Summary
This protocol uses endonuclease-dead, programmable RNA-guided RNA-targeting Cas13 RNases (d)Cas13 proteins fused with fluorescent proteins to visualize and track RNA dynamics in live cells. This protocol details several aspects of the procedure, including gRNA design, fluorescent protein selection, nuclear localization signal adjustment, raw data analysis, operation steps, and extended optional applications that have been successfully applied in the visualization of NEAT1, SatIII, MUC4, and GCN4 RNAs.
For complete information on the use and execution of this protocol, please refer to Yang et al. (2019).
Graphical Abstract
Highlights
-
•
CRISPR-dCas13b is a user-friendly system for RNA imaging
-
•
Tracking RNA with dPspCas13b and single gRNAs in living cells
-
•
RNA-RNA/DNA labeling by orthogonal dCas13bs or combined with dCas9
This protocol uses endonuclease-dead, programmable RNA-guided RNA-targeting Cas13 RNases (d)Cas13 proteins fused with fluorescent proteins to visualize and track RNA dynamics in live cells. This protocol details several aspects of the procedure including gRNA design, fluorescent protein selection, nuclear localization signal adjustment, raw data analysis, operation steps, and extended optional applications that have been successfully applied in the visualization of NEAT1, SatIII, MUC4, and GCN4 RNAs
BEFORE YOU BEGIN
Design and Clone gRNAs into Vectors
Timing: 3–4 days
-
1.
Design appropriate targeting gRNAs; we recommend using crispor.tefor.net for preliminary selection of gRNA sequences with high specificity. The gRNA used for dPspCas13b and dPguCas13b does not require a specific protospacer flanking site. But we recommend to select the gRNA spacer with a guanine (G) in the 5′ end that will avoid introduction of a mismatch while adding an extra G to ensure the transcription efficiency. Further, the length of gRNA spacer between 20–27 nucleotides is recommended. Of note, among all examined lengths of gRNAs for NEAT1, the best gRNA lengths for labeling depend on the targeted regions on NEAT1, while the shorter ones generally work better. We typically try three gRNAs for an abundant RNA of interest in HeLa cells.
Note: To avoid potential effect of CRISPR-dCas13 on targeting RNA localization and processing, do not design gRNAs that target functional sequence (such as known functional motifs or modules of an RNA of interest); perform RNA FISH of the same endogenous RNA in wild type cells as a reference; and perform co-staining of CRISPR dCas13-labeld RNA and FISH of the same endogenous RNA in cells to confirm signals by CRISPR-dCas13.
-
2.
Clone each gRNA into expressing vectors containing antibiotic-resistance open reading frame.
Note: Use dPspCas13b preferentially for single RNA visualization and tracking. There is no antibiotic-resistance open reading frame contained in the gRNA expressing vectors in our previous study (Yang et al., 2019), but an antibiotic-resistance containing vector will benefit the following antibiotic selection. Meanwhile, including several gRNAs in one expressing vectors as a multiplex is optional since co-expression of several gRNAs might benefit the targeting efficiency.
Generate Appropriate dCas13-FPs-NLS into Vectors
Timing: 3–4 days
-
3.
In the meantime, select appropriate copy numbers of fluorescent protein (FP) and nuclear localization signal (NLS) and clone them into expressing vectors containing antibiotic-resistance open reading frame. Based on the visualization of NEAT1, SatIII, MUC4 and GCN4 RNAs in live cells (Yang et al., 2019), we recommend dPspCas13b-3×EGFP-2×NLS for labeling nuclear RNAs, dPspCas13b-3×sfGFP-3×NLS or dPspCas13b-2×mNeonGreen-NLS for labeling cytoplasmic RNAs.
Note: It is not absolute necessary to use these three combinations of FPs and NLS to label nuclear or cytoplasmic RNAs. Different types and copies of FP and NLS could be tested for different RNAs and combinations of FP and NLS for dPguCas13b should be test separately. Meanwhile, plasmids including dPspCas13b and dPguCas13b proteins fused with FPs used in this protocol are capable of packaging lentivirus, which is optional to improve dCas13b-FP expression level.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine 3000 Reagent | Thermo | Cat#: 21341 |
| DPBS | Gibco | Cat#: 14190136 |
| FluoroBrite™ DMEM | Thermo | Cat#: A1896701 |
| Dulbecco’s Modified Eagle medium (DMEM) | Gibco | Cat#: 11965118 |
| Fetal Bovine Serum (FBS) | Gibco | Cat#: 10099141 |
| Opti-MEM | Gibco | Cat#: 31985070 |
| Experimental Models: Cell Lines | ||
| HeLa | ATCC | Cat#: CCL-2 |
| Recombinant DNA | ||
| pCMV-VSV-G | Addgene | Cat#: 8454 |
| pMD2.G | Addgene | Cat#: 12259 |
| PspCas13b crRNA backbone | Addgene | Cat#: 103854 |
| PguCas13b crRNA backbone | Addgene | Cat#: 103853 |
| dCas9 sgRNA backbone | Addgene | Cat#: 75390 |
| pHAGE-dPspCas13b-3×EGFP-2×NLS-IRES-puro | Addgene | Cat#: 132397 |
| pHAGE-dPspCas13b-3×sfGFP-3×NLS-IRES-puro | This paper | N/A |
| pHAGE-dPspCas13b-2×mNeonGreen-NLS-IRES-puro | Addgene | Cat#: 132403 |
| pHAGE-dPspCas13b-3×mRuby3-2×NLS | This paper | N/A |
| pHAGE-dPguCas13b-3×mRuby3-2×NLS | This paper | N/A |
| pHAGE-TO-dCas9-mEmerald | This paper | N/A |
| Software and Algorithms | ||
| Fiji/ImageJ | Fiji/ImageJ | https://imagej.net/Fiji/ |
| GraphPad Prism 8 | GraphPad Software | http://www.graphpad.com/scientific-software/prism/ |
| gRNAs Design Tool | CRISPR DESIGN | http://crispor.tefor.net |
| Office365 | Microsoft | https://www.office.com |
| MATLAB | MathWorks | https://www.mathworks.com |
| softWoRx 6.5 | GE Healthcare | N/A |
| Other | ||
| DeltaVision Elite imaging system | GE Healthcare | N/A |
| 35 mm no.1.5 glass-bottomed dishes | Cellvis | Cat#: D35-2001.5-N |
MATERIALS AND EQUIPMENT
Alternatives: We used Lipofectamine 3000 Reagent (Thermo) to perform transfection of plasmids, DeltaVision Elite imaging system (GE Healthcare) coupled with suites of live cells imaging system to obtain imaging data, but equivalent transfection method and imaging system can be used.
STEP-BY-STEP METHOD DETAILS
CRISPR-dCas13-FP/gRNA Delivery Method
Timing: 2 days
The purpose of this step is to deliver expression vectors including gRNA and dPspCas13b-FP into cells. The transfected cells followed by selection with antibiotics (puromycin in this protocol) is optional if the transfection level is low.
Note: A dozen gRNAs targeting NEAT1, SatIII, MUC4 and GCN4 have been tested with dPspCas13b (Yang et al., 2019).
-
1.
Plate cells in 12-well plates at approximately 30% confluency.
-
2.
Transfect dPspCas13b-FP and gRNA expressing vectors.
Note: Cell density should reach 70%–80% confluency.
CRITICAL: Plasmids transfection into cells should be performed within 24 h after cell plating to guarantee the cells are actively dividing.
-
3.
Recover Lipofectamine 3000 Reagent and Opti-MEM medium to 25°C.
-
4.
Dilute 2 μL Lipofectamine 3000 in 50 μL Opti-MEM and mix well with a pipette.
-
5.
Dilute 2 μL P3000, 0.7 μg gRNA and 0.3 μg dPspCas13b-FP plasmids in 50 μL Opti-MEM and mix well with a pipette.
CRITICAL: We recommend labeling NEAT1 with dPspCas13b in HeLa cells as a positive control due to the high expression and available proteins for positive signal confirmation (e.g., NONO, SFPQ, TDP43) when first set up CRISPR-dCas13b-FP labeling system in live cells. Additionally, mRNA MUC4 is another optional positive control for labeling cytoplasmic RNAs.
CRITICAL: At step 5, we suggest the ratio of each gRNA plasmid and dPspCas13b-FP plasmid above 2:1. In our previous study (Yang et al., 2019), we transfected 0.2 μg dPspCas13b-EGFP, 0.4 μg g5′-1 and 0.4 μg g5′-2 (targeting NEAT1) to label NEAT1 for cells in 12-well plate. Researchers could adjust the amount of transfected plasmids according to the requirements. In addition, to control artefactual and false-positive signal labeled by CRISPR-dPspCas13b, the positive gRNA control to label NEAT1 (available) and negative control (non-targeting gRNA) should be tested simultaneously.
Alternatives: Beyond single RNA labeling and tracking, application of orthogonal dPspCas13b and dPguCas13b on dual-color imaging of different transcripts as well as combination of dPspCas13b and nuclease-deficient Cas9 (dCas9, dSpCas9 used in this protocol) for simultaneous visualization of genomic DNA and RNA transcripts are optional.
-
•
At step 4, dilute 4 μL Lipofectamine 3000 in 50 μL Opti-MEM and mix well with a pipette.
-
•
At step 5, for dual-color RNA-RNA imaging, dilute 4 μL P3000, 0.3 μg dPspCas13b-FP, 0.7 μg gRNA (Psp), 0.3 μg dPguCas13b-FP and 0.7μg gRNA (Pgu) plasmids in 50 μL Opti-MEM and mix well with a pipette (dPspCas13b-3×EGFP-2×NLS-IRES-puro, gNEAT1 and dPguCas13b-3×mRuby3-2×NLS, gSatIII for NEAT1, SatIII labeling are used in this protocol); for dual-color DNA-RNA labeling, dilute 4 μL P3000, 0.3 μg dPspCas13b-FP, 0.7 μg gRNA, 0.3 μg dCas9-FP and 0.7 μg sgRNA (dPspCas13b-3×mRuby3-2×NLS, gMUC4/gSatIII and dCas9-Emerald, sgMUC4/sgSatIII for MUC4/SatIII DNA and RNA labeling are used in this protocol) in 50 μL Opti-MEM and mix well with a pipette.
-
6.
Add diluted plasmids to diluted P3000 reagent at 1:1 ratio and mix well with a pipette.
-
7.
Incubate for 15 min at 25°C (or room temperature).
-
8.
Add DNA-lipid complex dropwise and evenly into each well of the 12-well plates.
-
9.
Incubate cells for 8–12 h and refresh the medium (refresh the DNA-lipid containing medium will avoid the lipid toxicity), followed by plating into glass bottom dishes for microscopic observation.
Note: Summary of recommended plasmids usage for different applications of CRSIPR-dCas13b on living cell imaging is provided in Table 1.
Table 1.
Summary of Recommended Plasmids Usage for Different Applications of CRSIPR-dCas13b on Living Cell Imaging
| Assay | Plasmids Usage |
|---|---|
| Single RNA labeling and tracking | 0.3 μg dPspCas13b-FP, 0.7 μg gRNA (Psp) |
| Dual-color imaging of different RNA | 0.3 μg dPspCas13b-FP, 0.7 μg gRNA (Psp), 0.3 μg dPguCas13b-FP, 0.7μg gRNA (Pgu) |
| Dual-color imaging of genomic DNA and RNA | 0.3 μg dPspCas13b-FP, 0.7 μg gRNA (Psp), 0.3 μg dCas9-FP, 0.7 μg sgRNA |
Optional: In order to increase the proportion of cell transfection, antibiotic selection is optional after step 8.
Note: If take step of antibiotic selection, perform steps 10–12 listed below before transfection of dPspCas13b-FP and gRNA expressing vectors.
-
10.
Plating cells in 24-well plates at approximately 30% confluency.
-
11.
Add increasing amounts of antibiotics (e.g., 0–10 μg puromycin in this protocol) to duplicate wells of cells plated in culture medium.
-
12.
Observe the cells under a light microscope after 24 h and choose appropriate concentration of the antibiotic (the duration of culture may be extended, depending on the cell line used).
-
13.
Perform transfection of dCas13-FP and gRNA vectors according to steps 1–8.
-
14.
Incubate cells for 8–12 h and refresh the DNA-lipid containing medium.
-
15.
Change the medium to fresh culture medium supplemented with antibiotic (1 μg/mL puromycin for HeLa in this protocol) after 24 h.
-
16.
Select for 24–48 h.
-
17.
After transfection and selection, collect the cells and passage into glass bottom dishes for the following microscopic observation.
Note: Antibiotic selection should be performed when the transfection efficiency is low. In our previous study (Yang et al., 2019), gRNA expression vectors do not contain antibiotic resistance while dPspCas13b vectors do, thus, antibiotic selection is optional to select co-transfected cells. However, do not select for more than 2 days since transiently transfected plasmids are usually lost when cells divide. Additionally, package dPspCas13-FP into lentivirus for infection is also optional for some cell lines that are difficult to achieve desired level of protein expression by transfection.
Preparing Cells for Microscopy
Timing: 1 day
The purpose of this step is to prepare cells for observation under the microscope.
-
18.
After 24 h transfection (or antibiotic selection), cells are transferred into 35 mm No.1.5 glass-bottomed dishes (Cellvis) at 50%–70% confluency.
-
19.
Wash cells with DPBS and exchange the medium with fresh FluoroBrite™ DMEM containing with 10% FBS (prewarmed to 37°C before used) while the cells confluency reaches 80%–100%, and place back in the incubator for 1 h before observation by microscope.
-
20.
Equilibrate the microscope to 37°C and 5% CO2.
-
21.
Make sure cameras are aligned prior to image acquisition, if necessary, align using a multicolor calibration slide.
-
22.
Identify suitable cells for imaging using dPspCas13b-FP channel.
-
23.
Image cells in the dPspCas13b-FP channel using suitable parameters according to each used microscope of application, and 50%T, 0.2 second and 0.2 μm z-step are set on DeltaVision Elite imaging system (GE Healthcare) in this protocol.
-
24.
Image processing based on used microscope, deconvolution plugin with softWoRx 6.5 is used in this protocol.
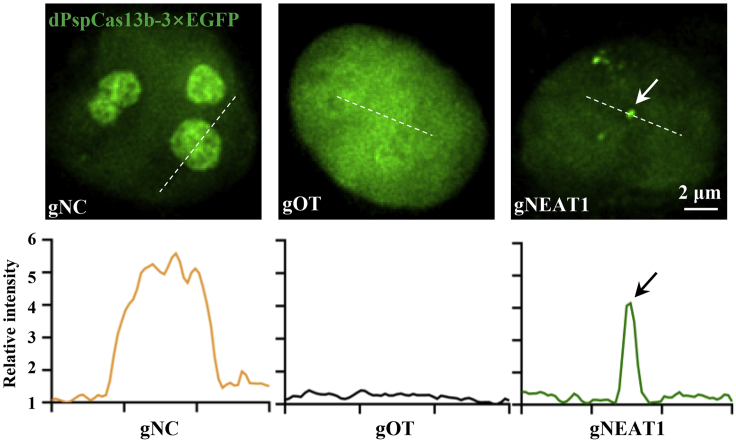
CRITICAL: In our experience, there are different FP expression and distribution patterns in cells under microscopic observation. For example, dPspCas13b-EGFP usually aggregates in the nucleolus with gNC (guide non-targeting control) (Yang et al., 2019). Importantly, however, off-target gRNA can lead to an evenly redistribution of dPspCas13b-EGFP in the nucleus, and effectual gRNA targeting NEAT1 produces reliable NEAT1 (paraspeckle) signals (Figure 1) (Yang et al., 2019).
Figure 1.
Examples of dPspCas13b-FP Distribution Patterns with Different gRNAs
Top, representative images of dPspCas13b-3×EGFP expressed cells with different gRNAs: gNC (guide non-targeting control), gOT (off-target gRNA), gNEAT1 (NEAT1 targeting gRNA). Bottom, line scans showing the relative intensity of the dotted lines in corresponding images. Arrows indicate the NEAT1 positive signal.
CRITICAL: A high level of dPspCas13b-EGFP expression generally introduces high background. Thus, the expression level of dPspCas13b-EGFP in the examined cells should be considered; its comparably lower expression as well as its uniform distribution in each nucleus (such as gNEAT1 panel in Figure1) should be observed preferentially.
EXPECTED OUTCOMES
In our experience, about 50% transfected HeLa cells show even distribution of dPspCas13b-EGFP. For positive signals of the interested RNAs (e.g., NEAT1, SatIII, MUC4), we set the criteria as signal to noise ratio (SNR) over 3. The RNAs of interest with an aggregation localization (e.g., NEAT1) or repeated sequence elements (e.g., SatIII, MUC4) are usually easy to be labeled by CRISPR-dPspCas13b. There is a positive correlation between the labeling efficiency/quality and the transfection efficiency.
QUANTIFICATION AND STATISTICAL ANALYSIS
All fluorescence imaging data is analyzed by Fiji/Image J.
There are two indicators to evaluate whether this CRISPR-dPspCas13b mediated RNA imaging in live cell work well: co-localization and signal to noise ratio (SNR) analysis. Co-localization is used to confirm the specific signal; SNR is used to evaluate the effectiveness of this method.
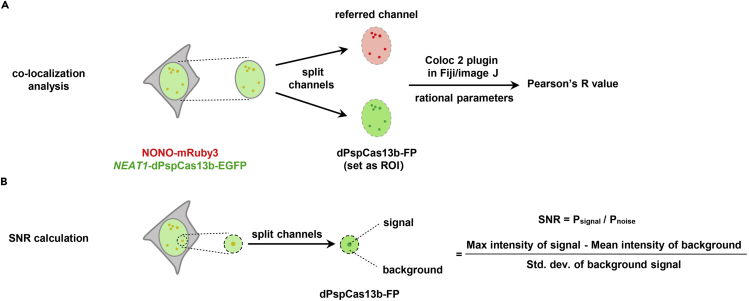
For co-localization analysis to ensure the positive signal of RNA, we suggest to co-express a known co-localized protein fused with another FP labeling in live cells or perform RNA Fluorescence in situ Hybridization (FISH) of the targeted RNA in fixed cells. Separate images into two channels of dPspCas13b-FP and the referred protein (or signals labeled by FISH). In the dPspCas13b-FP channel, select the nucleus region as the region of interest (ROI) for a nuclear RNA or the cytoplasmic region as the ROI for a cytoplasmic RNA. The parameters are set as below in Coloc 2 plugin: dPspCas13b-FP channel as channel 1, referred channel as channel 2, ROI in channel 1 as the ROI, threshold regression with bisection; PFS is set at 3.0 and costes randomisations is set at 10. After analyzed by Fiji/image J, the results are read out as Pearson’s R value with no threshold (Figure 2A).
Figure 2.
An Example of Quantification and Statistical Analysis
Schematic for co-localization analysis (A) and SNR calculation (B) of NEAT1-dPspCas13b-EGFP in NONO-mRuby3-KI HeLa cells. KI, knock in.
For SNR calculation of the labeled signals, select a circle with diameter of 3∼4 μm (about 10 folds over the resolution limit of used microscope) and the center of the spot (exclude spots of labeled signal) as background, calculate SNR with the formula: SNR=Psignal/Pbackground = (Max intensity of spots signal – Mean intensity of background GFP spot) / Std. dev. of background signal (Figure 2B).
LIMITATIONS
Although CRISPR-dPspCas13b provides a user-friendly tool to realize quick and robust tracking in live cells for abundant RNAs, it has limitations. No current guidelines to design efficient gRNAs targeting an RNA of interest and conformation of some structurally functional RNAs might limit the application of this approach. The non-repeat-containing RNAs limit the number of dCas13-FP to be recruited by gRNAs. Diffused localized RNAs often suffer from low SNR targeting by dCas13-FP and gRNAs. Finite fluorescence intensity of this method also limits its applications combined with fluorescence recovery after photobleaching (FRAP), Structured Illumination Microscopy (SIM) and long-term live-cell imaging.
TROUBLESHOOTING
Problem
Low transfection efficiency of dPspCas13b-FP and gRNA observed in step 22
Potential Solution
We recommend cloning several gRNAs into one vector as a multiplex assembly with separated U6 promoters, this will benefit the co-expression of gRNAs. Meanwhile, dPspCas13b-FP can be packaged as lentivirus for infection and antibiotic selection. Cells or single-cell clone with stable, uniformly distributed and appropriate level of expression of dPspCas13b-FP can be further selected from these infected cells.
Problem
No convincing signals in step 23
Potential Solution
The gRNA used might not work well. Sometimes gRNA designed by the prediction tool doesn’t work well in the dPspCas13b-FP expressing cells. We recommend designing the gRNA spacer sequences that target to repeated sequences in an RNA of interest, if any; or according to the antisense oligos (ASOs) or shRNA sequences that effectively targeting an RNA of interest.
Acknowledgments
This work was supported by the Chinese Academy of Sciences (XDB19020104), the Ministry of Science and Technology of China (2016YFA0100701), the National Natural Science Foundation of China (31725009, 31830108, and 31861143025), and the Howard Hughes Medical Institute (55008728).
Author Contributions
Y.W. and L.-Z.Y. wrote the manuscript, supervised by L.-L.C.
Declaration of Interests
L.-Z.Y., Y.W., and L.-L.C. have filed a patent for the dCas13b RNA labeling.
Contributor Information
Yang Wang, Email: wangyang2015@sibcb.ac.cn.
Ling-Ling Chen, Email: linglingchen@sibcb.ac.cn.
References
- Yang L.Z., Wang Y., Li S.Q., Yao R.W., Luan P.F., Wu H., Carmichael G.G., Chen L.L. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol. Cell. 2019;76:981–997.e7. doi: 10.1016/j.molcel.2019.10.024. [DOI] [PubMed] [Google Scholar]