.
Add 1.5 mL 0.05% Trypsin EDTA and leave at 18–23 ° C for 7–10 min until the cells are detached.
-
10.
Add 10.5 mL DMEM +/+ medium, take up all the cells and resuspend until a homogenous cell suspension is achieved.
-
11.Add 2 mL DMEM +/+ of the cell suspension to each new T175 flask and top up to 25–30 mL of total DMEM +/+ per flask:
-
→5 x T175 flask in DMEM +/+
-
→1 x T175 flask in DMEM +/+ with zeocin, use this flask for the next production batch when the cells are 90%–100% confluent again (usually after 3–4 days).
-
→
When the cells in the five T175 flasks containing DMEM +/+ are 90%–100% confluent (usually after 3–4 days), split the cells in 145 mm Petri dishes:
-
12.
Remove DMEM +/+ medium from the flasks and wash once with 5 mL PBS0.
-
13.
Add 0.05% Trypsin EDTA and leave at 18–23 ° C for 7–10 min.
-
14.
Add 3.5 mL of DMEM +/+ per flask, pool the cells from all five T175 flasks, and resuspend until a homogenous cell suspension is achieved (total volume = 25 mL DMEM +/+ medium with cells).
-
15.Measure the cell concentration per mL and dilute the cell suspension needed for seeding in 50 145 mm tissue culture Petri dishes to create one (the first) batch with 1100 ml of WCM. For 50 145 mm (2 x 106 cells per 22 ml ∗ 50=) 100 x 106 cells are needed in 1100 ml.
-
○Determine the cell concentration (with e.g. hemocytometer) [in #cells/mL] = Y
-
○Calculate 100/ Y = X, the volume of the cell suspension to dilute in 1100 ml.
-
○
-
16.
Make sure the cell suspension is homogeneous and add 22 mL of cell suspension per 145 mm tissue culture Petri dish.
Note: When more than the necessary amount of cells is counted, a part of the cell suspension may remain unused or extra 145 mm Petri dishes can be seeded.
-
17.
Incubate the cells at 37 °C, 5% CO2 for 8 days (do not harvest the WCM/ DMEM +/+ before day 8)
-
18.
Harvest the WCM from the Petri dishes in sterile 50 mL tubes and discard the Petri dishes with cells.
-
19.
Centrifuge for 5 min at 650 g to remove floating cells and debris.
-
20.
Filter medium through a 0.22 μm filter (stericups are preferable).
-
21.
Store the WCM in 50 mL tubes or in 0.5–1 L containers at 4 °C.
-
22.
Repeat the splitting of a 1 x T175 flask for the next batch as described above for 2–3 batches per WCM production.
-
23.
WCM can be stored at 4°C and used for 2 months. Testing and approving of the WCM batches: WCM quality can be tested by culturing organoids with this specific WCM batch. Differentiated, non- budding round, thick-walled organoid structures indicate low Wnt activity (also see Figure 10C). Running human colon organoid cultures often need time to adjust to a new batch of WCM. Judge organoid cultures after 2 weeks before making conclusions on the WCM quality.
Note: A running L-wnt3a cell line can be used for the production of multiple batches of WCM until passage 18–20.
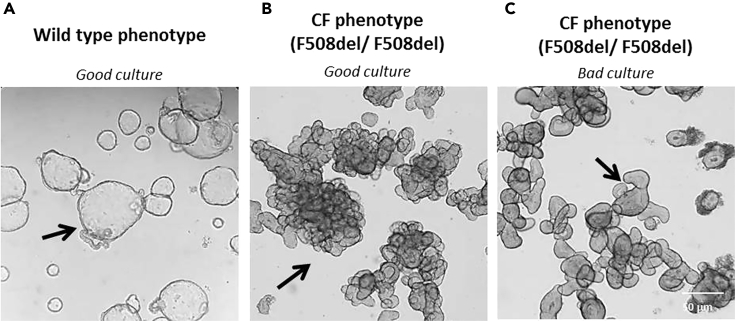
Figure 10.
Examples of 7-Day Old Human Colon Organoid Cultures (Bright Field, 4 x Magnification)
(A) Typical pre-swollen stem cell phenotype of a high quality, proliferated wild type (non-CF) human colon organoid culture.
(B) Non-swollen stem cell phenotype of a high quality, proliferated CF colon organoid culture.
(C) Example of a bad quality CF colon organoid culture with thick-walled, differentiated structures with decreased stem cell phenotype which is often due to low activity of Wnt-3A in WCM.
Noggin Conditioned Medium (NCM) Production
Note: HEK293T Noggin hFc cells are poorly adhesive cells (especially the first passage after thawing). At the first passage after thawing leave the cells for at least 5 days without touching. Make sure to handle the cells gently during washing with PBS0 to avoid detaching. An extra passaging step before starting the harvesting steps is strongly recommended to let the cells recover from the freeze-thawing step.
-
1.
Thaw a cryovial with HEK293T Noggin hFc cells by putting 13 mL of pre-warmed DMEM +/+ in a 15 mL tube (when thawing multiple cryovials, use a 50 mL tube with 30 mL DMEM +/+).
-
2.
Transfer the cells from the cryovial to the 15 mL tube by resuspending the cells with DMEM +/+.
-
3.
Centrifuge the 15 mL tube at 450 g for 5 min. Remove the supernatant and resuspend the cell pellet in 40 mL pre-warmed DMEM +/+ with additional G418 (final concentration 500 μg/mL).
-
4.
Make sure that a homogeneous cell suspension is achieved and transfer the cell suspension to a T175 flask.
-
5.
Incubate the T175 flask for 7 days at 37 °C and 5% CO2.
-
6.
Split the cells when 90-100% confluence is achieved into 6 x T175 flasks.
-
7.
Gently remove the DMEM +/+ from the T175 flask.
-
8.
Gently wash the cells once with 5 mL PBS0 and discard the PBS0.
-
9.
Add 1.5 mL 0.05% Trypsin EDTA, leave at 18–23 ° C for 7–10 min until the cells are detached.
-
10.
Add 10.5 mL DMEM +/+ medium, take up all the cells and resuspend until a homogenous cell suspension is achieved.
-
11.
Transfer the cell suspension to 15 mL tube
-
12.
Count the cells (with e.g. hemocytometer)
-
13.Add 5∗106 cells in 25 mL culture medium and transfer to a new T175 flask:
-
○1 x T175 in culture medium with G418 (500 μg/mL), use this flask for the next batch (usually cells are confluent after 3–4 days).
-
○X (depending on amount of cells) x T175 in culture medium without G418.
-
○
When the cells seeded in DMEM +/+ without G418 are ∼90% confluent (usually after 3–4 days):
-
14.
Remove the DMEM +/+ from the T175 flasks
-
15.
Gently (to avoid detachment of the cells) add 50 mL of Ad-DF+++ per flask.
-
16.
Incubate the flasks at 37 °C and 5% CO2 for 8–10 days.
-
17.
Harvest the Ad-DF+++ (NCM) on day 8–10. Never harvest NCM before day 8 or when cells are not at least 100% confluent.
-
18.
Pool the NCM from the T175 flasks in sterile 50 mL tubes and discard the T175 flasks.
-
19.
Centrifuge the NCM for 5 min at 650 g to remove floating cells and debris.
-
20.
Filter medium through a 0.22 μm filter (stericups are preferable).
-
21.
Store the NCM in 50 mL tubes or in 0.5–1 L containers at −20 °C.
Note: A running HEK293T-Noggin-hFc cell line can be used for the production of multiple batches of NCM until passage 18–20.
Note: No properly validated NCM quality/quantity assay has yet been developed. After testing the functionality (on human colon organoid cultures) the NCM batches can be thawed, pooled and refrozen in aliquots and are stable at −20 ºC for at least one year. Try to avoid unnecessary freeze-thaw cycles.
Note: Different NCM batches can show much variation in Noggin activit. It is advisable to produce multiple batches and mix an NCM pool to ensure a minimal efficiency level of active NCM.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides and Recombinant Proteins | ||
| 0.05% Trypsin EDTA | Thermo Fisher Scientific : Invitrogen | #25300-054 |
| Advanced Dulbecco’s Modified Eagles Medium with Nutrient Mixture F-12 Hams (Ad-DF) 500 mL | Thermo Fisher Scientific: Invitrogen | #12634 |
| B27 supplement | Thermo Fisher Scientific: Invitrogen | #17504-044 |
| Calcein, AM | Life Technologies: Gibco | #C3100MP |
| Dimethyl sulfoxide (DMSO) | Sigma Aldrich | #276855-1L |
| Dulbecco’s modified Eagle medium (DMEM) | Thermo Fisher Scientific: Invitrogen | #31966-021 |
| Fetal bovine serum (FBS, brand varies upon optimal batch selection) | Sigma/ Bovogen | |
| Forskolin | Sigma | #F3917-10mg |
| Gastrin | Sigma Aldrich | #G9145 |
| Gentamicin | Life Technologies: Gibco | #15710-049 |
| Glutamax | Thermo Fisher Scientific: Invitrogen | #35050 |
| G418 (100 mg/mL) | Invivogen | #ant-gn-5 |
| Human Epithelial Growth Factor (hEGF) | Peprotech | #AF-100-15 |
| HEPES | Thermo Fisher Scientific: Invitrogen | #15630-056 |
| Matrigel (important: protein concentration > 10 mg/mL) | Corning | #356231 |
| N-Acetylcysteine | Sigma Aldrich | #A9165 |
| Nicotinamide | Sigma Aldrich | #N0636 |
| p38 MAPK inhibitor (p38i) (SB202190) | Sigma Aldrich | #S7067 |
| Phosphate Buffered Saline 0 (without Ca and Mg) | Sigma/ Thermofisher/ sterile homemade | #D5652/#14190250 |
| Penicillin/ Streptomycin | Thermo Fisher Scientific: Invitrogen | #15140-122 |
| Primocin (50 mg/ mL) | InvivoGen | #ant-pm-1 |
| Recovery™ Cell Culture Freezing Medium | Thermo Fisher Scientific: Invitrogen | #12648010 |
| Recombinant Human R-Spondin 3 Protein (hR-spondin-3) | R&D | #3500 – RS/ CF |
| TGFb type I Receptor inhibitor (A83-01) | Tocris | #2939 |
| Ultrapure 0.5 M EDTA, pH 8 | Thermofischer | #15575020 |
| Vancomycin | Sigma Aldrich | #861987- 250mg |
| VX-661 | Selleckchem | #S7059 |
| VX-770 | Selleckchem | #S1144 |
| Y-27632 Dihydrochloride (ROCKi) | Abmole bioscience | #Y-27632 |
| Zeocin (100 mg/mL) 1g | Thermo Fisher Scientific: Invitrogen | #R25001 |
| Experimental Models: Cell Lines | ||
| HEK293T – Noggin hFc cell line | Optimal Noggin-hFc producing subclone obtainable through the HUB: http://hub4organoids.eu/ | n/a |
| L- Wnt 3A producing cell line | ATCC Optimal Wnt-3A producing subclone obtainable through the HUB: http://hub4organoids.eu/ |
#CRL-2647 |
| Human colon organoids genotype G542X / G542X | HUB: http://hub4organoids.eu/ | #HUB-02-D2-121 |
| Human colon organoids genotype F508del / R1162X | HUB: http://hub4organoids.eu/ | #HUB-02-D2-038 |
| Human colon organoids genotype F508del/ F508del | HUB: http://hub4organoids.eu/ | #HUB-02-D2-341 |
| Human colon organoids genotype F508del/ G551D | HUB: http://hub4organoids.eu/ | #HUB-02-D2-043 |
| Human colon organoids genotype F508del/S1251N | HUB: http://hub4organoids.eu/ | #HUB-02-D2-103 |
| Human colon organoids genotype F508del / R117H - 7T | HUB: http://hub4organoids.eu/ | #HUB-02-D2-004 |
| Software and Algorithms | ||
| Cell profiler (open- source) | McQuin et al., 2018 | https://cellprofiler.org/ |
| Zen blue® | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Graphpad Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Microsoft Excel | Microsoft Corporation | https://office.microsoft.com/excel |
| Other | ||
| 15 and 50 mL conical tubes | No recommended vendor | n/a |
| Microfuge tubes | No recommended vendor | n/a |
| 100 mL (plastic) bottles | No recommended vendor | n/a |
| 0.22 μm filters | No recommended vendor | n/a |
| Serological pipets | No recommended vendor | n/a |
| Micropipet filter tips + Micropipet tips without filter (200 μL) | No recommended vendor | n/a |
| Cryovials | No recommended vendor | n/a |
| 24-wells flat bottom tissue culture plates / 96-wells flat bottom tissue culture plates | No recommended vendor | n/a |
| T175 cell culture flasks | No recommended vendor | n/a |
| 145 mm Petri dishes | No recommended vendor | n/a |
| Cell Freezing Container (e.g. Mr Frosty) | No recommended vendor | n/a |
| Light/optical microscope appropriate for cell culture with standard ocular (10x) and 4x and 10x objective | No recommended vendor | n/a |
| Live cell imaging confocal microscope with 5x objective, incorporated incubator (allowing controlling temperature and CO2) and 488 nm laser | Recommended but not essential: Zeiss LSM800 | n/a |
| Liquid nitrogen tank | No recommended vendor | n/a |
Note: clinical reagents and materials mentioned in ‘obtaining human (colon) rectal biopsies’ (e.g. rectal suction biopsy device, forceps device, video endoscope) are not included in this list and are often hospital-specific. For information please contact your local gastroenterologist.
Materials and Equipment
| Culture Medium | Abbreviation | Description | Application |
|---|---|---|---|
| DMEM +/+ | DMEM +/+ | Dulbecco’s Modified Eagles Medium with additional 1% (v/v) pen- strep (10,000 U/mL) and 10 % (v/v) FBS | Preparation of conditioned media |
| Ad-DF+++ | Ad-DF+++ | Advanced Dulbecco’s Modified Eagles Medium with Nutrient Mixture F-12 Hams (Ad-DF 500 mL + 1% (v/v) glutamax, 1% (v/v) hepes buffer and 1% (v/v) pen-strep (see Table 3 for exact concentrations) | Preparation of conditioned media, handling of colon organoid cultures. |
| Human colon organoid medium −/− | 2x CM −/− | Colon organoid culture medium without WCM & R-spo3 (see stock preparation of human colon organoid medium components) | Preparation of 50 mL stock 2x human colon organoid medium saved at -20°C |
| Human colon organoid medium +/+ | CM +/+ | Colon organoid culture medium with WCM & R-spo3 (see stock preparation of human colon organoid medium components) | Human colon organoid medium used for regular culture and FIS |
| Noggin conditioned medium | NCM | Self-produced Noggin conditioned medium | 10% of human colon medium |
| Wnt-3A conditioned medium | WCM | Self-produced Wnt-3a conditioned medium | 50% of human colon medium +/+ |
Stock Preparation of Human Colon Organoid Medium Components
TIMING: 5–6 hours
Human colon organoid medium consists of several chemical components as well as self-produced conditioned media, such as Wnt-3A conditioned media (WCM) and Noggin conditioned media (NCM). In this protocol hR-spondin-3 (R-spo3) was preferred over self-produced R-spondin-1 conditioned media. However, a R-spondin-1 producing cell line is available and could be preferred due to cost reduction of media. R-spondin-1 conditioned media can also be stored for 1 year at -20 °C.
Preparing 2x Colon Organoid Medium −/−
Table 1 shows how to dissolve all individual components, whereas Table 2 shows how to prepare 2x colon organoid medium (2x CM) −/− by addition of all reagents except WCM and R-spo3. It is advisable to calculate the medium amounts that will be used within the following 2.5 months and adjust the volume of 2x CM −/− preparation.
Table 1.
Overview of the Stock Preparation of the Individual Medium Components with All Relevant Ordering and Dissolvent Information
| Compound | Amount | Concentration Stock | End Concentration Colon Medium +/+ | Storage |
|---|---|---|---|---|
| Ad-DF+++ | Add 5 mL Glutamax (100 x) | 40% | 8–12 weeks at 4 °C | |
| Add 5 mL HEPES (1M) | ||||
| Add 5 mL Pen/Strep (10,000 U/mL / 100 x) | ||||
| B27 | 10 mL direct use from bottle | 2% | 1 year at −20°C | |
| N-acetylcysteine | 815 mg Nac in 10 mL H20 | 500 mM | 1.25 mM | 1 year at −20°C |
| hEGF | Add 2 mL filter sterilized PBS0-BSA (0,1 %) to 1 vial hEGF | 0.5 mg/mL 10.000 x |
50 ng/mL | 1 year at −20°C |
| Nicotinamide | 6 gr nicotinamide in 50 mL PBS0 | 1 M 100 x |
10 mM | 4 months at −20°C |
| A83-01 | 10 mg A83-01 in 5 mL DMSO | 5 mM 10.000 x |
500 nM | 3 months at −20°C |
| P38 inhibitor (SB202190) | Add 500 μL DMSO to one bottle of 5 mg P38-inhibitor | 30 mM 3000 x |
3 months at −20°C | |
| Gastrin | Mix 0.5 mg gastrin to 2.383 mL PBS0 | 100 μM | 5 nM | 3 months at −20°C |
| R-spo3 | Reconstitute in PBS0 | Varies per batch | 250 ng/ mL | 3 months at −80°C |
| WCM | Collect from producing cell-line (see 2.1) | 2 x | 50% | 2 months at 4°C |
| NCM | Collect from producing cell-=line (see 2.2) | 10 x | 10% | 1 year at 20°C |
Table 2.
Preparation of 2x Colon Organoid Medium (CM) -/-
| Reagent | Final Conc. | 100 mL Total Medium (50 mL 2x CM −/−) |
|---|---|---|
| Ad-DF+++ | 36.8% | 36.8 mL |
| NCM | 10% | 10 mL |
| B27 | 2% | 2 mL |
| N-acetylcysteine | 1.25 mM | 250 μL |
| hEGF | 50 ng/mL | 10 μL |
| Nicotinamide | 10 mM | 1 mL |
| A83-01 | 500 nM | 10 μL |
| P38 inhibitor | 10 μM | 33 μL |
| Gastrin | 100 μM | 5 μL |
| Primocin | 100 μg/mL | 0.2 mL |
Store 2x CM −/− in aliquots at –20 °C for a maximum of 2.5 months. For example, aliquot 50 mL in 100 mL bottles so WCM can be added when preparing CM +/+ as described in the following section.
Human Colon Organoid Medium +/+ Preparation
When preparing fresh CM +/+ for direct use in human organoid cultures, thaw a bottle of frozen 2x CM −/− over 8–12 hours at 4 °C (preferably in the fridge) and add WCM and R-spo3 the following day. Table 3 shows the ratio of adding WCM and R-spo3 to create 100 mL total volume of CM +/+. CM +/+ can be used for 10 days if stored at 4 °C.
Table 3.
Preparation of Colon Organoid Medium (CM) +/+
| Reagent | Final Conc. | 100 mL CM +/+ | Storage |
|---|---|---|---|
| 2x CM -/ - | 50% | 50 mL | 10 days at 4 °C |
| WCM | 50% | 50mL | 2 months at 4 °C |
| R-spo3 | 250 ng/mL | Depends on LOT# | 7 days at 4 °C |
Step-by-Step Method Details
Obtaining Human (Colon) Rectal Biopsies
TIMING: ∼2 h
CRITICAL: Clinical procedures to isolate rectal or colon biopsies from patients either through forceps biopsy (A) or rectal suction biopsy (B) should ALWAYS be performed by trained medical staff in a medical clinic. Make sure all necessary precautions are in place for a safe procedure.
-
1.
Fill a 50 mL conical tube with +/− 20 mL of PBS0 at 4 °C
-
2.
If biopsies are transported over long distance or long duration (>8 h) before crypt isolation procedure can take place, biopsies should be stored at 4 °C in Ad-DF +++ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL).
Caution: Authorized personnel should wear gloves and a physician’s coat. This is the minimum protection that should be worn when working in the OR (operating room). Make sure all the apparatus are calibrated and sterilized.
CRITICAL: Procedures should not take place:
If the patient suffers from thrombocytopenia (less than 50 x 109 thrombocytes /L).
If the patient suffers from coagulation disorder.
If the patient is currently suffering from severe inflammation of the intestine.
In case of visibility problems during the endoscopy.
Possible Complications during and after the Procedure
Perforation of the intestinal wall.
Severe or persistent bleeding.
Attention Points
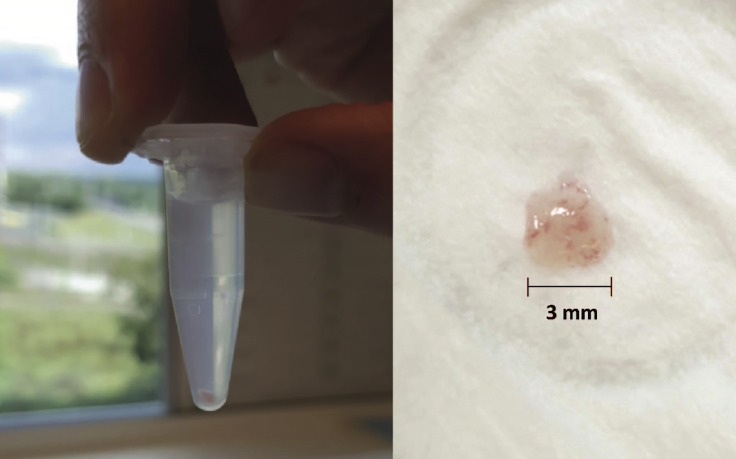
The nurse notifies the physician if the biopsies are of excellent, decent or inferior quality (see Figure 3).
Be considerate with regards to the position and signs of any pain or discomfort of the patient.
Figure 3.
Example of a Good Quality Biopsy
Note the pink colour and round shape.
Forceps Biopsy Procedure
Additional information on forceps biopsy procedure including video can be found elsewhere (Servidoni et al., 2013).
Note: A nurse/endoscopy-assistant guides the patient through the process and assists the physician throughout the procedure.
Caution: The (jumbo) forceps should be closed and pulled back when entering or exiting the endoscopic instrumentation channel.
Caution: The procedure should not take place in case there are visibility problems during the endoscopy.
Note: Before starting the procedure, be sure you have read and understood all the instructions detailed in this paragraph.
-
1.
Make sure the patient agrees to the procedure before starting and the appropriate informed consent form is signed.
-
2.
The nurse opens the 50 mL collection tube containing PBS0 or storage media (Ad+++ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL)) at 4°C and properly labelled for the rectal biopsies (with clean gloves).
-
3.
The nurse proceeds by opening a new sterile jumbo forceps packaging, removing the cover at the end of the forceps around the mouth of it and presents it to the physician.
-
4.
The physician enters the forceps inside the endoscopic instrumentation channel.
-
5.
The nurse/assistant opens or closes the forceps as instructed by the physician.
-
6.
When removing the forceps out of the endoscopic instrumentation channel it should be cleaned with a gauze to avoid blood spatters. The biopsies are collected in the collection tube.
Note: If the rectal/ colon biopsy is sticking to the jumbo forceps, use a sterile needle or sterile tweezers to remove the biopsy.
-
7.
Rinse the forceps in a cup of clean water before re-entering the jumbo forceps into the endoscope to collect more biopsies from the patient. Make sure the forceps are clean.
-
8.
Repeat steps 4–7 until a minimum of 4 good quality biopsies have been successfully isolated and collected (all biopsies can be collected in the same tube).
-
9.
After the procedure the nurse firmly closes the collection tube and takes off her gloves.
-
10.
If there is more than one biopsy collection tube, make sure it is marked and numbered by the nurses.
-
11.
Maintain the collection tube with the biopsies on ice at ∼4°C in a styrofoam box.
-
12.
After the procedure is finished the patient should remain in the hospital for an additional hour to check for potential rectal bleeding. In case of severe or persistent rectal bleeding a qualified physician should be consulted.
-
13.
When no complications are observed the patient can be discharged.
-
14.
Inform the patient about potential late onset rectal bleeding. In case of late onset rectal bleeding a qualified physician should be consulted.
Obtaining Biopsies by Rectal Suction Device
A nurse/endoscopy-assistant guides the patient through the process and assists the physician throughout the procedure.
Note: Before starting the procedure, make sure to have read and understood all the instructions detailed in this paragraph
-
1.
Make sure the patient agrees to the procedure before starting and the appropriate informed consent form is signed.
-
2.
Ensure an appropriate bowel preparation: apply an enema to the patient (Sodium phosphate enema (2.5 mL∗ (per) kg body weight; maximum dose 130 mL) by inserting the sodium phosphate solution rectally +/− 2–3 h before the biopsy procedure.
-
3.
Wait for the result of the enema.
-
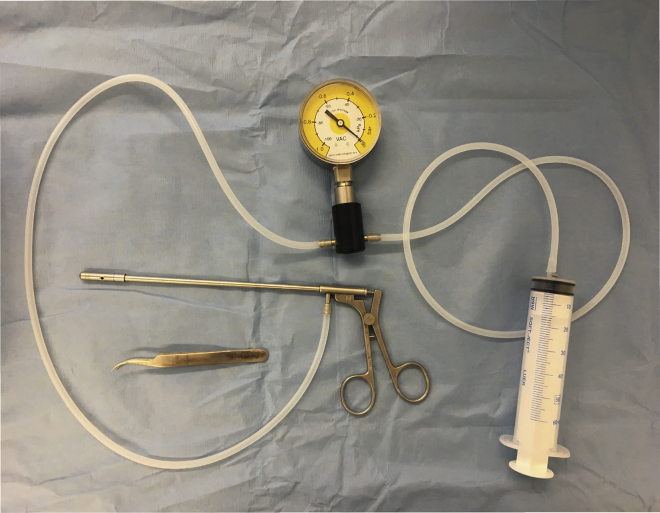
4.
Test the rectal suction device for vacuum pressure before using it in the patient (see Figure 1).
-
5.
The nurse opens the 50 mL collection tube containing PBS0 or storage media (Ad-DF+++ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL)) at 4°C and properly labelled for the rectal biopsies (with clean gloves).
-
6.
Apply lubricant (e.g. K-Y Jelly) to the rectal suction biopsy device.
-
7.
Carefully introduce the suction biopsy device rectally, approximately 5 cm distance from the anal verge.
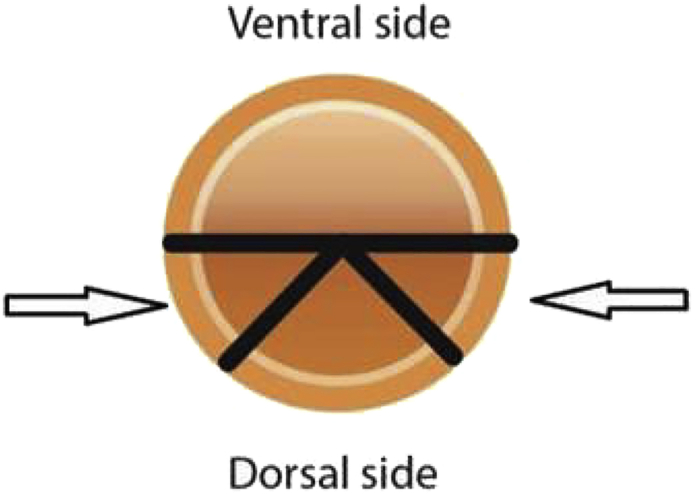
-
8.
Position the opening of the suction biopsy device between 45 and 90 degrees laterally from the dorsal side to avoid arteries (see Figure 2).
-
9.
Obtain a biopsy from the rectum/ colon, with a defined suction pressure of 30 kPa/0.3 Bar.
-
10.
Release the rectal (colon) biopsy from the device by twirling and shaking the frontal side in the collection tube containing 20 mL storage media at 4 °C (either PBS0 or Ad-DF+++ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL)) and until the biopsy is visible in the fluid.
-
11.
Repeat steps 5–10 until a minimum of 4 good quality biopsies have been successfully isolated and collected (all biopsies can be collected in the same tube).
-
12.
After the procedure the nurse firmLy closes the collection tube and takes off her gloves.
-
13.
Maintain the collection tube with the biopsies on ice at ∼4°C in a styrofoam box
-
14.
After the procedure is finished the patient should remain in the hospital for an additional hour to check for potential rectal bleeding. In case of severe or persistent rectal bleeding a qualified physician should be consulted.
-
15.
When no complications are observed the patient can be discharged.
-
16.
Inform the patient about potential late onset rectal bleeding. In case of late onset rectal bleeding a qualified physician should be consulted.
Figure 1.
Rectal Suction Biopsy Device with Manometer for Defined Suction Pressure and Syringe to Create Suction
Figure 2.
Cross-Sectional View of the Colon with Advised Positioning of the Rectal Suction Biopsy Device Opening
Transport of Biopsies to the Laboratory
CRITICAL: Intestinal crypts can be retrieved from a rectal/ colon biopsy (hereafter referred to as “colon biopsy”) with a high efficiency (> 95% success rate) within 48 h of the biopsy collection. However, tissue quality gradually decreases over time leading to a reduced chance of successfully isolating crypts from the biopsies. Therefore, immediate transport of the colon biopsies at 4 °C in AD-DF+++ is essential to ensure successful crypt isolation and establish efficient colon organoid cultures.
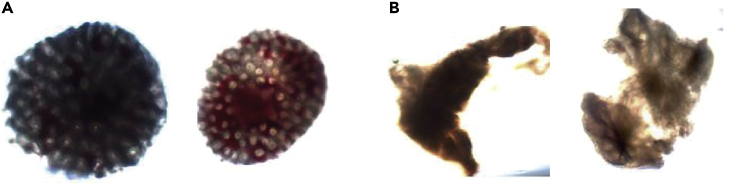
Note: Good quality colon biopsies should be round and have a slightly pink to red color (see Figures 3 and 4). When you are uncertain about the quality of the rectal biopsies, please check if intestinal crypts are visible under the light microscope. Good quality biopsies show a honeycomb- like structure in which the stem cell crypts reside. If this structure is not visible, the biopsy is of bad quality (Figure 4B).
Figure 4.
Examples of Biopsies as Viewed under a Microscope (Bright Field, 4 x Magnification)
(A) Good quality biopsies; note the honeycomb structure indicative of intestinal crypts.
(B) Bad quality biopsies, this can be due to lack of tissue depth of the biopsy leading to an absence of intestinal crypts.
-
1.
The colon biopsies should be stored on ice or at 4°C (fridge) in properly labelled 50 mL polypropylene conical tubes containing 30 mL of PBS0. Make sure the tissue is NOT frozen.
Note: If the biopsies are transported over long distances or over a long duration (>8 hs) before the crypt isolation procedure can be initiated, the biopsies should be stored in Ad-DF+++ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL) at 4°C.
-
2.
Make sure the 50 mL conical tube containing the colon biopsy in the storage medium is tightly closed and properly labelled. The date of the biopsy collection should be written on the tube (preferably on a sticker).
-
3.
During the biopsy transport, the temperature needs to be 4°C. Please use cold gel or ice packs during the shipment.
CRITICAL: Wet ice should not be used for the shipment, since thawing ice might damage the labelling of the tubes.
Organoid Crypt Isolation from Human Colon (Rectal) Biopsies
TIMING: ∼3 h
CRITICAL: All laboratory procedures related to colon organoid or cell cultures should be performed in a laminar flow to avoid contaminations and to protect the operator from biological risks.
CRITICAL: Always keep matrigel at 4°C or on ice to prevent polymerization above 10 °C.
CRITICAL: Pre-warm 24- and 96-wells plates used for organoid culturing to 37 °C for a minimum of >1 day. This is essential to ensure that after the colon organoids are plated, the matrigel polymerizes in the well efficiently and forms stable drops.
Preparations
-
1.
Prepare cold Ad-DF+++ and cold PBS0 (4 °C).
-
2.
Thaw (and keep) matrigel on ice or at 4 °C and dilute 1:1 with cold CM +/+ (= 50% matrigel).
-
3.
Prepare CM +/+ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL) to reduce the risk of infection.
EDTA Incubation
CRITICAL: In every step of this procedure, when handling the biopsies with pipettes, it is necessary to pre-coat the pipettes with Ad-DF+++ by aspirating media into the pipettes and discard the media. This covers the inside of the pipets with liquid and prevents biopsies from adhering to the plastic.
-
1.Wash the colon biopsies several times in cold PBS0 until the supernatant is clear using the following procedure:
-
a.Pipet the biopsies up and down ∼10 times in 10 mL with a 10 or 25 mL pipet.
-
b.Let the biopsies settle at the bottom for 30 seconds.
-
c.Remove the supernatant and add 10 mL of clean cold PBS0.
-
d.Repeat these steps 2–4 times until the biopsies and supernatant are clear of debris.
-
a.
-
2.
Discard the supernatant and add 10 mL of clean PBS0 to the biopsies and supplement it with 200 μL ultrapure EDTA pH 8, 0.5 M (final concentration 10 mM).
-
3.
Place the tube on a roller mixer for 90–120 min at 4 °C. The incubation time is dependent on the time between the biopsy collection and the start of the crypt isolation procedure. The longer the interval between biopsy collection and isolation procedures, the longer it is required to incubate. Crypts can be isolated from colon biopsies up until 48 h after biopsy collection but crypt yield decreases over time.
Isolate Intestinal Crypts from Biopsies
-
1.
Allow the colon biopsies to settle at the bottom of the tube.
-
2.
Discard the supernatant (PBS0 + EDTA).
-
3.
Add 3 mL cold PBS0 to the tube containing the biopsies and pipet the biopsies up and down vigorously 10–20 times. Crypts are released from the biopsies and float in the PBS0, which is visible by eye through the observation of the solution becoming more cloudy. If no crypts emerge from the colon biopsies after 90–120 min of EDTA incubation add new PBS0 + EDTA and incubate for another 60 min.
-
4.
Allow the biopsies to settle at the bottom and transfer the supernatant with the crypts to a clean 15 mL tube.
-
5.
Add fresh 3 mL of cold PBS0 to the biopsies again and repeat previous steps until no more crypts detach or until a sufficient amount of crypts are transferred to the clean tube.
-
6.
Add Ad-DF+++ to top up the solution containing the crypts in the 15 mL tube and centrifuge the crypts at 130 g for 5 min at 4 °C.
-
7.
Gently remove the supernatant, the crypt cell pellet is vulnerable and can easily be lost by aspiration.
-
8.
Add 10 mL of Ad-DF+++ to the crypt pellet for a second washing step and centrifuge at 130 g for 5 min at 4 ºC.
Plating Intestinal Crypts in Matrigel
-
1.
For the first passage (p.0) make sure to use a separate 24-wells plate per patient sample since isolated crypts from primary intestinal material during the first passage are most susceptible to infections.
-
2.
Keep the intestinal crypt pellet on ice.
-
3.Analyze the size of the pellet by eye to determine the appropriate volume of 50% matrigel.
-
○Usually, 4–6 wells per biopsy is sufficient for p.0. However, a higher number of wells can be used if the crypt yield is very high (7–12 wells) or a lower number when the yield is low (1–3 wells).
-
○
-
4.
To ensure the crypt density is not too low, first resuspend the crypt pellet in 100 μl 50% matrigel.
-
5.
Check the crypt density by plating a test droplet and view under the light microscope. Preferably do not plate out crypts too dense to allow for efficient (out)growth and proliferation of the structures. The ideal density is 15–20 crypts per 7.5 μL of matrigel drop.
Note: the crypt density can be adjusted according to the observations in the test droplet.
Too dense: If crypts are too densely seeded, dilute the sample in more matrigel and seed additional wells with the extra matrigel.
Too sparse: If crypts are too sparsely seeded, crypts can be centrifuged at 130 g 5 min at 4 °C and resuspended in smaller volumes of matrigel to increase the density.
Note: Prevent bubble formation during pipetting and plating.
-
6.
Plate 4∗ 7.5 μL of matrigel with about 15–20 crypts per drop of a (pre-warmed) 24-well plate.
-
7.
Resuspend the matrigel after plating out 3 wells to ensure the crypts have not settled at the bottom of the microcentrifuge tube.
-
8.
Incubate 30 min. at 37 °C.
-
9.
Add 500 μL of pre-warmed CM +/+ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL). An example of crypts directly after isolation can be found in Figure 5.
Note: gentamicin and vancomycin are only added to CM +/+ in the first week of culturing
-
10.
Incubate for 7 days at 37 °C, 5% CO2.
Figure 5.
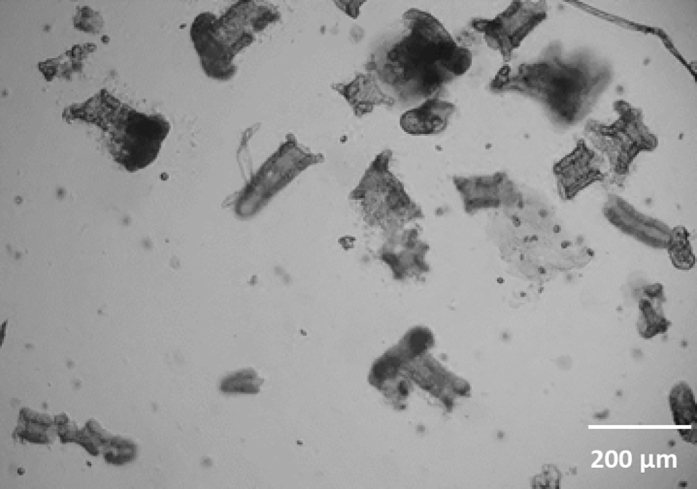
Crypts on Day 0 Directly after Isolation (Bright Field, 4 x Magnification)
First Week of Culture
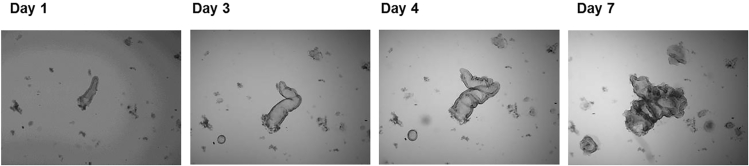
Note: Crypts close quickly and start to proliferate (visible as “budding”) after several days (see Figure 6).
Figure 6.
Development of Crypt into Budding Organoid Structure on Day 1; 3; 4 and 7 (Passage 0)
Arrows point to proliferating crypt and subsequent organoid structure (bright field, 4 x magnification).
-
1.
Often check wells for infections. When any of the wells shows clear signs of infection, aspirate the medium from the well and add 1 mL ethanol for 30 s. Aspirate ethanol and matrigel drops and re-add 1 mL of ethanol for 30 s. Aspirate and leave well empty (important: leaving ethanol creates ethanol vapor in the plate, which leads to cell death in the other wells). This approach prevents cross contamination of other wells from this patient sample.
-
2.
Refresh the wells with 500 μL colon organoid medium +/+ with additional gentamicin (50 μg/mL) and vancomycin (50 μg/mL) every 2–3 days (e.g. Monday, Wednesday, Friday). If crypts are isolated on a Friday, add 1 mL CM +/+ instead of 500 μL (with additional gentamicin and vancomycin) to ensure optimal growth during the weekend
Note: To remove the media, use a new tip for every well to decrease infection and well to well contamination risks.
-
3.
Once the crypt structures are budding, closed and proliferated (see Figure 6), passage them as described in 5.1. This can usually be performed around day 6–8.
Note: FIS experiments should only be performed after a minimum of two passages after isolation.
General Handling and Passaging of Human Colon Organoid Cultures
TIMING: ∼3 h
CRITICAL: Pre-warm 24- and 96-wells plates used for organoid culturing to 37 °C for a minimum of >1 day. This is essential to ensure that after the colon organoids are plated, the matrigel polymerizes in the well efficiently and forms stable drops.
Preparations
-
1.
Prepare cold Ad-DF+++ and warm CM +/+.
-
2.
Thaw (and keep) matrigel on ice or at 4 °C and dilute 1:1 with cold CM +/+ (= 50% matrigel).
Note: the splitting ratio should be usually 1:4–1:8 24-wells. If seven day old organoid cultures cannot be split according to this ratio because they have not proliferated sufficiently, check the medium quality or reseed organoids in 1:1 wells without disruption to allow more time to recover or proliferate until disruption.
Passaging Colon Organoids
-
1.
Discard the CM +/+ from the wells containing the colon organoids.
-
2.
Detach the matrigel with the organoids by aspirating 1 mL Ad-DF+++ and forcefully dispensing the medium directly onto the matrigel drops in the well several times with a p1000 filter tip. If necessary gently scrape the well with the end of the pipet tip to detach residue.
-
3.
Transfer the colon organoid suspension to a sterile 15 mL tube on ice. Several wells (up to 12) can be combined in the 1 mL.
-
4.
Put a p200 tip (without filter) on top of a p1000 filter tip (see Figure 7) and place the tip in the 1 mL organoid suspension on the bottom of the tube. Mechanically shear/disrupt the organoid structures by pipetting the 1 mL organoid suspension up and down 15–20 times through the p1000/ 200 tip combination. The organoids will be broken up into smaller parts. Resuspend with firm speed and fully take up and dispense the 1 mL suspension. Slow, mechanic shearing does not create efficient disruption. Minor foam formation is normal but try to prevent it by avoiding foam aspiration or dispension.
Note: steps 5 and 6 are relevant when colon organoid cultures are passaged before performing a FIS experiment or to clean an organoid culture that has many differentiated structures which need to be removed (see Figure 10C). Otherwise proceed with step 7.
-
5.
Add 5 mL of cold Ad-DF+++ to the 1 mL of disrupted organoids.
-
6.
Clean up the organoid suspension by resuspending the organoids and let the bigger, differentiated organoid structures sink down in a tube angles at 70° for 10 s (see Figure 8). Take up the smaller organoids by removing 1 mL from the top of the medium and transfer this to a sterile 15 mL tube on ice. Repeat this step until only 0.5–1 mL medium with bigger structures is left.
Note: if many large structures are still left in the remaining 1 mL after steps 5 and 6 (visual by eye), repeat steps 4–6 but during step 4 only disrupt 10 times.
-
7.
Top up the 15 mL tube containing the smaller, disrupted organoids with 10 mL cold Ad-DF+++.
-
8.
Centrifuge at 130 g for 5 min at 4 °C.
-
9.
Aspirate the supernatant and keep the organoid pellet.
CRITICAL: the colon organoid pellet is vulnerable and can be easily lost through aspiration of the supernatant. Be careful while aspirating the media: it is best to leave > 50 μL of supernatant on the organoids to avoid pellet aspiration. Remove the last supernatant with a p200 tip.
Figure 7.

Put a p200 Tip on Top of a p1000 Filter Tip to Disrupt the Organoid Structures
Figure 8.
Hold the 15 mL Tube Tilted at 70 ° and Transfer the Organoids from the Top of the Medium to a New Tube
Plating Colon Organoids in 24-Well Plates
-
1.
Resuspend the organoid pellet in an appropriate volume of 50% matrigel. The volume of matrigel needed is 30 μL per seeded well of a 24-wells plate assuming enough organoids have been passaged.
Example: 2 wells of a 7-day old, good quality organoid culture can be split into approximately 8–16 new wells of a 24-wells plate. To avoid too sparsely seeded wells, it is advisable to first resuspend in a matrigel volume sufficient to seed 6 wells. Therefore, the pellet can be resuspended in approximately 180 μL of 50% matrigel. In case the organoids are too densely seeded, the organoid suspension can be diluted by adding more matrigel.
-
2.
Check the number of organoids in a 10 μl test drop of matrigel by light microscopy, to decide if the organoid suspension should be further diluted (the density should be 15-30 structures per drop, see Figure 9B). Always recheck a new test drop after further dilution.
-
3.
While preventing bubbles, plate 4∗ 7.5 μL matrigel with a p20 tip per well in a (pre-warmed) 24-well plate (see Figure 9A). Slightly tilting the plate helps to deposit the drops in the desired positions.
Note: Organoid structures will quickly settle at the bottom of the tube within 30 s so frequently resuspend the organoid suspension with a p200. While resuspending keep the tube on ice to prevent the matrigel from solidifying in the tube.
-
4.
Incubate the plate for 15–20 min at 37 °C.
-
5.
Add 500 μL of pre-warmed of CM +/+ per well.
-
6.
Incubate the organoid cultures for 7 days at 37 °C, CO2 5% and refresh the wells with 500 μL of CM +/ + per well, every 2–3 days (Monday, Wednesday, Friday).
Note: Human colon organoids can usually be passaged every 7 days. Therefore, it is important to regularly check the quality of the organoid cultures. Good quality organoid cultures show a stem cell phenotype which is characterized by efficient budding structures. For wild type (or very mild CF genotype) colon organoids a clear pre-swollen lumen can be seen with bright field imaging compared to colon organoids with a CF genotype where no lumen is visible (see Figures 10A and 10B).When colon organoids lose their stem cell phenotype (usually due to insufficient Wnt-3A activity in the WCM) the organoids appear as thick-walled, differentiated structures (see Figure 10C). These bad quality organoid cultures are not appropriate for CFTR-related experiments because this leads to unreliable results.
Figure 9.
Organoid plating examples in 24-wells plates
(A and B) As shown in (A), add 4 drops of 7.5 μL containing organoids in a pre-warmed 24-wells plate. Slightly tilting the plate helps to deposit the drops in the designated location. Shown in (B) is an example of the ideal density of organoids in one 7.5 μl matrigel drop (bright field, 4 x magnification).
Freezing Human Colon Organoid Cultures
TIMING: ∼2 h
Two different methods of organoids freezing are described here. First is freezing trypsinized human colon organoids after 1–3 days replating (Freezing protocol A). This is an efficient method to store source colon organoid samples in a master cell bank. Second is freezing small organoid structures directly after disruption (Freezing protocol B); this is a fast, timesaving and efficient procedure appropriate for storing a local working cell bank.
-
1.
Disrupt and freeze organoids when well is full (7–10 days cultured organoids)
-
2.
Freeze one full well (of a 24-wells plate) per cryovial which should be sufficient to plate into 3–4 new wells after thawing.
-
3.
Prepare cold Recovery™ Cell Culture Freezing Medium.
-
4.
Prepare cold Ad-DF+++
-
5.
Prepare ROCKi 10 mM stock.
-
6.
Prepare warm CM +/+ with additional ROCKi 10 μM.
-
7.
For freezing protocol A: thaw (and keep) matrigel on ice and dilute 1:1 with cold CM +/+ (= 50% matrigel)
-
8.
Prepare 0.05% Trypsin EDTA to 18–23 ° C
Freezing Protocol A
-
1.
Perform step 1–4 and 7,8 of ‘General handling and passaging of human colon organoid cultures’, part B.
-
2.
Aspirate and discard the supernatant and add 4 mL of 0.05% Trypsin EDTA, and vortex for 30 s.
-
3.
Put the tube in a warm water bath at 37 °C for 1 min and vortex vigorously for 30 s.
-
4.
Inspect the solution in the tube by horizontally placing the tube on the light microscopy stage (4x objective), adjust focus so that organoids in solution are in visible. Observe the size of the organoid structures. If intact organoids are still visible put the tube in a warm water bath at 37 °C for an additional 1 min and vortex vigorously for 30 sec.
-
5.
When the organoids are sufficiently disrupted (∼100 x smaller), add 8 mL of Ad-DF+++ to neutralize the trypsin and resuspend 10 times the organoid suspension.
-
6.
Centrifuge the tube for 3 min at 450 g at 4 °C.
-
7.
Aspirate and discard the supernatant and add the required amount of medium and matrigel to the organoid pellet.
Note: For freezing, organoids must be seeded in a 1:1 ratio after trypsinization. Thus, 1 well of organoids from a 24-wells plate can be seeded into 1 new well of a 6-well tissue culture plate.
-
8.Mix the organoid suspension by resuspending without creating bubbles.
-
○Tip: Avoid bubbles by not fully aspirating and dispensing the pipet, but always keep a small amount of liquid in the tube.
-
○
-
9.
Seed 250 μL in a single well of a pre-warmed 6-well tissue culture plate by seeding 25 ∗ 10 μL matrigel drops .
-
10.
Place the plate in the incubator at 37 °C and leave the matrigel to solidify for 20–30 min.
-
11.
Add 2.5 mL of fresh CM +/+ + ROCKi in each 6-well plate well and transfer the plate to the incubator.
Note: 1–2 day old organoid cultures are ready to be frozen. This increases the efficiency of survival after thawing.
-
12.
Detach the matrigel and organoids by aspirating 1 mL Ad-DF+++ and forcefully dispensing the medium directly onto the matrigel drops in the well several times with a p1000. If necessary gently scrape well with end of pipet tip to detach everything.
-
13.
Transfer the organoid suspension to a sterile 15 mL tube.
-
14.
Wash the wells with another 1 mL of Ad-DF+++ and transfer to the same 15 mL tube.
-
15.
Fill up the 15 mL tube with with 12 mL of cold Ad-DF+++ and pipet up and down with a 5 mL pipet.
-
16.
Centrifuge the suspension for 3 min at 450 g at 4 °C, remove the supernatant and keep the pellet.
-
17.
Dissolve the organoid pellet with cold Recovery™ Cell Culture Freezing Medium and pipet up and down to properly resuspend all the organoid structures.
Note: one full well from a 6-well plate is frozen in 1 mL of Recovery™ Cell Culture Freezing Medium and is divided over 2 cryovials. Each vial should contain enough cells to be thawed into ≥ 4 wells of 24-well plate.
-
18.
Transfer 0.5 mL of organoids suspension in Recovery™ Cell Culture Freezing Medium to sterile cryovials.
-
19.
Place the cryovials at −80°C in a cell container that will freeze the organoids 1°C per minute (e.g. mr Frosty).
-
20.
After 24 h, transfer the cryovials for storage in liquid nitrogen.
Freezing Protocol B
-
1.
Perform step 1–4 and 7,8 of ‘General handling and passaging of human colon organoid cultures’, part B. However, shear/ disrupt the colon organoids 30–35 times to create even smaller organoid structures.
-
2.
Add cold Recovery™ Cell Culture Freezing Medium
- Use 1 mL of Recovery™ Cell Culture Freezing Medium in 1 cryovial per isolated well. 1 frozen well in 1 cryovial can be thawed into 3–4 wells of a 24 - well plate
-
→pellet of 1 full well + 1 mL Recovery™ Cell Culture Freezing Medium or FM → add to 1 cryovial
-
→pellet of 3 full wells + 3 mL Recovery™ Cell Culture Freezing Medium or FM → divide over 3 cryovials
-
→
-
3.
Place the cryovials at -80°C in a cell container that will freeze the organoids 1°C per minute (e.g. mr Frosty).
-
4.
After 24 h, transfer the cryovials for storage in liquid nitrogen.
Thawing Human Colon Organoids from Frozen Nitrogen Stock Vials
TIMING: ∼3 h
CRITICAL: Pre-warm 24- and 96-wells plates used for organoid culturing to 37 °C for a minimum of >1 day. This is essential to ensure that after the colon organoids are plated, the matrigel polymerizes in the well efficiently and forms stable drops.
Note: Human colon organoids cultures can be thawed from two different types of nitrogen stock vials: Thawing cultures that have been collected after 2-3 days in culture after trypsinization (freezing protocol A) or thawing cultures from vials with small organoid structures frozen directly after disruption (freezing protocol B).
-
1.
Prepare warm Ad-DF+++ (37 °C).
-
2.
Thaw (and keep) matrigel on ice or at 4 °C and dilute 1:1 with cold CM +/+ (= 50% matrigel)
-
3.
Prepare ROCKi 10 mM stock.
-
4.
Prepare warm CM +/+ with additional ROCKi 10 μM.
Note: Addition of ROCKi is only necessary during the first week after thawing
Colon Organoid Thawing Procedure
-
1.
Add 13 mL pre-warmed Ad-DF+++ in a 15 mL tube
-
2.
Rinse the outside of the cryovial with warm water or place in 37 °C water bath until the cell suspension in the vial is thawed. Clean the outside of the cryovial with 70% EtOH.
Alternative: first clean the outside of the cryovial with 70% EtOH and then add warm Ad-DF+++ to the organoid suspension to thaw it.
-
3.
Rapidly transfer organoids to 15 mL tube with 13 mL Ad-DF+++ (37 °C).
When thawing colon organoids frozen according to freezing protocol A
-
4.
Gently mix the suspension and centrifuge 450 g for 5 min at 4 °C.
-
5.
Discard supernatant and resuspend pellet in 100 μL of 50% matrigel.
-
6.
Check the organoid density under the microscope by seeding a 7.5 μL matrigel test drop. The matrigel drop should contain >100 structures of very small colon organoid structures. Organoids frozen according to freezing protocol A should be thawed and seeded with higher density due to the smaller sized structures compared to protocol B. To ensure optimal outgrowth and budding of the colon organoid structures it is suggested to seed multiple densities to cover the most optimal density for recovery and proliferation.
-
7.
Plate 4∗ 7.5 μL matrigel drops without bubbles to a single well of a (pre-warmed) 24-wells tissue culture plate. Usually one cryovial should contain enough organoids to seed in 3–4 wells.
-
8.
Dilute the remaining colon organoid suspension 1:1 with matrigel and plate two additional wells.
-
9.
Again, dilute the remaining colon organoid suspension 1:1 with matrigel and plate the fourth well.
When thawing colon organoids frozen according to freezing protocol B
-
10.
Gently mix the organoid suspension and centrifuge 130 g for 5 min at 4 °C.
-
11.
Discard the supernatant and resuspend the organoid pellet in 100 μL of 50% matrigel.
-
12.
Check the organoid density under the microscope: seed the organoid structures with a 10%–20% higher density compared to organoids already in culture, aiming for 50–70 structures per drop (10%–20% of the structures are usually not viable). Organoids frozen according to freezing protocol B should be thawed and seeded with lower density due to the larger sized structures compared to protocol A.
-
13.
Plate 4∗ 7.5 μL matrigel drops without bubbles per single well of a (pre-warmed) 24-wells tissue culture plate. Usually one cryovial should contain enough organoids to seed in 3–4 wells. Colon organoids will settle to the bottom quickly, so resuspend the organoids in the matrigel suspension frequently while plating.
-
14.
Incubate a maximum of 10 min at 37 °C. Thawed human intestinal organoids are sensitive to being left without warm CM +/+
-
15.
Add 500 μL of (pre-warmed) CM +/+ with additional ROCKi inhibitor (10 μM) to each well. Refresh the medium with 500 μL CM +/+, every 2–3 days (Monday, Wednesday, Friday).
-
16.
Incubate at 37 °C, 5% CO2 for 1 week.
CRITICAL: Monitor the progress of the thawed organoid structures daily.
Forskolin Induced Swelling (FIS) Assay
TIMING: ∼5 h
The FIS assay was developed to measure CFTR function in human colon organoids. Upon forskolin (fsk) stimulation (which indirectly activates CFTR through cyclic adenosine monophosphate (cAMP)), CFTR function can be assessed by observing the subsequent swelling of human colon organoids. Organoid swelling occurs through chloride excretion into the organoid lumen and osmosis. The assay can determine residual CFTR function and the effect of CFTR modulators (Dekkers et al., 2013, Boj et al., 2017).
CRITICAL: Pre-warm 24- and 96-wells plates used for organoid culturing to 37 °C for a minimum of >1 day. This is essential to ensure that after the colon organoids are plated, the matrigel polymerizes in the well efficiently and forms stable drops.
Preparations
-
1.
Incubate 96-well plates at 37 °C > 7 days. It is important to have warm plates to prevent dislodging of matrigel drops.
-
2.
Prepare cold Ad-DF+++.
-
3.
Thaw (and keep) matrigel on ice and dilute 1:1 with cold CM +/+ (= 50% matrigel).
-
4.
Prepare warm CM +/ +.
Plating Human Colon Organoids (Day 1)
Note: Some CFTR modulators (e.g. correctors) require longer periods incubation. Example: correctors VX-809 and VX-661 require 24 h incubation and are added directly to the CM +/+ on the day of the organoid plating, whereas potentiator (e.g. VX-770) should be added directly before the FIS measurement. Take this into account when CM +/+ is added. See ‘European FIS validation & standardization protocol’ for corrector preparations.
-
1.
Perform step 1-9 of ‘General handling and passaging of human colon organoid cultures’, part B.
-
2.Resuspend the organoid pellet in 50% matrigel (plating volume = 4 μL ∗ amount of wells + 50 μL)
-
○With good quality organoid cultures, one full well of organoids from a 24-wells plate can be seeded into 10–20 wells of a 96-wells plate
-
○
Example: 8 wells of a 7-day, good quality organoid culture can be split and seeded into ∼80–160 wells of a 96-wells flat bottom plate. To avoid too sparsely seeded wells, it is advisable to first resuspend in a matrigel volume sufficient to seed 80 wells. Therefore, the organoid pellet can be resuspended in approximately 370 (80∗4 + 50) μL of 50% matrigel. In case the organoids are too densely seeded, organoids can be diluted by adding more matrigel.
-
3.
Check the amount of organoids in a 4 μL test drop of matrigel under a light microscope, to determine whether the organoid suspension should be further diluted (the density should be 30–50 structures per drop). Always recheck a new test drop after further dilution.
Note: when the organoid density is too low, matrigel can be resuspended in 10 mL of Ad-DF+++ and centrifuged at 130 g for 5 min at 4 °C. Next, aspirate medium and resuspend the organoid pellet in less matrigel.
-
4.
Transfer the organoid suspension to a cold microcentrifuge tube.
-
5.
While preventing bubbles add a 4 μL drop to a (warm) flat-bottom 96-well plate (see Figure 11). This can be performed with a regular p20 pipet.
Note: Organoid structures will quickly settle at the bottom of the microcentrifuge tube within 30 s so resuspend the organoid suspension with a p200 regularly. While resuspending keep the tube on ice to prevent the matrigel from solidifying in the tube. It is advisable to use a repetitive pipet (like a Viaflo||, Integra) since the organoid seeding is performed more efficient, faster and homogeneously.
-
6.
Incubate the 96-wells plate for 5–10 min at 37 °C.
-
7.
Add 50 μL of CM +/+ to each well.
-
8.
Incubate 20–24 h (overnight) at 37 °C, CO2 5%.
Note: Regular organoid passaging and organoid seeding for FIS experiments can be combined. Guideline: prepare excess organoid suspension for seeding in a 96-well plate. Remaining organoid suspension can be diluted at least 1:1 and used for seeding in a 24-well plate for a following 7-day culture.
Figure 11.
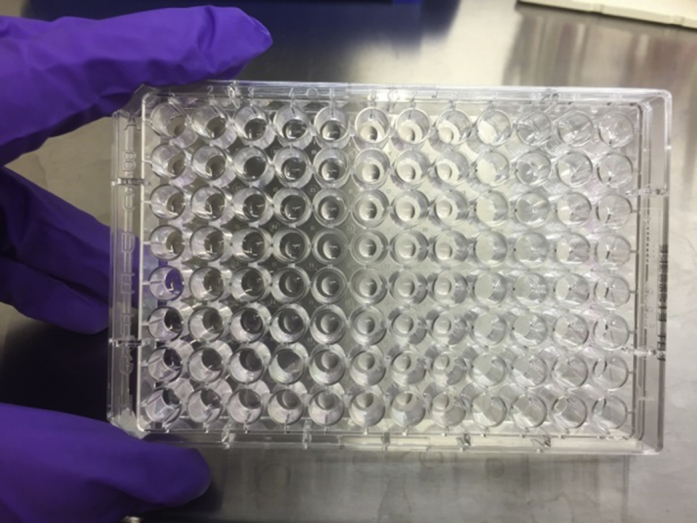
Add 4 μl Drops Containing 20–50 Organoids per Drop to Every Well in a Pre-warmed 96-Wells Plate
FIS Assay & Confocal Microscopy (Day 2)
-
1.
Incubate Ad-DF+++ at 18–37 °C.
-
2.
Turn on the live cell compartment of the confocal microscope and pre-incubate at 37 °C and 5% CO2 (Pre-heating the live cell compartment takes a minimum of 30–45 min).
-
3.
Prepare fsk and/or CFTR-modulators as desired. Prepare fsk and CFTR-modulator compounds in a 2 x concentration in Ad-DF+++ so when 50 μL is added to the organoids in a well of the 96-well plate containing already 50 μL of CM+/+, , the 1:1 dilution creates a 1 x final concentration of the desired fsk or compound concentration.
Example: if the desired final concentration of fsk within the well is 5 μM, prepare a fsk dilution of 10 μM. If 50 μL of 10 μM fsk is then added to the 50 μL CM +/+, the final concentration will be 5 μM.
-
4.
Prepare 8.4 mM stock of calcein green AM by adding 6 μL of DMSO to one 50 μg calcein green vial. For one full 96 well plate, add 1 μL of 8.4 mM calcein green solution in 1000 μL Ad-DF+++.
-
5.
Add 5 μL of calcein green solution to each well (final concentration is 0.84 μM)
-
6.
Gently resuspend the well 2–3 times with a multichannel for homogeneous staining and efficient uptake of calcein green by the organoid structures. Try tilting the plate and point tips of multichannel in the corner of each well to prevent touching the matrigel drop.
-
7.
Incubate the plate at 37 °C, CO2 5% for 15–30 min before starting the experiment.
-
8.Put the 96-well plate in the plate holder of the live cell imaging device and ensure the plate is in fixed position. Live cell imaging settings:
-
○Organoid structures with calcein green stain can be visualized upon emission at 488 nm and excitation at 515 nm (detection wavelength range 450–700 nm).
-
○Use 5 x objectives and ensure an overview of the full matrigel drop by adjusting the focus.
-
○Set the position (x, y) and focus (z) of the matrigel drops in the acquisition software.
-
○
Note: an autofocus option may be used when available.
-
9.Measurement settings for 60 min measurement:
-
○Interval = 10 min,
-
○Cycles = 7 (Cycle 1 is t = 0).
-
○
Note: the acquisition of images from the total 96-well plate in each time point should not exceed 5 min to ensure optimal comparison of time points between wells.
-
10.
Make sure the calcein green signal in the organoids structures is slightly oversaturated and the signal to noise ratio is optimized as much as possible (this is essential for optimal recognition of the organoid structures by the imaging software).
-
11.
Add 50 μL Ad-DF+++ containing fsk stimuli (and/or CFTR-modulators) to each well containing 50 μL of CM +/+. It is preferred to add the stimuli from a 96-well dummy plate with an 8-multichannel for accuracy and efficiency if the size of the live cell chamber allows this. If this is not possible, add the stimuli with a regular pipet.
-
12.
Start the acquisition immediately after stimuli are added.
European FIS Validation & Standardization Protocol
TIMING: ∼ 8 weeks
To ensure correct implementation of the human organoid technology and the application in the FIS assay, six reference human colon organoid lines (covering different classes of CFTR mutations from severe to mild -see Table 4). These organoids lines were tested for basal CFTR function and specific drug response to available CFTR modulators (VX-661 and VX-770). Based on the following protocol, swelling data (technical duplicates at three time points with weekly intervals (n=3)) were generated to compare the qualitative and quantitative results of the implemented organoid technology and FIS assay (not yet published data). These reference organoid lines were selected based on prevalent CFTR genotypes (see Table 4) and differences in CFTR basal function and response to CFTR modulators. Researchers that follow the detailed work instructions presented in this protocol should find identical or highly reproducible results (within 10% of AUC values, not yet published data) for these reference lines to confirm successful implementation of the human intestinal organoid technology and FIS assay.
Table 4.
Overview of the Different Organoid Cell Lines That Can Be Used to Validate FIS Assay Results
| Validation Organoid Cell-Lines | ||
|---|---|---|
| CF Class Mutation | Specific Genotype | HUB ID Code |
| Class I/ Class I | G542X / G542X | HUB-02-D2-121 |
| Class II/ Class I | F508del / R1162X | HUB-02-D2-038 |
| Class II/ Class II | F508del/ F508del | HUB-02-D2-341 |
| Class II/ Class III | F508del/ G551D | HUB-02-D2-043 |
| Class II/ Class III | F508del/S1251N | HUB-02-D2-103 |
| Class II/ Class IV (mild phenotype) | F508del / R117H - 7T | HUB-02-D2-004 |
Preparations for Validation Protocol
Medium amounts:
-
a)
1 liter CM +/+ (see ‘Stock preparation of human colon organoid medium components’ for instructions on media preparation)
-
b)
500 mL 2x CM −/− Prepare 10 x 100 mL bottles with 50 mL stock and store at −20 °C
-
c)
500 mL WCM Prepare 2 batches of > 375 mL (> 20 x 145 mm Petri dishes per batch) and pool. Store at 4 °C)
-
d)
0.25 mg R-spo3 Stock and aliquot volume dependent on LOT#. Store for 3 months at −80 °C.
-
e)
35 mL 50% Matrigel Aliquot 2 x 10 mL vials in 500 μL per microcentrifuge tube and store at −20 °C
General Experimental Set Up
The FIS validation assay is based on the experimental set up as previously performed and described (Dekkers et al., 2016, Science; Boj et al., 2017, Jove).
-
1.
Each reference organoid line was tested using four different conditions (basal CFTR function, VX-661; VX- 770 and VX-661 + VX-770, each in combination with 8 different fsk concentrations (0.008; 0.02; 0.05; 0.128; 0.32; 0.8; 2 or 5 μM) which results in 32 different experimental conditions.
-
2.
Biological replicates for each condition were measured in duplicate and experiments were repeated n=3.
-
3.
Human colon organoids were plated in a 96-well plate as schematically depicted in Table 5.
-
4.
VX-661 was added 24 h before the FIS experiment
-
5.
VX-770 was added acutely before the FIS experiment was initiated.
Table 5.
Experimental Plate Set Up for Measurement of Drug Response of Reference Organoid Cell Lines for Validation Data
| (Day 1) |
CM +/+ |
CM +/ + + VX-661 3 μM |
CM +/+ |
CM +/ + + VX-661 3 μM |
||||
|---|---|---|---|---|---|---|---|---|
| (Day 2) | Fsk stimuli | Fsk stimuli + VX-770 (3 μM) | ||||||
| Fsk concentration | 0.008 μM | 5 μM | 0.008 μM | 5 μM | 0.008 μM | 5 μM | 0.008 μM | 5 μM |
| 0.008 μM | 5 μM | 0.008 μM | 5 μM | 0.008 μM | 5 μM | 0.008 μM | 5 μM | |
| 0.02 μM | 2 μM | 0.02 μM | 2 μM | 0.02 μM | 2 μM | 0.02 μM | 2 μM | |
| 0.02 μM | 2 μM | 0.02 μM | 2 μM | 0.02 μM | 2 μM | 0.02 μM | 2 μM | |
| 0.05 μM | 0.8 μM | 0.05 μM | 0.8 μM | 0.05 μM | 0.8 μM | 0.05 μM | 0.8 μM | |
| 0.05 μM | 0.8 μM | 0.05 μM | 0.8 μM | 0.05 μM | 0.8 μM | 0.05 μM | 0.8 μM | |
| 0.128 μM | 0.32 μM | 0.128 μM | 0.32 μM | 0.128 μM | 0.32 μM | 0.128 μM | 0.32 μM | |
| 0.128 μM | 0.32 μM | 0.128 μM | 0.32 μM | 0.128 μM | 0.32 μM | 0.128 μM | 0.32 μM | |
| Per organoid reference line | ||||||||
Human Colon Organoid Culturing & Workflow (Also See Table 6)
Table 6.
Workflow and Timelines of FIS assay validation.
| Fis Assay Validation - Workflow & Timelines (Suggested) | ||
|---|---|---|
| Wk nr | Action | Extra comments |
| > Wk 0 |
|
|
| Wk 0 |
|
|
| Wk 1 |
|
|
| Wk 2 |
|
|
| Wk 3 |
|
|
| Wk 4 |
|
|
| Wk 5 |
|
|
| Wk 6 |
|
|
| Wk 7 |
|
|
| Wk 7–8 |
|
|
-
1.
When received, store cryovials with organoid reference lines in liquid nitrogen until ready for the FIS experiments.
-
2.
When ready, thaw a cryovial per reference organoid line as described in thawing protocol A
-
3.
Seed 3–4 wells of a pre- warmed 24- well plate for each organoid reference line and add CM +/+ with additional ROCKI.
-
4.
Check the organoid structures by light microscopy after 3 days and ensure there is enough space for the structures to expand. If not consider reseeding the cells with decreased organoid density to allow more space to proliferate and expand.
-
5.After 7 days of culture check the growth and quality of the organoid structures (compare to structures in Figure 10) :
-
a.When the organoids are not yet budding, but appear small and round, densely proliferating organoids: take up and wash the organoids, centrifuge at 130 g for 5 min at 4 °C without disruption/shearing and reseed 1 well into 3 new wells to create more space and allow for continuous proliferation of the structures.
-
b.When the organoids appear as already “budding” and healthy, large structures: disrupt 1 well according to ‘General handling and passaging of human colon organoid cultures’ (without the cleaning step) and seed into 3–4 new wells.
-
a.
-
6.After week 2 and a further 7 days in culture check the appearance of the organoid structures.
-
a.When grown into appropriate budding, proliferated organoid structures (see Figure 10B): Freeze two full wells into two cryovials according to freezing protocol B. Take the 3rd full well, disrupt and wash the organoids and seed into 3–4 new wells (without the cleaning step).
-
b.When not yet grown/proliferated into efficient budding organoid structures and the organoid structures still appear dense and small: repeat the washing and reseeding step without disruption/shearing and seed in 3 wells.
-
a.
-
7.
After week 3, all organoid lines should be of good culture quality, meaning they should appear as efficiently budding, proliferating structures and sufficient growth. Colon organoids are then ready to be frozen and to start experimental cultures to use in the FIS assay. Make sure a working cell bank is stored for every reference organoid cell line before any FIS experiments are started (see Table 6).
-
8.
To start the FIS experiment, expand each organoid reference line into 8 wells in a 24-well plate. This should be sufficient to perform a FIS experiment as well as to reseed organoids for the FIS experiment of the following week. After week 4, plate out organoids to perform the FIS assay. Use the experimental plate lay out as depicted in Table 5.
Preparation of CFTR Corrector VX-661 (Day 1)
-
1.
Prepare CM +/+ with VX-661 before plating out the organoids. VX-661 needs to be added 24 h in advance of the FIS measurement in CM+/+ with a final concentration of 3 μM. For example: from a 20 mM stock: add 95 μL of Ad-DF+++ to 5 μL of 20 mM stock creating 1 mM of VX-661 solution.
-
2.
Per 96-well plate add 9.6 μL of 1 mM VX-661 to 3 mL of CM+/+ creating a final concentration of 3.2 μM.
-
3.
Add 50 μL to each well that requires 24 h of VX-661 incubation (see Table 5 for plate lay out)
-
4.
The rest of the wells can be filled with regular CM +/+.
Preparation of CFTR Stimuli VX-770 & fsk (Day 2)
Per 96-well plate:
-
5.
Add 1 μL fsk (10 mM) to 1 mL Ad-DF+++ (= fsk (10 μM)), final concentration in assay will be 5 μM).
-
6.
Add 2 μL of VX-770 stock (20 mM) to 6 mL of Ad-DF+++ (= VX-770 6.4 μM, final concentration will be 3.2 μM.
-
7.
Take 1 mL of VX-770 6.4 μM solution and add 1 μL fsk (10 mM) (= fsk + VX-770 solution)
-
8.
Prepare the titrations (serial dilution) according to Table 7 below. Resuspend every suspension 10 times and use a new tip for every dilution step.
Table 7.
Preparation of the Fsk Titration with and without VX-770 Added Directly before the Start of the FIS Assay
| Order of Addition to Plate | Final Conc. (μM) | Ad-DF+++ (μL) | Fsk Titration (Column 1–4) | Ad-DF+++ (μL) + VX770 3 μM | Fsk Titration in 3 μM VX-770 (Column 5–8) |
|---|---|---|---|---|---|
| 8 | 5 | 1000 μL fsk (10 μM) | 1000 μL fsk / VX-770 mix | ||
| 7 | 2 | 600 μL Ad-DF+++ | 400 μL from 8 | 600 μL Ad-DF+++-770 | 400 μL fsk / VX-770 mix from 8 |
| 6 | 0.8 | 600 μL Ad-DF+++ | 400 μL from 7 | 600 μL Ad-DF+++-770 | 400 μL from 7 |
| 5 | 0.32 | 600 μL Ad-DF+++ | 400 μL from 6 | 600 μL Ad-DF+++-770 | 400 μL from 6 |
| 4 | 0.128 | 600 μL Ad-DF+++ | 400 μL from 5 | 600 μL Ad-DF+++-770 | 400 μL from 5 |
| 3 | 0.05 | 600 μL Ad-DF+++ | 400 μL from 4 | 600 μL Ad-DF+++-770 | 400 μL from 4 |
| 2 | 0.02 | 600 μL Ad-DF+++ | 400 μL from 3 | 600 μL Ad-DF+++-770 | 400 μL from 3 |
| 1 | 0.008 | 600 μL Ad-DF+++ | 400 μL from 2 | 600 μL Ad-DF+++-770 | 400 μL from 2 |
Expected Outcomes
Sometimes after 3–4 days the colon organoid concentration will be so dense that it is advisable to reseed the structures from 1 well into 3–4 new wells of a 24-wells plate. Take up the small organoids in 1 mL Ad-DF+++ in a microcentrifuge tube, centrifuge at a table top centrifuge and directly plate out in 100 μL fresh matrigel into 3–4 new wells of a 24-wells plate. This allows the small organoid structures to have more space to expand and start budding before they are ready for the first mechanical disruption/ shearing of the organoids.
After 7 days some organoid structures are already efficiently budding and ready for mechanical disruption for splitting. When organoid structures are still very small and not yet efficiently proliferating but the matrigel is fading: do not yet disrupt but take up the small organoids from four wells in 1 mL Ad-DF+++ in a microcentrifuge tube, centrifuge with a table top microcentrifuge and plate the organoids into 100–120 μL of fresh matrigel into 3-4 new wells of a 24-wells plate. This allows the small organoid structures to have more time to expand and start budding before they are ready for the first mechanical disruption/shearing of the organoids.
Quantification and Statistical Analysis
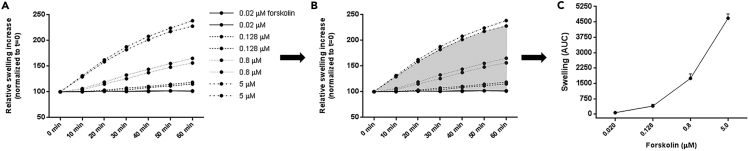
Raw Image Data Software Analysis
The goal of this procedure is to quantify the relative increase in total organoid area (organoid swelling) after stimulation of CFTR function by fsk. The raw data of the FIS assay consists of the confocal images of calcein green stained organoids that are generated at 10 min intervals, which are digitally stored as a time-series-clip of each well (see Figure 12). Image analysis software (e.g. Zen blue (Zeiss), Cell profiler (open-source)) is used to identify the perimeter of each closed organoid structure (X,Y plane), which is defined as a confined region-of-interest. The image software is set to ‘fill objects’ (region-of-interest). Each region-of-interest is expressed as area μm2 per time point per well. Total area μm2 associated with all regions-of-interest (organoids) per time point per well is calculated and compared to t = 0 min.
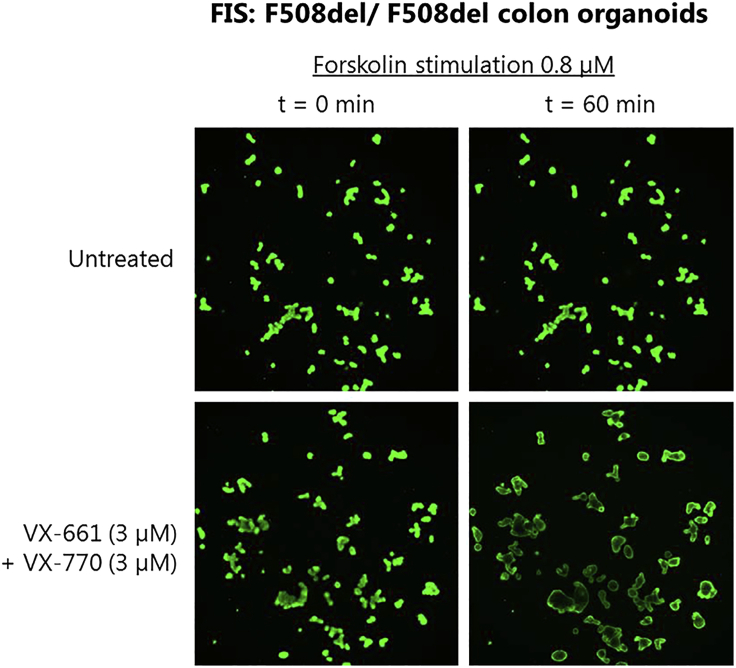
Figure 12.
Example of Organoid Swelling after 60 min FIS Assay of F508del/ F508del Calcein Green Stained Colon Organoids in the Absence or Presence of VX-661 (3 μM) & VX-770 (3 μM) and fsk (0.8 μM)
The area μm2 over a time series of 60 min (with 10 minute intervals creating 7 time points (t=0; 10; 20; 30; 40; 50; 60 min) represents the relative swelling increase from baseline. The total increase in area μm2 during the 7 time points is calculated as percent area increase per well. Hereby t=0 is set as a baseline of 100% and time point t=10 – t=60 min are normalized to time point 0 (see Table 8). This data can be used for further calculations with the following readouts:
-
1.
Percent increase of organoid area per fsk concentration and condition.
-
2.
The area under the curve (AUC) of the percent increase of organoid area per fsk concentration for all conditions.
Table 8.
Dissemination of Data Processing: From Raw Data to Area under the Curve (as Applied and Programmed in Excel).
| Calculations | Total Organoid Area μm2 (per Time Point, per Well) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Well nr 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| Time point (min) | 0.02 μM fsk (duplo) | 0.128 μM fsk (duplo) | 0.8 μM fsk (duplo) | 5 μM fsk (duplo) | ||||||
| 0 | (Raw data) | 1 | 281330 | 238453 | 263766 | 188102 | 331433 | 321666 | 300166 | 314690 |
| 10 | 2 | 283573 | 239998 | 263915 | 189347 | 345534 | 342520 | 388137 | 413799 | |
| 20 | 3 | 284395 | 241493 | 270094 | 194081 | 381535 | 382332 | 476159 | 510839 | |
| 30 | 4 | 283523 | 240770 | 274130 | 201630 | 416141 | 427078 | 546616 | 590889 | |
| 40 | 5 | 283548 | 243810 | 285890 | 207609 | 455630 | 463079 | 604865 | 654868 | |
| 50 | 6 | 287061 | 243062 | 294186 | 215582 | 490858 | 499504 | 653024 | 705319 | |
| 60 | 7 | 286687 | 241543 | 303803 | 223131 | 517691 | 531742 | 684018 | 750688 | |
| STEP A | Normalized to time point 0 per well (baseline set to 100%) | |||||||||
| (1/1) x 100 | 8 | 100,00 | 100,00 | 100,00 | 100,00 | 100,00 | 100,00 | 100,00 | 100,00 | |
| (2/1) x 100 | 9 | 100,80 | 100,65 | 100,06 | 100,66 | 104,25 | 106,48 | 129,31 | 131,49 | |
| (3/1) x 100 | 10 | 101,09 | 101,27 | 102,40 | 103,18 | 115,12 | 118,86 | 158,63 | 162,33 | |
| (4/1) x 100 | 11 | 100,78 | 100,97 | 103,93 | 107,19 | 125,56 | 132,77 | 182,10 | 187,77 | |
| (5/1) x 100 | 12 | 100,79 | 102,25 | 108,39 | 110,37 | 137,47 | 143,96 | 201,51 | 208,10 | |
| (6/1) x 100 | 13 | 102,04 | 101,93 | 111,53 | 114,61 | 148,10 | 155,29 | 217,55 | 224,13 | |
| (7/1) x 100 | 14 | 101,90 | 101,30 | 115,18 | 118,62 | 156,20 | 165,31 | 227,88 | 238,55 | |
| STEP B | Absolute baseline area | |||||||||
| 8-100 | 15 | 0,00 | 0,00 | 0,00 | 0,00 | 0,00 | 0,00 | 0,00 | 0,00 | |
| 9-100 | 16 | 0,80 | 0,65 | 0,06 | 0,66 | 4,25 | 6,48 | 29,31 | 31,49 | |
| 10-100 | 17 | 1,09 | 1,27 | 2,40 | 3,18 | 15,12 | 18,86 | 58,63 | 62,33 | |
| 11-100 | 18 | 0,78 | 0,97 | 3,93 | 7,19 | 25,56 | 32,77 | 82,10 | 87,77 | |
| 12-100 | 19 | 0,79 | 2,25 | 8,39 | 10,37 | 37,47 | 43,96 | 101,51 | 108,10 | |
| 13-100 | 20 | 2,04 | 1,93 | 11,53 | 14,61 | 48,10 | 55,29 | 117,55 | 124,13 | |
| 14-100 | 21 | 1,90 | 1,30 | 15,18 | 18,62 | 56,20 | 65,31 | 127,88 | 138,55 | |
| STEP C | Area difference between time points | |||||||||
| 16-15 | 22 | 0,80 | 0,65 | 0,06 | 0,66 | 4,25 | 6,48 | 29,31 | 31,49 | |
| 17-16 | 23 | 0,29 | 0,63 | 2,34 | 2,52 | 10,86 | 12,38 | 29,32 | 30,84 | |
| 18-17 | 24 | −0,31 | −0,30 | 1,53 | 4,01 | 10,44 | 13,91 | 23,47 | 25,44 | |
| 19-18 | 25 | 0,01 | 1,27 | 4,46 | 3,18 | 11,91 | 11,19 | 19,41 | 20,33 | |
| 20-19 | 26 | 1,25 | −0,31 | 3,15 | 4,24 | 10,63 | 11,32 | 16,04 | 16,03 | |
| 21-20 | 27 | −0,13 | −0,64 | 3,65 | 4,01 | 8,10 | 10,02 | 10,33 | 14,42 | |
| STEP D | Surface area per 10 minute interval | |||||||||
| 16 x10/2 | 28 | 3,99 | 3,24 | 0,28 | 3,31 | 21,27 | 32,42 | 146,54 | 157,47 | |
| 17 x 10 -(23 x 10/2) | 29 | 9,43 | 9,61 | 12,28 | 19,20 | 96,86 | 126,72 | 439,70 | 469,13 | |
| 18 x 10 - (24 x 10/2) | 30 | 9,34 | 11,23 | 31,64 | 51,85 | 203,37 | 258,15 | 703,68 | 750,50 | |
| 19 x 10 - (25 x 10/2) | 31 | 7,84 | 16,09 | 61,58 | 87,81 | 315,15 | 383,67 | 918,07 | 979,34 | |
| 20 x 10 - (26 x 10/2) | 32 | 14,13 | 20,90 | 99,60 | 124,90 | 427,87 | 496,25 | 1095,32 | 1161,15 | |
| 21 x 10 - (27 x 10/2) | 33 | 19,71 | 16,14 | 133,56 | 166,16 | 521,50 | 602,98 | 1227,17 | 1313,40 | |
| STEP E | Area under the curve (AUC) | |||||||||
| Total area | Sum of 28 - 33 | Y | 64,4 | 77,2 | 339 | 453,2 | 1586 | 1900,2 | 4530,5 | 4831 |
Calculations from Raw Analysis FIS Data in Excel
Exported data from imaging software is further processed and calculated in Excel. Data is transposed and calculated in a pre-set format according to the linear trapezoidal method that calculates the average increase over a given time window that is multiplied by the time units within the interval. First, the relative increase over time in area μm2 per well derived from the exported data is normalized per well to time point 0. Time point 0 is set to 100% and time point 10–60 min are normalized accordingly (see Table 8, example for 8 measured wells with 4 different conditions; time clips are presented as columns). The relative increase in swelling per well is now expressed in percentages creating 7 data points per well, which can be expressed to show swelling increase per well in time. To more condensely express swelling data (e.g. various forskolin concentrations) in one graph, the area under the curve (AUC) of the relative increase of swelling per well over 60 min can be calculated to create a single data point per well. By using the AUC values, the large datasets can be condensed into graphs capturing all data (see example data in Table 8 & Figure 13). A specific image analysis and data processing pipeline for the FIS assay in open source software is in development by Hagemeijer and Botelho (unpublished).
Figure 13.
Schematic: from raw data to area under the curve
(A) Raw data (area μm2 per well per time point) can be normalized to time point 0 min. Normalized data depicted in a graph shows relative increase in swelling per well over time (baseline is set to 100%). Here, duplicate wells are measured for swelling at 0.02; 0.128; 0.8 and 5 μM fsk for 60 min.
(B) The AUC of relative increase in swelling is calculated in Excel.
(C) Using AUC calculations for the different conditions, a condensed graph can be created which represents overall relative swelling data per condition over 60 min. Graph C shows the average AUC data of the duplicate wells (from B) with SD.
Limitations
This protocol aims to provide detailed descriptions on how to establish viable human colon organoid cultures and to reproducible FIS results. The quality and fast processing of rectal biopsies is crucial for successful isolation of stem cell crypts. Despite detailed protocols, quality of the colon organoid media can vary between batches and operators and is mainly caused by the variation in self-produced conditioned media like WCM and NCM. Currently, the activity of essential components such as Wnt-3A and Noggin can be assessed in quantitative assays but organoid responses to these factors may not fully be represented by cell-line based reporter assays which can affect the quality of the colon organoid cultures and outcome of FIS results. Therefore, pictures and examples are included in this protocol to provide guidance for the researcher and allow ongoing monitoring of human colon organoid culture quality and to compare FIS data. Various microscopy platforms can be used, but robustness of FIS data is highly dependent on the quality of the fluorescent images, and especially a low signal to noise ratio leads to unreliable data analysis and non-representative results. Therefore, optimized confocal microscopic procedures and efficient imaging software analysis are needed to ensure capturing high-quality images and calculation of FIS AUC.
Troubleshooting
Problem
When organoid structures are still very small and not yet efficiently proliferating but the matrigel is fading
Potential Solution
Do not yet disrupt the culture but take up the small organoids from four wells in 1 mL Ad-DF+++ in a microcentrifuge tube, centrifuge with a table top microcentrifuge and plate the organoids into 100 –120 μL of fresh matrigel into 3–4 new wells of a 24-wells plate. This allows the small organoid structures to have more time to expand and start budding before they are ready for the first mechanical disruption/ shearing of the organoids.
Problem
No crypts can be found after the total isolation procedure is performed on the biopsies.
Potential Solution
This is usually caused by a bad quality of biopsies (only superficial tissue removal) or too much time passing between the biopsy removal and the start of the crypt isolation procedure. In this case the colon biopsies can also be plated out directly in a 24-well in 250 μL 50% matrigel and addition of 1 mL CM +/+ (with additional genta- and vancomycin). Sometimes intestinal crypts might still emerge directly from the biopsy.
Problem
Organoids are too densely seeded
Potential Solution
Dilute the sample in more matrigel and seed additional wells
Problem
Organoids are too sparsely seeded
Potential solution
Add cold Ad-DF+++ to the organoids, centrifuged at 130 g for 5 min at 4 °C and resuspend in smaller volumes of matrigel with increased density.
Acknowledgments
This project has received funding from the European Union's Horizon 2020 research and innovation program under grant proposal number 755021-2. This project has been supported by the Dutch Cystic Fibrosis Foundation (NCFS). These protocols have been endorsed by the European CF Society (ECFS) and by the European Reference Network for Respiratory Diseases (ERN-LUNG) in 2019 and aim to standardize methods to ensure high reproducibility and correct interpretation of results among different laboratories.
Author Contributions
Conceptualization: A.M.V., P.M., C.K.E., J.M.B.; Methodology, A.M.V., S.W.F.S., M.S., F.P.V., R.H.J.H., E.K., J.F.D., S.F.B., J.M.; Validation; A.M.V., A.S.R., I.A.L.S., S.W.F.S., J.F.D.; Investigation, A.M.V., A.S.R., I.A.L.S., S.W.F.S.; Writing – Original Draft; A.M.V., P.M., M.S., A.S.R., I.A.L.S., M.S.; Writing – Review & Editing, J.M., S.F.B., R.V., M.D.A., K.B., J.M.B.; Funding Acquisition, P.M., J.M.B., C.K.E.; Supervision, C.K.E. , J.M.B.
Declaration of Interests
J.F.D. is an inventor on a patent related to the organoid technology. M.A.A. reports grants from Gilead Pharmaceuticals, Vertex Pharmceuticals, Proteostasis Therapeutics and personal fees from Vertex Pharmceuticals and PTC Pharmaceuticals outside the submitted work. K.B. reports consultancy work for CHIESI, grants from VERTEX, advisory board meetings for PROTEOSTASIS and ELOXX, grants from and advisory board meetings for GALAPAGOS, and advisory board meetings for TRANSLATE BIO outside the submitted work. R.V. is CEO of Hubrecht Organoid Technology, which provides organoid services to companies. C.K.E. reports grants from GSK, Nutricia, TEVA, Gilead, Vertex, ProQR, Proteostasis, Galapagos NV, and Eloxx outside the submitted work and has a patent 10006904 with royalties paid. J.M.B. is inventor on a patent related to CFTR function measurements in organoids and has received financial royalties. He also participates in industry-sponsored clinical trials (Galapagos NV, Vertex Pharmaceuticals, Proteostasis, Eloxx Pharmaceuticals) and has received travel reimbursements and speaker’s fees. The other authors have nothing to disclose.
References
- Boj S.F., Vonk A.M., Statia M., Su J., Vries R.R.G., Beekman J.M., Clevers H. Forskolin-induced swelling in intestinal organoids: an in vitro assay for assessing drug response in cystic fibrosis patients. J. Vis. Exp. 2017;120:1–12. doi: 10.3791/55159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers G., van Mourik P. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 2019;26:1701–1708. doi: 10.1016/j.celrep.2019.01.068. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., van de Graaf E.A., Nieuwenhuis E.E., Houwen R.H. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8:344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W., Bijvelds M.J., Scholte B.J. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Servidoni M.F., Sousa M., Vinagre A.M., Cardoso S.R., Ribeiro M.A., Meirelles L.R., de Carvalho R.B., Kunzelmann K., Ribeiro A.F., Ribeiro J.D., Amaral M.D. Rectal forceps biopsy procedure in cystic fibrosis: technical aspects and patients perspective for clinical trials feasibility. BMC Gastroenterology. 2013;13:91. doi: 10.1186/1471-230X-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]