SUMMARY
Organoids are three-dimensional (3D) constructs generated in stem cell cultures and are thought to mimic tissue and organ development in situ. However, until recently, they often exclusively recapitulated the development of the organ`s parenchyma without the major components of the organ stroma. Here, we describe a protocol to incorporate stromal components, first of all blood vessels, by co-culturing with induced pluripotent stem cell-derived mesodermal progenitor cells.
For complete details on the use and execution of this protocol, please refer to Wörsdörfer et al. (2019).
Graphical Abstract

Highlights
-
•
Generation of vascularized neural organoids
-
•
Analysis of tissue morphogenesis
-
•
3D reconstruction of vascular networks
Organoids are three-dimensional (3D) constructs generated in stem cell cultures and are thought to mimic tissue and organ development in situ. However, until recently, they often exclusively recapitulated the development of the organ`s parenchyma without the major components of the organ stroma. Here, we describe a protocol to incorporate stromal components, first of all blood vessels, by co-culturing with induced pluripotent stem cell-derived mesodermal progenitor cells.
BEFORE YOU BEGIN
Timing: 1–3 days
-
1.
Prepare the cell culture media (prepare reasonable amounts of medium and avoid storing for more than 7 days) (see Preparation of Medium)
-
2.
Prepare Matrigel-coated 6-well plates (coated plates can be stored for several days in the fridge, seal plates with Parafilm and make sure that plates don`t dry) (see Culturing iPS Cells)
-
3.
Prepare agarose-coated 96-well plates (we recommend to prepare these plates fresh at the day of use) (see Induction of Neural/Mesenchymal Organoids)
-
4.
Prepare solutions and buffers for paraffin sectioning, tissue clearing and immunofluorescence analyses (see Preparation of Buffers and Solutions)
-
5.
Cool the centrifuge to 4°C
-
6.
Before you start the experiment, take induced pluripotent stem cells (iPSCs) in cell culture and culture them for 2–3 passages. An 80% confluent well of a 6-well plate is sufficient to start the experiment.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sox1 | R&D Systems | AF3369 |
| NG2 | Millipore | AB5320 |
| CD31 | DAKO | M 0823 |
| MAP2 | Abcam | ab32454 |
| TUJ1 | Biolegend | MMS-435P |
| SMA | Santa Cruz | sc-53015 |
| Collagen I | Invitrogen | PA1-26204 |
| Collagen IV | Abcam | ab-6586 |
| Goat anti rabbit Cy2 | Jackson Immuno Research | 111-225-144 |
| Goat anti rabbit Cy3 | Jackson Immuno Research | 111-165-003 |
| Goat anti mouse Cy2 | Jackson Immuno Research | 115-225-146 |
| Goat anti mouse Cy3 | Jackson Immuno Research | 115-165-003 |
| DAPI | Roche | 2360276 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Advanced DMEM | Gibco | 12634028 |
| Neurobasal Medium | Gibco | 21103-049 |
| DMEM/F12 | Gibco | 11320-074 |
| StemMACS iPS Brew XF human | Miltenyi Biotec | 130-104-368 |
| N2-Supplement | Gibco | 17502-048 |
| B27-Supplement (without Vit. A) | Gibco | 12587010 |
| CHIR 99021 | Sigma | SML1046-25MG |
| L-Glutamine | Gibco | G7513 |
| SB 431542 | Miltenyi Biotec | 130-105-336 |
| Dorsomorphin | Miltenyi Biotec | 130-104-466 |
| Purmorphamine | Miltenyi Biotec | 130-104-465 |
| Penicillin/Streptomycin | Gibco | 11548876 |
| BMP4 | PeproTech | 120-05ET |
| Y27632 | Miltenyi Biotec | 130-103-922 |
| Ascorbic acid (Vit.C) | Sigma-Aldrich | A4545 |
| Accutase | Sigma-Aldrich | A6964 |
| BME | Biotechne | 3533-005-02 |
| hESC Matrigel | Corning | 354277 |
| Agarose | Biozym | 840006 |
| Hematoxylin | Chroma | 50837 |
| NaJO3 | Merck | 7412159 |
| KAl(SO4)2 | AppliChem | A2811 |
| C2H3Cl3O2 | AppliChem | A4431 |
| C6H8O7 | AppliChem | A1350 |
| Paraformaldehyde | Sigma-Aldrich | P6148 |
| Eosin Y | AppliChem | A0822 |
| DePeX | Serva | 18243.02 |
| 2-Propanol | Morphisto | REF 11365 |
| 1-Propanol (99.7%, anhydrous) | Sigma-Aldrich | 279544 |
| Ethanol 96% | Nordbrand | Sorte 641 |
| Ethanol 99% | Nordbrand | Sorte 642 |
| Ethyl cinnamate (Eci) | Sigma-Aldrich | W243000 |
| PBS | Sigma-Aldrich | D8537 |
| Mowiol | Sigma-Aldrich | 81381 |
| DAPI | Roche | 10236276001 |
| Experimental Models: Cell Lines | ||
| Induced pluripotent stem cells (iPSCs) | any human pluripotent stem cell line can be used | Kwok et al., 2018 |
| Software and Algorithms | ||
| Fiji | https://fiji.sc/ | n/a |
| NIS Elements Confocal | Nikon | n/a |
| Other | ||
| 6-well plate | Greiner | 657160 |
| 96-well plate, F-bottom | Greiner | 655180 |
| Cryomold | Tissue-Tek® | 4565 |
| PAP-Pen | Abcam | ab2601 |
| Tissue Cassette | Süsse | 100408 |
| Paraffin (Histosec pastilles) | Merck | 1.11609.2504 |
| 2D Rocker 2D series Model 2D basic | Roth | XT46.1 |
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Süleyman Ergün: sueleyman.erguen@uni-wuerzburg.de
Materials Availability
This study did not generate new unique materials or reagents.
Data and Code Availability
This study did not generate any unique datasets or code.
MATERIALS AND EQUIPMENT
Preparation of Medium
CRITICAL: Work sterile under a laminar flow cabinet. Thoroughly pipet all components, especially small molecules and cytokines. Wrong small molecule concentrations can lead to unwanted differentiation results. Use fresh small molecules and cytokines. We recommend to prepare small batches of medium and avoid storing of medium containing small molecules or cytokines for more than 7 days in the fridge. The amount of medium indicated below is enough for two 96-well plates. If you want to prepare less or more organoids adjust the amount of medium accordingly.
Prepare the following stock solutions for small molecules and cytokines and use them to prepare the medium:
BMP4: 50 μg/mL in 4 mM HCl, 0.1% BSA in d H2O
Chir99021: 10 mM in DMSO
Purmorphamine: 10 mM in DMSO
SB431542: 50 mM in DMSO
Dorsomorphin: 10 mM in DMSO
Vitamin C: 25 mg/mL in dH2O
Neural Induction Medium 1 (NIM1)__25 mL
| Ingredients | Final Concentration | Amount |
|---|---|---|
| DMEM- F12 | 50% | 12 mL |
| Neurobasal Medium | 50% | 12 mL |
| B27 without Vitamin A (50x) | 1x | 500 μL |
| N2-Supplement (100x) | 1x | 250 μL |
| L-Glutamine (200 mM) | 2 mM | 250 μL |
| CHIR 99210 (10 mM) | 3 μM | 7.5 μL |
| SB431542 (50 mM) | 10 μM | 5 μL |
| Dorsomorphin (10 mM) | 1 μM | 2.5 μL |
| Purmorphamine (10 mM) | 0.5 μM | 1.25 μL |
Neural Induction Medium 2 (NIM2)__25 mL
| Ingredients | Final Concentration | Amount |
|---|---|---|
| DMEM- F12 | 50% | 12 mL |
| Neurobasal Medium | 50% | 12 mL |
| B27 without Vitamin A (50x) | 1x | 500 μL |
| N2-Supplement (100x) | 1x | 250 μL |
| L-Glutamine (200 mM) | 2 mM | 250 μL |
| CHIR 99210 (10 mM) | 3 μM | 7.5 μL |
| Vitamin C (25 mg/mL) | 0.0625 mg/mL | 62.5 μL |
| Purmorphamine (10 mM) | 0.5 μM | 1.25 μL |
Neural Differentiation Medium (NDM)__200 mL
| Ingredients | Final Concentration | Amount |
|---|---|---|
| DMEM- F12 | 50% | 96 mL |
| Neurobasal Medium | 50% | 96 mL |
| B27 without Vitamin A (50x) | 1x | 4 mL |
| N2-Supplement (100x) | 1x | 2 mL |
| L-Glutamine (200 mM) | 2 mM | 2 mL |
| Vitamin C (25 mg/mL) | 0.0625 mg/mL | 500 μL |
| Penicillin/Streptomycin (100x) | 1x | 2 mL |
Mesodermal Induction Medium (MIM)__25 mL
| Ingredients | Final Concentration | Amount |
|---|---|---|
| Advanced DMEM-F12 | 100% | 25 mL |
| L-Glutamine (200 mM) | 2 mM | 250 μL |
| Vitamin C (25 mg/mL) | 0.0625 mg/mL | 62.5 μL |
| CHIR 99210 (10 mM) | 10 μM | 25 μL |
| BMP4 (50 μg/mL) | 25 ng/mL | 12.5 μL |
Preparation of Buffers and Solutions
Blocking Solution (Immunofluorescence)
| BSA | 4% |
|---|---|
| Triton X-100 | 0.2 % |
| in PBS |
Antibody Solution (Immunofluorescence)
Blocking solution + primary antibody (e.g. CD31 1:200; TUJ1 1:1000; MAP2 1:500; Sox1 1:100)
DAPI Stock Solution (Immunofluorescence)
| DAPI | 5 mg/mL |
|---|---|
| in dH2O |
Mayer`s Hemalum (HE Staining)
| Hematoxylin | 1 g/l |
|---|---|
| NaJO3 | 0.2 g/l |
| KAl(SO4)2 | 50 g/l |
| C2H3Cl3O2 | 50 g/l |
| C6H8O7 | 1 g/l |
| in ddH2O |
Eosin Staining Solution (HE Staining)
| Eosin Y | 1 g/l |
|---|---|
| in ddH2O |
Citrate Buffer (pH 6) (Antigen Retrieval)
| C6H8O7 | 1.8 mM |
|---|---|
| Na3C6H5O7 | 8.2 mM |
| In ddH2O |
Penetration Buffer (Tissue Clearing)
| DMSO | 20 % |
|---|---|
| Glycine | 300 mM |
| Triton X-100 | 0.2 % |
| in PBS |
Blocking Buffer (Tissue Clearing)
| DMSO | 10 % |
|---|---|
| BSA | 6% |
| Triton X-100 | 0.2 % |
| in PBS |
Wash Buffer (Tissue Clearing)
| Tween 20 | 0.2 % |
|---|---|
| in PBS |
Antibody Buffer (Tissue Clearing)
| DMSO | 5 % |
|---|---|
| BSA | 3% |
| Tween 20 | 0.2 % |
| in PBS |
STEP-BY-STEP METHOD DETAILS
Different strategies to achieve neural organoid vascularization were recently published by us and others. Vascularization was achieved by incorporation of endothelial cells (Pham et al., 2018), the transplantation into a mouse brain (Mansour et al., 2018, Daviaud et al., 2018) or the use of genetically altered induced pluripotent stem cells (iPSCs)(Cakir et al., 2019). Our protocol relies on the incorporation of mesodermal progenitor cells (MPCs), which have the potential to differentiate into all cell types of the vascular wall (Wörsdörfer et al., 2019).
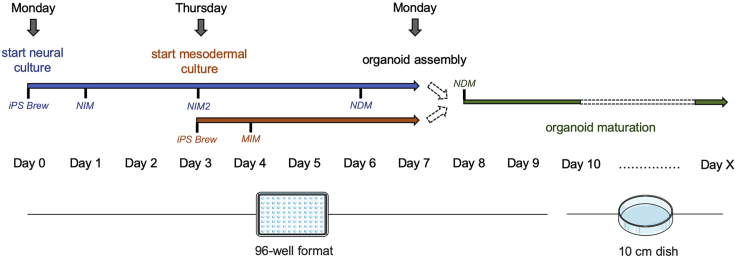
To generate vascularized neural organoids, iPSC-derived neural aggregates are co-cultured with aggregates of iPSC-derived MPCs. It takes 8 days to set up the neuro-mesenchymal co-cultures. These assembled multi-lineage organoid cultures can then be further cultivated for up to 200 days to achieve organoid maturation (Figures 1 and 2). First analyses are usually performed after 20 days in co-culture (day 28).
Figure 1.
Experimental Timing
The figure illustrates the timing of neural and mesenchymal aggregate cultures, neuro-mesenchymal organoid assembly and vascularized neural organoid maturation. It is feasible to start the experiment on a Monday.
Figure 2.
Schematic Depiction of the Workflow
(A) illustrates the generation of agarose-coated 96-well plates and the cell-seeding procedure to achieve iPSC aggregate formation for neural and mesodermal induction.
(B) depicts the induction, assembly, embedding, maturation and validation workflow.
Experimental Timing
-
1.
Generation of neuro-mesenchymal organoids (day 0 - day 7)
-
a)
Prepare neural organoids (day 0 - day 7)
-
b)
Prepare mesodermal organoids (day 3 - day 7)
-
c)
Assemble neural and mesodermal parts (day 7- day 8)
-
2.
Culture neuro-mesenchymal organoids until a vascular network is established (day 8 – day 28)
-
3.
Further culture vascularized organoids to achieve higher organoid maturation (day 28 – day x). Some glial cells (astrocytes, oligodendrocytes) or neuronal subtypes need up to 200 days to arise within the neural organoid.
Culturing iPS Cells
Timing: 3 days
-
4.
Prepare Matrigel-coated 6-well plates. For that purpose, dilute hESC Matrigel in DMEM-F12 using the dilution factor given by the distributor (this can vary from batch to batch depending on the protein concentration) and cover each well with 1 mL Matrigel solution. When working with Matrigel or Matrigel solution keep the Matrigel on ice and chill pipettes to avoid unwanted gelling. For coating, incubate the plates with Matrigel solution either for 17–20 h at 4°C or for 1 h at 20–23°C. Before using the plates for iPS cell culture discard Matrigel solution and cover the well with StemMACS iPS Brew medium. Do not allow the coated plate to dry.
-
5.
Seed 5x105 iPS cells per well in 2 mL StemMACS iPS Brew medium containing 10 μM ROCK inhibitor (Y27632).
-
6.
Culture cells at 5 % CO2 and 20 % O2 in a humidified incubator until they reach approx. 80% confluency (this takes usually 2 to 3 days) (Figure 3A).
Figure 3.
Generation of Neuro-Mesenchymal Organoids
(A) 80% confluent human iPS cells cultured on a Matrigel-coated well of a 6-well plate.
(B) iPSCs 2 h after seeding into an agarose-coated well of a 96-well plate. Note that cells already start to aggregate.
(C) Condensed iPS cell aggregate after 24 h in suspension culture.
(D) Neural organoid at day 7 of induction.
(E) Mesodermal organoid at day 4 of induction.
(F Neuro-mesenchymal organoid 1 day after assembly and prior to BME embedding.
(G) The neural part was generated using GFP-labeled iPSCs and is clearly detectable using fluorescence microscopy
(H) Neuro-mesenchymal organoid embedded in BME 2 days after assembly.
(I) Neuro-mesenchymal organoids after 20 days in co-culture (day 29 in total) in a non-adhesive 6-cm petri dish.
Note: A 80% confluent well of iPSCs is needed for the induction of neural (day 0) as well as mesodermal organoids (day 3)! If iPS cells grow overconfluent, this can influence their differentiation behavior and might negatively impact the outcome of the experiment. Change medium every day.
Induction of Neural Organoids
Timing: 8 days
-
7.
Prepare agarose-coated 96-well plates for suspension culture (Figure 2)
-
a.
Boil 1% agarose powder in water using e.g. a microwave oven
-
b.
Pipette 50 μL warm (approx. 60°C) liquid agarose gel into each well of a F-bottom 96-well plate
-
c.
Let cool down for 30 min until the agarose gel gets solid
Note: The intention of agarose coating is to create a non-adhesive conical mould. It supports the formation of a single cellular aggregate per 96-well. Proper mould formation works best with F-bottom 96-well plates.
-
8.
Prepare iPSC single cell suspension (Figure 2)
-
d.
Discard culture medium from iPSC culture
-
e.
Add 1 mL Accutase per well of a 6-well plate and incubate for 3 min at 37°C
-
f.
Prepare a 15 mL conical centrifuge tube with 5 mL DMEM
-
g.
Add 1 mL DMEM to each well with iPSCs treated with Accutase
-
h.
Mechanically detach the cells by gently pipetting the medium/Accutase solution and transfer the single cell suspension to the prepared 15 mL centrifuge tube.
-
i.
Pellet cells by centrifugation at 300 x g for 3 min at 4°C
-
j.
Count cell number/mL e.g. by using a Neubauer hemocytometer.
-
9.
Pipette 4000 cells/100 μL StemMACS iPS Brew medium containing 10 μM ROCK inhibitor (Y27632) into each well of the agarose-coated 96-well plate (Figure 3B) (day 0). To reduce the effort, prepare medium with cells sufficient for e.g. a complete 96-well plate (ca. 10 mL) and use a multichannel pipette (Figure 2).
-
10.
Culture cells in a humidified incubator (5 % CO2 and 20 % O2)
-
11.
After 24 h change medium to 100 μL NIM
-
12.
Culture for 48 h in NIM (day 1- day 3)
-
13.
Change NIM to 100 μL NIM2
-
14.
Culture for 72 h in NIM2 (day 4- day 6)
-
15.
Change NIM2 to 100 μL NDM
-
16.
Culture for 24 h in NDM (day 6- day 7)
Note: Daily medium changes are not required. Medium is only changed at the time points indicated in the protocol. It is difficult to remove all the cell culture medium from the 96-wells during medium change. Therefore, leave ca. 20 μL medium in the well and subsequently add 100 μL fresh medium. Use a 100 μL pipette or a vacuum aspirator to remove old medium and be careful not to aspirate the cell aggregates. This might need some practice.
CRITICAL: Prepare agarose plates fresh; do not store. The ideal amount of agarose needed (50–100 μL) may vary for 96-well plates from different distributers and should be tested. Use flat bottom 96-well plates. Use fresh small molecules and cytokines. We recommend to prepare small batches of medium and avoid storing of medium containing small molecules for more than 7 days in the fridge.
Note: It is feasible to start neural induction on a Monday.
Induction of Mesenchymal Organoids
Timing: 4 days
-
17.
Prepare agarose-coated 96-well plate as described above
-
18.
Prepare iPSC single cell suspension as described above
-
19.
Pipette 4000 cells/100 μL StemMACS iPS Brew medium containing 10 μM ROCK inhibitor (Y27632) into each well of the agarose-coated 96-well plate (Figure 3B) (day 3). To reduce the effort, prepare medium with cells sufficient for e.g. a complete 96-well plate (ca. 10 mL) and use a multichannel pipette (Figure 2).
-
20.
Culture cells in a humidified incubator (5 % CO2 and 20 % O2) (day 3 – day 4)
-
21.
After 24 h change StemMACS iPS Brew medium to 100 μL MIM (day 4)
-
22.
Culture for 72 h in MIM (day 4 – day 7)
Note: Daily medium changes are not required. Medium is only changed at the time points indicated in the protocol. It is impossible to remove all the cell culture medium from the 96-wells. It is OK to leave ca. 20 μL medium in the well and add 100 μL fresh medium. Use a 100 μL pipette to remove old medium and be careful not to aspirate the cell aggregates. This might need some practice.
CRITICAL: Prepare agarose plates fresh; do not store. The ideal amount of agarose may vary for 96-well plates from different distributers and should be tested. Use flat bottom 96-well plates. Use fresh small molecules and cytokines. We recommend to prepare small batches of medium and avoid storing of medium containing small molecules for more than 7 days in the fridge.
Assembling of Neural and Mesenchymal Aggregates
Timing: 2 days
-
23.
At day 7 of neural induction (Figure 3D) and day 4 of mesenchymal induction (Figure 2E) both organoid types are brought in co-culture (Figures 2B and 3F–3H) (day 7)
-
a.
Discard the NDM from neural organoids
-
b.
Add 100 μL of fresh NDM
-
c.
Discard most of the MIM from mesodermal organoids. Leave ca. 20 μL medium for organoid transfer
-
d.
Use a 1 mL pipette to transfer a single mesodermal organoid into a 96-well already containing a neural organoid
CRITICAL: Transfer the organoids cautiously, avoid damaging/breaking of the organoids. If organoids do not fit into the pipet tip, cut the tip using sterile scissors to create a wider opening
Note: We use ND medium for the culture of assembled organoids because vascular network formation is not negatively impacted under these conditions but neural differentiation is supported.
-
24.
At day 9 the combined organoids are embedded in 25 μL basement membrane extract (BME) to support neural tissue maturation.
-
a.
Thaw BME on ice (this takes approx. 2h)
-
b.
Discard the ND medium from the assembled organoid culture
-
c.
Add 25 μL BME carefully into each well of the 96-well plate to encapsulate the organoids
-
d.
Incubate for 30 min at 37°C (gelling of the BME)
-
e.
Add 100 μL ND medium
-
25.
Culture organoids for 24 h at 37°C in a humidified incubator (5 % CO2 and 20 % O2)
-
26.
Transfer assembled organoids into a 10 cm petri dish (day 10) (Figure 3I):
-
a.
Open the lid of the 96-well plate under the bench and quickly turn the plate upside-down.
-
b.
Tap gently to transfer the organoids into a 15 cm non-adhesive petri dish.
-
c.
Select organoids with clearly visible neural and mesodermal part directly attached to each other (Figure 3H). Use a 1 mL pipette to transfer those organoids to 10 cm petri dishes containing 10 mL fresh ND medium for long-term culture (10–15 organoids per dish). If organoids do not fit into the pipet tip, cut the tip using sterile scissors to create a wider opening
-
d.
Place petri dish with organoids for long-term culture on a 2D-rocking plate (we use a 2D rocker, tilting angle 8°, 15 rpm) in a humidified incubator at 37°C (5 % CO2 and 20 % O2).
CRITICAL: If more than 15 organoids are cultured per 10 cm petri dish, organoids tend to cluster together upon culturing.
-
27.
Culture organoids until they reach the desired maturation stage at 37°C (5 % CO2 and 20 % O2). A dense vascular network establishes already after 10–20 days in co-culture, neural organoid maturation can take more than 100 days (e.g. for the induction of astrocyte, oligodendrocytes or specific neuronal subtypes)
CRITICAL: Long culture time with regular medium changes is required to reach organoid maturation. Work as sterile as possible to avoid contaminations. We usually add Penicillin/Streptomyocin solution to the NDM to further reduce the risk of bacterial growth.
Note: During the maturation phase, a daily medium change is not necessary. We use 10 cm petri dishes with 10–15 organoids each and change medium every other day. The frequency of required medium changes depends on the number of organoids per petri dish and the organoid size (which depends on the time in culture).
CRITICAL: Change medium regularly. Otherwise, organoids start to die or develop large apoptotic core regions. Monitor the pH indicator in the cell culture medium daily. If necessary, increase the frequency of medium changes or reduce the number of organoids per petri dish accordingly.
Analyses of Vascular Network Formation and Organoid Morphology
Analyze vascularized neural organoids at different time points of differentiation (e.g. 20, 50 and 100 days in co-culture) to check for vascular network formation and tissue morphogenesis. In the following paragraph, we will present two suitable methods for organoid assessment: 1) paraffin sections and 2) tissue clearing analyses. Method 1 delivers detailed information about tissue morphogenesis and method 2 provides a 3-dimensional impression of the whole organoid structure and vascular network.
Paraffin Sections and Histological Analyses
Timing: 1 week
Sample Preparation
-
28.
Harvest organoids after the desired culture time (e.g. 20 days in co-culture)
-
29.
Transfer 5–10 organoids into a 2 mL reaction tube and wash 5 min with PBS
-
30.
Replace PBS by 4 % PFA in PBS and incubate over night at 4°C on a mini tube rotator or laboratory rocker
-
31.
Discard the PFA
-
32.
Wash again 3 x 5 min with PBS
-
33.
Whilst washing prepare 1 % agarose gel
-
a.
Boil 1% agarose in water using e.g. a microwave oven
-
b.
Let liquid agarose gel cool down to ca. 60°C
-
34.
Transfer organoids into Tissue-Tek Cryomolds and remove PBS (Figure 4A)
Figure 4.
Paraffin Embedding of Organoids
(A) Organoids placed into a cryomold.
(B) Organoids in cryomold embedded in 1% agarose.
(C) Agarose block with organoids removed from cryomold using a spatula.
(D) Tissue cassette.
(E) Agarose block with organoids embedded in paraffin ready for sectioning.
-
35.
Cover fixed organoids with 250- 500 μL liquid 1 % agarose gel (Figure 4B)
-
36.
Let agarose solution cool down until gel gets solid (ca. 15 min)
-
37.
Remove agarose block containing organoids from the Cryomold (Figure 4C) and place sample in a tissue cassette (Figure 4D).
-
38.
Store in 70% ethanol until further processing
Paraffin Embedding and Sectioning
-
39.
For paraffin embedding use a tissue processor (e.g. Leica TP1020) with the following program
-
a.
70% ethanol for 30 min
-
b.
70% ethanol for 30 min
-
c.
80% ethanol for 1 h
-
d.
96% ethanol for 1 h
-
e.
99% ethanol for 1 h
-
f.
100% 2-propanol for 1h
-
g.
100% 2-propanol for 1h
-
h.
100% 2-propanol for 1h
-
i.
99.9% Xylol for 1h
-
j.
99.9% Xylol for 1h
-
k.
99.9% Xylol for 1h
-
l.
Paraffin for 3h
-
m.
Paraffin for 3h
-
40.
Use a heated paraffin embedding station to embed samples into a paraffin block (Figure 4E)
-
41.
Cut 5 μm sections using a sliding microtome (we use a Leica SM2010 R)
-
42.
Transfer sections on microscope slides (we use SuperFrost Microscope Slides, Thermo Scientific)
-
43.
Let sections dry 17–20 h at 37°C in a lab oven
Note: Cryosectioning can be performed as well, however, we strongly recommend paraffin sectioning as it better preserves morphological details.
Deparaffination
-
44.
Place the specimen slides in a vertical staining cuvette.
-
45.
Deparaffination is achieved by incubation in a descending alcohol series
-
a.
2x 10 min 99.9% Xylol
-
b.
2x 5 min 99 % EtOH
-
c.
5 min 96 % EtOH
-
d.
5 min 80 % EtOH
-
e.
5 min 70 % EtOH
-
f.
5 min dH2O
-
46.
For heat-induced antigen retrieval, an acidic buffer (10 mM citric acid buffer (pH 6)) is used
-
g.
Place the slides in a vertical staining cuvette and cover with citrate buffer. Heat up using a microwave oven until the buffer is boiling, cool down for 1 min
-
h.
Repeat 6 times a 10 s boiling phase followed by 1 min cool down
-
i.
Last cool down for 30 min
-
47.
Wash 5 min with running dH2O
-
48.
Wash 3x 5 min with PBS
-
49.
Proceed with immunofluorecence analyses
Note: Antigen retrieval is required for immunofluorescence analyses but not for HE staining. Heat-induced antigen retrieval with citric acid buffer works well for the antibodies suggested in this protocol. However, if you use alternative antibodies these may require other antigen retrieval methods or buffers. Recommendations are usually provided by the distributor.
HE Staining and Immunofluorescence Analyses
To analyze organoid tissue morphology, histological stainings and immunofluorescence analyses can be performed. We recommend to perform HE staining to get a general idea of the tissue morphology (Figures 5A and 5B) and immunofluorescence analyses to detect specific structures (Figures 5C–5F). Recommended antibodies are CD31 for the detection of endothelial cells, SMA or NG2 marking peri-endothelial cells, Collagen I and Collagen IV antibodies detecting extracellular matrix, MAP2 or TUJ1 marking neurons and Sox1 for the detection of neural stem cells (Figures 5C–5F). A list of recommended antibodies can be found in the Key Resources Table.
Figure 5.
HE-Stainings and Immunofluorescence Analyses on Organoid Paraffin Sections
(A and B) HE-stainings. (A) provides an overview and (B) highlights details in higher magnification. The mesenchymal and the neural part are clearly distinguishable by their characteristic morphology.
(C–F) Immunofluorescence analyses of organoid sections (day 20 of neuro-mesenchymal co-culture) detecting (C) β3-Tubulin (TUJ1) and Collagen I (Col I), (D) CD31 and MAP2, (E) CD31 and smooth muscle actin (SMA), and (F) collagen IV (Col IV) and CD31.
HE Staining
-
50.
Place the specimen slides in a staining cuvette.
-
51.
Incubate 10 min in Mayer`s hemalum
-
52.
Rinse with dH2O
-
53.
Rinse 10 min under tap water
-
54.
Rinse with dH2O
-
55.
Incubate 10 min with Eosin stain
-
56.
Rinse with dH2O
-
57.
Incubate in 96% Ethanol for 2 min
-
58.
Incubate twice in 99% Ethanol for 5 min each
-
59.
Incubate twice in 99.9% Xylol for 5 min each
-
60.
Mount using an appropriate mounting medium (e.g. DePeX or Entellan)
Immunofluorescence Analyses
Optional: Encircle the organoid sections using a PAP-Pen to save antibodies
-
61.
Incubate specimens in blocking buffer for 1 h at 20–23°C
-
62.
Whilst blocking buffer is incubating prepare the antibody solution in blocking buffer (e.g. for the detection of endothelial cells and neurons we use CD31 antibody (DAKO, M0823, dilution: 1:200) and MAP2 antibody (abcam, ab32454, dilution: 1:1000))
Note: More recommended antibodies can be found in the Key Resources Table.
-
63.
Replace blocking buffer with antibody solution and incubate at 4°C for 17–20 h in a humidified chamber
-
64.
The next day, remove primary antibody solution and wash specimen 3x 5 min with PBS
-
65.
Meanwhile prepare the secondary antibody solution in PBS (e.g. goat anti mouse Cy3 (1:600) and goat anti rabbit Cy2 (1:300))
-
66.
Incubate with secondary antibody solution for 1 h at 20–23°C
-
67.
Discard secondary antibody solution and wash 3x with PBS
-
68.
Incubate specimen with DAPI staining solution (DAPI stock solution 1:5000 in PBS) for 10 min at 20–23°C
-
69.
Discard DAPI staining solution and wash 3x 5 min with PBS
-
70.
Mount the sections in Mowiol or another suitable mounting medium and cover with a coverslip
-
71.
Analyze and document specimens under the fluorescence microscope
Tissue Clearing to Visualize Vascular System
Timing: 1 week
Tissue clearing results in completely transparent organoids for microscopic analyses (Figure 6A). To highlight specific structures of interest, such as endothelial networks, whole-mount immunostaining with specific antibodies (e.g. CD31) is performed (Figures 6B–6D, Video S1).
-
72.
Harvest organoids after the desired culture time (e.g. 20 days in co-culture)
-
73.
Transfer organoids into a 2 mL reaction tube and wash 5 min with PBS
-
74.
Replace PBS by 4 % PFA and incubate 17–20 h at 4°C
-
75.
Wash organoids for 1 h in PBS
-
76.
Wash organoids 30 min in PBS
-
77.
Incubate in 50 % MeOH (in PBS) for 30 min
-
78.
Incubate in 80 % MeOH (in PBS) for 30 min
-
79.
Incubate in 100 % MeOH for 30 min
-
80.
Add fresh 100 % MeOH and incubate organoids 17–20 h at 4°C
-
81.
Make organoids accessible for antibodies
-
a.
Wash twice with 20 % DMSO (in MeOH) for 30 min
-
b.
Wash with 80 % MeOH (in PBS) for 30 min
-
c.
Wash with 50 % MeOH (in PBS) for 30 min
-
d.
Wash with PBS for 30 min
-
e.
Wash twice with 0.2 % Triton X-100 (in PBS) for 30 min
-
f.
Incubate organoids in penetration buffer 17–20 h at 37°C
-
82.
Blocking of non-specific antibody binding sites
-
a.
Incubate organoids in blocking buffer 17–20 h at 37°C
-
83.
Primary antibody treatment
-
a.
Wash twice with washing buffer for 1 h each
-
b.
Incubate organoids with 100–200 μL primary antibody (to detect vascular networks we use CD31 antibody (DAKO, M0823) in a 1:200 dilution) in antibody buffer for at least 24 h at 37°C
-
84.
Secondary antibody treatment and nuclear staining
-
a.
Wash 10 times with washing buffer for 30 min each
-
b.
Incubate organoids in 100–200 μL antibody buffer + secondary antibodies + DAPI (1 μg/mL) for 17–20 h at 37°C
-
85.
Dehydration
Note: Adjust pH of each solution to 9–9.5 with Triethylamine
-
a.
Wash 3 times with washing buffer for 30 min
-
b.
Wash 3 times with 30 % 1-Propanol (in dH2O, pH 9–9.5) for 30 min
-
c.
Wash 3 times with 50 % 1-Propanol (in dH2O, pH 9–9.5) for 30 min
-
d.
Wash 3 times with 70 % 1-Propanol (in dH2O, pH 9–9.5) for 30 min
-
e.
Wash 3 times with 100 % 1-Propanol (99.7 % anhydrous, pH 9–9.5) for 30 min
-
f.
Remove remaining 1-Propanol with a pipette and the rest with a filter paper
-
86.
Incubate organoids in ethyl cinnamate for at least 1 h before microscopy
-
87.
Store organoids in an Eppendorf tube with ethyl cinnamate at 4°C protected from light
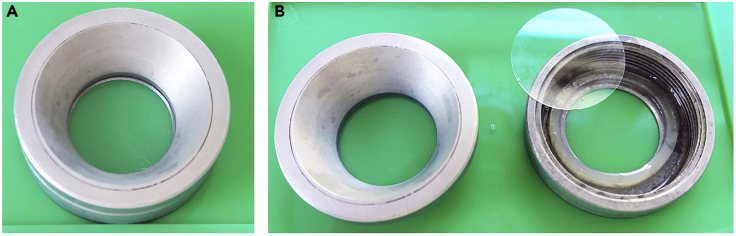
Note: All incubation steps are performed on a mini tube rotator or laboratory rocker. If not otherwise stated, incubation is at 20–23°C. Steps with 17–20 h incubation time can be also performed over the weekend, but always at 4°C. After starting the secondary antibody incubation, keep organoids protected from light. Once the organoids are transferred in ethyl cinnamate, they can be stored for several weeks at 4°C in the dark until microscopy. We use a custom-made glass bottom imaging chamber (Figure 7). Similar imaging chambers are also commercially available (e.g. Attofluor Cell Chamber, Invitrogen or CytoVista imaging chamber, Invitrogen). We use a confocal laser scanning microscope (Nikon Eclipse Ti) with long working distance air objectives (4x, 20x) for taking z-stack images. Fiji (ImageJ) or the Nikon NIS Elements Confocal software are used for 3-D reconstruction of imaged organoids.
Figure 6.
Tissue-Cleared Organoids for Vascular Network Assessment
(A) Representative organoids before and after tissue clearing.
(B) Maximum intensity projection of z-stack images from a cleared organoid (day 20 of neuro-mesenchymal co-culture) in low magnification. The endothelial network is detected using CD31 antibodies (red). Neural cells were derived from GFP-labeled iPSC line to highlight the neural part (green).
(C) This image shows only the CD31+ endothelial network.
(D) Maximum intensity projection of z-stack images of a cleared organoid in higher magnification highlighting a neuroepithelial structure with ventricle-like lumen surrounded by a CD31+ endothelial network.
Figure 7.
Custom-Made Imaging Chamber
(A) Assembled custom-made imaging chamber with glass bottom for imaging of cleared organoids under the confocal laser scanning fluorescence microscope.
(B) Disassembled imaging chamber consisting of top and bottom part (steel) with fine thread, seal ring and glass coverslip.
The endothelial network within a vascularized neural organoid was visualized using immunofluorescence analyses with a specific antibody targeted against CD31 (red). The neural aggregate was generated from a GFP-labeled iPSC line (green). A whole-mount staining was performed and the tissue was subsequently cleared using ethyl cinnamate. The video shows a 3D-reconstruction of a confocal z-stack.
EXPECTED OUTCOMES
This protocol should result in neuro-mesenchymal organoids. The organoids usually display a diameter of approximately 3 mm after 20 days in co-culture and are visible with the naked eye. Due to their size, apoptotic areas, especially within the neural part, are frequently observed as earlier reported for cerebral organoids (Lancaster et al., 2013). A vascular network should form throughout the mesenchymal part of the assembled organoid co-culture within the first 10–20 days. These vessels usually form a dense plexus at the neuro-mesenchymal interface reminiscent of the peri-neural plexus found during normal embryonic development (Hogan et al., 2004). From this plexus, endothelial sprouts start to infiltrate and vascularize the neural part of the organoid. This process starts around day 10–20 in co-culture. It is not necessary to add additional cytokines such as VEGF to the culture as neural stem cells intrinsically produce VEGF, even under normoxic conditions (Hogan et al., 2004, Roitbak et al., 2011). The forming vessel wall consist of an endothelial lining with typical ultra-structure, a Collagen IV-positive basement membrane as well as smooth muscle actin- and NG2-positive peri-endothelial cells (smooth muscle cells and/or pericytes) being recruited into the vascular wall (Wörsdörfer et al., 2019, Figures 5E and 5F).
LIMITATIONS
The protocol is robustly reproducible and delivers similar-sized neuro-mesenchymal organoids with a dense vascular network within the mesenchyme and a partial vascularization of the neural part. But, we observe a certain variability in the density and pattern of the vascular network and in the extent of endothelial sprouting into the neural part. The vessels show an endothelial layer, a basement membrane and peri-endothelial mural cells. However, the vascular network is not connected to a circulatory system. For that reason, the final maturation of the vessel wall cannot happen in this model, as it requires mechanical forces generated by the pulsatile blood flow. A possibility to overcome this issue would be the transplantation of these pre-vascularized organoids e.g. into a mouse brain to connect them to the hosts circulation (Cakir et al., 2019) or a connection of the organoid vessels to a microfluidic system (Sobrino et al., 2016).
TROUBLESHOOTING
Problem
Sometimes, iPSCs form several small aggregates per 96-well instead of a single aggregate.
Potential Solution
Optimize agarose coating. The amount of agarose and the type of 96-well plate determine the shape and depth of the forming agarose mould. We achieve best results with F-bottom 96-well plates (Greiner, 655180) and 50 μL agarose. For other 96-well plates the ideal amount of agarose should be tested. U-bottom plates did not work well in our hands and are not recommended.
Problem
Sometimes, several organoids cluster together during maturation culture.
Potential Solution
The BME embedding is gradually lost during maturation culture and different organoids can now cluster together. To avoid this, do not culture more than 10–15 organoids per 10 cm petri dish. Do not use an orbital shaker or rocker with three-dimensional tumbling motion as this collects all organoids in the center of the petri dish. We use a 2D cell culture rocker (Roth, XT46.1) with a tilting angle of 8° set to 15 rpm. If you use another device try to vary speed and/or tilting angle.
Problem
Sometimes, unwanted structures/tissues arise especially within the mesenchymal part during differentiation. These can be e.g. gastrointestinal epithelia.
Potential Solution
Always use fresh small molecules and avoid long-term storage of small molecules and cytokines in the fridge. Prepare medium fresh for each experiment and do not store medium containing small molecules for more than 1 week. These additives direct the differentiation process and ineffective small molecules/cytokines result in unwanted differentiation results. Moreover, high iPSC passage numbers or overconfluent cultures can lead to aberrant differentiation results. Check the used iPSC lines for pluripotency marker expression and correct karyotype before starting the experiment. Test frequently for possible mycoplasma contaminations.
Problem
A certain degree of apoptosis is normal as soon as organoids reach a diameter of more than 500 nm. However, sometimes organoids develop very large apoptotic areas, especially within the neural part.
Potential Solution
Change medium regularly to assure sufficient supply with nutrients and growth factor as well as ideal pH. Monitor the pH indicator in the medium and change medium as soon as it turns orange. When organoids are cultured for several weeks they get larger and consume more medium. For long-term culture, reduce the number of organoids per 10 cm dish (e.g. from 10 to 5 organoids/dish).
ACKNOWLEDGMENTS
We thank Heike Arthen, Doris Dettelbacher-Weber, Martina Gebhardt, Erna Kleinschroth, Ursula Roth, and Elke Varin for excellent technical assistance and all members of the Stem Cell and Regenerative Medicine Group for their support. We thank the Comprehensive Heart Failure Center Würzburg (CHFC Würzburg) and the German Research Foundation (DFG) for financial support by the Collaborative Research Center SFB688 TP A19 and TR225-B04. The work was supported by the IZKF-Würzburg (Interdisziplinäres Zentrum für Klinische Forschung der Universität Würzburg). This publication was funded by the German Research Foundation (DFG) and the University of Würzburg in the funding programme Open Access Publishing. Schematic representations of the workflow were produced using graphical elements taken from the image bank of Servier Medical Art (www.servier.com) licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0).
AUTHOR CONTRIBUTIONS
P.W. and S.E. conceived the original idea. P.W., A.R. and A.K. provided images and compiled figures. P.W., A.R., Y.A. and S.E. wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2020.100041.
Contributor Information
Philipp Wörsdörfer, Email: philipp.woersdoerfer@uni-wuerzburg.de.
Süleyman Ergün, Email: sueleyman.erguen@uni-wuerzburg.de.
References
- Cakir B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.J., Chapeton K., Patterson B., Yuan Y., He C.S. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviaud N., Friedel R.H., Zou H. Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro. 2018;5 doi: 10.1523/ENEURO.0219-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan K.A., Ambler C.A., Chapman D.L., Bautch V.L. The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development. 2004;131:1503–1513. doi: 10.1242/dev.01039. [DOI] [PubMed] [Google Scholar]
- Kwok C.K., Ueda Y., Kadari A., Günther K., Ergün S., Heron A., Schnitzler A.C., Rook M., Edenhofer F. Scalable stirred suspension culture for the generation of billions of human induced pluripotent stem cells using single-use bioreactors. J. Tissue Eng. Regen. Med. 2018;12:e1076–e1087. doi: 10.1002/term.2435. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A.A., Gonçalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M.T., Pollock K.M., Rose M.D., Cary W.A., Stewart H.R., Zhou P., Nolta J.A., Waldau B. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitbak T., Surviladze Z., Cunningham L.A. Continuous expression of HIF-1alpha in neural stem/progenitor cells. Cell. Mol. Neurobiol. 2011;31:119–133. doi: 10.1007/s10571-010-9561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrino A., Phan D.T., Datta R., Wang X., Hachey S.J., Romero-López M., Gratton E., Lee A.P., George S.C., Hughes C.C. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörsdörfer P., Dalda N., Kern A., Krüger S., Wagner N., Kwok C.K., Henke E., Ergün S. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci. Rep. 2019;9:15663. doi: 10.1038/s41598-019-52204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The endothelial network within a vascularized neural organoid was visualized using immunofluorescence analyses with a specific antibody targeted against CD31 (red). The neural aggregate was generated from a GFP-labeled iPSC line (green). A whole-mount staining was performed and the tissue was subsequently cleared using ethyl cinnamate. The video shows a 3D-reconstruction of a confocal z-stack.
Data Availability Statement
This study did not generate any unique datasets or code.