Summary
Fluorescent in situ hybridization (FISH) on the RNA moiety of human telomerase (hTR) with 50-mer probes detects hTR RNA accumulated in Cajal bodies. Using both live-cell imaging and single-molecule inexpensive FISH, our published work revealed that only a fraction of hTR localizes to Cajal bodies, with the majority of hTR molecules distributed throughout the nucleoplasm. This protocol is an application guide to the smiFISH method for the dual detection of hTR RNA and telomeres or Cajal bodies by immunofluorescence.
For complete details on the use and execution of this protocol, please refer to Laprade et al. (2020).
Graphical Abstract
Highlights
-
•
RNA smiFISH with multiple small probes reveals single molecules of hTR RNA in nucleus
-
•
The smiFISH technique is compatible with immunofluorescence for colocalization assay
-
•
Colocalized regions can be mapped in 3D images with the open source 3D ImageJ Suite
Fluorescent in situ hybridization (FISH) on the RNA moiety of human telomerase (hTR) with 50-mer probes detects hTR RNA accumulated in Cajal bodies. Using both live-cell imaging and single-molecule inexpensive FISH, our published work revealed that only a fraction of hTR localizes to Cajal bodies, with the majority of hTR molecules distributed throughout the nucleoplasm. This protocol is an application guide to the smiFISH method for the dual detection of hTR RNA and telomeres or Cajal bodies by immunofluorescence.
Before You Begin
Design and Order smiFISH Probes
Timing: 2–3 weeks to receive probes
Note: smiFISH is a single-molecule RNA FISH method developed by the labs of Florian Mueller and Edouard Bertrand, and is extensively described in their publication “smiFISH and FISH-quant – a flexible single RNA detection approach with super-resolution capability” by Tsanov et al. (2016). The user should refer to that paper for the complete guide to the smiFISH method and probeset design considerations.
Generate Probeset with Oligostan R Script
-
1.
Oligostan software was developed by Thierry Gostan of the SERANAD platform of IGMM France. Please refer to the Oligostan documentation for full details (link provided in the Key Resources Table).
-
a.
Save the hTR 451 bp sequence as a FASTA file.
-
b.
Run Oligostan script with the default settings (gene-specific probe length 26 to 32 nucleotides, probe GC content between 40%–60%, probe obeys PNAS rules 1, 2, and 4 as described in the Oligostan documentation). Only two probes are obtained for hTR.
-
c.
Change the Oligostan settings so that the PNAS filters are not applied (set to FALSE) and raise the maximum GC content to 70%. With these changes 8 probes are obtained for hTR, identified as TERC1 to TERC8 in Table 1.
Table 1.
Sequences of the smiFISH Probes for hTR and hGAPDH RNA, and the FLAP-Y Oligos
| Name | Sequence |
|---|---|
| FLAP-Y-Cy3 | /5Cy3/AATGCATGTCGACGAGGTCCGAGTGTAA/3Cy3Sp/ |
| FLAP-Y-Cy5 | /5Cy5/AATGCATGTCGACGAGGTCCGAGTGTAA/3Cy5Sp/ |
| TRProbe1 | GCATGTGTGAGCCGAGTCCTGGGTGCTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe2 | CGCGCGGGGACTCGCTCCGTTCCTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe3 | TTCCTGCGGCCTGAAAGGCCTGAACTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe4 | GGGCCAGCAGCTGACATTTTTTGTTTGCTTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe5 | GGCTTTTCCGCCCGCTGAAAGTCAGCTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe6 | GTCCCACAGCTCAGGGAATCGCGCTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe7 | GCCCAACTCTTCGCGGTGGCAGTGTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe8 | GCGGCCTCCAGGCGGGGTTCGGGTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe9 | CCGCAGGTCCCCGGGAGGGGCGATTACACTCGGACCTCGTCGACATGCATT |
| TRProbe10 | AGAATGAACGGTGGAAGGCGGCAGGCCTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe11 | CGCCTACGCCCTTCTCAGTTAGGGTTTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe12 | CCCCGAGAGACCCGCGGCTGACATTACACTCGGACCTCGTCGACATGCATT |
| TRProbe13 | CCTCCGGAGAAGCCCCGGGCCGATTACACTCGGACCTCGTCGACATGCATT |
| TRProbe14 | GAAAAACAGCGCGCGGGGAGCAAAAGCACTTACACTCGGACCTCGTCGACATGCATT |
| TRProbe15 | ACAAAAAATGGCCACCACCCCTCCCTTACACTCGGACCTCGTCGACATGCATT |
| TERC1 | GGCGAACGGGCCAGCAGCTGACATTTTTACACTCGGACCTCGTCGACATGCATT |
| TERC2 | TGTTTGCTCTAGAATGAACGGTGGAAGGCGGTTACACTCGGACCTCGTCGACATGCATT |
| TERC3 | CATGTGTGAGCCGAGTCCTGGGTGCATTACACTCGGACCTCGTCGACATGCATT |
| TERC4 | CGTTCCTCTTCCTGCGGCCTGAAAGGTTACACTCGGACCTCGTCGACATGCATT |
| TERC5 | CTGACAGAGCCCAACTCTTCGCGGTGTTACACTCGGACCTCGTCGACATGCATT |
| TERC6 | CCGAGGCTTTTCCGCCCGCTGAAAGTTTACACTCGGACCTCGTCGACATGCATT |
| TERC7 | CGCCTACGCCCTTCTCAGTTAGGGTTTTACACTCGGACCTCGTCGACATGCATT |
| TERC8 | GCGAGAAAAACAGCGCGCGGGGAGCAAAAGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _1 | GCTGGCGACGCAAAAGAAGATGCGGCTTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _2 | TGACCAGGCGCCCAATACGACCAAATTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _3 | CAATATCCACTTTACCAGAGTTAAAAGCAGCCTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _4 | AACCATGTAGTTGAGGTCAATGAAGGGGTCATTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _5 | CCATGGAATTTGCCATGGGTGGAATCATATTTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _6 | TGACAAGCTTCCCGTTCTCAGCCTTGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _7 | TCGCTCCTGGAAGATGGTGATGGGATTTCTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _8 | CCCAGCCTTCTCCATGGTGGTGAAGATTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _9 | GAGGGGGCAGAGATGATGACCCTTTTGGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _10 | ACACCCATGACGAACATGGGGGCATCTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _11 | GATCTTGAGGCTGTTGTCATACTTCTCATGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _12 | TAAGCAGTTGGTGGTGCAGGAGGCATTGCTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _13 | AGTTGTCATGGATGACCTTGGCCAGGGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _14 | GACTGTGGTCATGAGTCCTTCCACGATATTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _15 | CCACAGTCTTCTGGGTGGCAGTGATGGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _16 | TTGGCAGGTTTTTCTAGACGGCAGGTCAGGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _17 | CCTGCTTCACCACCTTCTTGATGTCATTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _18 | CTCAGTGTAGCCCAGGATGCCCTTGATTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _19 | TGGGTGTCGCTGTTGAAGTCAGAGGAGACCTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _20 | TGTCATACCAGGAAATGAGCTTGACAAAGTGGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _21 | ATGAGGTCCACCACCCTGTTGCTGTATTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _22 | CCAGCAGTGAGGGTCTCTCTCTTCCTTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _23 | CAGTAGAGGCAGGGATGATGTTCTGGTTACACTCGGACCTCGTCGACATGCATT |
| GAPDH _24 | GTTCAGCTCAGGGATGACCTTGCTTACACTCGGACCTCGTCGACATGCATT |
-
d.
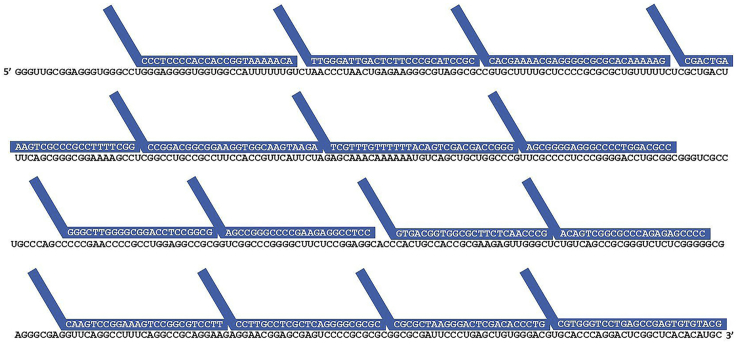
To obtain more probes, reduce minimum probe size to 23 and increase maximum GC percentage to 85%. With these permissive settings, 15 probes are obtained for hTR with delta G 37°C between −36 and −28 kcal/mol as recommended in Tsanov et al. 2016 (sequences TRProbe1 to TRProbe15 in Table 1). The 15 smiFISH probes are illustrated in Figure 1.
Figure 1.
Sequences of 15 smiFISH Probeset Aligned on hTR RNA
The hTR telomerase RNA sequence with the 15 smiFISH probes shown in blue. FLAP oligo-binding region of the probes are represented as diagonal tails.
-
e.
BLAST all the probes with blastn to ensure they meet criteria as described in the Oligostan documentation.
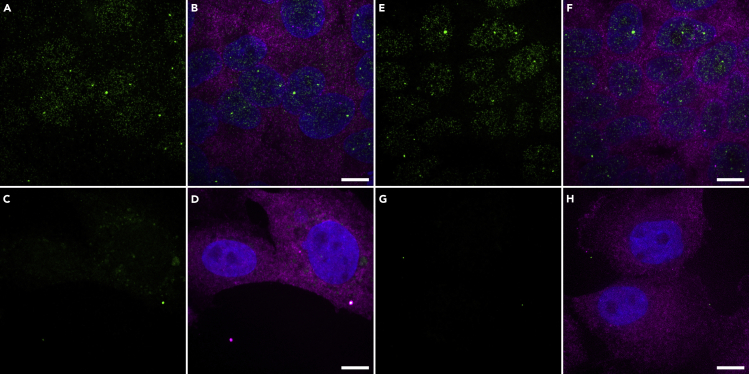
Note: We chose the FLAP-Y sequence for all the probes and ordered both the 8 probes and 15 probes sets from Integrated DNA Technologies Inc., along with the FLAP-Y-Cy3 and FLAP-Y-Cy5 oligos (listed in Table 1). This allowed us to test both sets to check if they generated specific signal for hTR RNA, as shown in Figure 2. The hTR smiFISH signal was nuclear in HeLa cells with both sets, while positive control GAPDH mRNA was mainly cytoplasmic. The WI38-VA13 clone cells were negative for hTR smiFISH signal with both probesets. With the smiFISH probes, hTR was detected all over the nucleus, unlike what is obtained with 50-mer probes for hTR FISH (Zhu et al., 2004). It is likely that the combination of multiple smiFISH probes for hTR and the lower formamide hybridization conditions favor probe binding to hTR RNA and thus enable single-molecule detection.
Figure 2.
Validation of the 8 Probes and 15 Probes Sets for hTR smiFISH
Dual-color smiFISH with 8 hTR smiFISH probes (A–D) or 15 hTR smiFISH probes (E–H), and 24 GAPDH smiFISH probes. smiFISH was performed on HeLa cells (A, B, E, and F) and hTR-negative WI38-VA13 cells (C, D, G, and H). hTR smiFISH probes labeled with FLAP-Y-Cy3 are shown in green. (B, D, F, and H) Merge with GAPDH smiFISH probes labeled with FLAP-Y-Cy5 (magenta) and DNA in blue. Maximum intensity projection of Z stacks acquired on a Zeiss Axio-Observer Z1 Yokogawa CSU-X1 spinning disk confocal microscope with a Photometrics Evolve EMCCD camera (resolution x, y = 0.133 μm). Scale bars, 10 μm.
CRITICAL: To validate a new set of smiFISH RNA probes, perform a negative control FISH on a cell line that does not express the target RNA. If such cells are not available for the RNA of interest, you can deplete the target by RNA interference as shown in Querido et al. (2017). RNAse-treated cells are not a sufficient control since they will be negative even if many probes in the new set cross-react with other RNAs.
Set Up the Immunofluorescence Detection
Timing: 2 days or more
-
2.
Before combining smiFISH and immunofluorescence (IF), the antibody detection step should be validated. This step should be as short as possible to minimize smiFISH signal loss.
-
a.
Test several concentrations of your primary antibody with a 2-h incubation time at 20°C–22°C (using the permeabilization, blocking, and IF antibody buffer conditions listed in this protocol).
-
b.
Test higher dilution of the secondary antibody to minimize background. This is especially important for super-resolution microscopy.
-
c.
Once antibody incubation is optimized, test smiFISH and IF separately, and combined. Compare the signals obtained with each condition.
CRITICAL: Always include controls with no antibody and secondary antibody alone.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal Anti-TRF2 antibody produced in rabbit | Novus Biologicals | Cat# NB110-57130; RRID: AB_844199 |
| Monoclonal Anti-Coilin antibody produced in mouse, Clone pdelta | Sigma-Aldrich | Cat# C1862; RRID: AB_476827 |
| Alexa Fluor®488 AffiniPure Donkey Anti-Mouse IgG(H+L) | Jackson ImmunoResearch | Cat# 715-545-150; RRID: AB_2340846 |
| Alexa Fluor® 594 AffiniPure Donkey Anti-Rabbit IgG(H+L) | Jackson ImmunoResearch | Cat# 711-585-152; RRID:AB_2340621 |
| Alexa Fluor® 647 Anti-Coilin mouse monoclonal antibody, Clone pdelta | Novus Biologicals (Discontinued) | Cat# NB600-1275AF647; RRID: AB_608469 |
| Alternative: Alexa Fluor® 647 Anti-Coilin mouse monoclonal antibody, Clone pdelta | Santa Cruz | Cat# sc-56298 AF647 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 32% Paraformaldehyde (formaldehyde) aqueous solution | Electron Microscopy Sciences | 15714(EM) |
| Formamide (certified ACS) | Thermo Fisher Scientific | F84-1 |
| Dextran Sulfate | Thermo Fisher Scientific | BP1585 |
| Ribonucleoside-Vanadyl Complex (VRC) | New England Biolabs | S1402S |
| NEBuffer 3 | New England Biolabs | B7003S |
| Triton™ X-100 BioXtra | Sigma-Aldrich | T9284 |
| Bovine Serum Albumin 98% | Sigma-Aldrich | A3059 |
| tRNA from E. coli MRE 600 (Roche) | Sigma-Aldrich | 10109541001 |
| Gelatin from porcine skin, Type A Cell culture tested | Sigma-Aldrich | G1890 |
| Hoechst 33342 – FluoroPure™ Grade | Thermo Fisher Scientific | P10144 |
| ProLong™ Glass Antifade Mountant | Thermo Fisher Scientific | P36982 |
| VECTASHIELD® Antifade Mounting Medium with DAPI | Thermo Fisher Scientific | H1200 |
| Experimental Models: Cell Lines | ||
| HeLa 1.3 (HeLa 1.2.11 clone 1.3) | De Lange Laboratory, Rockefeller University | N/A |
| HeLa 1.3 hTR-5′-3xMS2 | Laprade et al. 2020 | N/A |
| WI-38 VA13 | ATCC | CCL-75.1 |
| Oligonucleotides | ||
| Probes for smiFISH | See Table 1 | N/A |
| Software and Algorithms | ||
| Fiji ImageJ software | Schneider et al., 2012 | https://imagej.net/Fiji |
| 3D ImageJ Suite with RoiManager 3D | Ollion et al., 2013 | https://imagejdocu.tudor.lu/plugin/stacks/3d_ij_suite/start |
| FigureJ ImageJ plugin | N/A | https://imagejdocu.tudor.lu/plugin/utilities/Figurej/start |
| BLASTN suite | National Center for Biotechnology Information | https://blast.ncbi.nlm.nih.gov/Blast.cgi?LINK_LOC=blasthome&PAGE_TYPE=BlastSearch&PROGRAM=blastn |
| Oligostan R script | Tsanov et al., 2016 | https://bitbucket.org/muellerflorian/fish_quant/src/master/Oligostan/Oligostan.r |
| Oligostan documentation | Tsanov et al., 2016 | https://bitbucket.org/muellerflorian/fish_quant/src/master/Oligostan/Documentation/ |
| R software | The R Project for Statistical Computing | https://www.r-project.org/ |
| R Studio Desktop software | R Studio | https://rstudio.com/ |
| Zen Microscopy software | Carl Zeiss Microscopy GmbH | https://www.zeiss.com/microscopy/int/products/microscope-software.html |
| Other | ||
| TetraSpeck™ Microspheres, 0.1 μm, fluorescent blue/green/orange/dark red | ThermoFisher Scientific | T7279 |
| SSC buffer, 20×, Sterile, Mol. grade | Wisent | 880-040-LL |
| PBS 10×, pH 7.4 without Ca2+ and Mg2+ | Wisent | 311-012-CL |
| Fisherbrand Cover glasses 22 × 22 mm no1.5 | ThermoFisher Scientific | 12-541B |
| Pkg of 2, 16 × 20 cm, glass plates | Bio-Rad | 1651821 |
| PARAFILM® M 4 in. wide roll | Bemis | PM999 |
| RNaseZAP™ cleaning agent | Sigma-Aldrich | R2020 |
Materials and Equipment
FLAP hybridization reaction mix for PCR cycler
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| MilliQ water | n/a | 6.5 |
| 10× NEBuffer 3 | 1× | 1 |
| 20 μM hTR 15 probeset | 4 μM | 2 |
| 100 μM FLAP-Y-Cy5 oligo | 5 μM | 0.5 |
| Total | n/a | 10 |
CRITICAL: The FLAP oligo must be in excess of the smiFISH probeset molecules. Throughout this protocol molecular grade nuclease-free ddH2O is used to prepare solutions and reaction mixes. Freshly filtered MilliQ or commercial nuclease-free water are good options.
Fixation buffer
| Reagent | Final Concentration | Volume (mL) |
|---|---|---|
| MilliQ water | n/a | 31 |
| 10× PBS nuclease-free | 1× | 4 |
| 32% Paraformaldehyde (formaldehyde) | 4% | 5 |
| Total | n/a | 40 |
CRITICAL: Prepare fresh. Formaldehyde is a carcinogen. Use in a chemical fume hood. We recommend the Electron Microscopy Sciences 10 mL ampoules sealed under inert gas to minimize artifacts.
smiFISH wash buffer
| Reagent | Final Concentration | Volume (mL) |
|---|---|---|
| MilliQ water | n/a | 80 |
| 20× SSC nuclease-free | 1× | 5 |
| Formamide | 15% | 15 |
| Total | n/a | 100 |
Prepare fresh. Formamide is toxic and should be used in a chemical fume hood. The 6-well plate can be wrapped with plastic film to prevent exposure when smiFISH washes are performed.
smiFISH hybridization mix
| Reagent | Final Concentration | Volume (μL) |
|---|---|---|
| MilliQ water | n/a | 26.3 |
| 20× SSC nuclease-free | 1× | 5 |
| Formamide | 15% | 15 |
| 2% BSA nuclease-free∗ | 4.5 mg/mL | 22.5 |
| 40% dextran sulfate∗∗ | 10.6% | 26.5 |
| VRC (200 mM) | 2 mM | 1 |
| tRNA (25 mg/mL)∗∗∗ | 0.425 mg/mL | 1.7 |
| FLAP-annealed probeset | 0.08 μM | 2 |
| Total | n/a | 100 |
Prepare on ice and mix well by vortexing, spin down, and keep on ice. Try to minimize bubble formation.
Note: A rocking platform adjusted to a low angle and speed should be used to provide gentle agitation of the 6-well plates with the coverslips during incubation and washing steps.
Prepare BSA 20 mg/mL (2%) solution in nuclease-free water, filter with 0.2 μm, aliquot and store at −20°C. Note that a higher BSA concentration than Tsanov et al. 2016 is used in the hybridization mix with the goal of reducing non-specific probe sticking.
Heat 40% Dextran in nuclease-free water to 65°C to dissolve fully, 0.2-μm filter, and keep at 4°C. Dextran sulfate concentration in the hybridization solution is critical to successful RNA in situ hybridization.
Prepare tRNA 25 mg/mL solution in sterile nuclease-free water, aliquot and keep at −20°C.
IF antibody buffer
| Reagent | Final Concentration | Volume (mL) |
|---|---|---|
| MilliQ water | n/a | to 100 |
| 10× PBS nuc-free | 1× | 10 |
| Triton X-100 | 0.1% | 0.1 |
| BSA nuclease-free | 2% | n/a |
| Total | n/a | 100 |
Prepare fresh. Dissolve well and pass through a 0.2-μm filter to sterilize and remove undissolved BSA. Keep on ice.
Alternatives: Triton™ X-100 is included in European REACH Annex XIV. TERGITOL™ 15-S-9 is a suggested alternative, but we have not tested it.
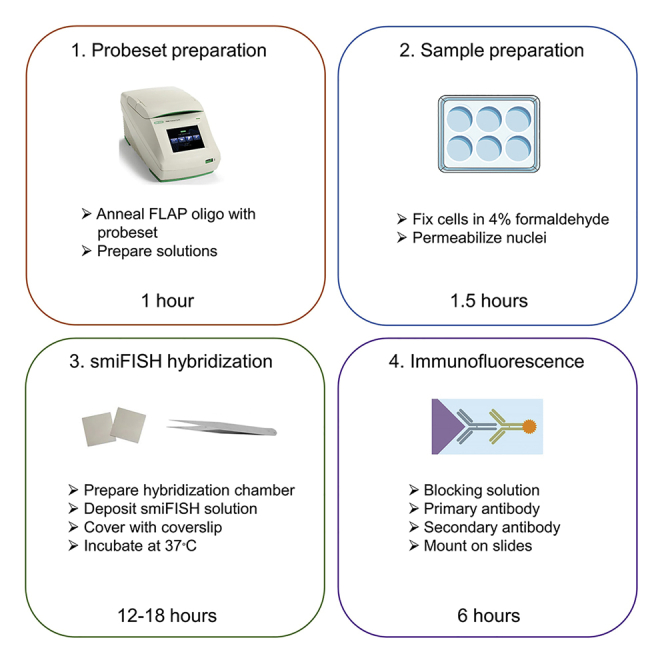
Step-By-Step Method Details
Anneal smiFISH Probeset with FLAP Oligo
Timing: ∼1 h
The principle of smiFISH is that the oligo marked with the two dye molecules (the FLAP) is complementary to the 3′ end of each probeset oligo. They must be annealed together before using the probeset.
-
1.
Anneal FLAP oligo to the probeset in a PCR machine
-
a.
The smiFISH probeset is prepared as a 20 μM equimolar mixture of the 15 hTR probes in Tris-EDTA pH8.0 (TE).
-
b.
Anneal in PCR cycler then spin down and store on ice protected from light.
| PCR conditions | ||
|---|---|---|
| Steps | Temperature | Time |
| Denaturation | 85°C | 3 min |
| Annealing | 65°C | 3 min |
| Annealing | 25°C | 5 min |
| Lid: 99°C | ||
Note: For dual-color FISH as shown in Figure 2, the same FLAP sequence can be used since each probeset is annealed with its FLAP-dye oligo in a separate tube. Once annealed, we have not seen any exchanges between FLAP-dye oligos in dual-color FISH.
Prepare Samples
Timing: ∼1.5 h
To detect RNA and proteins by microscopy, the cells must be grown on glass coverslips. Once they reach an appropriate density, the cells are fixed by formaldehyde cross-linking, then permeabilized to create openings for the fluorescent probes and antibodies to enter.
Note: Prepare gelatin-coated coverslips fresh for every experiment. Autoclave acid-washed coverslips in a beaker of water with 0.5% gelatin. Allow the gelatin beaker to cool then place coverslips in a 6-well plate, rinse with 1× PBS, replace with media and cells. Do not let the gelatin-coated coverslips dry. Alternative coverslip preparation methods appropriate to the cell type are also compatible with smiFISH.
-
2.
Cells grown in a 6-well plate on 22 × 22 mm no. 1.5 coverslips are fixed in 4% formaldehyde (fixation buffer).
-
a.
Bring 6-well plate to the chemical fume hood. Remove media and replace with 2 mL fixation buffer per well. Wrap plate in plastic film and incubate for 20 min at 20°C–22°C with gentle agitation.
-
b.
Rinse twice with 2 mL per well of 1× PBS.
Pause Point: Fixed cells can be kept in 1× PBS at 4°C for several days. However, proceeding with smiFISH the same day is optimal in order to minimize RNA degradation.
-
c.
Permeabilize with 2 mL per well of 0.5% Triton X-100 in 1× PBS for 5 min at 20°C–22°C with gentle agitation.
-
d.
Rinse twice with 2 mL per well of 1× PBS.
-
e.
Rinse once with 2 mL per well of 1× SSC 15% formamide.
-
f.
Incubate 15 min or more with 2 mL per well of 1× SCC 15% formamide at 20°C–22°C.
CRITICAL: Do not let the coverslips dry out at any step.
smiFISH Hybridization
Timing: 12–18 h
Hybridization with the probeset oligos occurs at 37°C overnight with a high concentration of probes in a small volume. The hybridization solution must not evaporate, and RNAse contamination should be avoided.
-
3.
Prepare smiFISH hybridization mix and store on ice protected from light.
-
4.
Prepare a nuclease-free airtight hybridization chamber as shown in Figure 3.
Figure 3.
Setup of an Airtight Hybridization Chamber on a Glass Plate
(A) Start with a clean 16 × 20 cm glass plate.
(B) Fix Parafilm to glass by pressing lightly with a rounded pen or PCR film paddle tool.
(C) Pipet 50 μL of smiFISH hybridization mix on Parafilm.
(D) Invert coverslip cell-side down on hybridization mix.
(E and F) (E) Cover with second sheet of Parafilm and (F) seal four sides.
(G and H) (G) Wrap in plastic film and (H) aluminum foil.
-
a.
Fix a length of Parafilm (paper side on top) on a large glass plate by rubbing the edges with a rounded pen cap. Remove paper.
-
b.
Pipet 50 μL of hybridization mix on the Parafilm and invert coverslip cell-side down on the drop. Try not to trap bubbles.
-
c.
Remove paper from a second length of Parafilm, deposit on top, and seal the four sides. Wrap glass plate in plastic film.
-
d.
Incubate at 37°C overnight protected from light.
-
5.
Unwrap and remove the top sheet. Pinch up the bottom Parafilm sheet near each coverslip to break the liquid tension. Carefully pick up the coverslip and place cell-side up in a clean 6-well dish containing 2 mL 1× SSC 15% formamide per well.
-
6.
Wash twice with 1× SSC 15% formamide at 37°C for 30 min.
-
7.
Wash twice with 1× PBS at 20°C–22°C.
Note: Clean needle-tip tweezers with RNAseZAP or similar solution, rinse in 70% ethanol.
Pause Point: If you are only performing smiFISH, the coverslips are now ready for DNA stain and mounting.
Immunofluorescence
Timing: ∼6 h
Before detecting the protein of interest with antibody, the cells are incubated with high protein solution to reduce non-specific sticking. After the primary antibody incubation, secondary antibody cross-linked to dye molecules is used to detect the primary antibody. The nuclear boundary is marked by staining the DNA with a fluorescent dye.
-
8.
Incubate with 2 mL per well of IF antibody buffer for 1 to 1.5 h at 20°C–22°C with gentle agitation to block non-specific binding.
CRITICAL: Protect from light during all steps.
-
9.
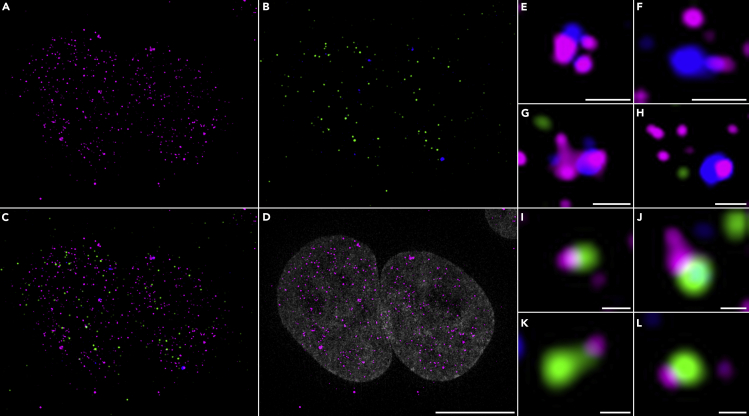
Dilute primary antibodies in IF antibody buffer. For Figure 4, we combined anti-coilin mouse mAb 1: 500 and anti-TRF2 rabbit pAb 1:200.
Figure 4.
Super-Resolution Imaging of hTR smiFISH Combined with IF against Coilin and TRF2
(A) HeLa 1.3 cells hybridized with the 15 probes set for hTR smiFISH labeled with FLAP-Y-Cy5 shown in magenta.
(B) Coilin (in blue) and TRF2 (in green) were detected by immunofluorescence.
(C) Overlay of coilin, TRF2, and hTR smiFISH.
(D) Overlay of hTR with DNA stain in gray, scale bar, 10 μm.
(E–L) Zoom on regions of image C showing Cajal bodies (E–H) or telomeres (I–L) with partially overlapped hTR signal. Panels (E–H) scale bars, 0.5 μm. Panels (I–L) scale bars, 0.2 μm. Maximum intensity projection of Z stacks acquired on a Zeiss Elyra PS.1 super-resolution microscope by structured illumination using an Andor iXon3 EMCCD camera (resolution after SR-SIM processing x, y = 0.040 μm).
-
10.
Prepare antibody incubation chamber
-
a.
Use a clean 15 cm plastic dish with lid. Cut out a Parafilm circle and fix to the bottom of the dish with the paper side up.
-
b.
Remove paper. Pick up coverslip and gently blot the edge on a Kimwipe to drain excess liquid. Deposit on Parafilm cell-side up.
-
c.
Pipet 150 μL of 1st antibody solution on top of the coverslip. Replace dishcover without moving the dish.
-
11.
Incubate primary antibody for 2 h at 20°C–22°C.
-
12.
Pick up coverslip and place in 6-well plate containing 1× PBS 0.1% Triton X-100 wash buffer
-
13.
Wash 4 times (5 min each with gentle agitation) in 2 mL per well 1× PBS 0.1% Triton X-100 wash buffer at 20°C–22°C.
-
14.
Wash twice in 2 mL per well of IF antibody buffer.
-
15.
Dilute secondary antibodies in IF antibody buffer. For Figure 4, we combined anti-mouse AF488 1: 6500 and anti-rabbit AF594 1:6500.
-
16.
Prepare antibody incubation chamber as in step 10. Incubate 2nd antibody for 50 to 60 min at 20°C–22°C.
-
17.
Wash twice in 2 mL per well of IF antibody buffer.
-
18.
Wash 4 times in 2 mL per well of 1× PBS 0.1% Triton X-100 wash buffer.
-
19.
Incubate with H33342 staining solution (0.5 μg/mL in 1× PBS) for 5–10 min at 20°C–22°C.
-
20.
Wash twice with 1 mL/well 1× PBS.
-
21.
Pick up coverslip and gently blot the edge on a Kimwipe to drain excess liquid. Mount one coverlip per slide with Prolong mounting media. Let it harden at 20°C–22°C protected from light for the recommended 24–72 h. Seal the four corners of the coverslip with a drop of nail polish and let dry completely before imaging.
Alternatives: Use other antifade mounting medium. We have also had good results with Vectashield (skip step 19 if mountant contains DAPI).
Expected Outcomes
Single-molecule RNA FISH typically has a low signal-to-noise ratio. Negative and positive controls should be included, as they are essential to determine appropriate settings for image acquisition and image analysis steps. The control cells should be negative or depleted for the RNA target. Highly expressed mRNAs are good positive controls for smiFISH, although they may not be resolved as single molecules. The sequences for a 24 probes set for human GAPDH are provided in Table 1. Figure 2 shows GAPDH smiFISH results in HeLa and VA13 cells photographed with a spinning disk confocal microscope.
Quantification and Statistical Analysis
Several software packages are available for image analysis of single-molecule RNA FISH but many work only on 2D images, usually by applying a maximum intensity projection (MIP) on the 3D Z stack. For the quantification of hTR single-molecule FISH, analysis of the image stacks in 3D was necessary as the nuclear volume contained many spots that could appear to falsely overlap in a MIP image. The 3D ImageJ Suite created by Thomas Boudier has a variety of segmentation options with the needed flexibility for this application (Ollion et al., 2013). A brief outline of the steps to segment the nuclei, hTR spots, and Cajal body regions with the 3D Suite, is given below. Values for background subtraction and seeds threshold will need to be set experimentally dependent on the signal-to-noise ratio of the smiFISH and IF spots.
Steps for nuclei segmentation:
-
1
Save a copy of the DAPI or H33342 single-channel 16-bit Tiff Z stack
-
2
Process>Filters>GaussianBlur3D
-
3
Adjust Threshold - Li method
-
4
Plugin>3D>3D Simple Segmentation
Save as segmented nuclei image
-
5
If necessary, separate nuclei that touch by drawing on the image, save it as a seed image, then apply seeded watershed (Plugin>3D>3D Watershed)
-
6
Import segmented nuclei into 3D RoiManager
Steps for hTR spot segmentation:
-
1
Save a copy of the hTR smiFISH single-channel 16-bit Tiff Z stack
-
2
Process>Math>Subtract background
-
3
Plugin>3D>3DFastFilters - MaximumLocal
Save as seed image
-
4
Plugin>3D>3D Watershed
Save as segmented spots image
-
5
Import segmented spots from one nucleus by selecting it into 3D RoiManager
-
6
Select hTR spots in 3D RoiManager and press 3D Quantif to measure on unmodified hTR 16-bit Tiff
Steps for Cajal body (CB) segmentation:
-
1
Save a copy of the coilin single-channel 16-bit Tiff Z stack
-
2
Process>Math>Subtract background
-
3
Plugin>3D>3DFastFilters - MaximumLocal
Save as seed image
-
4
Plugin>3D>3D Spot segmentation-Gaussian fit classical
Save as segmented CB image
-
5
Import segmented CB from one nucleus by selecting it into 3D RoiManager
-
6
Select CB and hTR spots in 3D RoiManager and press Colocalization to list the hTR spots and CB that overlap
Note: Prior to colocalization image analysis, the hTR, TRF2 and coilin image stacks should be aligned using reference multicolor bead images (Tetraspeck 0.1 μm beads listed in the Key Resources Table). Prepare beads slide in the same mounting media used for the cells, and photograph in the same imaging session.
Limitations
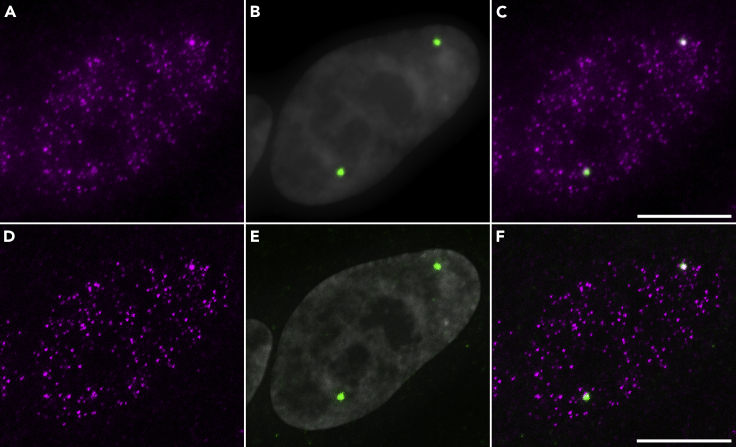
The length and sequence of the RNA could make probeset design challenging for some targets. The main limitation of single-molecule RNA FISH however is the microscope system itself. Epifluorescence microscopes have been shown to work well for single-molecule RNA FISH, as shown in Raj et al. 2008 and Tsanov et al., 2016, but the images might require deconvolution, as illustrated in Figure 5. Scanning confocal microscopes may not be sufficiently sensitive for the weak single-molecule RNA FISH signal, but the spinning disk microscope has worked in our hands (Figure 2). We recommend lasers as the lightsource and an ultra-sensitive camera to capture the emission with a high magnification high NA objective. Structured illumination super-resolution microscopy (SR-SIM) has been particularly successful in resolving single RNA molecules as unique spots (Figure 4), as shown in Adivarahan et al. 2018 and Laprade et al. 2020.
Figure 5.
IF against Coilin Followed by hTR smiFISH Imaged by Epifluorescence Microscopy
For this experiment, HeLa 1.3 hTR-5′-3xMS2 cells cells were incubated with coilin antibody labeled with AF647 for 1 h, followed by overnight smiFISH hybridization with the 15 hTR probeset plus 4 probes for the MS2 region (as described in Laprade et al. 2020) labeled with FLAP-Y-Cy3.
(A and D) hTR smiFISH shown in magenta.
(B and E) Coilin IF in green merged with DAPI in gray.
(C and F) Overlay of coilin and hTR.
(D–F) Images were deconvolved with the ZEN Fast Iterative algorithm default settings (Zeiss software). Maximum intensity projection of Z stacks acquired on a Zeiss Axio Imager Z2 epifluorescence microscope with a Photometrics sCMOS Prime camera (resolution x, y = 0.065 μm). Scale bars, 10 μm.
Troubleshooting
Problem 1
RNA smiFISH signal indistinguishable from background.
Potential Solutions
Change reagent(s) that might be contaminated with RNAse. Bovine serum albumin (BSA) is a likely suspect. Collaborators have reported that they omitted the BSA from the smiFISH hybridization buffer and this solved the low signal problem. Use filter tips when pipetting.
If there is high endogenous fluorescence in your cell type in one channel, changing fluorophores for the FLAP oligo may help. In addition, if the lightsource is a Metal Halide lamp, there may not be enough power to excite Cy5 sufficiently for single-molecule imaging, Cy3 or AF488 could work much better.
If your RNA length permits, try adding more probes.
Problem 2
Immunofluorescence detection no longer works when combined with smiFISH.
Potential Solutions
A solution of 2× SSC 10% formamide can be used for smiFISH incubation and washing, this lower formamide concentration might help to maintain the epitope for some antibodies.
The order of steps can be reversed, i.e., permeabilization, blocking, and antibody incubation can be performed on the first day, followed by smiFISH incubation overnight. However, the risk of RNA degradation is greater if antibody incubation is done first. Figure 5 shows the results of an IF first-smiFISH second experiment.
The use of a primary antibody directly tagged with the dye of choice can sometimes resolve a high background problem and be ideal for smiFISH combined with IF.
If the protein of interest is GFP or mCherry-tagged, the fluorescence of these tags can be conserved during fixation and smiFISH procedure. Make sure that the pH of the fixation solution is close to pH 7.4 in order to preserve fluorescence.
Resource Availability
Lead Contact
Pascal Chartrand (p.chartrand@umontreal.ca).
Materials Availability
For cell lines, please refer to Laprade et al., 2020. All other materials are available commercially.
Data and Code Availability
The 16 bit TIFF files for the microscopy images in Figures 2, 4, and 5 are available on a Mendeley archive created for this paper. The ImageJ 3D Suite analysis files are included for Figure 4. Software is listed in the Key Resources Table.
Acknowledgments
This work was funded by the Canadian Institutes of Health Research (PJT-162156) and the Fonds de Recherche du Québec-Santé (FRQS). We acknowledge the microscopy core facility in Biochemistry and Molecular Medicine at Université de Montréal.
Author Contributions
E.Q. developed the protocol. A.S. and P.C. supervised and provided comments on the text.
Declaration of Interests
A.S. is a co-founder of Repare Therapeutics and a member of its scientific advisory board.
Contributor Information
Emmanuelle Querido, Email: e.querido@umontreal.ca.
Pascal Chartrand, Email: p.chartrand@umontreal.ca.
References
- Adivarahan S., Livingston N., Nicholson B., Rahman S., Wu B., Rissland O.S., Zenklusen D. Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol. Cell. 2018;72:727–738. doi: 10.1016/j.molcel.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade H., Querido E., Smith M.J., Guérit D., Crimmins H., Conomos D., Pourret E., Chartrand P., Sfeir A. Single-molecule imaging of telomerase RNA reveals a Recruitment – Retention model for telomere elongation. Mol. Cell. 2020;79:115–126. doi: 10.1016/j.molcel.2020.05.005. [DOI] [PubMed] [Google Scholar]
- Ollion J., Cochennec J., Loll F., Escudé C., Boudier T. TANGO: A Generic Tool for High-throughput 3D Image Analysis for Studying Nuclear Organization. Bioinformatics. 2013;29:1840–1841. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E., Dekakra-Bellili L., Chartrand P. RNA fluorescence in situ hybridization for high-content screening. Methods. 2017;126:149–155. doi: 10.1016/j.ymeth.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to Image J: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov N., Samacoits A., Chouaib R., Traboulsi A.M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44:e165. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tomlinson R.L., Lukowiak A.A., Terns R.M., Terns M.P. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 16 bit TIFF files for the microscopy images in Figures 2, 4, and 5 are available on a Mendeley archive created for this paper. The ImageJ 3D Suite analysis files are included for Figure 4. Software is listed in the Key Resources Table.