Summary
Mammalian hematopoietic stem cells (HSCs) maintain life-long hematopoiesis in the bone marrow. HSCs remain quiescent in vivo, unlike more differentiated progenitors, and enter the cell cycle rapidly after bone marrow injury or in vitro culture. We have recently demonstrated the ability to maintain HSC quiescence in vitro by mimicking the bone marrow microenvironment. Here, we provide a detailed protocol for keeping functional HSCs in the quiescent state in vitro.
For complete details on the use and execution of this protocol, please refer to Kobayashi et al. (2019).
Graphical Abstract
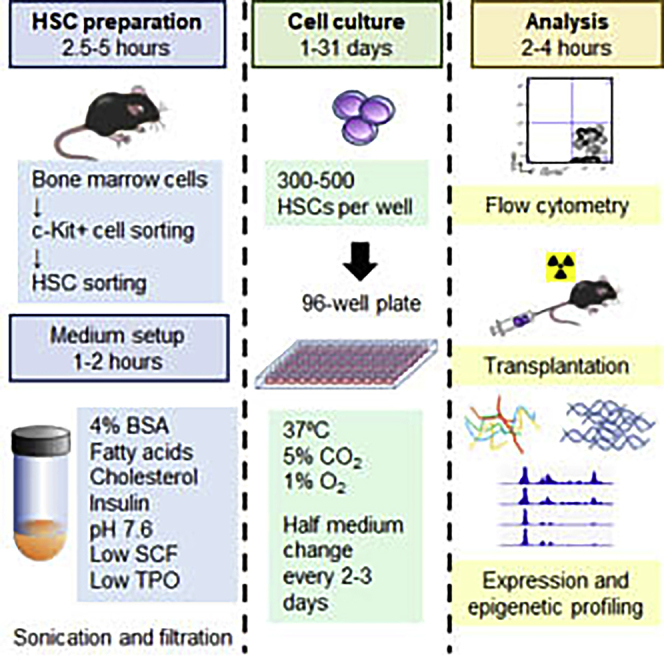
Highlights
-
•
A protocol for the maintenance of HSC quiescence in vitro is described
-
•
Oxygen levels as well as fatty acid and cytokine concentrations are key factors
-
•
The bone marrow microenvironment during steady state is effectively mimicked
Mammalian hematopoietic stem cells (HSCs) maintain life-long hematopoiesis in the bone marrow. HSCs remain quiescent in vivo, unlike more differentiated progenitors, and enter the cell cycle rapidly after bone marrow injury or in vitro culture. We have recently demonstrated the ability to maintain HSC quiescence in vitro by mimicking the bone marrow microenvironment. Here, we provide a detailed protocol for keeping functional HSCs in the quiescent state in vitro.
Before You Begin
Medium Set Up
Timing: 1–2 h
-
1.
Prepare DMEM/F12 medium with HEPES and glutamine (Thermo Fisher Scientific)
-
2.
Add 4% w/v BSA (Sigma Aldrich) to the medium.
-
3.
Dissolve fatty acid sodium salt (palmitate, oleate) and cholesterol in methanol separately at 16 mg/mL (palmitate), 30 mg/mL (oleate), and 4 mg/mL (cholesterol) (Tokyo Chemical Industry Co., Ltd.) in 15 mL or 50 mL glass tubes with a cap (N-16 or NX-50 Maruemu Corporation). The fatty acid solution can be stored at −30°C and repeatedly thawed upon use.
-
4.
Turn on a heater of a water bath to keep 37°C.
-
5.
Mix fatty acid and/or cholesterol solutions in another 50 mL glass tube.
-
6.
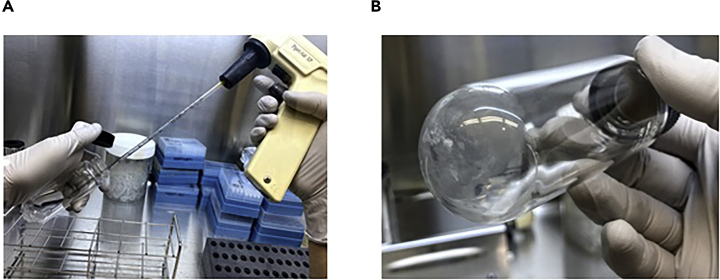
Blow air into the solution by using a 1-mL pipette and pipette-aid. If an N2 cylinder is available, blow nitrogen gas into the solution to prevent the oxidation of unsaturated fatty acids (Figure 1A).
-
7.
Heat the glass tube with fatty acid salt in a water bath at 37°C (up to 50°C is acceptable) with the cap off until the methanol is evaporated (Figure 1B).
-
8.
Adjust pH of the medium within a range of 7.6 by adding 1 M NaOH solution.
-
9.
Add medium containing 4% BSA or 4% fatty acid-free BSA directly to the glass tube to resuspend fatty acids and/or cholesterol. We usually dissolve at a final concentration of 100 μg/mL sodium palmitate, 100 μg/mL sodium oleate, and 20 μg/mL cholesterol. Three to 15 mL of medium is appropriate for sonication at step 10.
-
10.
Sonicate the medium until it becomes clear. It takes 20 min. Avoid overheating the medium.
-
11.
Add 2-mercaptoethanol at a final concentration of 55 μM and 1/1,000 (which is 1/10 of recommended concentration) of insulin, transferrin, and sodium selenite (IST) mixture (Thermo Fisher Scientific). Then, filter the medium using a polyethersulfone 0.22-μm filter (Millipore). Cytokines should be added before use at the following concentrations: 1.5 ng/mL SCF and 1 ng/mL thrombopoietin (TPO) for flat-bottom plate culture, and 3 ng/mL SCF and 0.1 ng/mL TPO for round-bottom plate culture. See Materials and Equipment for further details.
Figure 1.
Procedure to Evaporate Methanol Away from the Lipid Solution
(A) Blowing the air to the lipid solution by a pipette aid.
(B) After evaporation, lipid salts adhere to the bottom of the tube.
Optional Steps
-
12.
Fatty acids can be used instead of fatty acid salts but require a longer sonication time to dissolve.
-
13.
The BSA concentration in the medium can be reduced to 2% with little effect on the capacity to maintain the HSC number.
-
14.
BSA can be replaced with 4% fatty acid-free BSA (Sigma Aldrich or other providers), but may result in less effective maintenance of HSCs presumably due to contaminant or deficiency in essential factors bound to BSA.
-
15.
The use of lipid-rich BSA including AlbuMAX (Thermo Fisher Scientific #11020039) instead of BSA increases the total cell count and decreases the SCF requirement to as low as 1.5 ng/mL on day 7 of culture, with differentiation into megakaryocytes and increased expression levels of megakaryocyte-related genes.
-
16.
For the culture of progenitors other than HSCs in a slow cycling state, change the combination of cytokines as described in (Kobayashi et al., 2019) and (Watanuki et al., 2019). In general, Lin-Sca1+c-Kit+ LSK cells other than HSCs require a higher concentration of SCF (up to 10 ng/mL) and the addition of IL6, Flt3L, and/or IL7 is essential for MPP4.
CRITICAL: Since fatty acid concentrations and other unidentified contaminants vary among batches of BSA, test the efficacy of the culture conditions for every new batch of BSA. In our experience, the pH and lipid concentration bound to BSA affect the experimental results. Adjustment of the final pH from 7.4 to 7.8 by sodium hydroxide and the addition of lipids (e.g., 100 μg/mL sodium palmitate, 100 μg/mL sodium oleate, and 20 μg/mL cholesterol) may improve the maintenance of HSCs.
CRITICAL: Cytokines should be added immediately before use.
Note: The cytokine concentration is a key determinant of the success of the culture. Before animal experiments, determine the optimal cytokine concentration able to maintain the initial cell number after a 7-day culture period. Concentrations of 1.5–5 ng/mL SCF and 0.1–4 ng/mL TPO are sufficient to maintain the surface marker phenotype of HSCs from 8- to 14-week-old mice after a 7-day culture period. A lower cytokine concentration yields a higher frequency of phenotypic HSCs. To achieve a stringent quiescent state in HSCs, SCF < 2ng/mL when using a flat-bottom plate and SCF <5ng/mL when using a round-bottom plate are essential.
Note: In general, the SCF concentration required for HSC maintenance is inversely related to the age of a mouse (i.e., younger animals require a higher concentration of SCF). Additionally, the efficiency of HSC maintenance is expected to increase when older mice are used (e.g., >24 weeks old).
Note: To dissolve cholesterol quickly at concentrations exceeding 10 μg/mL, the addition of at least 100 μg/mL fatty acid sodium salt is required.
Note: Adding unsaturated fatty acids (i.e., oleic acid or linoleic acid) without saturated fatty acids (i.e., palmitic acid or stearic acid) results in rapid cell death in cultured HSCs.
Note: BSA conjugation is required to deliver fatty acids into HSCs. Fatty acid supplements and LDL cholesterol are less efficient than fatty acids conjugated with BSA.
Note: The addition of a very high concentration of fatty acids (>800 μg/mL) results in a loss of HSCs.
Pause Point: The medium can be stored before the addition of 4% BSA at 4°C for at least 2 months. After 4% BSA is added, the medium should be stored at −80°C and thawed immediately before use. The frozen medium should be used within 2 months.
Preparation of Mice
Timing: 30 min
1. Prepare adequate number of C57BL/6 mice to obtain the target amount of HSCs. Approximately 2,000 HSCs can be obtained from two femurs and two tibias of a 10-week-old male mouse when a strict gating strategy is applied for cell sorting. A total of 300 to 500 HSCs per well in a 96-well culture plate is suitable for analyses.
Optional Steps
-
2.
CD45.1 congenic mice, GFP-expressing transgenic mice, or other genetically modified mice can be used according to the purpose of the experiment.
CRITICAL: All procedures involving experimental animals should be performed in accordance with relevant national and institutional regulations and within dedicated experimental animal facilities. Please follow internal guidelines related to the purchase, housing, and breeding of experimental mice.
CRITICAL: All materials and reagents should be kept sterile.
CRITICAL: The protocols are optimized for mice over 7 weeks of age. For mice younger than 4 weeks, cytokine concentrations should be adjusted.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD4-PerCP-Cy5.5 (clone: RM4-5) | TONBO Biosciences | Cat# 65-0042-U100; RRID: AB_2621876 |
| Anti-mouse CD8a-PerCP-Cy5.5 (clone: 53-6.7) | TONBO Biosciences | Cat# 65-0081-U100; RRID: AB_2621882 |
| Anti-mouse B220-PerCP-Cy5.5 (clone: RA3-6B2) | TONBO Biosciences | Cat# 65-0452-U100; RRID: AB_2621892 |
| Anti-mouse B220-APC (clone: RA3-6B2) | BioLegend | Cat# 103212; RRID: AB_312997 |
| Anti-mouse Ter-119-PerCP-Cy5.5 (clone: TER-119) | TONBO Biosciences | Cat# 65-5921-U100 |
| Anti-mouse Gr1 (Ly-6G/6C)-PerCP-Cy5.5 (clone: RB6-8C5) | BioLegend | Cat# 108428; RRID: AB_893558 |
| Anti-mouse Gr1-PE-Cy7 (clone: RB6-8C5) | TONBO Biosciences | Cat# 60-5931-U100; RRID: AB_2621870 |
| Anti-mouse Mac1 (CD11b)-PerCP-Cy5.5 (clone: M1/70) | TONBO Biosciences | Cat# 65-0112-U100; RRID: AB_2621885 |
| Anti-mouse Sca-1 (Ly-6A/E)-PE-Cy7 (clone: E13-161.7) | BioLegend | Cat# 122514; RRID: AB_756199 |
| Anti-mouse c-Kit (CD117)-APC-Cy7 (clone: 2B8) | BioLegend | Cat# 105826; RRID: AB_1626278 |
| c-Kit (CD117) MicroBeads Mouse | Miltenyi Biotec | Cat# 130-091-224 |
| Anti-mouse CD150-PE (clone: TC15-12F12.2) | BioLegend | Cat# 115904; RRID: AB_313683 |
| Anti-mouse CD150-BV421 (clone: TC15-12F12.2) | BioLegend | Cat# 115926; RRID: AB_2562190 |
| Anti-mouse CD48-FITC (clone: HM48-1) | BioLegend | Cat# 103404; RRID: AB_313019 |
| Anti-mouse CD48-PE (clone: HM48-1) | BioLegend | Cat# 103406; RRID: AB_313021 |
| Anti-mouse CD41-FITC (clone: MWReg30) | BD Biosciences | Cat# 553848; RRID: AB_395085 |
| Anti-mouse CD41-PE (clone: MWReg30) | BD Biosciences | Cat# 558040; RRID: AB_397004 |
| Anti-mouse CD41-APC (clone: MWReg30) | BioLegend | Cat# 133914; RRID: AB_11125581 |
| Anti-P-selectin (CD62P)-BV421 (clone: RB40.34) | BD Biosciences | Cat# 564289 |
| Anti-mouse EPCR (CD201)-PE (clone: RCR-16) | BioLegend | Cat#141504 |
| Anti-CD34-BV421 (clone: RAM34) | BD Biosciences | Cat# 562608; RRID: AB_11154576 |
| Anti-Flt3 (CD135)-APC (clone: A2F10) | BioLegend | Cat# 135310; RRID: AB_2107050 |
| Anti-mouse CD45-PE-Cy7 (clone: 30-F11) | BioLegend | Cat# 103114; RRID: AB_312979 |
| Anti-mouse CD45-APC-Cy7 (clone: 30-F11) | BD Biosciences | Cat# 557659; RRID: AB_396774 |
| Anti-mouse CD45-FITC (clone: 30-F11) | BD Biosciences | Cat# 553080; RRID: AB_394610 |
| Anti-mouse CD45-PE (clone: 30-F11) | BD Biosciences | Cat# 553081; RRID: AB_394611 |
| Anti-mouse CD45-BV421 (clone: 30-F11) | BioLegend | Cat# 103133; RRID: AB_10899570 |
| Anti-mouse CD45-APC (clone: 30-F11) | BD Biosciences | Cat# 559864; RRID: AB_398672 |
| Anti-mouse CD45-PerCP-Cy5.5 (clone: 30-F11) | BD Biosciences | Cat# 550994; RRID: AB_394003 |
| Fc-block (anti-mouse CD16/32) (clone: 2.4-G2) | BD Biosciences | Cat# 553142; RRID: AB_394657 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PBS | Nacalai Tesque | Cat# 14249-24 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat# 26140079 |
| DMEM/F-12 medium | Thermo Fisher Scientific | Cat# 11320-033 |
| IST | Thermo Fisher Scientific | Cat# 41400-045 |
| Penicillin | Meiji Seika | PGLD755 |
| Streptomycin sulfate | Meiji Seika | SSDN1013 |
| Sodium selenite | Nacalai Tesque | Cat# 11707-04 |
| Bovine serum albumin | Sigma Aldrich | Cat# A4503 |
| Fatty-acid free bovine serum albumin | Sigma Aldrich | Cat# A7030-10G |
| 2-Mercaptoethanol (2-ME) 1000× | Life Technologies | Cat# 21985-023 |
| Sodium palmitate | Tokyo Chemical Industry Co., Ltd. | Cat# P0007 |
| Sodium oleate | Tokyo Chemical Industry Co., Ltd. | Cat# O0057 |
| Cholesterol | Tokyo Chemical Industry Co., Ltd. | Cat# C0318 |
| Ammonium Chloride | Fujifilm | Cat# 017-2995 |
| Sodium Hydrogen Carbonate | Fujifilm | Cat# 191-01305 |
| EDTA 2Na | Fujifilm | Cat# 345-01865 |
| Recombinant Murine SCF | PeproTech | Cat# 250-03 |
| Recombinant Human TPO | PeproTech | Cat# 300-18 |
| Recombinant Murine Flt3 ligand | PeproTech | Cat# 250-31L |
| Recombinant Murine IL6 | PeproTech | Cat# 216-16 |
| Recombinant human Flt3 ligand | PeproTech | Cat# 300-19 |
| Recombinant human insulin | Nacalai Tesque | Cat# 12878-86 |
| Recombinant human holo-transferrin | Nacalai Tesque | Cat# 34443-44 |
| Propidium iodide | Life Technologies | Cat# P3566 |
| Flow-Check Fluorospheres | Beckman Coulter | Cat# 7547053 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6JJmsSlc | Japan SLC, Inc. | http://www.jslc.co.jp/english/index2.htm |
| Software and Algorithms | ||
| FlowJo version 10 | BD Biosciences | https://www.flowjo.com/solutions/flowjo |
| Other | ||
| AutoMACS Pro | Miltenyi Biotec | N/A |
| FACS Aria3u | BD Biosciences | N/A |
| VELVO-CLEAR VS-25 (sonicator) | VELVO-CLEAR | N/A |
Materials and Equipment
Recipe for HSC Maintenance Media
| Components | Final Concentration |
|---|---|
| DMEM/F-12 (HEPES and glutamine added) | NA |
| BSA | 4% w/v |
| Sodium palmitate | 100 μg/mL |
| Sodium oleate | 100 μg/mL |
| Cholesterol | 20 μg/mL |
| IST mixture | 1/1,000 volume of the medium |
| NaOH solution (1 M) | Adjust pH to 7.6 |
| 2-mercaptoethanol (55 mM) | 1/1,000 volume of the medium |
| SCF (20 μg/mL) stock solution | 1.5 ng/mL (flat-bottom plate) 3 ng/mL (round-bottom plate) |
| TPO (20 μg/mL) stock solution | 1 ng/mL (flatbottom plate) 0.1 ng/mL (round-bottom plate) |
Alternatives: A flat-bottom tissue culture plate (Corning #353072) can be used instead of a round-bottom plate. A flat-bottom tissue culture plate reduces the cytokine requirement for HSC maintenance for unknown reasons.
Alternatives: The single-cell sorting mode of FACS Aria or other sorters can be used to directly sort HSCs in the culture well.
Alternatives: Using an 8-channel pipet when dispensing HSCs may reduce variation in cell counts.
Alternatives: For hypoxic culture, multi-gas incubator equipped with CO2 and O2 sensor is needed. We culture cells with APM-30DR (astec), but other models such as Tri-gas CO2 Incubators (Thermo Fisher Scientific) can be used.
Alternatives: For long-term culture (>7 days), half of the medium should be replaced every 3 to 4 days throughout the culture period. Carefully aspirate 100 μL of medium using a 200-μL pipette without disrupting the colony and add the same volume of pre-warmed fresh medium gently to each well.
Step-by-Step Method Details:
Mouse Bone Marrow Collection
Timing: ~10 min per mouse
This step describes the procedure for mouse dissection followed by collection of bone marrow.
-
1.
Euthanize the mouse by cervical dislocation.
-
2.
Sterilize the dissection area, dissection kit, and the fur of the mouse with 70% ethanol.
-
3.
Cut the skin of the abdomen and strip off the skin to the ankles.
-
4.
Cut off the foot from the tibia just above the ankle and strip off the triceps surae along with the Achilles tendon.
-
5.
Cut the patellar tendon, remove the quadriceps femoris and the biceps femoris muscles from the femur, and dislocate the hind limb from the hip joint.
-
6.
Dislocate femurs and tibiae at the patella, remove residual muscles and connective tissues entirely using scissors (on paper towel), and place the bones in 20–30 mL of ice-cold PBS + 2% FCS in a 10-cm Petri dish (Greiner).
-
7.
Flush the femurs and tibiae with PBS + 2% FCS using a 21-gauge needle (Terumo) and a 10-mL syringe (Terumo) to collect the bone marrow plug and disperse the plug by refluxing through the needle.
-
8.
Transfer the cell suspension to a 50-mL conical tube.
-
9.
Centrifuge the suspension at 680 × g for 5 min at 4°C.
Collection of the c-Kit-Positive Fraction
Timing: ~60 min
This step describes the procedure for sorting of c-Kit+ cells with AutoMACS. This step is required in order to reduce the sorting time by FACS as well as to reduce the volume of antibodies used.
-
10.
Discard the supernatant by inverting the tube quickly and remove droplets on the rim using a paper towel.
-
11.
Add 5 mL of red cell lysis buffer (0.17 M NH4Cl, 1 mM EDTA, 10 mM NaHCO3) per mouse and incubate on ice for 1 min.
-
12.
Add 2 volumes of PBS + 2% FCS and centrifuge at 680 × g for 5 min at 4°C.
-
13.
Discard the supernatant and resuspend cells in 10–20 mL of PBS + 2% FCS
-
14.
Filter the cell suspension through a 40-μm nylon mesh (BD Biosciences) and centrifuge at 680 × g for 5 min at 4°C.
-
15.
Discard the supernatant and resuspend cells in 80 μL/mouse PBS + 2% FCS.
-
16.
Transfer the cell suspension to a 1.5-mL tube.
-
17.
Add an anti-CD16/32 antibody to block Fc-receptor (2 μL/mouse) and incubate for 5 min at 4°C
-
18.
Add anti-c-Kit magnetic beads (Miltenyi) at a 1/5 v/v ratio and incubate for 15 min at 4°C in the dark.
-
19.
Add 0.5 to 1 mL of PBS + 2% FCS and centrifuge at 680 × g for 5 min at 4°C. Discard the supernatant.
-
20.
Repeat step 19.
-
21.
Resuspend cells in 1 mL of PBS +2% FCS, filter the suspension with a 40-μm filter, and transfer to a 5-mL tube. Then rinse the 1.5-mL tube with another 1 mL of PBS +2%FCS, and transfer to the 5-mL tube.
-
22.
Enrich c-Kit-positive cells using Auto-MACS Pro (Miltenyi) and the Possel-S program.
-
23.
Transfer the cell suspension of the positive fraction (2 mL) into two 1.5-mL tubes separately, centrifuge at 340 × g for 5 min at 4°C, and discard the supernatant.
Antibody Staining
Timing: ~45 min
This step describes the procedure for staining c-Kit+ cells with fluorescent-dye conjugated antibodies. This step is required in order to sort HSCs by FACS.
-
24.
Label cells with an antibody cocktail as follows: lineage markers (CD4, CD8a, Gr-1, Mac-1, Ter-119, B220)-PerCP-Cy5.5, c-Kit-APC-Cy7, Sca-1-PE-Cy7, CD150-PE, CD41-FITC, CD48-FITC, Flt3-APC, CD34-BV421. Alternatively, lineage markers (CD4, CD8a, Gr-1, Mac-1, Ter-119, B220)-PerCP-Cy5.5, c-Kit-APC-Cy7, Sca-1-PE-Cy7, CD150-BV421, CD48-FITC, Flt3-APC, EPCR-PE. Use all antibodies at 0.5 μL per mouse and mixed with 20 μL of PBS + 2% FCS. Resuspend the cell pellet to the antibody cocktail and combine the sample into one tube. Incubate cells for 30 min at 4°C in the dark.
-
25.
Add 1 mL of PBS +2% FCS and centrifuge at 340 × g for 5 min at 4°C. Discard the supernatant.
-
26.
Resuspend cells in 0.5–2 mL of PBS + 2% FCS + 0.1% propidium iodide, filter the suspension with a 40-μm filter, and transfer to a 5-mL tube.
Note: If old mice (> 1 year old) are used, the CD41 antibody should be excluded to account for the high expression of CD41 in HSCs derived from old mice.
Optional Steps
If compensation of fluorochrome is needed for the flow cytometer, prepare single-color stained bone marrow cells as follows. This step is mandatory when the configuration of the flow cytometer is altered. Following steps are branched from step 16.
-
27.
Split bone marrow cells to the number of colors +1 (for no-stain control) you use and dispense them to 1.5-mL tubes. Cell number of around 5 × 105 in 50 μL of PBS + 2% FCS is optimal.
-
28.
Label each of the compensation sample with 1μL of fluorochrome-conjugated anti-CD45 antibodies for 30 min at 4°C. For instance, following samples are prepared: no-stain control, anti-CD45-BV421, anti-CD45-FITC, anti-CD45-PE, anti-CD45-PerCP-Cy5.5, anti-CD45-PE-Cy7, anti-CD45-APC, anti- anti-CD45-APC-Cy7.
-
29.
Add 1 mL of PBS +2% FCS to each compensation sample and centrifuge at 340 × g for 5 min at 4°C. Discard the supernatant.
-
30.
Resuspend cells in 0.5 mL of PBS + 2% FCS, filter the suspension with a 40-μm filter, and transfer to a 5-mL tube.
-
31.
Perform fluorescent compensation for the flow cytometer you use.
Cell Sorting
Timing: 10 min per mouse
This step describes the procedure for sorting purified HSCs by FACS.
-
32.
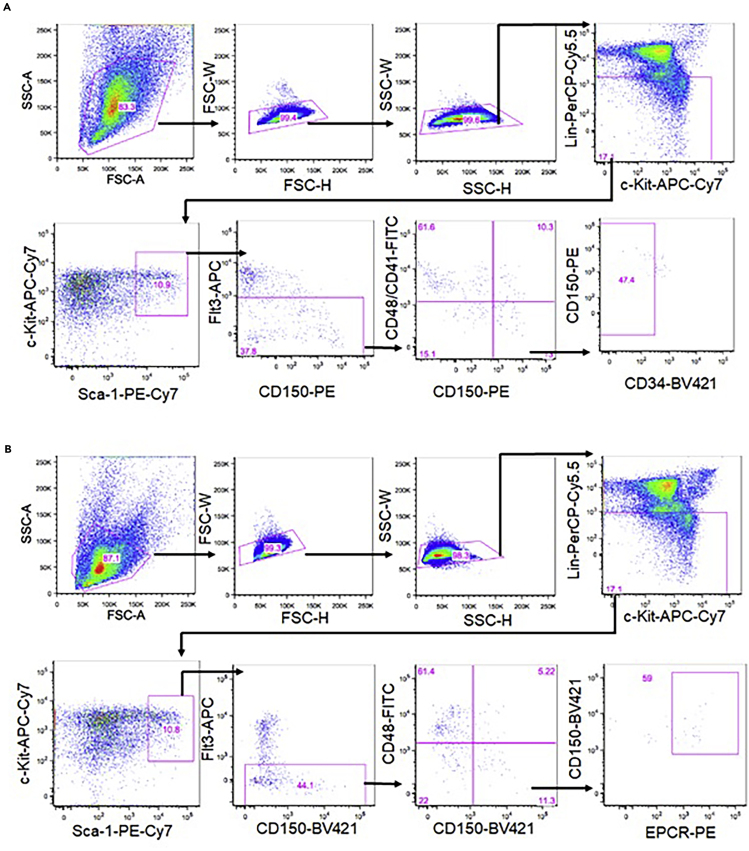
Sort HSCs with marker phenotype of CD150+CD41-CD48-Flt3-CD34- LSK or CD150+CD48-Flt3-EPCR+ LSK using the FACS AriaIIIu in a 1.5-mL tube containing 500 μL of culture media with 4% w/v BSA. Gating strategy we use is presented on Figure 2.
-
33.
Centrifuge sorted cells at 340 × g for 5 min at 4°C and discard the supernatant, and carefully aspirate the supernatant to avoid disturbing the pellet.
-
34.
Keep the cell pellet on ice until culture
Note: When cultures are conducted in fatty acid-free conditions, use fatty acid-free BSA as a sorting medium to avoid fatty acid contamination in the culture medium.
Figure 2.
Gating Strategy for Sorting HSC
(A) Gating strategy for sorting CD150+CD41-CD48-Flt3-CD34- LSK cells.
(B) Gating strategy for sorting CD150+CD48-Flt3-EPCR+ LSK cells
Cell Culture
Timing: 10–30 min
This step describes the procedure for culturing HSCs sorted by FACS.
-
35.
Transfer 200 μL of the medium prepared as described in the Medium Set Up section into each well of a 96-well round-bottom plate (Corning #353077).
-
36.
Fill all unused wells with 100–200 μL of PBS to avoid medium evaporation.
-
37.
Resuspend sorted HSCs in the culture medium at 30–100 cells/μL.
-
38.
Add 10 μL of the HSC suspension to each well (300–1,000 cells per well). Considering the balance between technical variation and efficient use of cells, we usually culture 400 cells per well.
-
39.
Place the culture plate in a humidified multi-gas incubator (Astec) at 37°C under 1% O2 and 5% CO2 conditions.
Note: Culturing less than 300 cells per well would lead to the larger technical variation thus more wells per condition is needed to detect the biological difference. Although we have not experimentally tested, more than 1,000 cells would potentially compromise the maintenance rate due to cytokine/nutrient deprivation or accumulation of unfavorable cytokines/chemokines, so that we avoid culturing more than 1,000 cells in a single well.
Optional Steps
-
40.
A flat-bottom tissue culture plate (Corning #353072) can be used instead of a round-bottom plate. A flat-bottom tissue culture plate reduces the cytokine requirement for HSC maintenance for unknown reasons.
-
41.
The single-cell sorting mode of FACS Aria or other sorters can be used to directly sort HSCs in the culture well.
-
42.
Using an 8-channel pipet when dispensing HSCs may reduce variation in cell counts.
-
43.
For long-term culture (>7 days), half of the medium should be replaced every 3 to 4 days throughout the culture period. Carefully aspirate 100 μL of medium using a 200-μL pipette without disrupting the colony and add the same volume of pre-warmed fresh medium gently to each well.
Expected Outcomes
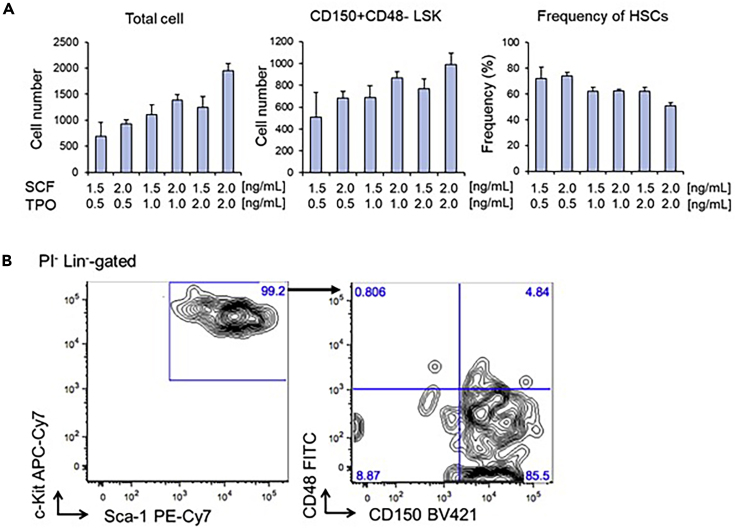
Following 6 to 7-day culture of purified HSCs, the frequency of 50 to 80% of phenotypic HSCs characterized by the marker phenotype of CD150+CD48-LSK (Figure 3) with less than 10% of PI+ cells among total cells is expected. The total cell number depends on the cytokine concentration you added. Higher concentrations of SCF and TPO induce cell cycle entry and differentiation. SCF decreases the expression of either CD150 or Sca-1. TPO induces the expression of CD48 and P-selectin. TPO also induces megakaryocytic differentiation.
Figure 3.
A Representative FACS Plot Following Culture
A total of 400 HSCs (CD150+CD48-EPCR+LSK cells) were sorted and cultured for 6 days with the indicated cytokine concentration.
(A) Shown are the total cell number, the number of CD150+CD48-LSK cells, and the frequency of CD150+CD48-LSK cells among total cells.
(B) A representative FACS plot of cells cultured with a condition of SCF 1.5ng/mL and TPO 1.0ng/mL.
Statistics and Quantification
Analysis of Cells by MACSQuant
Timing: ~2 h
We test the HSC maintenance capacity of culture media by examining the surface marker phenotype of cultured HSCs after 7 days. To minimize time and reduce sampling variation, we utilize the MACSQuant equipped with an automated sampler.
-
1.
Aspirate and discard 170 μL of the medium in wells of a 96-well plate using an 8-channel pipette not to disturb cells softly attaching to the bottom of the wells.
-
2.
Stain samples by adding 10 μL of an antibody cocktail to the residual medium (~30 μL) in wells of the 96-well plate for 30 min at 4°C. The antibody cocktail includes anti-lineage markers (CD4, CD8a, Gr-1, Mac-1, B220, Ter-119)-PerCP-Cy5.5, anti-c-Kit-APC-Cy7, anti-Sca-1-PE-Cy7, anti-CD150-PE, anti-CD48-FITC, anti-CD41-APC. Use all antibodies at 0.1 μL/well and diluted in PBS + 2% FCS up to 10 μL/well.
-
3.
After incubation, add 100 μL of PBS + 2% FCS to wells and centrifuge plates for 5 min at 4°C at 400 × g with low acceleration and medium deceleration.
-
4.
Aspirate 100 μL of the supernatant and resuspend the cell pellet in 200 μL of PBS + 2% FCS + 0.1% PI + 1% Flow-Check Fluorospheres (Beckman Coulter).
-
5.
Using the MACSQuant instrument, acquire samples in fast mode, check the mix sample check box, and analyze volumes of 100 μL.
-
6.
Export data as FCS files and analyze results using FlowJo. Determine relative cell numbers using the bead count (Flow-Check; ~1,000/μL). Megakaryocytes can be identified as cells with high forward scatter and side scatter as well as high CD150 and CD41 expression levels.
Optional Steps
-
1.
We have observed an aging-like phenotype of HSCs (i.e., an increase in megakaryocyte-related genes, including CD41 (Gekas and Graf, 2013; Grover et al., 2016) ) after the culture procedure. To monitor the aging phenotype, we often add 0.1 μL of an anti-P-selectin-BV421 antibody to the antibody cocktail.
-
2.
Analyses other than flow cytometry can be performed. We have successfully performed transplantation assays, qPCR, cDNA microarray, RNA-sequencing, and ATAC-sequencing analyses well after culture. To collect cells, transfer all culture media to 1.5-mL tubes, followed by centrifugation at 2,000 × g for 5 min. Aspirate the supernatant, and store the cell pellet on ice for up to 30 min for subsequent analyses.
-
3.
If the configuration of MACSQuant is changed, prepare single-color stained bone marrow cells for compensating fluorescence spillover before use just as steps 27–31 in the Methods in detail section. Briefly, label 5 × 105 of bone marrow cells resuspended in 50 μL of PBS + 2% FCS with 1μL of fluorochrome-conjugated anti-CD45 antibodies for 30 min at 4°C. For instance, following samples are prepared: no-stain control, anti-CD45-BV421, anti-CD45-FITC, anti-CD45-PE, anti-CD45-PerCP-Cy5.5, anti-CD45-PE-Cy7, anti-CD45-APC, anti- anti-CD45-APC-Cy7. Following staining, wash once and resuspend cells in 0.5 mL of PBS + 2% FCS. Then perform auto-compensation according to the MACSQuant instruction.
CRITICAL: Flow-Check Fluorospheres contain formaldehyde and thus are potentially toxic to cultured cells. Analyzing more than 50 wells at the same time results in reduced cell survival in the latter part of wells, compromising the experiment.
Limitations
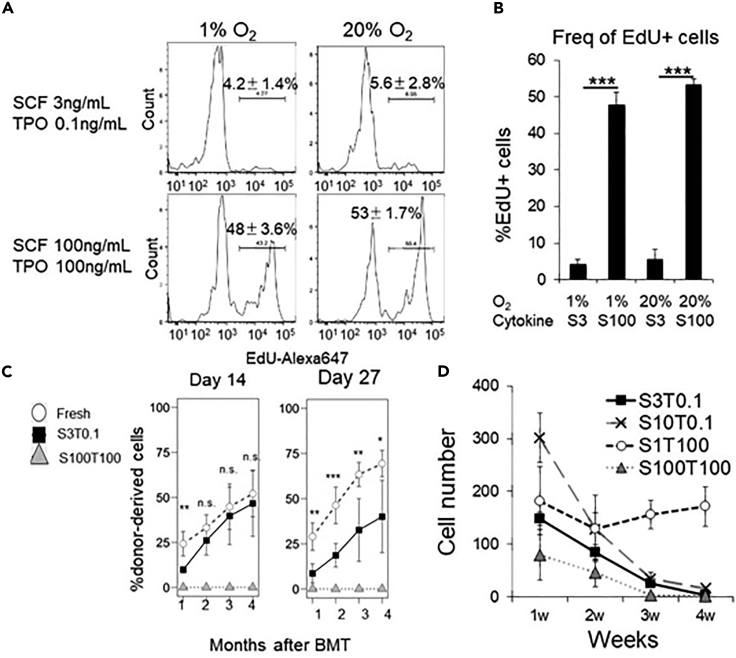
Although the aim of this protocol is to recapitulate the physiological bone marrow microenvironment for the maintenance of HSCs in an undifferentiated and quiescent state in vitro, HSCs gradually lose reconstitution potential as well as the surface marker phenotype within 1 month (Figures 4C and 4D). We recommend verifying the function of cultured HSCs by transplantation assays.
Figure 4.
Features of Cultured HSCs
(A) EdU incorporation assay to determine the cell cycle status either under quiescent culture condition (SCF 3 ng/mL and TPO 0.1 ng/mL) or under the high cytokine concentration (SCF 100 ng/mL and TPO 100 ng/mL). Shown are representative plots (n=4, mean ±SD).
(B) Bar graphs of the frequency of EdU+ cells as determined in (A).
(C) The frequency of donor-derived cells in peripheral blood transplanted with cultured HSCs following 14-day (left) or 27-day (right) culture under indicated cytokine concentrations (S: SCF, T: TPO). A total of 500 freshly isolated HSCs is transplanted as control (n=6 for each group, mean ±SD).
(D) The total cell number of a surface marker phenotype of CD150+CD48- LSK cells after 1, 2, 3, and 4 week-culture in the indicated conditions (n=4 for each group, mean ±SD).
The physiological state is not simply determined by the cell cycle status. We used an EdU incorporation assay to determine the cell cycle status and confirmed the slow cycling rate following culture (5% of cells incorporated EdU, Figures 4A and 4B). However, some genes associated with cell cycle progression (e.g., Cdk6) were upregulated, while stem cell genes (e.g., Mecom and Hoxb5) are downregulated.
The culture media still contains supraphysiological levels of insulin, amino acids, and pyruvic acid compared with serum concentrations (and possibly bone marrow concentrations). In addition, lipid and cytokine concentrations in the bone marrow environment have not been determined. Further medium optimization may be required to recapitulate the precise physiological bone marrow environment.
Troubleshooting
Problem
Fatty acids and cholesterol are not fully dissolved in the medium
Potential Solution
Extend the sonication time or strengthen the power of sonication. Of note, cholesterol does not dissolve without fatty acids. Fatty acids at >1,000 μg/mL are also difficult to dissolve.
Problem
The number of cells after culture is lower than the initial count
Potential Solution
1. Low Survival Rate
The cytokine concentration, lipid concentration, pH, and bottom shape (i.e., flat or round) affect the survival rate. The lipid concentration and pH vary among BSA batches; thus, it is necessary to optimize these parameters when a new BSA batch is used. Alternatively, increase the cytokine concentration.
2. Low Sorting Rate
Ensure that cells are properly sorted in the collection tube.
3. Accidental Aspiration of Cells
Aspirate media immediately after centrifugation to avoid loosening of the cell pellet. Carefully aspirate the medium to avoid disturbing the cell pellet.
Problem
The number of cells after culture is higher than the initial count
Potential Solution
Decrease the cytokine concentration.
Problem
The frequency of phenotypic HSCs is low after culture
Potential Solution
The cytokine concentration, purity of sorted HSCs, oxygen concentration, and contaminants in BSA affect the frequency of phenotypic HSCs after culture.
1. Inappropriate Cytokine Concentration
Higher concentrations of SCF and TPO induce cell cycle entry and differentiation. SCF decreases the expression of either CD150 or Sca-1. TPO induces the expression of CD48 and P-selectin. TPO also induces megakaryocytic differentiation. Optimize the concentrations of SCF and TPO according to your purpose.
2. Low Purity of Sorted HSCs
Pure HSCs can maintain the marker phenotype CD150+CD48-LSK at a rate of 50% following a 7-day culture period. If anti-CD41 and CD34 antibodies are excluded from the antibody cocktail, HSC purity is reduced. Sorting Sca-1-high cells and/or EPCR+ cells (Wilson et al., 2015) may improve the purity of HSCs.
3. Inappropriate Oxygen Concentration
Make sure that the O2 sensor of the incubator is functioning properly. Avoid frequent opening and closing of the door of the incubator or alternatively, use a CO2 incubator with multiple separate compartments to minimize the effect of door openings.
4. Contaminants in BSA
Removing BSA contaminants is difficult. Test several batches of BSA if contamination is the suspected cause of a low frequency of HSCs.
Resource Availability
Lead Contact
Keiyo Takubo; keiyot@gmail.com
Materials Availability
We did not generate any new materials.
Data and Code Availability
We did not generate a dataset or code.
Acknowledgements
We thank M. Haraguchi and S. Tamaki for technical support and laboratory management. H.K. was supported in part by a KAKENHI grant from MEXT/JSPS (19K17847) and a grant from the National Center for Global Health and Medicine. K.T. was supported in part by KAKENHI Grants from MEXT/JSPS (18H02845, 18K19570), grants from the National Center for Global Health and Medicine (26-001, 19A2002), AMED grants (JP18ck0106444, JP18ae0201014, JP20bm0704042) and grants from the Ono Medical Research Foundation, the Kanzawa Medical Research Foundation, and the Takeda Science Foundation.
Author Contributions
Conceptualization, H.K. and K.T.; Investigation, H.K.; Writing—original draft, H.K. and K.T.
Declaration of Interests
The authors, HK and KT, have a patent related to this work (JP-A-P2019-201599A).
References
- Gekas C., Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood. 2013;121:4463–4472. doi: 10.1182/blood-2012-09-457929. [DOI] [PubMed] [Google Scholar]
- Grover A., Sanjuan-Pla A., Thongjuea S., Carrelha J., Giustacchini A., Gambardella A., Macaulay I., Mancini E., Luis T.C., Mead A. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 2016;7:11075. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Morikawa T., Okinaga A., Hamano F., Hashidate-Yoshida T., Watanuki S., Hishikawa D., Shindou H., Arai F., Kabe Y. Environmental optimization enables maintenance of quiescent hematopoietic stem cells ex vivo. Cell Rep. 2019;28:145–158.e9. doi: 10.1016/j.celrep.2019.06.008. [DOI] [PubMed] [Google Scholar]
- Watanuki S., Kobayashi H., Sorimachi Y., Yamamoto M., Okamoto S., Takubo K. ATP turnover and glucose dependency in hematopoietic stem/progenitor cells are increased by proliferation and differentiation. Biochem. Biophys. Res. Commun. 2019;514:287–294. doi: 10.1016/j.bbrc.2019.04.123. [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Kent D.G., Buettner F., Shehata M., Macaulay I.C., Calero-Nieto F.J., Sanchez Castillo M., Oedekoven C.A., Diamanti E., Schulte R. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We did not generate a dataset or code.