Abstract
Chemoresistance is a major cause of treatment failure and poor outcome in neuroblastoma. In this study, we investigated the expression and function of dual-specificity phosphatase 26 (DUSP26), also known as mitogen-activated protein kinase phophatase-8, in human neuroblastoma. We found that DUSP26 was expressed in a majority of neuroblastoma cell lines and tissue specimens. Importantly, we found that DUSP26 promotes the resistance of human neuroblastoma to doxorubicin-induced apoptosis by acting as a p53 phosphatase to downregulate p53 tumor suppressor function in neuroblastoma cells. Inhibiting DUSP26 expression in the IMR-32 neuroblastoma cell line enhanced doxorubicin-induced p53 phosphorylation at Ser20 and Ser37, p21, Puma, Bax expression as well as apoptosis. In contrast, DUSP26 overexpression in the SK-N-SH cell line inhibited doxorubicin-induced p53 phosphorylation at Ser20 and Ser37, p21, Puma, Bax expression and apoptosis. Using in vitro and in vivo assays, we found that DUSP26 binds to p53 and dephosphorylates p53 at Ser20 and Ser37. In this report, we show that DUSP26 functions as a p53 phosphatase, which suppresses downstream p53 activity in response to genotoxic stress. This suggests that inhibition of this phosphatase may increase neuroblastoma chemosensitivity and DUSP26 is a novel therapeutic target for this aggressive pediatric malignancy.
Keywords: neuroblastoma, DUSP26, MKP8, doxorubicin, p53, phosphatase
Introduction
Neuroblastoma is the most common extracranial solid tumor in children and is responsible for 15% of all cancer-related deaths within this population. Multidisciplinary therapeutic approaches including chemotherapy, bone marrow transplantation, radical tumor resections and radiation therapy have affected the outcomes in low-risk neuroblastoma. However, o20% of patients with high-risk neuroblastoma live past 5 years (Brodeur, 2003). These high-risk patients are the most difficult to treat because of the highly resistant nature of these tumors. Many clinical trials are underway to target these therapy resistance pathways using small molecule inhibitors in hopes of increasing the efficacy of current treatment modalities.
The tumor suppressor p53 is a nuclear transcription factor that has a central role in the pathogenesis of human cancers (Levine, 1997; Hupp et al., 2000; Vogelstein et al., 2000). The p53 protein is normally expressed at low levels in a latent form in the cytoplasm. Many stress signals and oncogenic changes activate p53 protein and lead to cell cycle arrest or apoptotic cell death. The regulation of p53 tumor suppressor activity is complex and includes posttranslational events such as phosphorylation and acetylation. Phosphorylation at several serine and threonine residues in p53 has been shown to occur after cells are exposed to DNA-damaging agents and is critical for p53 to function as an effective tumor suppressor (Kruse and Gu, 2009). Aberrant regulation of the p53 tumor suppressor pathway has been suggested as an important mechanism of chemoresistance in neuroblastoma (Tweddle et al., 2003).
Available evidence suggests that neuroblastoma maintains functional p53; however, the downstream apoptotic mechanisms are suppressed in the majority of primary and relapsed cases (Moll et al., 1995; Goldman et al., 1996; Chen et al., 2007). More than 50% of all human cancers have p53 gene mutations or deletions (Vousden and Lu, 2002). The p53 protein is mostly wild type (WT) in neuroblastoma, suggesting that inhibition of p53 tumor suppressor function contributes to chemoresistance, tumor metastasis and poor patient survival (Moll et al, 1995; Tweddle et al., 2001). Thus, the strategy of restoring p53 function represents an attractive therapeutic approach to this cancer (Tweddle et al., 2003).
Dual-specificity phosphatases (DUSPs) are a subset of protein tyrosine phosphatases, many of which dephosphorylate serine, threonine, and tyrosine residues on mitogen-activated protein kinases, and hence are also referred to as mitogen-activated protein kinase phosphatases. This group of phosphatases has been shown to have an important role in the regulation of intracellular signaling cascades governing cell growth, differentiation and apoptosis (Jeffrey et al., 2007; Patterson et al., 2009). In some types of cancer cells, the expression level of a DUSP with oncogenic potential is elevated. For example, overexpression of the Cdc25 phosphatase in many tumors is associated with a poor prognosis (Pestell et al., 2000). In other cancers, a DUSP with tumor suppression potential is inactivated. For example, the DUSP6/mitogen-activated protein kinase phophatase-3 protein has been suggested to be a strong candidate tumor suppressor gene at 12q22 locus in pancreatic cancer (Furukawa et al., 2003). In our previous report, we cloned and identified DUSP26, also known as mitogen-activated protein kinase phophatase-8, as a p38 phosphatase (Vasudevan et al., 2005). Moreover, recent studies suggest DUSP26 could promote cell survival of anaplastic thyroid cancer cells by inhibiting p38 kinase phosphorylation (Yu et al., 2007).
In this study, we examine the role of DUSP26 in the resistance of neuroblastoma cells to doxorubicin-induced apoptosis. We provide experimental evidence that DUSP26 inhibits p53 tumor suppressor function by dephosphorylating p53 at Ser20 and Ser37 and contributes to the resistance of neuroblastoma cells to doxorubicin-induced apoptosis.
Results
DUSP26 is overexpressed in majority of human neuroblastoma cell lines and primary tissue specimens
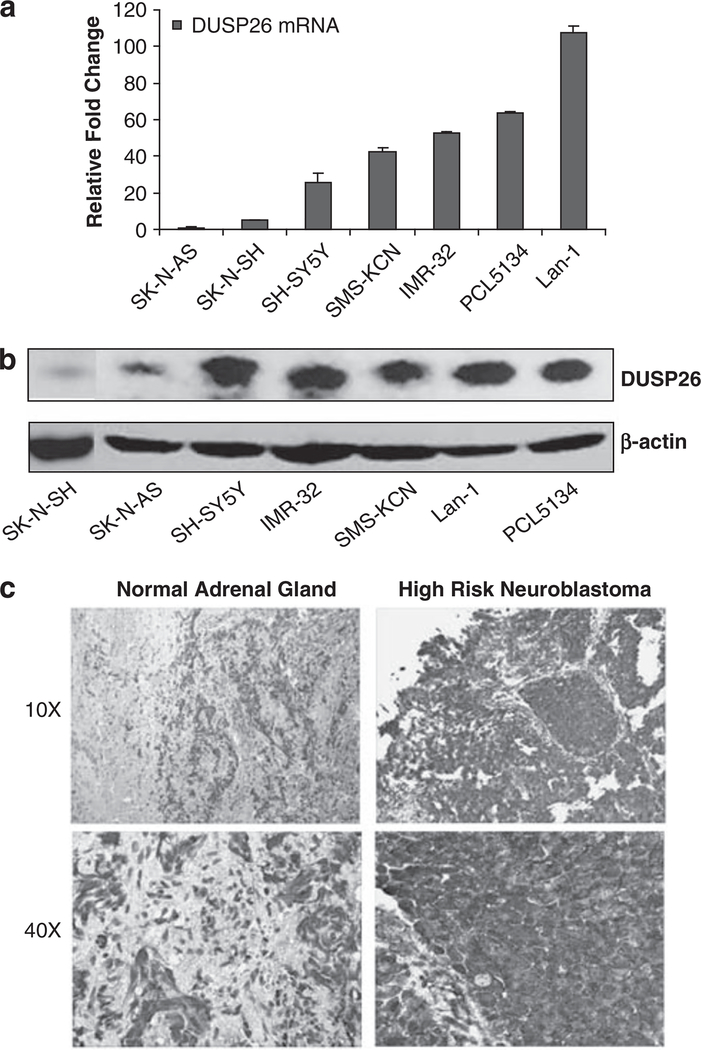
To test the prevalence of DUSP26 expression in neuroblastoma, we first measured the protein and mRNA levels of DUSP26 in seven cultured neuroblastoma cell lines. As shown in Figure 1a, five of the seven neuroblastoma cell lines have high levels of DUSP26 mRNA expression, ranging from 25.8 to 107.5-fold higher than the low expressing SK-N-AS and SK-N-SH cell lines. In addition, these five neuroblastoma cell lines have high DUSP26 protein expression (Figure 1b).
Figure 1.
DUSP26 expression in neuroblastoma cell lines and primary tumor specimens. (a) DUSP26 mRNA level in human neuroblastoma cell lines determined by quantitative reverse transcriptase (RT)–PCR. (b) DUSP26 protein level in human neuroblastoma cell lines determined by immunoblotting analysis. (c) Immunohistochemistry analysis of DUSP26 expression in primary neuroblastoma tumor specimens. Neuroblastoma paraffin-embedded tissues were stained with anti-DUSP26 antibodies and developed with DAB substrate and hematoxylin counterstain. Slides were viewed at × 10 and × 40 magnification.
Using a rabbit-polyclonal antibody against DUSP26, we stained paraffin-embedded tissue samples for DUSP26 expression. A total of 17 samples were stained and read in a blinded manner by an attending pediatric pathologist. Figure 1c shows the specificity of DUSP26 staining in the neuroblastoma cells; whereas, the adjacent normal adrenal tissue shows minimal to no staining. We analyzed DUSP26 expression with an immunohistochemistry score determined by adding the % cells stained score (scale of 1–4) and staining intensity score (scale 1–3) for a total score scale of 1–7. The high-risk neuroblastoma samples (categorized according to the Children’s Oncology Group risk stratification system) showed the highest levels of DUSP26 expression with a mean score of 5.14. This was statistically significant compared with the low-risk group when analyzed with Student’s t-test (Supplementary Table 1).
As DUSP26 was previously shown to be amplified at the DNA level in anaplastic thyroid cancer (Yu et al., 2007), we performed fluorescence in situ hybridization analysis of DUSP26 with six neuroblastoma cell lines and found that DUSP26 is not amplified in these neuroblastoma cell lines (data not shown).
DUSP26 inhibits doxorubicin-induced apoptosis in neuroblastoma
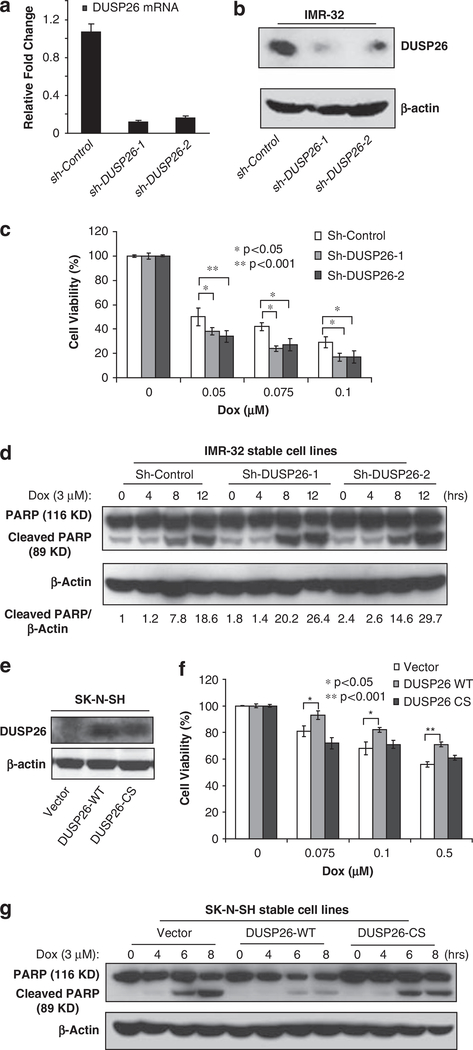
As DUSP26 is overexpressed in human NB cell lines, DUSP26 may have an important role in the chemoresistance of the cancer cells. To test this hypothesis, we first examined the effect of DUSP26 knockdown on doxorubicin-induced cellular toxicity in p53 wild-type neuroblastoma cells. As shown in Figures 2a and b, DUSP26 expression is effectively suppressed by two independent, retrovirally delivered short hairpin RNA sequences directed against DUSP26 (sh-DUSP26–1 and sh-DUSP26–2) compared with the short hairpin RNA control (sh-Control) in the IMR-32 human neuroblastoma cell line. After a treatment period with various concentrations of doxorubicin, there is a significant enhancement of doxorubicin-induced apoptosis in the two sh-DUSP26-transduced cell lines compared with the sh-Control-transduced cells (Figure 2c). To confirm this result, we examined the effect of DUSP26 knockdown on doxorubicin-induced PARP cleavage. In this assay, we found that both sh-DUSP26 cell lines show more doxorubicin-induced PARP cleavage compared with sh-Control cells (Figure 2d).
Figure 2.
DUSP26 promotes the resistance of neuroblastoma cells to doxorubincin-induced cytotoxicicity. Knockdown of DUSP26 expression in IMR-32 cells stably transduced with sh-Control and two sh-DUSP26 constructs detected by quantitative reverse transcriptase (RT)-PCR (a) and immunoblotting analysis (b) after 10 days of puromycin (2 μg/ml) selection. (c) The IMR-32 sh-Control, sh-DUSP26–1 and sh-DUSP26–2 cell lines were plated in 96-well plates at 1 × 104 cells per well. After 24 h of growth, all of the cell lines were treated with the indicated μM concentration of doxorubicin for 24 h. Cell viability was determined with the CCK-8 cell viability assay relative to the 0 μM group. All experiments were performed in triplicate and statistical significance was determined by Student’s t-test comparing each sh-DUSP26 with sh-Control group where *P<0.05 and **P<0.001. (d) Knockdown of DUSP26 expression enhances doxorubicin-induced PARP cleavage in neuroblastoma cells. Cells were treated with or without doxorubicin (3 μM) for indicated time points, harvested and subjected to immunoblotting analysis with anti-PARP antibodies to detect the presence of full-length (116 kDa) and cleaved (89 kDa) PARP proteins. The relative level of the cleaved PARP was indicated. (e) DUSP26 expression in SK-N-SH cells stably transduced with pBabe-vector control, DUSP26-WT and DUSP26-CS were analyzed by immunoblotting analysis with anti-DUSP26 antibodies after 10 days of puromycin (2 μg/ml) selection. (f) The SK-N-SH pBabe-vector control, DUSP26-WT and DUSP26-CS cell lines were plated in 96-well plates at 1 × 104 cells per well. After 24 h of growth, all of the cell lines were treated with the indicated μm concentration of doxorubicin for 24 h. Cell viability was determined with the CCK-8 cell viability assay relative to the 0 μm group. All experiments were performed in triplicate and statistical significance was determined by Student’s t-test comparing DUSP26-WT with the vector control group where *P<0.05 and **P<0.001. (g) The SK-N-SH pBabe-vector control, DUSP26-WT and DUSP26-CS cells were seeded into 6-cm dishes for two days. The cells were either left untreated or stimulated with 3 μM doxorubicin for 4, 6 or 8 h. After that, cells were harvested and subjected to immunoblotting analysis with anti-PARP antibodies.
To further validate our results, we also overexpressed DUSP26-WT and -C152S (CS) phosphatase-deficient mutant in p53 WT SK-N-SH cells with a low baseline expression of DUSP26. In this assay, we found that overexpression of DUSP26-WT promoted the resistance of the cells to doxorubicin-induced cellular toxicity and PARP cleavage compared with the vector control and DUSP26-CS mutant (Figures 2e–g).
DUSP26 modulates p53 phosphorylation and function in neuroblastoma cells
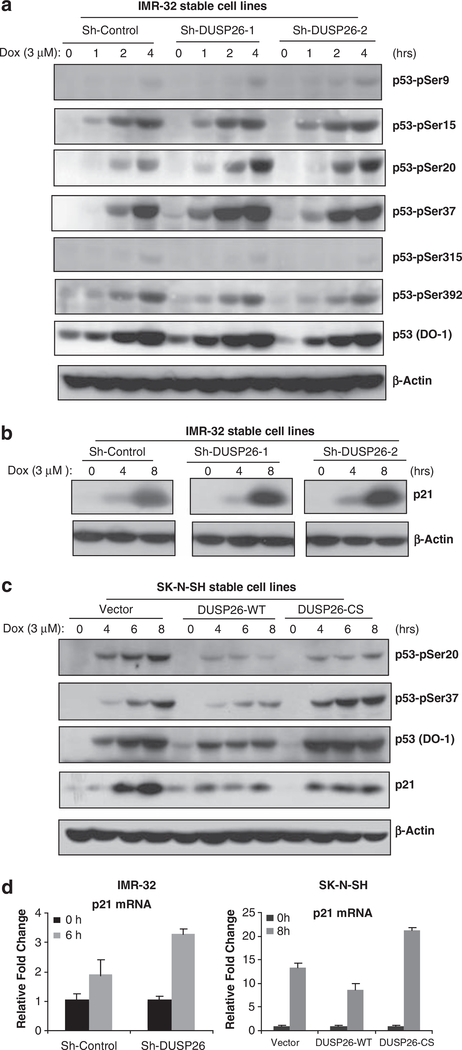
To determine the mechanism of DUSP26 function in chemoresistance of neuroblatoma cells, we tested the effects of DUSP26 suppression and overexpression on doxorubicin-induced p53 phosphorylation and activation in neuroblastoma cells. In this assay, we used a group of anti-phospho-p53 antibodies to screen the possible phospho-p53 residues that are modulated by DUSP26 in neuroblastoma cells. Interestingly, we found that knockdown of DUSP26 expression significantly enhanced doxorubicin-induced p53 phosphorylation at Ser20 and Ser37 residues in IMR-32 neuroblastoma cell line. In addition, overexpression of DUSP26-WT inhibited p53 phosphorylation at Ser20 and Ser37 residues compared with vector control and DUSP26-CS mutant in the transduced SK-N-SH cells (Figures 3a and c). Consistent with this result, doxorubicin induced a higher level of p21 expression in IMR-32 cells with DUSP26 knockdown whereas overexpression of DUSP26-WT but not -CS mutant inhibited doxorubicin-induced p21 expression in SK-N-SH cells (Figures 3b–d). Furthermore, we also found that doxorubicin induced a higher level of p53 target genes such as Bax, p53AIP1 and Puma in DUSP26 knockdown IMR-32 cells whereas overexpression of DUSP26-WT but not -CS mutant inhibited the expression of Bax, p53AIP1 and Puma in SK-N-SH cells (Supplementary Figure 1). We also found that knockdown of DUSP26 expression enhanced doxorubicin-induced p53 phosphorylation, p53 target gene expression and apoptosis in SH-SY5Y—another neuroblastoma cell line with higher DUSP26 expression (Supplementary Figure 2). In addition, we found that DUSP26 expression negatively modulated etoposide (VP-16)-induced p53 phosphorylation, p53 target gene expression and apoptosis in a similar manner to the effect observed with doxorubicin (Supplementary Figures 3a and b).
Figure 3.
DUSP26 inhibits doxorubicin-induced p53 phosphorylation at Ser-20 and Ser-37 residues and downstream p53 activity. Sh-Control, sh-DUSP26–1 and sh-DUSP26–2 IMR-32 cells were seeded into 6-cm dishes for 2 days and then were either left untreated or treated with 3 μm of DOX for 1, 2, 4 or 8 h. Then cells were harvested and subjected to immunoblotting analysis with an array of (a) anti-phospho-p53 and (b) anti-p21 antibodies. (c) Vector control, DUSP26-WT and -CS mutant SK-N-SH cell lines were seeded into 6-cm dishes for 2 days and then were either left untreated or treated with 3 μm of DOX for 4, 6 or 8 h. Cells were then harvested and subjected to immunoblotting analysis with anti-p53-pSer20, -pSer37, p53 (DO-1) and anti-p21 antibodies. (d) The p21 transcript level was detected by quantitative reverse transcriptase (RT)–PCR in IMR-32 cells and SK-N-SH cells.
DUSP26 modulates p53 function by binding to and dephosphorylating p53 at Ser20 and Ser37
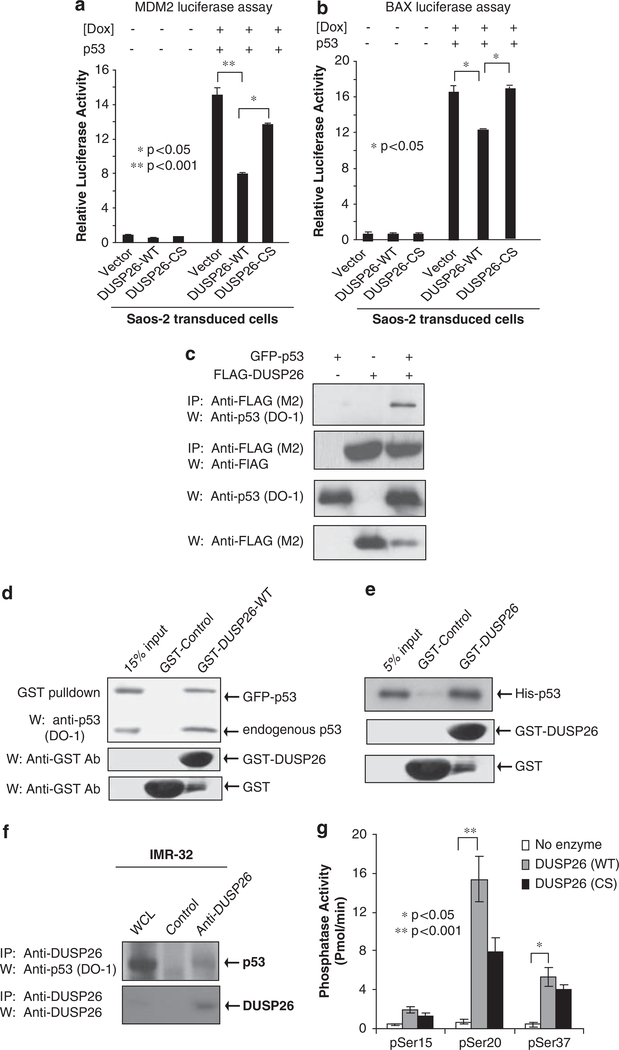
Having established a connection between DUSP26 phosphatase activity and p53 phosphorylation and downstream function, we further examined the functional interactions between DUSP26 and p53. The p53-dependent MDM2-luciferase reporter plasmid was first transfected with or without the p53 expression plasmid into Saos-2 cells stably transduced with empty vector, DUSP26-WT or -CS mutant. Then cells were treated with or without doxorubicin. In this assay, we found that DUSP26-WT strongly inhibits p53-dependent MDM2-luciferase reporter activity compared with vector control and DUSP26-CS mutant (Figure 4a). A similar result was obtained when the p53-dependent Bax-luciferase reporter plasmid was used in this assay (Figure 4b).
Figure 4.
DUSP26 functionally and physically interacts with p53 and acts as a p53 phosphatase in vitro. (a, b) One microgram of MDM2 (a) or Bax (b) firefly-luciferase reporter and 20 ng of Renilla-Luc plasmids were co-transfected into Saos-2 cells transduced with vector, DUSP26-WT, DUSP26-CS along with p53 as indicated. At 48 h after transfection, cells were exposed to doxorubicin (1 μm) for 24 h as indicated. The relative luciferase activity was measured and normalized with the Renilla activity. Error bars indicate±s.d. in duplicate experiments. Statistical significance was determined by Student’s t-test where Po0.05 was considered significant. (c) FLAG-DUSP26 and green fluorescent protein (GFP)-p53 expression plasmids were cotransfected into HEK293T cells. After 48 h of incubation, cells were lysed and immunoprecipitation was performed with anti-FLAG Ab. Immunoblotting analysis was then performed with anti-p53 antibodies (DO-1). (d) GST-control and GST-DUSP26-WT full length (FL) were purified from Escherichia coli and bound to glutathione sepharose resin. This resin was then incubated with lysate from GFP-p53-transfected HEK293T cells. Immunoblotting analysis was then performed with anti-p53 antibodies (DO-1). (e) The same resin was co-incubated with purified recombinant His-p53. Resin was then washed, resolved on SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotting analysis was performed with anti-p53 Ab (DO-1). (f) Lysates from parental IMR32 cells after doxorubicin stimulation were immunoprecipitated with anti-DUSP26 antibodies and subjected to immunoblotting analysis with anti-p53 antibodies (DO-1). (g) The p53 phosphopeptides indicated were co-incubated with purified recombinant GST-DUSP26-WT or -CS mutant in an in vitro phosphatase assay.
To determine whether DUSP26 physically binds to p53, we co-transfected the green fluorescent protein-p53 expression plasmid with vector control or FLAG-DUSP26 into HEK293T cells. The FLAG-DUSP26 proteins were immunoprecipitated from the cell lysates with anti-FLAG antibodies and immunoblotted with antibodies specific for p53 (DO-1). As shown in Figure 4c, we found that FLAG-DUSP26 pulls down green fluorescent protein-p53. Consistently, we found that immunoprecipitating 53 with the anti-p53 (DO-1) antibodies pulls down FLAG-DUSP26 (Supplementary Figure 4a). Using an in vitro assay with recombinant protein, we found that recombinant glutathione S-transferase (GST)-DUSP26-WT binds to both endogenous and overexpressed p53 from HEK293T cell lysates (Figure 4d). We also found that the GST-DUSP26-CS mutant binds overexpressed p53 in HEK293T cells suggesting that this binding occur independent of the phosphatase activity (Supplementary Figure 4b). Furthermore, recombinant GST-DUSP26-WT binds to recombinant His-p53 in vitro, suggesting that that these two proteins directly bind without any intermediary proteins (Figure 4e).
The anti-DUSP26 antibodies were tested for DUSP26 immunoprecipitation using the FLAG-DUSP26 expression plasmid in HEK293T cells. We found effective immunoprecipitation of FLAG-DUSP26 as confirmed by immunoblotting with anti-FLAG antibodies (Supplementary Figure 5a). To further confirm the interaction between DUSP26 and p53 in neuroblastoma cells, neuroblastoma cells were first treated with doxorubicin to induce p53 expression, and then endogenous DUSP26 was immunoprecipitated from IMR-32 cell lysate with anti-DUSP26 antibodies and immunoblotted for p53. As shown in Figure 4f, endogenous DUSP26 co-immunoprecipitates with endogenous p53. Effective p53 immunoprecipitation also found in an additional neuroblastoma cell line SH-SY5Y with a high endogenous DUSP26 expression (Supplmentary Figure 5b).
To further confirm the role of DUSP26 as a p53 phosphatase targeting on p53 Ser20 and Ser37 residues, we performed an in vitro phosphatase assay to address whether DUSP26 directly dephosphorylates p53 at these two sites. In this assay, p53 Ser15, Ser20 and Ser37 phosphopeptides were co-incubated with recombinant GST-DUSP26-WT and -CS mutant proteins in an in vitro phosphatase reaction. DUSP26 WT showed a higher level phosphatase activity toward p53 phosphoSer20 and phospho-Ser37 compared with phosphoSer15 (Figure 4g). These results strongly suggest that DUSP26 physically interacts with p53 and functionally inhibits p53 function by modulating p53 phosphorylation at Ser20 and Ser37 residues in neuroblastoma cells.
Discussion
Neuroblastoma is a p53 WT tumor for which relapse with chemoresistant disease is the primay cause of death (Brodeur, 2003). The mechanism for this chemoresistance in the presence of intact apoptotic machinery remains unclear. In this study, we examine the role of DUSP26 in promoting neuroblastoma chemoresistance. As has been shown with other important DUSPs, we present evidence that DUSP26 is overexpressed in neuroblastoma cell lines and in high-risk neuroblastoma tissue samples. The high-risk tissue samples represent patients with high-risk features such as age >1 year of age, MYCN amplification, poorly differentiated histopathologic characteristics and poor prognosis (London et al., 2005).
Our studies show that DUSP26 overexpression may be one of the mechanisms by which neuroblastoma develops chemoresistance. We found that overexpression of DUSP26 in a low DUSP26 expressing cell line increased cell viability in the setting of doxorubicin treatment by both cell viability assay and PARP cleavage. With DUSP26 inhibition with short hairpin RNA, the cells became more chemosensitive to doxorubicin treatment showing that p53 is essential for chemotherapy-related cell death in neuroblastoma. Doxorubicin-induced genotoxic stress is an accepted model of p53 transactivation and has been a mainstay of therapy for neuroblastoma for many years (Lowe et al., 1994; Matthay et al., 1998).
In neuroblastoma, the p53 protein is most commonly in the WT form and is functional. p53 has an essential role in tumor suppression and in the cellular response to stress (Toledo and Wahl, 2006). Phosphorylation of p53 protein is involved in the regulation of p53 protein stability and biochemical activities. Many serine residues within the N- and C-terminal regions of p53 are phosphorylated in response to stress conditions such as hypoxia and DNA damage (Kruse and Gu, 2009). Both Ser20 and Ser37 phosphorylation on the p53 protein has been shown to have an important role in p53 stability and downstream apoptotic function (Chehnab et al., 1999; Unger et al., 1999; Li et al., 2006; Amano et al., 2009).
Previously, human Cdc14 phosphatase has been reported to interact with and dephosphorylate p53 at Ser-315 while PPM1D dephosphorylates p53 at Ser15 without binding (Li et al., 2000; Lu et al., 2005). The results from our in vitro phosphatase assays suggest that p53 Ser20 and Ser37 are specifically targeted by DUSP26 compared with Ser15. In addition, our DUSP26 knockdown and overexpression data further suggest that p53 Ser20 and Ser37 are targeted in vivo by DUSP26. These data indicate that phosphorylation of Ser20 and Ser37 facilitate p53 tumor suppressor function in neuroblastoma. Overexpression of DUSP26 in neuroblastoma results in physical binding and dephosphorylation of p53 in vivo which downregulates p53 tumor suppressor activity in neuroblastoma cells in response to doxorubicin. Interestingly, we found that doxorubincin and VP-16-induced p21 expression was partially inhibited by DUSP26-CS mutant (Figure 3c, Supplementary Figure 3b). One explanation is that DUSP26-CS mutant may still have residual phosphatase activity. However, we can not exclude the possibility that binding of DUSP26-CS mutant to p53 causes some level of inhibition. It is also possible that the physical interaction of DUSP26-p53 and the enzymatic activity of DUSP26 both have inhibitory effects on p53 phosphorylation.
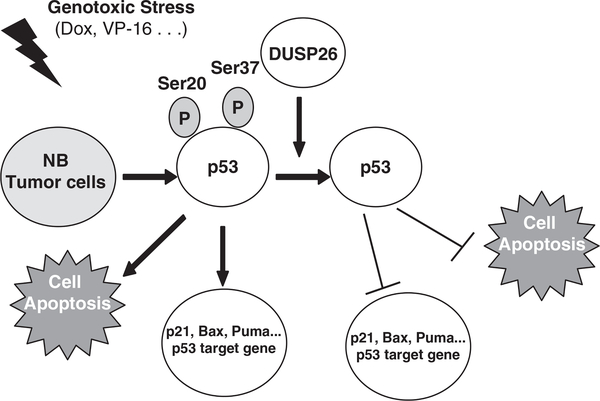
Our data, in conjunction with previous studies, suggest a model in which genotoxic stress/DNA damage causes p53 induction and phosphorylation in neuroblastoma. DUSP26 binds to and dephosphorylates p53 at Ser 20 and Ser37 to inhibit p53 tumor suppressor function in neuroblastoma cells (Figure 5). We suggest that overexpression of DUSP26 has an important role in the capacity of neuroblastoma cells to overcome the mechanisms that promote apoptosis and inhibit tumor progression.
Figure 5.
A working model for DUSP26-mediated p53 dephosphorylation and inactivation. Genotoxic stress such as doxorubicin and VP-16 induces p53 expression and phosphorylation in neuroblastoma cells. DUSP26 binds to and dephosphorylates p53 at Ser20 and Ser37 residues to inhibit p53 tumor suppressor function and promotes the resistance of tumor cells to doxorubicin-induced cell death.
Thus, overexpression of DUSP26 may be one of the mechanisms in neuroblastoma to escape doxorubincin-induced cell death and gain chemoresistence. Therefore, therapies designed to interfere with the DUSP26-p53 functional interaction would increase chemosensitivity by reactivating cellular apoptosis pathways to promote tumor cell death and better response to therapy.
Materials and methods
Patients and tumor tissue collection
The neuroblastoma patients participating in this study were recruited from the Texas Children’s Cancer Center at Texas Children’s Hospital in Houston, TX, USA from 1995 through 2006. All of the procedures were approved by an institutional review board. Fresh tumor tissues were collected from patients with pathologically and clinically confirmed neuroblastoma. A portion of tumor specimens were kept in −80 °C and sectioned for total RNA and protein extraction. Children’s Oncology Group risk group and INSS stage assignments were conducted by reference to the medical record.
Cell culture and compound treatment
Human neuroblastoma tumor cell lines (IMR-32, SK-N-SH, SH-SY5Y, SK-N-AS, LAN-1) and HEK293T cells, were obtained from the American Type Culture Collection, (Manassas, VA, USA), and the neuroblastoma cell lines SMS-KCN and PCL-5134 were kindly provided by Dr A Davidoff at the St Jude’s Children’s Hospital, Memphis, TN, USA. Briefly, cell lines were maintained in minimum essential medium (IMR-32, SK-N-SH, SH-SY5Y, Lan-1), and in Dulbecco’s modified Eagle’s medium (SK-N-AS, HEK293T). All of media were supplemented with 10% heat-inactivated fetal calf serum, 2 mm glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin (all from Invitrogen, Gaithersburg, MD, USA). Doxorubicin and VP-16 (Sigma-Aldrich) were diluted in Hanks’ balanced salt solution buffer and dimethylsulphoxide, respectively, and added to the cell culture medium at a final concentration of 0.05–3 μm.
Quantitative reverse transcription–PCR
Total RNA was extracted from tumor cell lines using TRIZOL(R) reagent (Invitrogen, Carlsbad, CA, USA), and the purity of RNA was determined by measuring the absorbance at 260/280 nm (A260/A280) in a spectrophotometer. The following primer pairs were used: DUSP26 mRNA (forward, 5′-AACCTGTCTTGGGCAGAAAC-3′; reverse 5′-ATGGGTTCAGTTGCCAGGTA-3′); p21WAF1/CIP1 mRNA (forward, 5′-CGAAAACGGCGGCAGACCAGCATGA-3′; reverse, 5′-TGAGGCCCTCGCGCTTCCAGGAC-3′); Bax mRNA (forward, 5′-AACTGGTGCTCAAGGCCCTG-3′; reverse, 5′-GGGTGAGGAGGCTTGAGGAG-3′); p53 AIP1 mRNA (forward, 5′-TCTTCCTCTGAGGCGAGCT-3′; reverse, 5′-AGGTGTGTGTGTCTGAGCCC-3′); Puma mRNA (forward, 5′-GACGACCTCAACGCACAGTA-3′; reverse, 5′-GG AGTCCCATGATGAGATTGT-3′) and glyceraldehyde 3-phosphate dehydrogenase mRNA (forward, 5′-CCACATCGCTCA GACACCAT-3′; reverse 5′-GGCAACAATATCCACTTTACC AGAGT-3′). Glyceraldehyde 3-phosphate dehydrogenase was used as internal control.
Antibodies
Antibodies specific for anti-p53-pSer9, anti-p53-pSer15, anti-p53-pSer20, anti-p53-pSer37, anti-p53-pSer315, anti-p53-pSer392, anti-Puma, horseradish peroxidase-linked anti-rabbit immunoglobulin secondary antibodies and horseradish peroxidase-linked anti-mouse immunoglobulin were from Cell Signaling Technology. Anti-b-actin, anti-FLAG-M2 were from Sigma-Aldrich. Anti-DUSP26 was from MBL International Corporation. Anti-p21 (C-19), anti-Bax (N-20), anti-p53AIP1 (G-20), anti-p53 (DO-1), and horseradish peroxidase-linked anti-goat immunoglobulin secondary antibodies were from Santa Cruz Biotechnology.
Immunohistochemical staining
Immunohistochemical was performed to detect DUSP26 protein expression on sections of paraffin-embedded neuroblastoma tumor tissue. Immunohistochemical staining was performed using Two-Step Histostaining Reagent (Zymed Laboratories Inc., South San Francisco, CA, USA) according to the manufacturer’s instructions. The anti-DUSP26 antibody (1:50) was used as primary antibody. Diaminobenzidine tetrahydrochloride was then used as a chromogen, followed by counterstaining with hematoxylin solution.
Immunoblotting and immunoprecipitation
Cells were harvested in ice-cold phosphate-buffered saline (pH 7.4) and spun down. The pellets were dissolved in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% IGEPAL, 0.25% Na-deoxycholate, 1 mm phenylmethyl-sulfonyl fluoride, 1 mm dithiothreitol, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm benzamidine, 20 mm disodium p-nitrophenylphosphate, 0.1 mm sodium orthovanadate, 10 mm sodium fluoride, phosphatase inhibitor cocktail A and B (Sigma-Aldrich)). The cell lysates were either subjected directly to 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis for immunoblotting analysis or immunoprecipitated for 3 h with the indicated antibodies. Immune complexes were recovered with protein A/G-agarose (Santa Cruz Biotechnology) for 3 h, then washed three times with wash buffer containing 20 mm HEPES (pH 7.4), 50 mm NaCl, 2.5 mm MgCl2, 0.1 mm EDTA, and 0.05% Triton X-100. Then the immunoprecipitates or 10% whole cell lysates were resolved on SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes followed by immunobloting analysis using the ECL-Plus Western blotting system (GE Healthcare Biosciences Corp., Pittsburgh, PA, USA) according to the manufacturer’s instruction.
For DUSP26 immunoblotting cell pellets were lysed in a different lysis buffer (62.5 mm Tris-HCl, 10% SDS, 10% glycerol and 50 mm dithiothreitol). Samples were then sonicated and centrifuged at 13 000 r.p.m. One hundred micrograms of protein were mixed with 0.1% bromophenol blue, boiled and resolved by SDS–polyacrylamide gel electrophoresis.
Fluorescence in situ hybridization
Metaphase chromosomes were prepared from six neuroblastoma cell lines (IMR-32, SK-N-SH, SH-SY5Y, SK-N-AS, LAN-1 and JF). Fluorescence in situ hybridization analyses were performed as described previously (Yu et al., 2007), using BAC RP11–258M15 clone located in the region of interest as probe.
Purification of recombinant GST-DUSP26 proteins
This bacterial expression plasmid (pGEX-DUSP26) was used to purify GST GST-DUSP26-WT or GST-DUSP26-CS from Escherichia coli as described before (Vasudevan et al., 2005). A modification to the sonication step was made in which 1% Sarcosyl (Sigma-Aldrich) was added to the lysis buffer before sonication, and Triton X-100 (1%), 5 μg/ml DNase, and 5 μg/ml RNase (Roche) were added after sonication. The recombinant proteins were then eluted with 20 mm glutathione in 50 mm Tris (pH = 8.0) and dialyzed in a buffer containing 20 mm HEPES, 150 mm KCl, 0.2 mm EDTA, 20% glycerol and protease inhibitors. The protein concentrations were then assessed with a Bradford Protein Assay (Bio-Rad). The proteins were visualized by 10% SDS–polyacrylamide gel electrophoresis and Coomassie blue staining of the gel.
RNA interference and overexpression constructs and transduction
DUSP26 short hairpin RNA expression was achieved by cloning the following ligated sequences into the pSuper-PURO retrovirus expression vectors per manufacturer’s protocol (Oligoengine). The RNA interference target sequences used were as follows: pSuper-Scrambled control (sh-Control), 5′-CGTCTTTTCGGACTTAGAGAG-3′; pSuper-DUSP26–1 (sh-DUSP26–1), 5′-AAGACAGCCTGTAACCATGCC-3′, and pSuper-DUSP26–2 (sh-DUSP26–2), 5′-AAGATGTGCCC TGGTAACTGG-3′. For retroviral transduction, we used a previously published protocol (Yu et al., 2008). Stable cell lines were established in IMR-32 cells after 10 days of puromycin (2 μg/ml) selection.
The stable pBabe-vector control, DUSP26-WT and DUSP26-CS cell lines were established in SK-N-SH neuroblastoma cells. A full-length complementary DNA clone containing the DUSP26 gene was obtained from American Type Culture Collection (IMAGE Consortium ID 3535215). DUSP26 ORF was amplified with the following primers: 5′-CACCAAGCTTACCATGTGCCCTGGTAACTGG-3′ and 5′-GAGCCAGGGACCTACCCA-3′, and cloned into the pcDNA3.1 vector (Invitrogen) (DUSP26-WT). To create a catalytically inactive DUSP26, a serine was substituted for the cysteine at position 152 using a PCR-directed mutagenesis technique as described previously (Maruta et al., 1991). The DUSP26-CS mutant gene was then cloned into the pcDNA3.1 vector (DUSP26-CS). From this primary construct, DUSP26 was subcloned into p3xFLAG-CMV-10 (FLAG-DUSP26) (Sigma) using HindIII/XbaI. The following primer set, 5-CA CCGGATCCATGTGCCCTGGTAACTG-3 and 5-GAGCC AGGGACCTACCCA-3, was used to subclone DUSP26 into pGEX-KG (Pharmacia) using BamHI/XbaI. This bacterial expression plasmid (pGEX-DUSP26) was used to purify GST GST-DUSP26-WT or GST-DUSP26-CS from E. coli as described before (Vasudevan et al., 2005).
Luciferase reporter assay
The MDM2-Luc-luciferase reporter plasmid was previously described (Slack et al., 2005). The Bax-Luc-luciferase reporter plasmid was kindly provided by Dr Lawrence A Donehower (Baylor College of Medicine, Houston, TX, USA). Saos-2 cells were seeded at a concentration of 3 × 105 cells per well and cultured overnight in six-well plates. The p53 expression plasmid, p53-dependent MDM2-firefly-luciferase reporter and the control Renilla luciferase plasmid were co-transfected with empty vector, DUSP26-WT or -CS expression plasmids into the cells. At 24 h after transfection, cells were treated with or without doxorubincin (1 μm) for 24 h and then harvested in lysis buffer (Promega, Madison, WI, USA), luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity was calculated by dividing the firefly luciferase activity by the Renilla luciferase activity. Data represent three independent experiments performed in duplicate.
CCK-8 cell proliferation assay
Cell proliferation was measured by the tetrazolium salt-based proliferation assay (CCK-8 assay; Wako Chemicals, Osaka, Japan) following the manufacturer’s instructions and as described previously (Shang et al., 2009).
In vitro protein phosphatase assays
For in vitro phosphatase assays on phosphopeptides, recombinant DUSP26-WT or -CS mutant proteins prepared from bacterial lysates was diluted in buffer (50 mm Tris-HCl [pH 7.5], 0.1 mm ethyleneglycol tetraacetic acid and 0.02% 2-mercaptoethanol) and incubated with bovine serum albumin (1 mg/ml) and 30 mm MgCl2 containing 100 μm phosphopeptide for 20 min at 30 °C. Free phosphate was determined using a malachite green/molybdate-based assay following protocols provided by the manufacturer (Promega). Absorbance was measured at 600 nm. The amount of phosphate released was calculated by a phosphate standard curve. The p53-pSer15 (Ac-VEPPL(pS)QETFS-amide), p53-pSer20 (Ac-SQETF(pS) DLWKL-amide), p53-pSer37 (Ac-LSPLP(pS)QAMDD-amide) phosphopeptides were synthesized by New England Peptides (Gardner, MA, USA).
Statistical analyses
A two-tailed Student’s t-test was used for statistical analysis of comparative data using Microsoft Excel software (Microsoft Corp., Redmond, WA, USA). Values of P<0.05 were considered as significant and indicated by asterisks in the figures. All statistical tests were two-sided and a P-value of <0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Dr Xiangwei Wu for providing GFP-p53 expression plasmid and Mdm2-Luc reporter. The work was supported in part by the grants from the Hope Street Kids Foundation (to JY), the Bear Necessities Pediatric Cancer Foundation (to JY), the American Cancer Society Grant RSG-06-070-01-TBE (to JY), the Society of University Surgeons Ethicon Resident Scholarship (SAV) and the NIH/NCI-NRSA training grant 1F32CA113059-01A1 (SAV).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Amano T, Nakamizo A, Mishra SK, Gumin J, Shinojima N, Sawaya R et al. (2009). Simultaneous phosphorylation of p53 at serine 15 and 20 induces apoptosis in human glioma cells by increasing expression of pro-apoptotic genes. J Neurooncol 92: 357–371. [DOI] [PubMed] [Google Scholar]
- Brodeur GM. (2003). Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 3: 203–216. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. (2003). Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol 15: 164–171. [DOI] [PubMed] [Google Scholar]
- Chehnab NH, Malikazy A, Stravridi ES, Halazonetis TD. (1999). Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA 96: 13777–13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Malcolm AJ, Wood KM, Cole M, Variend S, Cullinane C et al. (2007). p53 is nuclear and functional in both undifferentiated and differentiated neuroblastoma. Cell Cycle 6: 2685–2696. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Lozano G. (2009). p53 and MDM2: antagonists or partners in crime? Cancer Cell 15: 161–162. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Sunamura M, Motoi F, Matsuno S, Horii A. (2003). Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol 162: 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SC, Chen CY, Lowsing TJ, Gilmer TM, Kastan MB. (1996). The p53 signal transduction pathway is intact in human neuroblastoma despite cytoplasmic localization. Am J Pathol 148: 1381–1385. [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Lane DP, Ball KL. (2000). Strategies for manipulating the p53 pathway in the treatment of human cancer. Biochem J 352 (Part 1): 1–17. [PMC free article] [PubMed] [Google Scholar]
- Jeffrey KL, Camps M, Rommel C, Mackay CR. (2007). Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov 6: 391–403. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. (2009). Modes of p53 regulation. Cell 137: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ. (1997). p53, the cellular gatekeeper for growth and division. Cell 88: 323–331. [DOI] [PubMed] [Google Scholar]
- Li DW, Liu JP, Schmid PC, Schlosser R, Feng H, Liu WB et al. (2006). Protein serine/threonine phosphatase-1 dephosphorylates p53 at Ser-15 and Ser-37 to modulate its transcriptional and apoptotic activities. Oncogene 25: 3006–3022. [DOI] [PubMed] [Google Scholar]
- Li L, Ljungman M, Dixon JE. (2000). The human Cdc14 phosphatases interact with and dephosphorylate the tumor suppressor protein p53. J Biol Chem 275: 2410–2414. [DOI] [PubMed] [Google Scholar]
- London WB, Castleberry RP, Matthay KK, Look AT, Seeger RC, Shimada H et al. (2005). Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children’s Oncology Group. J Clin Oncol 23: 6459–6465. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE et al. (1994). p53 status and the efficacy of cancer therapy in vivo. Science 266: 807–810. [DOI] [PubMed] [Google Scholar]
- Lu X, Nannenga B, Donehower LA. (2005). PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 19: 1162–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta H, Holden J, Sizeland A, D’Abaco G. (1991). The residues of Ras and Rap proteins that determine their GAP specificities. J Biol Chem 266: 11661–11668. [PubMed] [Google Scholar]
- Matthay KK, Perez C, Seeger RC, Brodeur GM, Shimada H, Atkinson JB et al. (1998). Successful treatment of stage III neuroblastoma based on prospective biologic staging: a Children’s Cancer Group study. J Clin Oncol 16: 1256–1264. [DOI] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. (1995). Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci USA 92: 4407–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson KI, Brummer T, O’Brien PM, Daly RJ. (2009). Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J 418: 475–489. [DOI] [PubMed] [Google Scholar]
- Pestell KE, Ducruet AP, Wipf P, Lazo JS. (2000). Small molecule inhibitors of dual specificity protein phosphatases. Oncogene 19: 6607–6612. [DOI] [PubMed] [Google Scholar]
- Shang X, Burlingame SM, Okcu MF, Ge N, Russell HV, Egler RA et al. (2009). Aurora A is a negative prognostic factor and a new therapeutic target in human neuroblastoma. Mol Cancer Ther 8: 2461–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A et al. (2005). The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci USA 102: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. (2006). Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923. [DOI] [PubMed] [Google Scholar]
- Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. (2001). Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res 61: 8–13. [PubMed] [Google Scholar]
- Tweddle DA, Pearson AD, Haber M, Norris MD, Xue C, Flemming C et al. (2003). The p53 pathway and its inactivation in neuroblastoma. Cancer Lett 197: 93–98. [DOI] [PubMed] [Google Scholar]
- Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G et al. (1999). Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J 18: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan SA, Skoko J, Wang K, Burlingame SM, Patel PN, Lazo JS et al. (2005). MKP-8, a novel MAPK phosphatase that inhibits p38 kinase. Biochem Biophys Res Commun 330: 511–518. [DOI] [PubMed] [Google Scholar]
- Vasudevan SA, Shang X, Chang S, Ge N, Diaz-Miron JL, Russel HV et al. (2009). Neuroblastoma derived secretory protein is a novel secreted factor overexpressed in neuroblastoma. Mol Cancer Ther 8: 2478–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. (2000). Surfing the p53 network. Nature 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. (2002). Live or let die: the cell’s response to p53. Nat Rev Cancer 2: 594–604. [DOI] [PubMed] [Google Scholar]
- Yu W, Imoto I, Inoue J, Onda M, Emi M, Inazawa J. (2007). A novel amplification target, DUSP26, promotes anaplastic thyroid cancer cell growth by inhibiting p38 MAPK activity. Oncogene 26: 1178–1187. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ge N, Xie M, Sun W, Burlingame S, Pass AK et al. (2008). Phosphorylation of Thr-178 and Thr- 184 in the TAK1 T-loop is required for interleukin (IL)-1-mediated optimal NFkappaB and AP-1 activation as well as IL-6 gene expression. J Biol Chem 283: 24497–24505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.