Summary
DNA-FISH remains the method of choice to visualize genomic regions in situ ranging from a single locus to entire chromosomes. Current methods to generate probes rely on expensive kits that vary in labeling efficiency and are limited by the size and/or amount of starting material and by the choice of fluorophores. Here we describe a protocol to prepare inexpensive ($20) DNA-FISH probes using an isothermal polymerase, incorporating labeled nucleotides while amplifying minute amounts of any template (PCR fragments/BAC/YAC/fosmids).
For complete details on the use and execution of this protocol, please refer to Grosmaire et al. (2019) and Sharma et al. (2014).
Graphical Abstract

Highlights
-
•
Direct incorporation of modified nucleotides during isothermal template amplification
-
•
Adaptable to the size, amount, type of template DNA, and choice of fluorophores
-
•
Inexpensive probes and easily scalable from single locus to multiple chromosomes
-
•
Method consistently yields good quantity of highly labeled probes (∼1 dye/100 bases)
DNA-FISH remains the method of choice to visualize genomic regions in situ ranging from a single locus to entire chromosomes. Current methods to generate probes rely on expensive kits that vary in labeling efficiency and are limited by the size and/or amount of starting material and by the choice of fluorophores. Here we describe a protocol to prepare inexpensive ($20) DNA-FISH probes using an isothermal polymerase, incorporating labeled nucleotides while amplifying minute amounts of any template (PCR fragments/BAC/YAC/fosmids).
Before You Begin
DNA Template Preparation
The amount of total starting material required is between 10-100 ng. This amount can be easily recovered from a single miniprep (for plasmids/cosmids/fosmids) or a PCR reaction.
A variety of methods are available to find suitable starting material. For example, the UCSC genome browser provides a track with BACs and YACs covering the mouse and the human genome. Additional methods have been published elsewhere (Nedbal et al., 2012) and are available online (https://github.com/webfish/).
The isothermal polymerase uses a rolling circle amplification mechanism (RCA) which works optimally with a circular template. If starting from shorter (< 2 kb) PCR fragments, use the following method to circularize the fragments:
| Reagents | Volume in microliter (ul) |
|---|---|
| DNA | up to 1 ug |
| T4 ligase buffer (10x) | 5 ul |
| Water | to 42 ul |
| T4 ligase | 2 ul |
| T4 PNK | 1 ul |
Incubate at 37°C for one h.
Purify either with a PCR purification kit (e.g., QIAGEN minElute) or with SPRI beads (2x Ampure) according to the manufacturer’s instructions.
Note: Although not tested here, purified genomic DNA can also be directly used for amplification. Please refer to Pan et al. (2008).
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phi29 DNA polymerase | NEB | M0269S |
| Phi29 DNA polymerase 10X reaction buffer | NEB | Included in M0269S |
| BSA | NEB | Included in M0269S |
| Deoxynucleotide (dNTP) Solution Set | NEB | N0446S |
| Aminoallyl-dUTP | Fisher Scientific | R0091 |
| ATTO-488 NHS ester | Attotec | AD 488-3 |
| ATTO-647N NHS ester | Attotec | AD 647N-3 |
| ATTO-565 NHS ester | Attotec | AD 565-3 |
| Glycogen from oyster | Millipore Sigma | G8751 |
| Ribonucleic acid, transfer (tRNA) | Millipore Sigma | R9001 |
| Formamide | Millipore Sigma | 47670 |
| T4 DNA Ligase | NEB | M0202 |
| T4 PNK | NEB | M0201 |
| Phenol solution | Millipore sigma | P4557 |
| Ethanol 100%, 95% and 70% | Lab Stock | N/A |
| 3M Sodium acetate pH 5.2 | Lab Stock | N/A |
| 0.2M Sodium bicarbonate solution pH 9 | Lab Stock | N/A |
| AG 501-X8 mixed bed resin | Biorad | 1426425 |
| MilliQ or Nuclease Free Water | Lab Stock | N/A |
| Critical Commercial Assays | ||
| MinElute PCR Purification Kit | QIAGEN | 2800a4 |
| Oligonucleotides | ||
| Random Nonamer NNNNNNN*N*N (* phosphorothiolate bond) | Most oligonucleotides providers | N/A |
| Recombinant DNA | ||
| DNA template (e.g., purified fosmid minipreps, PCR amplicons and plasmids) | User defined | N/A |
| Other | ||
| Heat block | N/A | N/A |
| Minicentrifuge | N/A | N/A |
| −80 freezer | N/A | N/A |
| Branson Ultrasonics 250 Digital Sonifier or similar Thermocycler (Optional for PCR amplification and incubations) | N/A | N/A |
| Magnetic stand (Optional if using magnetic beads for DNA purification) | N/A | N/A |
Materials and Equipment
Deoxynucleotide Master Mix (dAGC) Preparation
Prepare the following mix of dATP, dGTP, dCTP to obtain a final concentration of 10 mM:
| Reagent | Volume in microliter (ul) |
|---|---|
| 100 mM dATP | 10 ul |
| 100 mM dGTP | 10 ul |
| 100 mM dCTP | 10 ul |
| Nuclease free water | 70 ul |
| Final Volume | 100 ul |
Prepare 10 ul aliquots and store at −20°C.
Alternatives: A master mix with all nucleotides, including dTTP (1 mM final concentration) and amino-dUTP (1mM final concentration can be prepared by replacing the 70 ul of nuclease free water with 0.5 ul of dTTP 100 mM, 5 ul of amino-dUTP 10 mM and 64.5 ul of nuclease free water). In any case, we recommend aliquoting the premixed nucleotides in 3 ul volume/vial to avoid multiple freeze-thaw cycles.
Sodium Bicarbonate Buffer Preparation
Prepare 0.2 M sodium bicarbonate solution and adjust pH to 9 using sodium hydroxide.
CRITICAL: Maintaining the pH of this buffer is critical. Frequent use might change the pH. Therefore, make aliquots and store at −20°C.
ATTO NHS Ester Dye Preparation
ATTO dyes come as a lyophilized powder. Reconstitute the dye by adding DMSO to a final concentration of 1 uM, prepare 5-10 ul aliquots and store at −20°C indefinitely.
CRITICAL: NHS ester groups are highly reactive to moisture and amine groups; this negatively affects dye coupling to DNA. Therefore, it is advisable to use the aliquots only once.
Random Nonamer Preparation (100 uM)
Most primer providers produce these random primers. Order NNNNNNNNN. In case the GC/AT content of the target organism genome is more than 20% away from 50%, the ratio between the different nucleotides can be adjusted. Reconstitute the lyophilized primers to a final concentration of 100 uM in water.
CRITICAL: The phi29 polymerase has 3′ exonuclease activity, therefore it is essential to use phosphorothioate bonds for the last two bonds of the random nonamer to avoid digestion of these by the polymerase.
Purified Glycogen Preparation
TIMING: 1 h
Glycogen is used an inert carrier to significantly improve nucleic acid precipitation in ethanol. Once precipitated, it forms a visible pellet, facilitating the localization of the co-pelleted DNA. While commercial alternatives exist, glycogen can be quickly prepared from powder at much lower costs and stored indefinitely at −20°C as frozen aliquots. (This protocol is adapted from a protocol for glycogen preparation available on the website of the Molecular Oncology laboratory, University of Chicago Medical Center)
-
1.
Dissolve 5 g of glycogen (from Oyster) in 10 mL of MilliQ water. Transfer 0.5-1ml to a 2ml microcentrifuge tube.
-
2.
Add an equal volume of aqueous phenol (Millipore Sigma P4557; this is not the phenol/chloroform/isoamyl alcohol mixture used for DNA extraction). Mix vigorously by shaking the tubes or vortexing. Handle phenol carefully by using proper personal protective equipment and working in a chemical hood.
-
3.
Spin tube at 13,000 rpm for 10 min at 4C. Recover the aqueous upper phase and transfer to a fresh tube.
-
4.
Add an equal volume of chloroform and mix thoroughly by vortexing.
-
5.
Spin tube at 13,000 rpm for 10 min at 4°C. Recover the aqueous phase and transfer to a fresh tube.
-
6.
Add an equal volume of 100% ethanol to the obtained supernatant. This will precipitate the glycogen.
-
7.
Spin at 13,000 rpm for 10 min at 4°C and remove supernatant.
-
8.
Air dry pellet. Weigh and resuspend in MilliQ water to obtain a final concentration of 20 mg/mL. Make aliquots and store at −20°C.
Deionized Formamide Preparation
TIMING: 1-2 h
CRITICAL: Formamide is a hazardous material. Use proper personal protective equipment while handling and work under a chemical hood.
Add 5 g of AG 501-X8 mixed bed resin (Biorad 1426425) to 100 mL of Formamide (Sigma 47670). Stir for 1 h and filter through Whatman paper. Prepare 1 mL aliquots and store at −20°C indefinitely.
Alternatives: Cost for commercial kits that apply similar chemistry begins at about $500 for the preparation of 10 probes ($50/probe). Our cost estimate for all reagents required to setup our protocol is about a $1000 for 50 reactions ($20/probe).
Step-by-Step Method Details
Set Up Amplification Reaction
TIMING: 16 h
To generate high quality FISH probes, our approach relies on efficient incorporation of an amine-modified nucleotide (dUTP) during the amplification of template DNA. Nick translation only moderately amplifies DNA while PCR based probe preparation methods are not possible with large fragments. Therefore, we used the phi29 DNA polymerase, an isothermal high fidelity DNA replicator capable of whole genome amplification from single cells (Pan et al., 2008). Using random nonamer primers, phi29 DNA polymerase generates large concatemers of template DNA in an unbiased rolling circle amplification reaction. phi29 is able to incorporate aminoallyl-dUTP instead of dTTP at a ratio of up to 1:1 without compromising the amplification yield.
-
1.
Put 50 ng of purified template DNA in a final volume of 2 ul in an eppendorf tube.
Note: The amount of initial template DNA is important. It is reported that even 10 ng of total DNA template is sufficient for amplification and more than 50 ng does not increase the final yield (Pan et al., 2008).
-
2.
Denature the DNA at 95°C for 3 min and immediately chill on ice for 5 min.
-
3.
Add the following to the DNA in the following order (or make a master mix for multiple tubes):
| Reagent | Volume in microliter (ul) |
|---|---|
| Nuclease-free water | 15.4 ul |
| 10X reaction buffer | 3 ul |
| BSA 10 mg/mL | 0.6 ul |
| Nonamer 100 uM | 2 ul |
| dAGC (10 mM) mixture | 3 ul (final concentration 1 mM) |
| dTTP (1 mM) | 1.5 ul (final concentration 50 uM) |
| Aminoallyl-dUTP (1 mM) | 1.5 ul (final concentration 50 uM) |
| Phi29 DNA polymerase | 1 ul |
| Final Volume | 30 ul |
Alternatively, when using a nucleotide mixture with all nucleotides including amino-dUTP, use 3 ul of this mixture and add an additional 3 ul of nuclease free water (total volume of water 18.4 ul).
CRITICAL: The dTTP:aminoallyl-dUTP ratio is a critical variable. We tested a variety of ratios of dUTP:dTTP, keeping the final concentration at 100 uM. Based on the final amplification yield and labeling efficiency, a 1:1 ratio gave the best results.
CRITICAL: Change the Phi29 10x reaction buffer and BSA after 20-25 reactions as the amplification yield decreases with the number of freeze-thaw cycles. Alternatively, make aliquots of the reaction buffer. According to the manufacturer, older reaction buffers can be supplemented with 4 mM DTT to restore activity.
-
4.
Vortex briefly, spin and incubate the tube at 30°C for 12-16 h. Immediately proceed to the next step.
-
5.
Heat inactivate the enzyme at 65°C for 10 min. Cool on ice.
-
6.
Run 1-2 ul of the reaction on 1% agarose gel. A single faint band is visible with a high molecular weight that runs above the 10 kb band of the DNA ladder (Figure 1). Given the rolling circle amplification process, the size of the band will be independent of the starting material size.
Figure 1.

Detection of Amplified Template DNA
Agarose gel picture shows the size of the amplified product (lane 2 and 3) compared to DNA ladder (lane 1).
Alternate Option for Multiple Fosmid Amplification
-
1.If more than one template is to be amplified together (example: chromosome paints made of multiple fosmids), separate master mixes can be prepared, as follows:
-
a.Pool 100 ng of each template
 CRITICAL: Empirical testing showed that mixing more than 10 fosmids/plasmids per tube led to uneven staining, likely due to small differences in template concentrations leading to larger differences between probes after amplification. We therefore recommend to make another master mix if more than 10 different templates have to be amplified simultaneously.
CRITICAL: Empirical testing showed that mixing more than 10 fosmids/plasmids per tube led to uneven staining, likely due to small differences in template concentrations leading to larger differences between probes after amplification. We therefore recommend to make another master mix if more than 10 different templates have to be amplified simultaneously. -
b.Calculate the concentration (ng/ul) of the new master mix of fosmids since the corresponding volumes are not the same for the individual purified fosmid stocks.
-
c.Take a volume corresponding to 50 ng of this new mix (should not be more than 2 ul) in a separate tube and amplify using the protocol above.
-
d.Independently amplified master mixes can then be pooled together for further downstream processing.
-
a.
The idea is to maintain equal stoichiometry between the different fosmids present in the same tube thereby providing an equal chance for each independent fosmid to be amplified in the mix and achieve homogeneity.
Sonication and Purification of the Amplification Products
TIMING: 1-2 h
DNA fragments obtained by RCA are concatemers of hundreds of copies of the original template. To facilitate entry into the nucleus and efficient hybridization, the DNA probe needs to be sheared into smaller fragments of 300-700 bp (average 500bp). For this purpose, we use a Branson sonicator to shear DNA into fragments of desirable probe length.
-
1.
Add 370 ul of MilliQ water to the amplification reaction.
-
2.
Sonicate the sample in an ice box using a Branson sonifier with the following parameters:
Amplitude: 10%
Time: 80 s
Pulse: 0.5 s (ON and OFF)
-
3.
Purify the reaction using a PCR clean up kit or SPRI beads (1.8x Ampure) according to the manufacturer’s instructions. Elute in 80 ul MilliQ water. Estimate the DNA concentration using Nanodrop. (Under ideal conditions, the reaction would yield a total of 1.5 to 2.4 ug of DNA)
Note: Sonication time should be modified to achieve an average fragment length of 500 base pairs (Figure 2). It is desirable to check the sonicated product on a gel for the first time to ensure fragment length.
Figure 2.

Size Distribution of Fragmented DNA
Agarose gel picture shows the size of the fragmented DNA (lane 2) post sonication compared to the DNA ladder (lane 1).
Note: The reaction volume should be raised to 400 ul in order to avoid frothing of the sample.
Note: If using a different sonicator, it is recommended to optimize conditions for efficient shearing.
DNA Precipitation
TIMING: 2-18 h
DNA fragments obtained post sonication are precipitated before fluorophore labeling for two reasons: a) to remove any unincorporated amine labeled dUTP that could possibly react with the amine reactive dyes leading to inefficient labeling; and b) to concentrate DNA for buffer exchange in the labeling step. We recommend using purified glycogen as a carrier to efficiently precipitate DNA.
-
1.
Add the following to the reaction:
| Reagent | Volume in microliter (ul) |
|---|---|
| 3M sodium acetate (pH 5.2) | 10 ul (or 0.1 volume of the total sample volume) |
| Glycogen (purified, 20 mg/mL) | 1.5 ul (see ‘Protocol to prepare purified glycogen’) |
| Chilled −20°C 100% ethanol | 250 ul |
Alternatives: 1 ul of tRNA (10 mg/ml) can be added instead of glycogen as a precipitation carrier.
-
2.
Vortex and spin briefly and incubate the reaction at −20°C for at least 1 h.
PAUSE POINT: The reaction can be left up to 16 hours at −20°C. Longer incubation times improve precipitation.
-
3.
Spin the tube at 16000 g for 12 min at 4°C.
-
4.
Remove supernatant and wash pellet with 500 ul of 70% Ethanol.
-
5.
Spin again at 16000 g for 5 min at 4°C.
-
6.
Air dry the pellet.
-
7.
Add 6 ul of nuclease free water to the tube and incubate at 37°C for 15 min. Pipette gently to dissolve the pellet.
PAUSE POINT: Tubes can be stored at −20°C for days to weeks.
Labeling the DNA Fragments with Fluorescent Dye
TIMING: 2-16 h
The aminoallyl group linked to dUTP reacts readily with the N-hydroxysuccinimidyl (NHS)-ester group of fluorescent dyes to form a stable amide bond, thereby coupling the dye to the modified nucleotide (Wiederschain 2011). To maintain the NHS ester in its reactive state, the pH of the buffer has to be adjusted to be between 7 and 9 while the buffer cannot contain any other amine groups (e.g., Tris). At this stage, the DNA fragments can be coupled to virtually any fluorophore of choice.
-
1.
Denature the DNA by incubating the tube at 95°C for 3 min and immediately cool on ice.
-
2.
Spin briefly and add in the following order:
| Reagent | Volume in microliter (ul) |
|---|---|
| Sodium bicarbonate solution | 3 ul |
| Fluorescent dye (in DMSO) | 1-1.5 ul (1000-1500 picomoles) |
Note: The ideal amplification yields ∼500 picomoles of 1:1 dUTP/dTTP incorporated DNA. Therefore, 1000 picomoles of the fluorescent dye are in large excess for the coupling reaction.
-
3.
Vortex for 15 s, spin briefly and incubate at 18-24°C in the dark for at least one h.
PAUSE POINT: Reaction can be left up to 3 days in the dark at 18-24°C. Increased incubation times do not improve dramatically the labeling yield.
Purification and Precipitation of the Labeled DNA
TIMING: 3-20 h
DNA fragments coupled to fluorescent dyes as described in section IV, need to be purified to remove excess dye. These fragments can then be precipitated individually or pooled together with other probes depending on the experiment. For example, probes targeting different regions of single/multiple chromosomes can be pooled together at this step to generate probes for chromosome painting.
-
1.
Add 340ul of MilliQ water to the reaction and purify using the PCR purification kit or SPRI beads (1.8x Ampure) according to the manufacturer’s instructions. Elute in 20 ul of MilliQ water.
-
2.
Check the probe concentration and absorption of the fluorochrome at the specific wavelength using the ‘microarray’ function of Nanodrop (Figure 3).
Figure 3.
Quantitative Analysis of the FISH Probe
Snapshot of a nanodrop screen showing the absorbance spectrum of the final probe. The readings have been generated using the microarray function on the Nanodrop. Nucleotides absorb at 260 nm whereas the dye (ATTO-488) absorbs at 488nm. The spectrum shows a clear absorbance peak at the latter wavelength. The ratio between the two absorbances is used to calculate the labeling efficiency (see text for details).
-
3.
Labeling efficiency can be calculated using a Dye:Base ratio calculator (http://www.genelink.com/tools/gl-BDratiores.asp)
-
4.
Transfer a volume corresponding to 400 ng of DNA of each probe (in case there is more than one probe required for simultaneous hybridization) into a separate tube.
-
5.
To precipitate, add:
| Reagent | Volume in microliter (ul) |
|---|---|
| Sodium acetate | 10 ul (for every 90 ul) |
| tRNA (10mg/mL) | 1 ul |
| Chilled 95% ethanol | 400 ul |
-
6.
Vortex vigorously and incubate at −20°C for at least 2 h
PAUSE POINT: Reaction can be left up to 16 hours at −20°C. Longer precipitation improves purification yield.
-
7.
Spin the reaction at 11,000 rpm for 20 min at 4°C.
-
8.
Decant the supernatant and wash with cold 70% ethanol. Spin for 5 min with the same conditions.
-
9.
Remove the supernatant carefully and dry the pellet in the dark. At this point, the pellet should be colored and fluoresce under a dissecting scope (as in Figure 4).
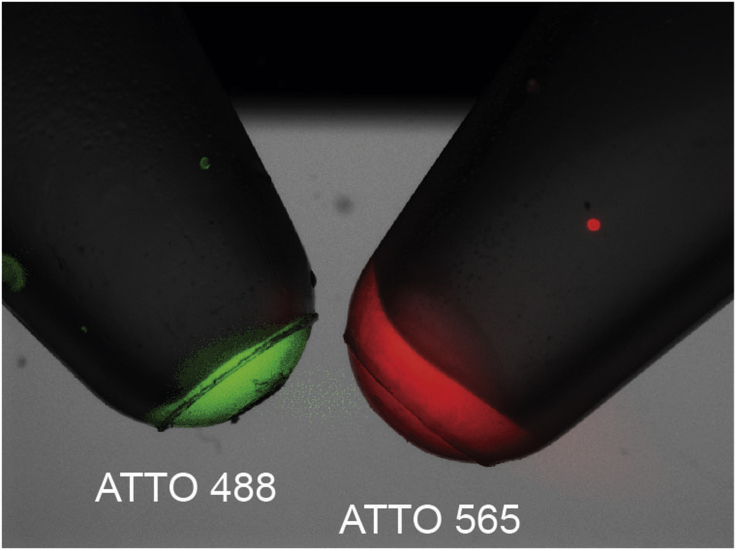
Figure 4.
Visual Assessment of the FISH Probe
Images of microtubes containing final dissolved probes labeled with ATTO-488 and ATTO-565 under a fluorescent dissecting scope (monochrome pictures acquired sequentially, displayed in false colors).
-
10.
Dissolve the obtained pellet in 20 ul deionised formamide and store at −20°C indefinitely (Figure 4).
CRITICAL: Do not use regular formamide. Probes resuspended in deionised formamide hybridize optimally in FISH experiments. See ‘before you begin’ section for the preparation of deionised formamide.
If probes of different concentration have to be mixed before being applied to the sample, the precipitated probes can be reconstituted in nuclease free water before adding deionized formamide to the hybridization mixture. The efficacy and long term storage of probes suspended in water should be determined by the user.
Expected Outcomes
Expected labeling varies with the kind of dye being used. In our experience, ATTO-488 gives a labeling of 1-1.5 dye molecules/100 bases, on average. On the other hand, ATTO-647N gives 0.5-1 dye molecules/100 bases and ATTO-565 gives 0.8-1.2 dye molecules/100 bases. These ranges of probe labels provide good signals upon hybridization under a standard confocal microscope (Figure 5).
Figure 5.
Probes Generated Using This Protocol Efficiently Label a Single Locus or Entire Chromosomes
Partial z stacks of C. elegans embryos with 2-color DNA-FISH performed with probes of different sizes. Top panel shows simultaneous hybridization of (i) a probe generated from a plasmid stably integrated as a single copy in the worm genome (red, ATTO-488) and (ii) a locus on chromosome X (green, ATTO-647N). Bottom panel shows chromosome X (red, ATTO-488) and V (green, ATTO-647N) painting probes generated from a mixture of 50 fosmids spread across each chromosome. Blue scale bar is 5 μm.
Limitations
We tested directly incorporating fluorophore labeled dUTP during DNA amplification but in our hands this strategy did not work. We failed to obtain any product suggesting that phi29 is unable to incorporate directly labeled nucleotides.
Our protocol generates double stranded probes that are used for DNA-FISH but cannot be used for RNA-FISH.
Troubleshooting
Problem 1
Low yield of amplified product
Potential Solution A
Change phi29 reaction buffer.
Potential Solution B
Check if random nonamers have phosphorothioate bonds.
Problem 2
Poor fragmentation of DNA post sonication.
Potential Solution
Increase sonication time and amplitude on test reaction.
Problem 3
Poor labeling efficiency
Potential Solution A
Check pH of sodium bicarbonate buffer and make sure it is 9.
Potential Solution B
Use fresh aliquot of NHS-ester dye.
Acknowledgments
We thank the past and present members of the Meister laboratory for their support during the development of this technique and for careful reading of this manuscript. The Meister laboratory is supported by the Swiss National Foundation (PP00P3_133744/159320, 31003A_176226), the Novartis Biomedical Research Foundation, and the University of Bern.
Author Contributions
P.M. conceived the idea. R.S. optimized the conditions, standardized the protocol, and performed the FISH and imaging experiments. P.M. and R.S. analyzed the data and wrote the manuscript.
Declaration of Interests
The authors declare no conflicts of interest.
Contributor Information
Rahul Sharma, Email: rsharma@salk.edu.
Peter Meister, Email: peter.meister@izb.unibe.ch.
References
- Grosmaire M., Launay C., Siegwald M., Brugière T., Estrada-Virrueta L., Berger D., Burny C., Modolo L., Blaxter M., Meister P. Males as somatic investment in a parthenogenetic nematode. Science. 2019;363:1210–1213. doi: 10.1126/science.aau0099. [DOI] [PubMed] [Google Scholar]
- Nedbal J., Hobson P.S., Fear D.J., Heintzmann R., Gould H.J. Comprehensive FISH probe design tool applied to imaging human immunoglobulin class switch recombination. PLoS One. 2012;7:e51675. doi: 10.1371/journal.pone.0051675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Urban A.E., Palejev D., Schulz V., Grubert F., Hu Y., Snyder M., Weissman S.M. A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc. Natl. Acad. Sci. USA. 2008;105:15499–15504. doi: 10.1073/pnas.0808028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., Jost D., Kind J., Gómez-Saldivar G., van Steensel B., Askjaer P., Vaillant C., Meister P. Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev. 2014;28:2591–2596. doi: 10.1101/gad.248864.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederschain G.Y. Molecular probes handbook. A guide to fluorescent probes and labeling technologies. Biochemistry Moscow. 2011;76:1276. [Google Scholar]