Summary
Pulmonary neuroendocrine cells (PNECs) are sensory cells within the lung airway epithelia. Here, we provide a detailed protocol for generating induced PNECs (iPNECs) from human induced pluripotent stem cells (iPSCs). The cellular and molecular profile of iPNECs resembles primary human PNECs. Primary human PNECs are exceedingly rare, comprising only 1% of the adult lung. Therefore, a self-renewing source of patient-specific iPNECs facilitates the creation of reproducible human cellular models to study lung diseases characterized by PNEC dysfunction.
For complete details on the use and execution of this protocol, please refer to Hor et al. (2020).
Graphical Abstract
Highlights
-
•
Efficient generation of iPNECs from human iPSCs
-
•
Air-liquid-interface culture augments iPNEC specification
-
•
Continuous Notch inhibition increases iPNEC specification
-
•
iPNEC gene expression signature is highly concordant with primary human fetal PNECs
Pulmonary neuroendocrine cells (PNECs) are sensory cells within the lung airway epithelia. Here, we provide a detailed protocol for generating induced PNECs (iPNECs) from human induced pluripotent stem cells (iPSCs). The cellular and molecular profile of iPNECs resembles primary human PNECs. Primary human PNECs are exceedingly rare, comprising only 1% of the adult lung. Therefore, a self-renewing source of patient-specific iPNECs facilitates the creation of reproducible human cellular models to study lung diseases characterized by PNEC dysfunction.
Before You Begin
General Laboratory Preparations
Timing: 60–120 min
-
1.
Prepare all stock solutions.
-
2.
Prepare media.
-
3.
Set water bath to 37oC.
-
4.
Warm media to 37oC.
-
5.
All procedures are performed in a Class II biological hood with standard aseptic technique and cells are cultured in a humidified 37oC incubator with 5% CO2.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NKX2.1 (1:100) | Abcam | ab76013 |
| FOXA2 (1:200) | Cell Signaling Technology | 8186T |
| SOX17 (1:200) | Thermo Fisher | AF1924-SP |
| SOX2 (1:200) | Cell Signaling Technology | 2748 |
| SOX9 (1:500) | Abcam | ab185230 |
| ZO1 (1:200) | Thermo Fisher | 40-2200 |
| SYP (1:500) | Abcam | ab32127 |
| SYP (1:100) | Santa Cruz Biotechnology | sc-17750: SYP (D-4) |
| ENO2 (1:100) | Santa Cruz Biotechnology | sc-271384: Enolase (A-5) |
| CHGA (1:100) | Proteintech | 60135-1-Ig |
| ROBO2 (1:100) | R&D Systems | AF3147 |
| PGP9.5 (1:100) | Santa Cruz Biotechnology | sc-271639: UCH-L1 (C-4) |
| EPCAM (1:500) | R&D Systems | AF960 |
| VIM (1:500) | Sigma-Aldrich | V6630 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-21206 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-31572 |
| Donkey anti-rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | R37119 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-21432 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | A-21202 |
| Donkey anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | A-31570 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| mTeSR1 media | STEMCELL Technologies | 85850 |
| Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12) | Life Technologies | 11330032 |
| Roswell Park Memorial Institute (RPMI) 1640 Medium | Life Technologies | 11875-093 |
| FBS | Genesee Scientific | 25-514 |
| Geltrex™ LDEV-Free Reduced Growth Factor Basement Membrane Matrix | Thermo Fisher Scientific | A1413202 |
| ReLeSR | STEMCELL Technologies | 05782 |
| Accutase | STEMCELL Technologies | 7920 |
| Y-27632 (Rock inhibitor) | Selleckchem | S1049 |
| SB431542 | Selleckchem | S1067 |
| Recombinant Human FGF7/KGF | Peprotech | 100-19 |
| Recombinant Human FGF2 | Peprotech | 100-28 |
| Recombinant Human FGF10 | Peprotech | 100-26 |
| Activin A | Humanzyme | HZ-1140 |
| Recombinant Human Wnt-3α | R&D Systems | 5036-WN-010 |
| Recombinant Human Noggin | Peprotech | 120-10C |
| Recombinant Human BMP4 | Humanzyme | HZ-1045 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A2058 |
| Phosphate-buffered saline (PBS), 1× | Genesee Scientific | 25-507 |
| DAPT | Cayman Chemical | 208255-80-5 |
| Laminin | Life Technologies | 23017015 |
| Fibronectin | R&D Systems | 1918-FN-02M |
| Collagen IV | Sigma-Aldrich | C7521-5MG |
| Iscove's Modified Dulbecco's Media (IMDM) | Life Technologies | 12440053 |
| Ham’s F12 Media | Life Technologies | 11765-054 |
| B-27 with RA supplement (50×) | Life Technologies | 17504-044 |
| N-2 supplement (100×) | Life Technologies | 17502-048 |
| Glutamax | Life Technologies | 35050 |
| Ascorbic Acid | Sigma-Aldrich | A8960-25G |
| α-Monothioglycerol (MTG) | Sigma-Aldrich | M1753-100ML |
| Bovine Brain Extract | Lonza | CC-4098 |
| Epinephrine | Sigma-Aldrich | E4642 |
| Ethanolamine | Sigma-Aldrich | E0135 |
| Fetal Clone II | Hyclone | SH30066 |
| Hydrocortisone | Sigma-Aldrich | H4001 |
| Insulin | Sigma-Aldrich | I9278 |
| Phosphoryl ethanolamine | Sigma-Aldrich | P0503 |
| Retinoic Acid | Sigma-Aldrich | R2625 |
| 3,3′,5-Triiodo-L-thyronine sodium salt | Sigma-Aldrich | T6397 |
| Transferrin | Sigma-Aldrich | T8158 |
| Ultroser G serum substitute | Crescent chemicals | 67042.1/S |
| Paraformaldehyde (PFA) | VWR | 100496-496 |
| CAS Block | Thermo Fisher Scientific | 008120 |
| Triton X-100 | Sigma-Aldrich | T8787 |
| Phosphate Buffered Saline (PBS) | Genesee Scientific | 25-507 |
| Immunomount | Thermo Fisher Scientific | 9990402 |
| Vectashield with DAPI | Vector Laboratories | H-1500 |
| DAPI | Sigma-Aldrich | D9542 |
| DNase | Worthington Biochemical Corporation | LS003172 |
| SYBR Green | Bio-rad | 1725125 |
| Critical Commercial Assays | ||
| Qiagen RNA Easy Mini Kit | Qiagen | 74004, 74134 |
| Protoscript first strand synthesis kit | New England Biolabs | E6560S |
| Enolase Quantikine ELISA Kit | R&D | DENL20 |
| Experimental Models: Cell Lines | ||
| Healthy human lymphoblastoid cell line-derived iPSCs (iPSC 1). Lymphoblastoid cell line reprogrammed by Ichida Lab USC | National Institute of Neurological Disorders and Stroke (NINDS) Biorepository, Coriell Institute | ND00184 (original lymphoblastoid cell line) |
| Healthy human lymphoblastoid cell-derived iPSCs (iPSC 2). Lymphoblastoid cell line reprogrammed by Ichida Lab USC | National Institute of Neurological Disorders and Stroke (NINDS) Biorepository, Coriell Institute | ND03719 (original lymphoblastoid cell line) |
| Human foreskin fibroblast-derived iPSC line (FiPSC #3C15) | Ryan Laboratory, USC | Derived from BJ fibroblasts (ATCC, CRL-2522) |
| Deposited Data | ||
| Raw and analyzed iPNEC data | Hor et al., 2020 | GEO: GSE146990 |
| Oligonucleotides (qPCR Primers) | ||
| Gene | Forward Primer | Reverse Primer |
| β-Actin | 5′-CATGTACGTTGGTATCGAGGC-3′ | 5′-CTCCTTAATGTCACGCACGAT-3′ |
| HES1 | 5′-ACGTGCGAGGGCGTTAATAC-3′ | 5′-GGGGTAGGTCATGGCATTGA-3′ |
| HEY1 | 5′-AGAGTGCGGACGAGAATGGAAACT-3′ | 5′-CGTCGGCGCTTCTCAATTATTCCT-3′ |
| ASCL1 | 5′-CCCAAGCAAGTCAAGCGACA-3′ | 5′-AAGCCGCTGAAGTTGAGCC-3′ |
| GRP | 5′-AAAGAGCACAGGGGAGTCTTC-3′ | 5′-TCCTTTGCTTCTATGAGACCCA-3′ |
| DLL3 | 5′-CGTCCGTAGATTGGAATCGCC-3′ | 5′-TCCCGAGCGTAGATGGAAGG-3′ |
| GAD67 | 5′-GCTTCCGGCTAAGAACGGT-3′ | 5′-TTGCGGACATAGTTGAGGAGT-3′ |
| ENO2 | 5′-CTGATGCTGGAGTTGGATGG-3′ | 5′-CCATTGATCACGTTGAAGGC-3′ |
| ROBO2 | 5′-GTTTGTGTTGCGAGGAACTATCT-3′ | 5′-GTTTTGTCGGAAGTCATCTCGTA-3′ |
| SYP | 5′-CTCAGCATCGAGGTCGAGTTC-3′ | 5′-GAGGAGTAGTCCCCAACTAAGAA-3′ |
| SST | 5′-CCCAGACTCCGTCAGTTTCT-3′ | 5′-AAGTACTTGGCCAGTTCCTGC-3′ |
| SV2 | 5′-TAGACCAGGCACTCATTGGC-3′ | 5′-ACCCCTCCCCACAGTTACTT-3′ |
| PENK | 5′-TCTGAACCCGGCTTTTCCAA-3′ | 5′-TACGCAAGCCAGGAAGTTGAT-3′ |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Quantitative RT-PCR software | Thermo Fisher Scientific | 7900HT SDS software v2.1 |
| Other | ||
| 6-Well Cell Culture Plates | Genesee Scientific | 25-105 |
| Transwell® Inserts, Sterile, Corning® (3470), 24 well plate | VWR | 29442-082 |
| Vacuum Filter Systems, 500 mL | Genesee Scientific | 25-227 |
| 0.45 μm pore filter | Genesee Scientific | 25-246 |
| BD Disposable Syringes with Luer-Lok™ Tips (1 and 5 mL) | VWR | BD309646, BD328438 |
Note: General laboratory consumables like serological pipettes (1 mL, 5 mL, 10 mL, 25 mL, 50 mL), p10, p20, p200 and p1000 pipet tips, aspirating pipettes, centrifuge tubes (15 mL, 50 mL), and hemocytometer (for cell counting) are also required.
Materials and Equipment
Growth Factor/Cytokine Stock Solutions
Reconstitute growth factors as per the manufacturer's recommendations and aliquot for storage.
Activin A Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Activin A | 100 μg/mL | 100 μg |
| PBS 1× | n/a | 1 mL |
| BSA | 0.1% (w/v) | 1 mg |
| Total | n/a | 1 mL |
n/a – not applicable
CRITICAL: Prepare 50 μL aliquots of the stock solutions and store them at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
Wnt-3a Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Wnt-3a | 25 μg/mL | 10 μg |
| PBS 1× | n/a | 400 μL |
| BSA | 0.1% (w/v) | 400 μg |
| Total | n/a | 400 μL |
n/a – not applicable
CRITICAL: Prepare 50 μL aliquots of the stock solutions and store them at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
SB431524 Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| SB431524 | 50 mM | 1 mg |
| DMSO | n/a | 0.5203 mL |
| Total | n/a | 0.5203 mL |
n/a – not applicable
Use the 50 mM stock to further dilute 1:100 to obtain 500 μM stock solution.
CRITICAL: Prepare 100 μL aliquots of the stock solutions and store at −20°C for up to 1 year.
Noggin Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Noggin | 20 μg/mL | 20 μg |
| PBS 1× | n/a | 1 mL |
| Total | n/a | 1 mL |
n/a – not applicable
CRITICAL: Prepare 50 μL aliquots of the stock solutions and store at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
BMP4 Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| BMP4 | 50 μg/mL | 10 μg |
| PBS 1× | n/a | 200 μL |
| BSA | 0.1% (w/v) | 200 μg |
| Total | n/a | 200 μL |
n/a – not applicable
CRITICAL: Prepare 10 μL aliquots of the stock solutions and store at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
FGF-2 Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| FGF-2 | 10 μg/mL | 10 μg |
| PBS 1× | n/a | 1 mL |
| BSA | 0.1% (w/v) | 1 mg |
| Total | n/a | 1 mL |
n/a – not applicable
CRITICAL: Prepare 50 μL aliquots of the stock solutions and store at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
FGF-7 (KGF) Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| FGF-7 | 10 μg/mL | 10 μg |
| PBS 1× | n/a | 1 mL |
| BSA | 0.1% (w/v) | 1 mg |
| Total | n/a | 1 mL |
n/a – not applicable
CRITICAL: Prepare 50 μL aliquots of the stock solutions and store at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
FGF-10 Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| FGF-10 | 10 μg/mL | 25 μg |
| PBS 1× | n/a | 2.5 mL |
| BSA | 0.1% (w/v) | 2.5 mg |
| Total | n/a | 2.5 mL |
n/a – not applicable
CRITICAL: Prepare 50 μL aliquots of the stock solutions and store at −80°C for up to 6 months. Working stocks can be stored at −20°C for a 1–2 weeks. Avoid repeated freeze-thaw cycles of the stock solutions.
Ascorbic Acid Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Ascorbic Acid | 50 mg/mL | 50 mg |
| Milli-Q H2O (autoclaved) | n/a | 1 mL |
| Total | n/a | 1 mL |
n/a – not applicable
CRITICAL: Do not store this solution. Prepare fresh stock solution for every use.
Y-27632 (ROCK inhibitor) Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Y-27632 (ROCK inhibitor) | 40 mM | 5 g |
| DMSO | n/a | 390.3 μL |
| Total | n/a | 390.3 μL |
n/a – not applicable
CRITICAL: Prepare 10 μL aliquots of the stock solutions and store at −80°C for up to 2 years. Working stocks can be stored at −20°C for a 1–2 weeks.
DAPT Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| DAPT | 1 mg/mL | 5 mg |
| DMSO | n/a | 5 mL |
| Total | n/a | 5 mL |
n/a – not applicable
CRITICAL: Prepare 100 μL aliquots of the stock solutions and store at −20°C for up to 2 years. Avoid repeated freeze-thaw cycles of the stock solutions.
Bovine Brain Extract
| Reagent | Final Concentration | Amount |
|---|---|---|
| Bovine Brain Extract | 9 mg/mL | 5 mL |
Prepare 250 μL aliquots and preserve at −20°C for up to 6 months.
Epinephrine Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Epinephrine | 15 mM | 6.5 mg |
| Milli-Q H2O (autoclaved) | n/a | 2 mL |
| Total | n/a | 2 mL |
n/a – not applicable
Prepare 50 μL aliquots and store at −20°C for up to 1 year.
Ethanolamine Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Ethanolamine | 250 μM | 1.527 mg |
| Milli-Q H2O (autoclaved) | n/a | 100 mL |
| Total | n/a | 5 mL |
n/a – not applicable
Prepare 1 mL aliquots and store at −20°C for up to 1 year.
Hydrocortisone Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Hydrocortisone | 200 μM | 1 g |
| Ethanol | n/a | 1 mL |
| Milli-Q H2O (autoclaved) | n/a | 12.8 mL |
| Total | n/a | 13.8 mL |
n/a – not applicable
CRITICAL: First, dissolve the hydrocortisone at 1 mg/mL in absolute EtOH and then add 12.8 mL H2O.
Filter the solution using a 0.45 μm-pore filter. Prepare 1 mL aliquots and store at −20°C for up to 6 months.
Insulin Stock Solution
Prepare 100-μL aliquots of the commercial Insulin solution (10.2mg/mL, check lot concentration) and freeze them at −20°C for up to 1 year.
Phosphoryl ethanolamine Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Phosphoryl ethanolamine | 50 mM | 15 mg |
| PBS, 1× | n/a | 2 mL |
| Total | n/a | 5 mL |
n/a – not applicable
Prepare 50 μL aliquots and store at −20°C for up to 1 year.
Retinoic Acid Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Retinoic Acid | 100 mM | 1 mg |
| Milli-Q H2O (autoclaved) | n/a | 33.28 mL |
| Total | n/a | 33.28 mL |
n/a – not applicable
Prepare 1 mL aliquots and store at −80°C for up to 1 year.
Note: When preparing Media G further dilute this stock at 1:1,000 dilution with water to prepare a 100 μM stock for ease of media preparation.
Transferrin Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| Transferrin | 2.5 mg/mL | 100 mg |
| Milli-Q H2O (autoclaved) | n/a | 40 mL |
| Total | n/a | 40 mL |
n/a – not applicable
Prepare 100 μL aliquots and store at −20°C for up to 1 year.
3,3′,5-Triiodo-L-thyronine sodium Stock Solution
| Reagent | Final Concentration | Amount |
|---|---|---|
| 3,3′,5-Triiodo-L-thyronine sodium | 15 mM | 100 mg |
| DMEM/F-12 Media | n/a | 10 mL |
| Total | n/a | 10 mL |
n/a – not applicable
Prepare 50 μL aliquots and store at −20°C for up to 1 year.
Extracellular Matrix (ECM) Stock Solution
| Reagent | Stock Concentration | Final Concentration | Amount |
|---|---|---|---|
| Fibronectin | 1 mg/mL | 5 μg/mL | 25 μL |
| Laminin | 500 μg/mL | 5 μg/mL | 50 μL |
| Collagen IV | 600 μg/mL | 60 μg/mL | 500 μL |
| PBS, 1× | n/a | n/a | 4.5 mL |
| Total | n/a | n/a | 5 mL |
n/a – not applicable
Note: This should be prepared immediately before coating the Transwells and kept on ice throughout the preparation.
Differentiation Media Compositions
Complete Serum-free Differentiation Medium (cSFDM)
| Reagent | Final Concentration | Amount |
|---|---|---|
| Iscove's Modified Dulbecco's Media (IMDM) | 75% | 375 mL |
| Ham’s F-12 | 25% | 125 mL |
| BSA | 0.75% (w/v) | 3.75 mg |
| B-27 with RA | 0.5× | 5 mL |
| N-2 | 0.5× | 2.5 mL |
| Glutamax | 1× | 5 mL |
| Ascorbic Acid | 50 μg/mL | 500 μL |
| MTG | 4.5 × 10−4 M | 19.55 μL |
| Total | n/a | 500 mL |
n/a – not applicable
Filter the solution using vacuum filter units. This medium can be prepared and stored at 4°C for 1 week. Pre-warm only the volume needed for that day in a water bath at 37°C before adding to cells.
Note: This media composition is based on previously published endoderm-derivation protocols (Longmire et al., 2012; Gouon-Evans et al., 2006).
Media A. mTeSR1 (commercially available)
| Reagent | Final Concentration | Amount |
|---|---|---|
| Basal medium | 400 mL | |
| 5× Supplement | 1× | 100 mL |
Store at 4oC.
This may be substituted for any suitable iPSC maintenance media however only mTeSR1 was validated in this protocol.
Medium B
| Reagent | Final Concentration | Amount |
|---|---|---|
| RPMI 1640 | 100 mL | |
| Wnt-3a | 25 ng/mL | 100 μL |
| Activin A | 100 ng/mL | 100 μL |
Make fresh for every use and filter through a 0.45 μm pore filter.
Medium C
| Reagent | Final Concentration | Amount |
|---|---|---|
| RPMI | 100 mL | |
| FBS | 1% | 1 mL |
| Activin A | 100 ng/mL | 100 μL |
Make fresh for every use and filter through a 0.45 μm pore filter.
Medium D
| Reagent | Final Concentration | Amount |
|---|---|---|
| cSFDM | 100 mL | |
| SB431542 | 500 nM | 100 μL |
| Noggin | 20 ng/mL | 100 μL |
Make fresh for every use and filter through a 0.45 μm pore filter.
Medium E
| Reagent | Final Concentration | Amount |
|---|---|---|
| cSFDM | 100 mL | |
| SB431542 | 500 nM | 100 μL |
| BMP-4 | 5 ng/mL | 10 μL |
Make fresh for every use and filter through a 0.45 μm pore filter.
Medium F
| Reagent | Final Concentration | Amount |
|---|---|---|
| cSFDM | 100 mL | |
| BMP-4 | 5 ng/mL | 10 μL |
| FGF-2 | 10 ng/mL | 100 μL |
| FGF-10 | 10 ng/mL | 100 μL |
| KGF | 10 ng/mL | 100 μL |
This medium can be prepared for 3–5 days use and stored at 4°C. Filter through a 0.45 μm pore filter. Pre-warm the amount needed for daily use only.
Medium G
This medium can be prepared for the week and stored at 4°C. Filter through a 0.45 μm pore filter. Pre-warm only the volume needed for cell culture on a daily basis. This media can be prepared in a larger batch, aliquoted, protected from light and stored at −20°C for up to 6 months.
Note: This media composition is based on previously published protocol to culture human bronchial epithelial cells (Neuberger et al., 2011).
| Composition | Final Concentration | Amount |
|---|---|---|
| DMEM/F-12 | 480 mL | |
| Ultroser G | 2% | 10 mL |
| Fetal Clone II | 2% | 10 mL |
| Insulin | 2.5 μg/mL | 122.5 μL |
| Bovine Brain Extract | 22.5 μg/mL | 1.25 μL |
| Transferrin | 2.5 μg/mL | 500 μL |
| Hydrocortisone | 20 nM | 50 μL |
| 3,3′,5-Triiodo-L-thyronine | 500 nM | 16.7 μL |
| Epinephrine | 1.5 μM | 50 μL |
| Retinoic Acid | 10 nM | 50 μL |
| Phosphoryl ethanolamine | 250 nM | 2.5 μL |
| Ethanolamine | 250 nM | 500 μL |
| Total | n/a | 500 mL |
Step-by-Step Method Details
Preparation of iPSC for Differentiation
Timing: 2–4 days
-
1.
Coating plates for iPSC culture.
Note: Before you begin thaw Geltrex™ matrix in a refrigerator for 16–20 h at 2 to 8°C and carefully pipet up and down to avoid introducing air bubbles. It is best to aliquot and work with small volumes of Geltrex™ matrix solution to avoid multiple freeze/thaw cycles.
-
a.
Dilute Geltrex™ matrix at 1:100 in pre-chilled (4°C) DMEM/F-12 (for example: 1 mL Geltrex™ matrix in 99 mL DMEM/F-12).
Note: Add sufficient diluted Geltrex™ matrix solution to cover the entire growth surface area. For example, add 2 mL per one well of a 6-well plate.
-
b.
Incubate the coated plates at 37°C for a minimum of 60 min.
Note: The coated dish is stable for two weeks when wrapped with parafilm and stored at 4°C.
CRITICAL: Do not allow the coated surface to dry out. Add enough media (1.5–2 mL volume for 1 well of a 6 well-plate) to avoid drying. Keep Geltrex on ice throughout all aliquotting and plating steps to prevent premature gelling
-
c.
At the time of use, we recommend keeping the plates at room temperature (20–25°C) for 1 h before aspirating. Carefully aspirate off the supernatant above the Geltrex™ matrix coating, then immediately plate cells in the pre-equilibrated cell culture medium.
-
2.
Expansion of iPSCs.
-
a.
iPSCs are maintained in mTeSR1 medium in 6-well tissue culture-treated plates on Geltrex™ matrix.
-
b.
iPSCs are split routinely at 1:3 or 1:4 when cells have formed tight colonies and are no more than 70% confluent (generally every 4–5 days).
-
c.
To split iPSCs, aspirate media, and add 1 mL ReLeSR™ for 30 s. Remove ReLeSR™ and incubate at 37oC for 5 min. Add 2 mL mTESR1 media and tap the dish gently to release the iPSC colonies as cell aggregates.
Note: Incubation time for ReLeSR™ depends on the individual iPSC line and should be optimized to ensure selective detachment of undifferentiated iPSC colonies only.
-
d.
Replate the aggregates in 2 mL fresh mTESR1 per well in a new Geltrex™-coated well of a 6- well plate.
Note: 1 well of iPSC colonies was split at a 1:3 or 1:4 ratio for routine maintenance.
Timing: 2 h
-
3.
Preparation of Transwells for Differentiation
-
a.
Day 1. The day prior to preparing cells for differentiation, prepare extracellular matrix (ECM) to coat the Transwells in PBS on ice. This is comprised of fibronectin (5 μg/mL), laminin (5 μg/mL), and collagen IV (60 μg/mL).
-
b.
Add 150 μL of ECM solution to the apical side of each Transwell filter.
-
c.
Incubate the ECM-coated Transwell filters at room temperature (20–25°C) for 16–20 h inside the cell culture hood.
-
d.
After a minimum of 16 h, aspirate the ECM solution and gently wash twice with 1× PBS followed by a single wash with autoclaved Milli-Q water.
-
e.
Transwells can be stored at this point at room temperature (20–25°C) or 4°C, sealed with parafilm, for up to a week.
CRITICAL: The quality of coating is a major factor for successful cell adherence to the membrane. Sub-optimal coating can lead to cells peeling off at early stages of differentiation.
Timing: 2 h
-
4.
Plate cells for differentiation
-
a.
Day 0. Remove mTeSR from the iPSC colonies.
-
b.
Wash the cells gently with PBS. Aspirate the PBS.
-
c.
Add 500 μL of room temperature (20–25°C) Accutase.
-
d.
Incubate at 37°C for 2–3 min (max).
-
e.
Collect cells by gently dissociating with a 5 mL pipette into a 15 mL Falcon tube and centrifuge at 180 × g (1,000 rpm) for 5 min.
-
f.
Resuspend in mTeSR1 containing 10 μM ROCK inhibitor and count the cells (1 μL per 4 mL mTESR1)
-
g.
Prepare Transwells by adding mTeSR1 containing ROCK inhibitor to the desired number of wells. For 24 well inserts use 350 μL in the bottom chamber and 150 μL in the Transwell.
-
h.
Count cells and add 150K live cells to each 6.5 mm Transwell.
-
i.
Incubate at 37°C for 20–24 h.
Differentiation to Definitive Endoderm (DE)
Timing: 5 days
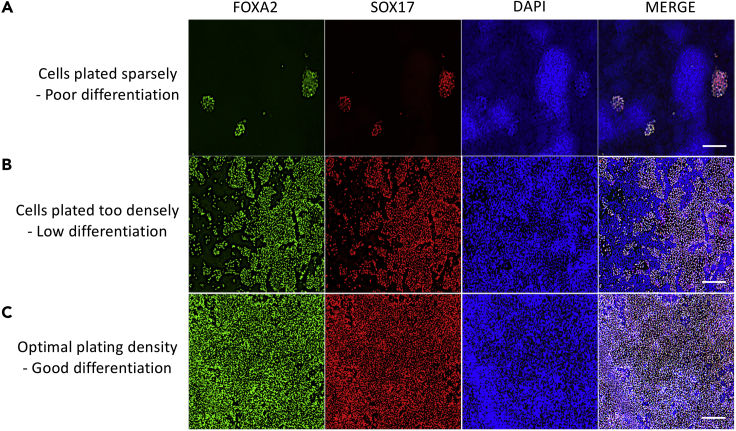
CRITICAL: Cells must be a minimum of 80% confluent before adding Medium B. If the cells are plated sparsely or are too confluent at the beginning of day 1 media change then differentiation efficiency is severely affected. Figure 1 provides an example of the ideal plating density required to move forward (Figure 1). See Troubleshooting 1.
Figure 1.
Effect of Plating Density on Differentiation Efficiency
(A–C) Efficiency of definitive endoderm (DE) derivation at day 5 varies based on iPSC confluency before starting the differentiation. Representative immunofluorescence (IF) images of cells show expression of FOXA2 (green) and SOX17 (red). Nuclei are counterstained with DAPI (blue). Differentiation started with too few cells (<70% confluency) or too many cells results in poor (A) or low (B) DE differentiation. Differentiation started with optimal plating density of 70%–80% confluency at the time of starting the experiment results in best DE differentiation (C). Scale bars, 200 μm.
-
5.
Day 1. Prepare Medium B by adding Wnt-3a (25ng/mL) and Activin A (100 ng/mL) to pre-warmed RPMI medium.
-
6.
Remove and replace the mTeSR1 medium from the basal side of the Transwells with 350 μL of Medium B.
CRITICAL: Avoid disrupting the cells on the insert by using a 200 μL pipette to aspirate the media very gently.
-
7.
Carefully aspirate mTesR1 medium from the apical side of the Transwells and add 150 μL of Medium B into each 6.5 mm Transwell.
CRITICAL: It is essential to add and remove media gently and not to touch/disturb the cells.
CRITICAL: Add media to the edge of the Transwell.
-
8.
Incubate at 37°C for 24h.
-
9.
Day 2. Prepare Medium C by adding FBS (1%) and Activin A (100 ng/mL) to pre-warmed RPMI medium.
-
10.
Remove and replace the Medium B from the basal side of the Transwells with 350 μL of Medium C.
-
11.
Carefully aspirate Medium B from the apical side of the Transwells and add 150 μL of Medium C into each 6.5 mm Transwells.
-
12.
Incubate at 37°C for 48h.
CRITICAL: Significant cell death can be observed during days 1–3. To move to day 4 cells should be near 100% confluent. Observe closely on a microscope and only proceed if cells are >80% confluent. See Troubleshooting 2.
-
13.
Day 4. Prepare Medium D by adding FBS (2%), SB431524 (500 nM) and Noggin (20 ng/mL) to prewarmed cSFDM medium.
-
14.
Remove and replace the Medium C from the basal side of the Transwells with 350 μL of Medium D.
-
15.
Carefully aspirate Medium C from the apical side of the Transwells and add 150 μL of Medium D into each 6.5 mm Transwells.
-
16.
Incubate at 37°C for 24h.
-
17.
Day 5. Quality Control Analysis DE differentiation. Take 1 well of cell culture to assay for definitive endoderm (DE) markers like FOXA2 and SOX17 with immunofluorescence.
Immunofluorescence Validation of DE
Timing: 2–3 days
-
18.
Fix Transwells in 4% (vol/vol) paraformaldehyde (PFA) for 1 h at 4°C.
-
19.
Wash 3× with PBS.
-
20.
Permeabilize in 0.5% Triton X-100 for 1 h at 4oC.
-
21.
Block for 1 h with commercial CAS block.
-
22.
Incubate in primary antibody (See Table 1) diluted in CAS block at the required dilution (provided in Key Resources Table) at 4°C for 16–20 h.
-
23.
Wash 3× in 0.1% Triton X-100 in PBS.
-
24.
Incubate for 1 h in secondary antibodies diluted in CAS block at 1:500 dilution.
-
25.
Wash 3× in 0.1% Triton X-100 in PBS.
-
26.
Add DAPI (1 μg/mL) and incubate for 5 min.
-
27.
Wash 3× with PBS.
-
28.
Mount on glass microscope slides in Immunomount.
Alternatives: Transwells can be washed after secondary antibody incubation and directly mounted on the glass microscope slide using Vectashield with DAPI.
Note: By day 5 of differentiation >85% of the cells must express FOXA2+ and SOX17+ endoderm (Figure 1C).
CRITICAL: Cultures with poor DE conversion showing <50% FOXA2+ and SOX17+ cells will have higher heterogeneity as the differentiation proceeds, affecting the ratio of different airway cell types present in the cultures at later stages. Discontinue these cultures as subsequent differentiation will be inadequate.
Table 1.
Markers for Characterizing Different Stages of Human iPNEC Differentiation
| Cell types | Markers | Assays |
|---|---|---|
| Definitive endoderm (DE) | FOXA2, SOX17 (CXCR4) | Immunofluorescence |
| Anterior foregut endoderm (AFE) | FOXA2, SOX17 | Immunofluorescence |
| Lung progenitors | NKX2.1 (lung endoderm marker), ZO1 (tight junction marker), EPCAM (epithelial marker) | Immunofluorescence |
| Induced PNECs (iPNECs) | SYP, PGP9.5, ENO2, CHGA, ROBO2 | Immunofluorescence, qPCR |
Differentiation to AFE
Timing: 4 days
-
29.
Day 5. Prepare Medium E by adding FBS (2%), SB431524 (500 nM) and BMP-4 (5 ng/mL) to pre-warmed cSFDM medium.
-
30.
Remove and replace the Medium D from the basal side of the Transwells with 350 μL of Medium E.
-
31.
Carefully aspirate Medium D from the apical side of the Transwells and add 150 μL of Medium E into each 6.5 mm Transwell.
-
32.
Incubate at 37°C for 48h.
-
33.
Day 7. Repeat steps 30–32 until day 9.
-
34.
Incubate at 37°C for 48h.
-
35.
Day 9. Quality Control Analysis AFE differentiation. Take 1 well of cell culture and assay for continued presence of FOXA2 and SOX17 with immunofluorescence (See Table 1) (protocol described in step 18).
Note: By day 9 of differentiation >85% of the cultures should express FOXA2+ and SOX17+ endoderm.
CRITICAL: Cultures with poor AFE conversion showing <50% FOXA2+ and SOX17+ will have higher heterogeneity as the differentiation proceeds affecting the ratio of different airway cell types present in the cultures at later stages. Discontinue these cultures as subsequent differentiation will be inadequate.
Differentiation toward NKX2.1+ Lung Progenitors (LP)
Timing: 8 days
Note: Medium F can be prepared for up to 5 days, but needs to be stored at 4°C, and only the volume needed for that day should be pre-warmed at a time.
-
36.
Day 9. Prepare Medium F by adding FBS (2%), BMP-4 (5 ng/mL), FGF-2 (10 ng/mL), FGF-10 (10 ng/mL) and KGF (10 ng/mL) to cSFDM medium.
-
37.
Aspirate and replace the Medium E from the basal side of the Transwells with 350 μL of Medium F.
-
38.
Carefully aspirate Medium E from the apical side of the Transwells and add 150 μL of Medium F into each 6.5 mm Transwell.
-
39.
Incubate at 37°C for 48h.
-
40.
Day 11. Remove and Replace Medium F (350 μL in basal chamber, 150 μL in apical chamber).
-
41.
Incubate at 37°C for 48h.
-
42.
Repeat steps 40–41 until day 17.
-
43.
Day 17. Quality Control Analysis of LP differentiation. Take 1 well of cell culture and assay for LP markers (See Table 1) including NKX2.1, SOX2 and SOX9 with immunofluorescence (protocol described in step 18).
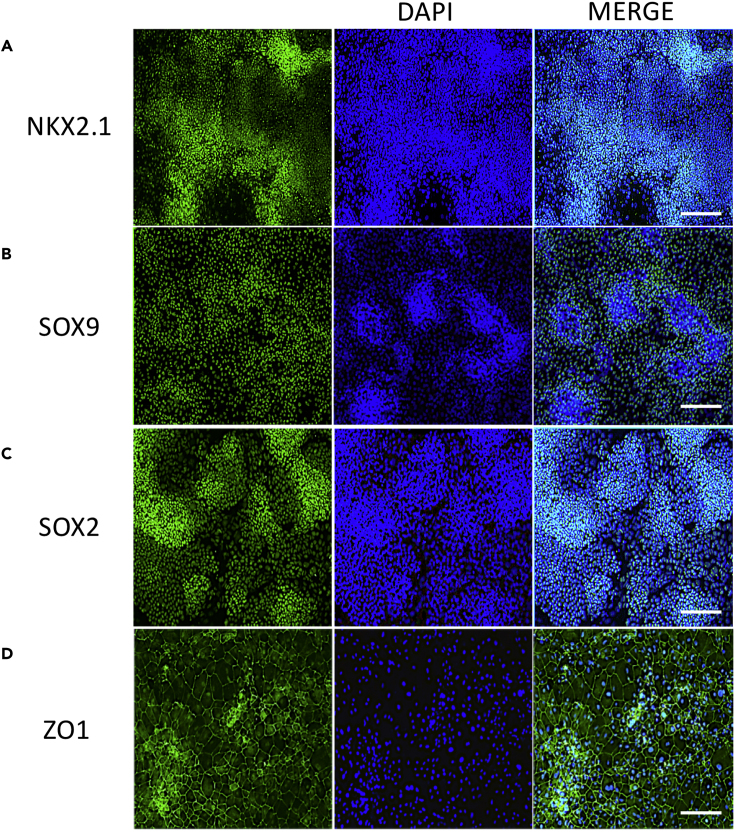
Note: By day 17 cultures should contain a population of NKX2.1+, SOX2+ and SOX9+ lung progenitors. Appearance of tight junction protein ZO1 shows the formation of an epithelium with tight junctions (Figure 2).
Figure 2.
Generation of Lung Progenitors during iPNEC Differentiation from Human iPSCs
(A–D) Cultures stained for (A) lung endoderm marker NKX2.1 (green), (B) distal progenitor marker SOX9 (green), (C) proximal airway progenitor marker SOX2 (green) and (D) tight junction marker ZO1 (green) at day 17 of differentiation. Nuclei are counterstained with DAPI (blue). Scale bars, 200 μm.
Differentiation and Maturation of PNEC at Air-Liquid Interface (ALI)
Timing: 14–75 days
Note: The end point depends on the experiments to be performed. We have maintained cultures for up to 75 days after transfer to the ALI (day 91).
Note: Medium G can be prepared in advance and frozen (−80°C) in aliquots for use up to 2–3 months, once thawed needs to be stored at 4°C, and only the volume needed for that day should be pre-warmed at a time.
-
44.
Day 17. Prepare Medium G complete with 10 μM DAPT.
Note: DAPT should be added to Medium G on day of use only.
-
45.
Aspirate and replace Medium F from the basal side of the cultures and add 350 μL of Medium G.
-
46.
Aspirate Medium F from the apical side of the cultures.
CRITICAL: avoid disrupting the epithelium by using mechanical suction to aspirate the media. Use a 200 μL pipette tip to do it manually.
-
47.
Gently wash the apical side with 100 μL PBS and aspirate.
-
48.
Incubate at 37°C for 48h.
-
49.
Day 19. Aspirate and replace Medium G from the basal side of the cultures.
-
50.
Incubate at 37°C for 48h.
-
51.
Repeat steps 49–50 for as long as the cultures need to be maintained.
Quality Control Analysis of iPNECs
Timing: 3–5 days
-
52.Analyzing mRNA expression changes using Quantitative RT-PCR (qRT-PCR)
-
a.Take 1 well of cell culture from each of these time points at days 5, 10, 13, 17, 21, 25 and 31 for checking the mRNA expression levels of different PNEC markers (See Table 1) during differentiation by performing a qRT-PCR.
-
a.
-
53.Quantitative RT-PCR protocol: Isolate total RNA using the Qiagen RNA Easy Mini Kit as per the manufacturer’s protocol.
-
a.Aliquot 500 ng of RNA and proceed to DNase-treatment as per the manufacturer’s protocol.
-
b.Use the DNase-treated RNA aliquot to prepare cDNA using a Protoscript first strand synthesis kit.
-
c.Set up 5 μL qRT-PCR reactions with SYBR Green using the following cycling parameters.
-
a.
| PCR Cycling Conditions | |||
|---|---|---|---|
| Steps | Temperature | Time | Cycles |
| Initial Denaturation | 95°C | 2 min | 1 |
| Denaturation | 95°C | 15 s | 40 cycles |
| Annealing | 60°C | 1 min | |
| Extension | 72°C | 1 min | |
| Hold | 4°C | Forever | |
Note: qRT-PCR is conducted and analyzed using 7900HT SDS v2.1 software (Thermo Fisher Scientific). Each run consists of a triplicate of technical repeats per sample/gene. Beta-actin was used as an internal control.
Note: Data shown in Figures 3D, 5A, S2D-G of Hor et al., (2020).
Note: Day 0 iPSCs express OCT4, NANOG and SOX2 genes. They should be expressed highly at day 0 and by DE stage (day 4–5) should be significantly lower.
IF Analysis of PNEC Markers
Timing: 2–3 days
-
54.
Take 1 wells of cell culture at days 31, 60 and 91 for immunofluorescence staining (protocol described in step 18). Stain for all the PNEC markers (See Table 1) – SYP, CHGA, ROBO2, PGP9.5 and ENO2. Co-staining with VIM (vimentin, a mesenchymal marker) and EPCAM (epithelial marker) is recommended to characterize iPNECs.
Note: iPNECs should stain for SYP, CHGA, ROBO2, PGP9.5 and ENO2 and should not co-localize with VIM and EPCAM.
Note: All mean fluorescent intensity (MFI) percentage quantifications and Sholl analysis measurements of number of intersections and branching density of iPNECs can be performed using ImageJ.
Note: Data shown in Figures 1A, 1B, 2, 3, S1A-B, S2A-B, S4 of Hor et al. (2020).
Analyzing Neuropeptide Secretion
Timing: 1–2 days
-
55.
Collect media from the basal chamber of the Transwells of day 91 cultures to perform enzyme-linked immunosorbent assay (ELISA) in order to measure secreted levels of neuron-specific enolase (ENO2). Follow the manufacturer’s instructions to perform ELISA using Human Enolase 2/Neuron-specific Enolase Quantikine ELISA Kit (R&D systems).
Note: Data shown in Figures 5B of Hor et al. (2020).
Expected Outcomes
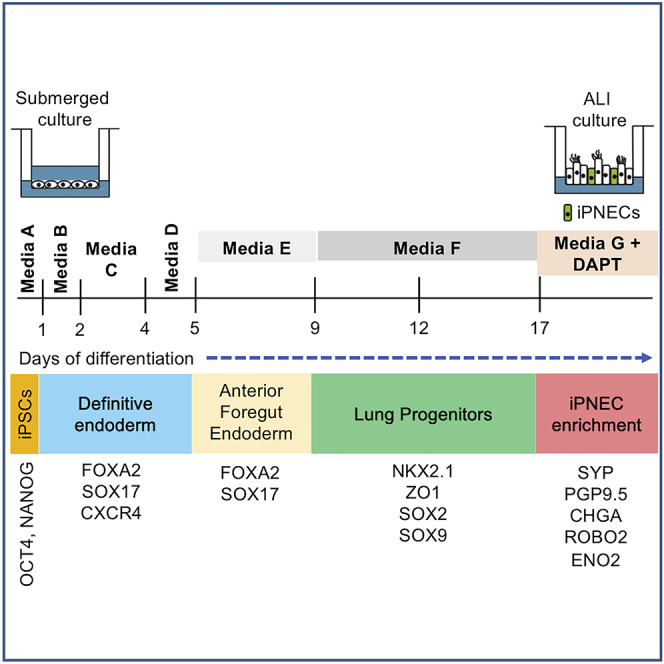
This protocol describes the generation of iPNECs that have potential utility in modeling lung disorders characterized by PNEC dysfunction. The first stage of differentiation towards DE is initiated by supplementing RPMI media with Activin A (days 1–3) along with rhWnt-3a protein (day 1). These conditions promote the exit from the pluripotency state and progression towards DE. This is indicated by cells undergoing an epithelial to mesenchymal (EMT), down-regulation of pluripotency markers (OCT4, NANOG, SOX2) and the transient upregulation of pan-endodermal markers such as FOXA2 and SOX17. At day 4, a combination of Noggin and SB141524 successfully inhibits BMP and TGF-β signaling pathways, respectively, pushing cells toward AFE from DE. From day 9 onwards, a cocktail of FGFs and BMP4 ensures patterning of lung endoderm from AFE, thereby increasing NKX2.1 expression during anteriorization. It is critical that the NKX2.1+ primed airway epithelial progenitors induced by day 17 are maintained at an air liquid interface (ALI) in Media G on the basolateral side of the Transwell filters (Neuberger et al., 2011) for the differentiation of iPNECs. This method, adapted from human primary bronchial epithelial cells in vitro culture technique, essentially reflects a mature lung epithelial niche and helps promote maturation and polarization of the epithelial layer (Van Haute et al., 2009; Wong et al., 2012; Yamaya et al., 1992). Maintaining lung progenitors persistently in a submerged condition result in iPNEC loss (Hor et al., 2020). This is possibly due to the hypoxic microenvironment created around the cells and consequential Notch upregulation inhibiting PNEC fate specification (Gerovac et al., 2014; Yao et al., 2018). ALI alleviates these issues by mimicking a niche similar to a breathing lung. Continued Notch inhibition with DAPT augments iPNEC fate specification and yield during the differentiation protocol (Henke et al., 2009; Linnoila, 2006; Nelson et al., 2009). Approximately 40%–60% of the cultures show PNEC marker expression by day 31 which is continued over a period of >70 days at ALI (Hor et al., 2020). Increased arborization of membrane markers is observed over time, possibly reflecting maturation of iPNECs similar to that observed in the NEBs of adult lungs (Pan et al., 2006, 2004).
Limitations
The predominant limitation of the protocol is a current inability to purify the iPNECs due to a lack of a cell surface markers that is specific and selective for PNECs. We are continuing to work to identify such a marker which will substantially increase the utility for this protocol for studying disorders involving PNECs. We demonstrate neuropeptide expression using qRT-PCR and ELISA; however additional functional experiments, including a more comprehensive analysis of neuropeptide marker release under hypoxic conditions and mechanical stress or in response to allergic stimulants would be beneficial for establishing iPNEC functional competency for disease modeling.
Troubleshooting
Problem 1
At day 1, the cells do not reach 70%–80% confluence (step 5).
Potential Solution
Leave the cells for another 24 hrs to reach the optimal confluence before starting the differentiation.
Problem 2
At day 3, the cells do not reach 80%–100% confluence (steps 12 and 13).
Potential Solution
If differentiation does not reach the desired >80%, do not continue the differentiation. This is usually an indicator that there was an issue with the quality of the starting iPSC and will result in poor efficiency of differentiation. Ensure that iPSC maintain pluripotency, are karyotypically stable and that all differentiated cells are removed prior to starting the protocol.
Problem 3
Low differentiation at days 5 and 9 as shown by staining for DE and AFE markers (FOXA2 and/or SOX17).
Potential Solution
If the percent DE/AFE conversion is really poor (<60%), do not continue the differentiation as it will not give rise to enough lung progenitors and thereby end up with mixed cell types alongside non-pulmonary lineage contamination. Optimization of plating may be needed or additional days to differentiate to DE.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Amy L. Ryan, afirth@usc.edu.
Materials Availability
Human iPSC lines used in this study are available upon request from Dr. Ichida ichida@usc.edu and the Lead Contact, Dr. Ryan, afirth@usc.edu
Data and Code Availability
We have uploaded and made available our single-cell RNA sequencing data generated from our day 91 differentiation cultures on Gene Expression Omnibus (GEO) database under accession number: GSE146990.
Acknowledgments
We acknowledge the use of the services at the USC Optical Imaging Facility. This work was supported by NIH grants R35HL135747, R00NS077435, and R01NS097850; the New York Stem Cell Foundation; the USC Broad Innovation Award; and the Hastings Foundation. Z.B. is Ralph Edgington Chair in Medicine.
Author Contributions
Conceptualization, P.H., A.L.R., and Z.B.; Methodology, P.H., J.K.I., A.L.R., and Z.B.; Writing – Original Draft, P.H. and A.L.R.; Writing – Review & Editing, P.H., A.L.R., J.K.I., and Z.B.; Funding Acquisition, J.K.I. and Z.B.; Resources, J.K.I., A.L.R., and Z.B.; Supervision, J.K.I., A.L.R., and Z.B.
Declaration of Interests
J.K.I. is a co-founder of AcuraStem Incorporated. J.K.I. declares that he is bound by confidentiality agreements that prevent him from disclosing details of his financial interests in this work.
Contributor Information
Zea Borok, Email: zborok@usc.edu.
Amy L. Ryan, Email: afirth@usc.edu.
References
- Gerovac B.J., Valencia M., Baumlin N., Salathe M., Conner G.E., Fregien N.L. Submersion and hypoxia inhibit ciliated cell differentiation in a notch-dependent manner. Am. J. Respir. Cell Mol. Biol. 2014;51:516–525. doi: 10.1165/rcmb.2013-0237OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke R.M., Meredith D.M., Borromeo M.D., Savage T.K., Johnson J.E. Ascl1 and Neurog2 form novel complexes and regulate delta-like3 (Dll3) expression in the neural tube. Dev. Biol. 2009;328:529–540. doi: 10.1016/j.ydbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor P., Punj V., Calvert B.A., Castaldi A., Miller A.J., Carraro G., Stripp B.R., Brody S.L., Spence J.R., Ichida J.K., Ryan Firth A.L., Borok Z. Efficient generation and transcriptomic profiling of human iPSC-derived pulmonary neuroendocrine cells. iScience. 2020;23:101083. doi: 10.1016/j.isci.2020.101083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnoila R.I. Functional facets of the pulmonary neuroendocrine system. Lab. Invest. 2006;86:425–444. doi: 10.1038/labinvest.3700412. [DOI] [PubMed] [Google Scholar]
- Longmire T.A., Ikonomou L., Hawkins F., Christodoulou C., Cao Y., Jean J., Kwok L.W., Mou H., Rajagopal J., Shen S.S., Dowton A.A., Serra M., Weiss D.J., Green M.D., Snoeck H.-W., Ramirez M.I., Kotton D.N. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.R., Hartman B.H., Ray C.A., Hayashi T., Bermingham-McDonogh O., Reh T.A. Acheate-scute like 1 (Ascl1) is required for normal Delta-like (Dll) gene expression and Notch signaling during retinal development. Dev. Dyn. 2009;238:2163–2178. doi: 10.1002/dvdy.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger T., Burton B., Clark H., Van Goor F. Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre-clinical testing of CFTR modulators. Methods Mol. Biol. 2011;741:39–54. doi: 10.1007/978-1-61779-117-8_4. [DOI] [PubMed] [Google Scholar]
- Pan J., Luk C., Kent G., Cutz E., Yeger H. Pulmonary neuroendocrine cells, airway innervation, and smooth muscle are altered in Cftr null mice. Am. J. Respir. Cell Mol. Biol. 2006;35:320–326. doi: 10.1165/rcmb.2005-0468OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Yeger H., Cutz E. Innervation of pulmonary neuroendocrine cells and neuroepithelial bodies in developing rabbit lung. J. Histochem. Cytochem. 2004;52:379–389. doi: 10.1177/002215540405200309. [DOI] [PubMed] [Google Scholar]
- Gouon-Evans V., Boussemart L., Gadue P., Nierhoff D., Koehler C.I., Kubo A., Shafritz D.A., Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat. Biotechnol. 2006 doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- Van Haute L., De Block G., Liebaers I., Sermon K., De Rycke M. Generation of lung epithelial-like tissue from human embryonic stem cells. Respir. Res. 2009;10:105. doi: 10.1186/1465-9921-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Bear C.E., Chin S., Pasceri P., Thompson T.O., Huan L.-J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao E., Lin C., Wu Q., Zhang K., Song H., Chuang P. Notch signaling controls transdifferentiation of pulmonary neuroendocrine cells in response to lung injury. Stem Cells. 2018 doi: 10.1002/stem.2744. [DOI] [PubMed] [Google Scholar]
- Yamaya M., Finkbeiner W.E., Chun S.Y., Widdicombe J.H. Differentiated structure and function of cultures from human tracheal epithelium. Am. J. Physiol. 1992;262 doi: 10.1152/ajplung.1992.262.6.L713. L713–724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have uploaded and made available our single-cell RNA sequencing data generated from our day 91 differentiation cultures on Gene Expression Omnibus (GEO) database under accession number: GSE146990.