Abstract
INTRODUCTION
Antibiotic resistance in carbapenemase-producing Klebsiella pneumoniae is acquired and disseminated mainly by plasmids. Therefore, we aimed to investigate the occurrence of carbapenemase genes, analyze the genetic diversity by ERIC-PCR, and examine the most common plasmid incompatibility groups (Incs) in clinical isolates of K. pneumoniae from colonization and infection in patients from a hospital in Brazil.
METHODS
Twenty-seven isolates of carbapenem-resistant K. pneumoniae were selected and screened for the presence of carbapenemase genes and Incs by PCR, followed by amplicon sequencing.
RESULTS
The bla KPC and bla NDM genes were detected in 24 (88.8 %) and 16 (59.2 %) of the isolates, respectively. Thirteen isolates (48.1 %) were positive for both genes. The IncFIB (92.6 %) and IncQ (88.8 %) were the most frequent plasmids, followed by IncA/C, IncHI1B, and IncL/M, indicating that plasmid variability existed in these isolates. To our knowledge, this is the first report of IncHI1B in Brazil. We found eight isolates with clonal relationship distributed in different sectors of the hospital.
CONCLUSIONS
The accumulation of resistance determinants, the variability of plasmid Incs, and the clonal dissemination detected in K. pneumoniae isolates demonstrate their potential for infection, colonization, and the dissemination of different resistance genes and plasmids.
Keywords: Klebsiella pneumoniae, Antimicrobial resistance, Plasmids, Incompatibility groups
INTRODUCTION
Klebsiella pneumoniae is clinically important because it is involved in a variety of healthcare-association infections (HAI) such as in the urinary and respiratory tracts, wounds, endocarditis, and sepsis 1 , 2 , 3 . Intestinal colonization is one of the main factors favoring infection by K. pneumoniae, as colonized carriers can serve as important reservoirs for the spread of bacteria in the hospital environment. Further, K. pneumoniae can harbor resistance genes and spread them through conjugative plasmids and transposons 4 .
The excessive and indiscriminate use of beta-lactam antimicrobials has culminated in the emergence of antibiotic-resistant K. pneumoniae strains and other carbapenem-resistant Enterobacteria 1 , 5 , 6 , 7 . The development of resistance is related to the production of beta-lactamases which is mediated by conjugative plasmids 8 , 9 , 10 , 11 , 12 .
Klebsiella pneumoniae carbapenemase (KPC) has become endemic in several countries and is frequently detected in K. pneumoniae isolates from Brazilian hospitals 1 , 13 , 14 , 15 , 16 , 17 . The bla KPC gene is often located on the transposon Tn4401 (Eilertson et al. 18 ), which has been found in several transferable plasmids 4 , 19 , 20 , which ensures its dispersion among Klebsiella species and other genera of Gram-negative bacteria (Belder et al. 21 ). The bla KPC gene can also be found in non-Tn4401 elements (NTEKPC) 16 , 22 .
Additionally, resistance to carbapenems can occur due to the production of other enzymes, such as metallo-beta-lactamases, e.g., the New Delhi metallo-beta-lactamase-1 (NDM-1). Since its detection in 2008 in New Delhi, India 23 strains producing NDM-1 have been reported in many countries, including Brazil 6 , 8 , 9 , 24 , 25 . Considering the importance of knowing which resistance genes and plasmids are circulating among multi-drug resistant clinical isolates in hospitals in Brazil, we investigated the bla KPC, bla GES, bla NDM, bla VIM, and bla IMP genes, and the most common plasmid Incs in K. pneumoniae (FIB, Q, A/C, L/M, N, HI2, and HI1B) to analyze the clonal relationship between KPC resistant clinical isolates obtained from a public hospital in Recife-PE, Brazil.
METHODS
Bacterial isolates
Twenty-seven isolates of K. pneumoniae selected for being resistant to one or more carbapenems were isolated from different patients and sites of infection or colonization. The patients were admitted to a public hospital in the city of Recife-PE, Brazil, between 2017 and 2018. The isolates were kept as frozen stocks at -80ºC in 15 % glycerol.
Antimicrobial susceptibility
The Minimum Inhibitory Concentration (MIC) for the antimicrobials Amikacin (AMI); Amoxicillin-clavulanic acid (AMC); Ampicillin (AMP); Cefazolin (CFZ); Cefepime (CPM); Cephalothin (CFL); Cefotaxime (CTX); Cefoxitin (CFO); Ceftazidime (CAZ); Ceftriaxone (CRO); Cefuroxime (CRX); Ciprofloxacin (CIP); Colistin (COL); Ertapenem (ERT); Gentamicin (GEN); Imipenem (IMI); Levofloxacin (LEV); Meropenem (MER); Piperacillin-tazobactam (PIPT); Trimethoprim-sulfamethoxazole (TRIS) was determined using automated equipment from BD Phoenix 100. The susceptibility profile was interpreted according to the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) 26 .
DNA extraction and PCR conditions for resistance genes
Genomic DNA was extracted using a commercial kit as per the manufacturer’s instructions (Wizard Genomic DNA Purification kit, Promega). After extraction, the DNA was quantified using the NanoDrop 2000c UV-Vis spectrophotometer. For PCR amplification of the bla KPC, bla NDM, bla GES, bla VIM, and bla IMP genes, the primers described in Table 1 were used. The amplification reactions were prepared in a total volume of 25 μL per tube, comprising a final concentration of 25 mM MgCl2, 8 mM dNTPs, 1U Taq DNA Polymerase (Promega), 10 μM of each primer, 5×buffer, and 1 ng of DNA.
TABLE 1: Primers used in the PCR and sequencing of the amplicons in isolates of K. pneumoniae.
| Gene | Primer | Sequence (5`- 3`) | Temp.a | Amplicon size (base pair) | Reference |
|---|---|---|---|---|---|
| bla KPC | KPC-1a | TGTCACTGTATCGCCGTC | 63°C | 800 | Yigit et al. (2001) 42 |
| KPC-1b | CTCAGTGCTCTACAGAAAACC | ||||
| bla GES | GES-F | GAAACCAAACGGGAGACGC | 60ºC | 207 | Nordmann (2011) 43 |
| GES-R | CTTGACCGACAGAGGCAACT | ||||
| bla NDM | NDM-F | TAAAATACCTTGAGCGGGC | 52ºC | 439 | Nordmann (2011) 43 |
| NDM-R | AAATGGAAACTGGCGACC | ||||
| bla VIM | VIM-F | CAGATTGCCGATGGTGTTTGG | 62°C | 600 | Dong et al. (2008) 44 |
| VIM-R | AGG TGGGCCATTCAGCCAGA | ||||
| bla IMP | IMP-F | GGAATAGAGTGGCTTAATTCTC | 60°C | 232 | Dong et al. (2008) 44 |
| IMP-R | GTGATGCGTCYCCAAYTTCACT | ||||
| HI-2 | IncHI-2-F | GGAGCGATGGATTACTTCAGTAC | 64ºC | 644 | Caratolli et al. (2005) 28 |
| IncHI-2-R | GGCTCACTACCGTTGTCATCCT | ||||
| L/M | IncL/M-F | GGATGAAAACTATCAGCATCTGAAG | 62ºC | 758 | Caratolli et al. (2005) 28 |
| IncL/M-R | CTGCAGGGGCGATTCTTTAGG | ||||
| A/C | IncA/C-F | GAGAACCAAAGACAAAGACCTGGA | 62ºC | 465 | Caratolli et al. (2005) 28 |
| IncA/C-R | ACGACAAACCTGAATTGCCTCCTT | ||||
| N | IncN-F | GTCTAACGAGCTTACCGAAG | 62ºC | 559 | Caratolli et al. (2005) 28 |
| IncN-R | GTTTCAACTCTGCCAAGTTC | ||||
| HI1B | HI1B-Fw | CAA AAC GAG AGA TAT TCAACCC CTG ATT | 63ºC | 900 | Caratolli et al. (2005) 28 |
| HI1B-Rw | CTT GAT GAT ACA GGG | ||||
| FIB | IncFIB-F | GGAGTTCTGACACACGATTTTCTG | 62ºC | 702 | Caratolli et al. (2005) 28 |
| IncFIB-R | CTCCCGTCGCTTCAGGGCATT | ||||
| Q | Ori-V-F | CTCCCGTACTAACTGTCACG | 61ºC | 436 | Smalla et al. (2001) 27 |
| Ori-V-R | ATCGACCGAGACAGGCCCTGC | ||||
| Rep-B-F | TCGTGGTCGCGTTCAAGGTACG | 64ºC | 1.160 | ||
| Rep-B-R | CTGTAAGTCGATGATCTGGGCGTT | ||||
| Ori-T-F | TTCGCGCTCGTTGTTCTTCGAGC | 63ºC | 191 | ||
| Ori-T-R | GCCGTTAGGCCAGTTTCTCG | ||||
| NA | ERIC-1 | ATGTAAGCTCCTGGGGATTAAC | 36°C | NA | Duan et al., (2009) 29 |
| ERIC-2 | AAGTAAGTGACTGGGGTGAGCG |
NA: not applicable; Temp.a: the annealing temperature of the primers
The following thermal cycling conditions were used for bla KPC amplification: initial denaturation for 5 min at 95ºC, followed by 30 cycles of 1 min at 95ºC for denaturation, 1 min at 63ºC for primer annealing, and 1 min at 72ºC for the extension step. Subsequently, a final elongation step of 10 min was performed at 72ºC. For amplification of the bla GES gene, the following conditions were used: 3 min at 93°C, followed by 40 cycles of 1 min at 93°C, 1 min at 55°C and 1 min at 72°C, and a final extension for 7 min at 72°C. For the amplification of the bla NDM gene, the conditions used were 10 min at 94°C, followed by 36 cycles of 30 s at 94°C, 40 s at 52°C, 50 s at 72°C, and a final extension of 5 min at 72°C. For the bla VIM and bla IMP genes, we used PCR cycling of 5 min at 95ºC, followed by 30 cycles of 1 min at 95ºC, 1 min at 60oC and 62oC for the bla IMP and bla VIM genes, respectively, and an extension step for 1 min at 68ºC. Subsequently, a 5-minute final elongation step was performed at 68ºC.
Incs PCR
To detect the plasmid Incs, the primers for IncA/C, IncL/M, IncN, IncHI2, IncFIB, IncHI1B, and IncQ were selected and used 27 , 28 , 29 , as these plasmid incompatibility groups are more frequently described in the literature for K. pneumoniae. The cycling conditions used were 5 min at 94°C, followed by 35 cycles of 1 min denaturation at 94°C, 1 min of annealing with the specific temperature of each initiator used in the reaction (Table 1), and 1 min of extension at 72°C. The final extension was performed for 10 min at a temperature of 72°C.
Electrophoresis and sequencing of resistance genes
The PCR products were analyzed via electrophoresis using a 1 % agarose gel in TBE buffer (0.089 M Tris-Borate and 0.002 M EDTA) at a constant voltage of 100 V. The gels were visualized under ultraviolet light using a transilluminator (Bio Rad) and photographed with a photo-documentation system (Photocap, Vilber Lourmat). The amplicons for each gene were purified using the SV Total DNA Isolation System (Promega) and the DNA was sequenced by the method of Sanger et al. (1997). The nucleotide sequences were analyzed using the BLAST program (http://www.ncbi.nlm.nih.gov/).
Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR)
To assess the clonal relationship between the isolates, ERIC-PCR was performed as described by Duan et al. 2009 29 and Cabral et al. 2012 14 (Table 1). The DARWIN 6.0 software was used to generate the dendrogram.
RESULTS
Antimicrobial resistance profile
Although isolates resistant to at least one carbapenem were selected, we observed that most isolates were resistant to three carbapenems tested (i.e., ertapenem, imipenem and meropenem), except for isolate K5-A3 which was sensitive to imipenem and showed intermediate sensitivity to meropenem, and the K6-A3 isolate which had intermediate sensitivity to imipenem (Table 2). The antimicrobials that showed the best activity against carbapenem-resistant K. pneumoniae isolates were amikacin and colistin, showing that 96.2% and 88.9% were sensitive to these antimicrobials, respectively.
TABLE 2: The source of isolation; minimum inhibitory concentration (MIC) for ertapenem (ERT), imipenem (IMI), and meropenem (MER); genes for carbapenemases (Resistance genes); plasmid Incs (Incs); and the ERIC-PCR profile of K. pneumoniae isolates from a public hospital in Recife-PE, Brazil.
| Isolates | Sector | Clinical sample | MIC(ERT) | MIC(IMI) | MIC(MER) | Resistance Genes | Incs | ERIC-PCR |
|---|---|---|---|---|---|---|---|---|
| K5-A3 | ICU | Surgical drain | >4(R) | <=1(S) | 2(I) | bla KPC | Q, FIB | E7 |
| K6-A3 | CU | Blood | >4(R) | 2(I) | 4(R) | bla KPC | Q, FIB | E2 |
| K8-A3 | ICU | Urine | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E2 |
| K9-A3 | GE | Blood | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E7 |
| K10-A3 | ICU | Blood | >4(R) | >8(R) | >8(R) | bla KPC | Q, FIB, HI1B | E4 |
| K11-A3 | CU1 | Blood | >4(R) | 8(R) | 4(R) | bla KPC | Q, HI1B | E12 |
| K12-A3 | ICU | Blood | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E1 |
| K16-A3 | Cardiology | Urine | >4(R) | >8(R) | >8(R) | bla NDM | FIB | E3 |
| K24-A3 | ICU | Urine | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB, A/C | E1 |
| K26-A3 | GE | Urine | >4(R) | 8(R) | 4(R) | bla KPC, bla NDM | Q, FIB | E11 |
| K27-A3 | MC | Urine | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E1 |
| K30-A3 | CU1 | Blood | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E1 |
| K31-A3 | ICU | Urine | >4(R) | >8(R) | >8(R) | bla KPC | FIB, A/C | E1a |
| K2-A3 | Cardiology | Rectal Swab | >4(R) | (R) | (R) | bla KPC | Q, FIB, L/M | E5 |
| K3-A3 | CU2 | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E7 |
| K13-A3 | CU1 | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC | Q, FIB, A/C | E10 |
| K14-A3 | CU2 | Rectal Swab | >1(R) | >8(R) | >32(R) | bla KPC | Q, FIB, HI1B | E4 |
| K15-A3 | Cardiology | Rectal Swab | >1(R) | >8(R) | >32(R) | bla KPC | Q, FIB | E6 |
| K17-A3 | ICU | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q | E2 |
| K18-A3 | CU1 | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC | Q, FIB | E1 |
| K20-A3 | CU1 | Rectal Swab | >4(R) | >8(R) | >8(R) | bla NDM | FIB | E1 |
| K21-A3 | Cardiology | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC | Q, FIB, A/C | E1a |
| K29-A3 | CU | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E1 |
| K32-A3 | ICU | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E8 |
| K34-A3 | ICU | Rectal Swab | >4(R) | >8(R) | >8(R) | bla NDM | Q, FIB | E1a |
| K36-A3 | CU2 | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB, L/M | E9 |
| K37-A3 | ICU | Rectal Swab | >4(R) | >8(R) | >8(R) | bla KPC, bla NDM | Q, FIB | E1 |
K: Klebsiella pneumoniae; GE: General Emergency; MC: Medical Clinic; ICU: Intensive care unit; CU: Coronary Unit; MIC: Minimal Inhibitory Concentration; ERT: Ertapenem, IMI: Imipenem, MER: Meropenem, R: Resistant; I: Intermediate; S: Sensitive; A3: public hospital; +, presence of the gene; - absence of the gene; Shaded text: clinical isolates from colonization.
Beta-lactamase genes
The bla KPC and bla NDM genes were detected in 24 (88.8 %) and 16 (59.2 %) isolates of K. pneumoniae, respectively (Table 2), by amplifying the expected 800 bp and 621 bp genes for bla KPC and bla NDM respectively. The bla GES, bla VIM, and bla IMP genes were not detected. The bla KPC and bla NDM genes were detected in 13 (48.1 %) of the isolates analyzed. The bla KPC-2 and bla NDM-1 variants were confirmed by sequencing the PCR product from representative isolates.
Plasmid Incompatibility Groups (Incs)
The FIB (n=25; 92.6 %) and Q (n=24; 88.8 %) Incs were the most frequent in the K. pneumoniae isolates analyzed in this study, followed by the Incs A/C, HI1B, and L/M that were detected in 4 (14.8 %), 3 (11.1 %) and 2 (7.4 %) isolates, respectively. The Incs N and HI2 were not detected. Two isolates (K16-A3 and K20-A3) showed the presence of the IncFIB alone, and the isolate K17-A3 had only IncQ. The remaining 24 isolates had more than one Inc that were investigated in this study (Table 2).
Molecular typing by ERIC-PCR
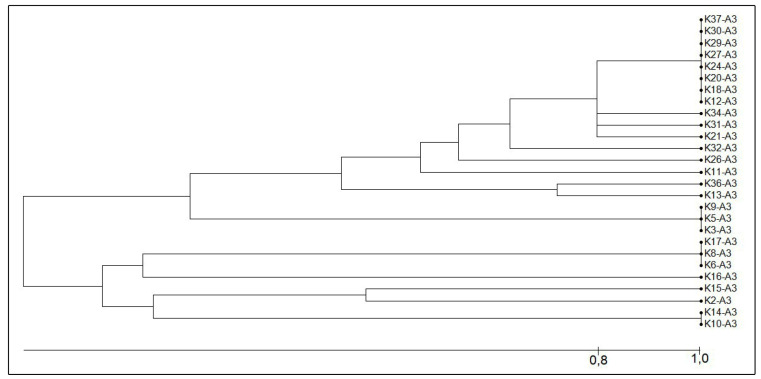
Of the 27 isolates of K. pneumoniae analyzed, 12 different genetic profiles were identified (Figure 1 and Figure 2) by ERIC-PCR. Eleven isolates showed an 80 % similarity (E1a profile), of which eight isolates showed a 100 % similarity (E1 profile). Four isolates showed an identical resistance profile (presence of the bla KPC and bla NDM genes) and the presence of the same plasmid Incs (Q and FIB). The E2 profile grouped three isolates that were different for their resistance profile and the presence of Incs. The E4 profile grouped two isolates that showed an identical resistance profile (only the bla KPC gene was detected) and Incs (Q, FIB and HI1B). The E7 profile grouped three isolates, of which two isolates were identical for their resistance profile (bla KPC and bla NDM gene) and in the presence of Incs (Q and FIB), whereas one isolate in this profile had the bla KPC gene alone and the same plasmid Incs. The isolates that were not clonally related had a different resistance profile and the presence of distinct plasmid Incs.
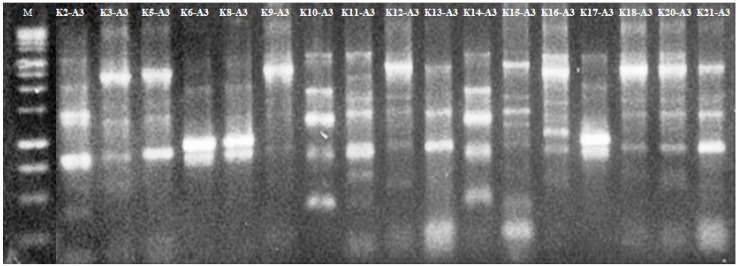
FIGURE 1: Electrophoresis using a 1.5 % agarose gel for ERIC-PCR from representative isolates of K. pneumoniae. Lane 1: 1 Kb molecular weight marker (Promega), lanes 2-17: K. pneumoniae isolates.
FIGURE 2: Dendrogram generated from the results of the ERIC-PCR using the Darwin 6.0 software, illustrating the relationship between the profiles of the 27 isolates of K. pneumoniae that were resistant to carbapenems and obtained from Recife-PE, Brazil.
Comparative analysis of the presence of genes for carbapenemases, plasmid Incs, and ERIC-PCR, based on colonization or infection as sources for isolation.
Of the 27 carbapenem-resistant K. pneumoniae isolates, 12 were from infected samples (blood or urine), 14 were from colonization samples (rectal swab), and 1 was from a cavity drain (Table 2). The majority (58.3 %) of the isolates from the infected samples that had the bla NDM gene also harbored the bla KPC gene, except for the K16-A3 isolate that harbored the bla NDM gene alone. On analyzing the isolates from colonization samples, we observed that six isolates were positive for the bla KPC and bla NDM genes. The Incs FIB, Q, A/C, and HI1B were detected in isolates from infection and colonization, and IncL/M was detected in only two isolates from colonization.
DISCUSSION
Most of the K. pneumoniae isolates evaluated in this study were resistant to all beta-lactams tested. These data justify the alert published by the CDC in 2013, which states that carbapenemase-producing enterobacteria are a global threat due to the high rates of resistance to antimicrobials, and urgent and effective actions are required to control them 30 . Lorenzoni et al. (2017) 31 performed studies with carbapenem-resistant enterobacteria isolated from a hospital in the Rio Grande do Sul, Brazil, and detected high rates of sensitivity to colistin and amikacin in K. pneumoniae isolates. Further, the authors found that the bla KPC gene was detected in 80 % of the isolates, corroborating the data in this study, where we found an occurrence of 88.8 % for the bla KPC gene. These occurrence rates of the bla KPC gene identified in this study highlight the persistence of this gene in K. pneumoniae, and since its detection in 2006 from Recife, Brazil 13 , it remains the main carbapenemase associated with carbapenem-resistant K. pneumoniae samples in several Brazilian states, including Recife-PE 1 , 5 , 32 .
The occurrence of the bla NDM gene in 59.2 % of the isolates analyzed in this study is worrying and highlights the emergence of another carbapenemase that, in addition to KPC, can also hydrolyze carbapenems. Barberino et al. (2018) 33 were the first to report the presence of the bla NDM gene in clinical isolates of K. pneumoniae and Citrobacter in two patients admitted to a public hospital in Salvador, Bahia, in northeastern Brazil. Da Silva et al. (2019) 34 detected the bla NDM-1 gene in different species of Gram-negative bacteria isolated from nine Brazilian states but did not include the state of Pernambuco. Scavuzzi et al. (2019) 35 detected an isolate of K. pneumoniae that in addition to harboring the bla NDM gene, also harbored bacterial virulence genes, and were the first to report strains carrying the bla NDM gene in bacterial isolates from Recife-PE. Additionally, Firmo et al. (2019) 17 detected the occurrence of bla NDM in 25 % of K. pneumoniae isolates, and also in isolates from Recife-PE. In our study, we detected a greater number of isolates with the bla NDM gene (n=16; 59.2 %), and these results indicated the rapid dissemination of this gene.
The rate of occurrence of the bla NDM and the bla KPC genes deserves to be highlighted because of the accumulation of these genetic mechanisms of resistance in the same bacterial species. In Brazil, the accumulation of resistance determinants in K. pneumoniae has been described by other authors. Nava et al. (2018) 7 detected the occurrence of the bla NDM , bla KPC, and bla TEM genes in clinical isolates of K. pneumoniae from a university hospital in Londrina-PR. The concomitant presence of the bla KPC and bla NDM genes in colonization isolates examined in this study is worrying and reinforces the need for surveillance cultures, as patients showing bacterial colonization are an important reservoir for the spread of resistance mechanisms within the hospital environment and are the main gateway to the development of infection.
We observed that K. pneumoniae isolates, despite being clonally related as indicated by an ERIC-PCR assay, presented different types of plasmids and different resistance genes. This may occur because the ERIC-PCR technique amplifies repetitive intergenic regions of the bacterial chromosome, yet does not necessarily amplify plasmid regions, where most of the resistance genes are located 36 . Clonal dissemination of the isolates was observed in different sectors of the hospital under study and among colonized and infected patients. Therefore, our results indicate that K. pneumoniae can potentially spread in the hospital environment.
The persistence of genes that confer resistance to carbapenems results due to the clonal dissemination of the isolates and via the dispersion of these genes through conjugative or mobile plasmids 8 , 9 , 10 , 11 . In this study, we detected five types of Incs which are described in the literature as being potentially responsible for the spread of bla KPC and bla NDM genes in K. pneumoniae isolates. Additionally, all Incs identified in this study already harbor resistance genes, including bla KPC and bla NDM 4 , 16 , 19 , 20 , 37 , 38 , and the Incs FIB and Q were the most frequently detected in this study. This study is the first report of the IncFIB in bacterial isolates from Recife-PE, Brazil. IncFIB is a conjugative plasmid that has been associated with the dissemination of the bla IMP gene in E. cloacae in Japan 39 , and was also reported in Africa in E. coli isolates carrying the bla TEM gene 40 . In Europe, it was responsible for the spread of the bla NDM-1, bla SHV-12, bla CTXM-15, and bla OXA-1 genes in K. pneumoniae 11 .
The second most frequently detected Inc in K. pneumoniae isolates was IncQ, a plasmid that harbors carbapenem resistance genes and had been gaining prominence in some regions of Brazil. Nicoletti et al. (2015) 41 identified the bla KPC gene inserted into an IncQ plasmid in K. pneumoniae. Cerdeira et al. (2019) 16 detected two isolates of K. pneumoniae that were resistant to carbapenems and had the bla KPC gene inserted into IncQ plasmids. According to Smalla et al. (2000) 27 , IncQ is a small plasmid that can vary between 5.1-14.2 kb in size, and it can be found in several host bacterial cells. IncQ is not conjugative, but is mobilizable and promiscuous, and can be transferred from one bacterium to another by conjugative plasmids, which are present in the same bacterial cell. Given the variability in plasmids, including for conjugative plasmids, which were detected in this study in K. pneumoniae isolates, IncQ can probably be disseminated to other species. Additionally, it plays an important role in enhancing the dissemination of the bla KPC gene in Brazil 16 .
Incs A/ C and L/M, despite being detected in a smaller number of isolates in this study, have been described in previous studies as carrying resistance genes in different species of enterobacteria in Brazil, including K. pneumoniae 20 . Pereira et al. (2015) 37 detected the simultaneous presence of the bla KPC and bla NDM genes in E. hormaechei in Rio de Janeiro, with bla NDM inserted into IncA/C. Only one study has investigated Incs in K. pneumoniae isolates from Recife-PE, Brazil; however, the plasmids were not typed, and the small number of isolates analyzed (only four) may be a limitation of the study 19 .
We also detected the presence of the conjugative plasmid IncHI1B in this study, and to our knowledge, this is the first report of this Inc in Brazil. This plasmid has been reported in clinical isolates of E. cloacae, K. pneumoniae, E. coli, and C. freundii as carrying the bla NDM gene in hospitals in the United States of America 42 . Additionally, Al Baloushi et al. (2018) 10 detected isolates of K. pneumoniae carrying the bla NDM gene in IncHI1B in Saudi Arabia. Matsumura et al. (2018) 38 performed a conjugation and transformation experiment in isolates from surveillance programs and identified the bla VIM gene in plasmids IncL/M, IncN2, IncHI1B, and IncFIB in K. pneumoniae isolates from Greece and Spain. These studies show the ability of IncHI1B to host genes for carbapenemases in different species.
We conclude that the accumulation of resistance determinants, the variability of plasmid Incs, and the clonal dissemination of these in K. pneumoniae isolates from infection and colonization, demonstrate the ability of this species to acquire genes for resistance and disseminate them via conjugative and mobilizable plasmids. The importance of the early phenotypic and genetic identification of resistance mechanisms in bacterial isolates from infection and colonization samples needs to be highlighted to prevent and halt the development of infection in patients hospitalized due to immuno-depression.
ACKNOWLEDGMENTS
We are grateful to the Bioinformatics and Evolutionary Biology Group - LABBE UFPE, in particular Dr. Heidi Lacerda and Dr. Valdir Queiroz Balbino for their promptness in sequencing the amplicons. We thank Dra. Josineide Ferreira Barros for the help in obtaining the analyzed isolates from the study, as well as in their automated identification.
Footnotes
Financial Support: CAPES-Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and FACEPE Fundação de Amparo à Ciência e Tecnologia de PE (PPSUS 2017 APQ-0837-2.12/17).
Limitations of the study: It was not possible to identify all known plasmid Incs, as well as to sequence the plasmids to determine the specific location of each resistance gene.
REFERENCES
- 1.Scavuzzi AM, Alves LC, Veras DL, Brayner FA, Lopes ACS. Ultrastructural changes caused by polymyxin B and meropenem in multiresistant Klebsiella pneumoniae carrying blaKPC-2 gene. J Med Microbiol. 2017 doi: 10.1099/jmm.0.000367. [DOI] [PubMed] [Google Scholar]
- 2.Kuntaman K, Shigemura K, Osawa K, Kitawa K, Sato K, Yamada N, et al. Occurrence and characterization of carbapenem-resistant Gram-negative bacilli: A collaborative study of antibiotic-resistant bacteria between Indonesia and Japan. Int J of Urol. 2018;25(11):966–972. doi: 10.1111/iju.13787. [DOI] [PubMed] [Google Scholar]
- 3.Henriksen AS, Smart JI, Hamed K. Susceptibility to ceftobiprole of respiratory-tract pathogens collected in the United Kingdom and Ireland during 2014-2015. Infec Drug Resist. 2018;11:1309–1320. doi: 10.2147/IDR.S176369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalmolin TV, Martins AF, Zavascki AP, Morales DL, Barth AL. Acquisition of the mcr-1 gene by a high-risk clone of KPC-2- producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn Micr and Infec Dis. 2018;90(2):132–133. doi: 10.1016/j.diagmicrobio.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Alves DP, Carvalho-Assef APDA, Conceição-neto OC, Aires CAM, Albano RM, Folescu TW, et al. Enterobacter cloacae harbouring blakpc-2 and qnrb-1 isolated from a cystic fibrosis patient: a case report. New Microbes New Infect. 2018;25:49–51. doi: 10.1016/j.nmni.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melgarejo JL, Cardoso MH, Pinto IB, Júnior FL, Mendo S, Oliveira CL, et al. Identification, molecular characterization, and structural analysis of the blaNDM-1 gene/enzyme from NDM-1-producing Klebsiella pneumoniae isolates. J Antibiot Res. 2018;72(3):155–163. doi: 10.1038/s41429-018-0126-z. [DOI] [PubMed] [Google Scholar]
- 7.Nava RG, Oliveira-Silva M, Nakamura-Silva R, Pitondo-Silva A, Vespero EC. New sequence type in multidrug-resistant Klebsiella pneumoniae harboring the blaNDM-1-encoding gene in Brazil. Int J Infect Dis. 2018;79:101–103. doi: 10.1016/j.ijid.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Rozales FP, Ribeiro VB, Ribeiro VB, Magagnin CM, Pagano M, Lutz L, et al. Emergence of NDM-1-producing Enterobacteriaceae in Porto Alegre, Brazil. Int J Infect Dis. 2014;25:79–81. doi: 10.1016/j.ijid.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Nodari SC, Siebert M, Matte SU, Barth LA. Draft genome sequence of a GES-5- producing Serratia marcescens isolated in southern Brazil. Braz J Microbiol. 2017;48(2):191–192. doi: 10.1016/j.bjm.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Baloushi AE, Pál T, Ghazarawi A, Sonnevend A. Genetic support of carbapenemases in double carbapenemase producer Klebsiella pneumonia e isolated in the Arabian Peninsula. Acta Mirobiol Imm H. 2018;23:1–16. doi: 10.1556/030.65.2018.005. [DOI] [PubMed] [Google Scholar]
- 11.Paskova V, Medvecky M, Shalova A, Chudejova K, Bitar I, Jakubu V, et al. Characterization of NDM-Encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front Microbiol. 2018;9:1–12. doi: 10.3389/fmicb.2018.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmieri M, D'andrea MM, Pelegrin AC, Mirande C, Brkic S, Cirkovic I, et al. Genomic Epidemiology of Carbapenem- and Colistin-Resistant Klebsiella pneumoniae Isolates From Serbia: Predominance of ST101 Strains Carrying a Novel OXA-48 Plasmid. Front Microbiol. 2020;11:1–10. doi: 10.3389/fmicb.2020.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteiro J, Santos AF. First report of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53(1):333–334. doi: 10.1128/AAC.00736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabral AB, Melo RCA, Maciel MAV, Lopes ACS. Multidrug resistance genes, including blaKPC and blaCTX-M-2, among Klebsiella pneumoniae isolated in Recife, Brazil. Rev Soc Bras Med Trop. 2012;45(5):572–578. doi: 10.1590/s0037-86822012000500007. [DOI] [PubMed] [Google Scholar]
- 15.Melo RCA, Barros EMR, Loureiro NG, Melo HRL, Maciel MAV, Lopes ACS. Presence of fimH, mrkD and irp2 virulence genes in KPC-2-producing Klebsiella pneumoniae isolates in Recife-PE, Brazil. Curr Microbiol. 2014;69(6):824–831. doi: 10.1007/s00284-014-0662-0. [DOI] [PubMed] [Google Scholar]
- 16.Cerdeira LT, Lam MMC, Wyres KL, Wick RR, Judd LM, Lopes R, et al. Small IncQ-1 and Col-like Plasmids Harboring blaKPC-2 and non-Tn4401 Elements (NTEkpc-lld) in high-risk lineages of Klebsiella pneumoniae CG258. Antimicrob Agents Chemother. 2019;63(3):1–4. doi: 10.1128/AAC.02140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firmo EF, Beltrão EMB, Silva FRF, Alves LC, Brayner FA, et al. Association of blaNDM−1 with blaKPC−2 and aminoglycoside-modifying enzymes genes among Klebsiella pneumoniae, Proteus mirabilis and Serratia marcescens clinical isolates in Brazil. J Glob Antimicrob Resist. 2019;21:255–261. doi: 10.1016/j.jgar.2019.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Eilertson B, Chen L, Li A, Chavda KD, Chavda B, Kreiswirth BN, et al. CG258 Klebsiella pneumoniae isolates without β-lactam resistance at the onset of the carbapenem-resistant Enterobacteriaceae epidemic in New York City. Antimicrob Agents Chemother. 2019;74(11):17–21. doi: 10.1093/jac/dky394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira PS, Araujo CF, Seki LM, Zahner V, Carvalho-Assef APD, Asensi MD. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340) Antimicrob Agents Chemother. 2013;68:312–316. doi: 10.1093/jac/dks396. [DOI] [PubMed] [Google Scholar]
- 20.Andrade LN, Curião T, Ferreira J, Longo JM, Clímaco EC, Martinez R, et al. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniaeclonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother. 2011;55(7):3579–3583. doi: 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belder D, Lucero C, Rapoport M, Rosato A, Faccone D, Petroni A, et al. Genetic diversity of KPC-producing Escherichia coli, Klebsiella oxytoca, Serratia marcescens and Citrobacter freundii isolates from Argentina. Microb Drug Resist. 2018;24(7):958–965. doi: 10.1089/mdr.2017.0213. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Mathemal B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho-Assef APD, Pereira PS, Albano RM, Berião GC, Chagas TPG, Timm LN, et al. Isolation of NDM-producing Providencia rettgeri in Brazil. J Antimicrob Chemother. 2013;68:2956–2957. doi: 10.1093/jac/dkt298. [DOI] [PubMed] [Google Scholar]
- 25.Carvalho-Assef APD, Pereira PS, Albano RM, Berião GC, Tavares CP, Chagas TPG, et al. Detection of NDM-1-, CTX-M-15-, and qnrB4-producing Enterobacter hormaechei isolates in Brazil. Antimicrob Agents Chemother. 2014;58:2475–2476. doi: 10.1128/AAC.02804-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 29th ed. Wayne, PA: CLSI; 2019. [Google Scholar]
- 27.Smalla K, Heuer H, Gotz A, Niemeyer D, Krogerrecklenfort E, Tietze E., et al. Exogenous Isolation of Antibiotic Resistance Plasmids from Piggery Manure Slurries Reveals a High Prevalence and Diversity of IncQ-Like Plasmids. Appl Environ Microbiol. 2000;66(11):4854–4862. doi: 10.1128/aem.66.11.4854-4862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caratolli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Duan H, Chai T, Liu J, Zhang X, Qi C, Gao J, et al. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ Res. 2009;109(5):511–517. doi: 10.1016/j.envres.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CDC . Antibiotics resistance threats in the United States. U.S department of health and human services centers for disease control and prevention. Centers for Disease Control and Prevention, Office of Infectious Disease Antibiotic resistance threats in the United States, 2013. 2013. http://www.cdc.gov/drugresistance/threat-report-2013 [Google Scholar]
- 31.Lorenzoni VV, Silva DC, Rampelotto RF, Brites PC, Villa B, Horner R, et al. Evaluation of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Rio Grande do Sul, Brazil. Rev Soc Bras Med Trop. 2017;50(5):685–688. doi: 10.1590/0037-8682-0209-2017. [DOI] [PubMed] [Google Scholar]
- 32.Cabral AB, Maciel MA, Barros JF, Antunes MM, Lopes ACS. Detection of blaKPC-2 in Proteus mirabilis in Brazil. Rev Soc Bras Med Trop. 2015;48(1):94–95. doi: 10.1590/0037-8682-0152-2014. [DOI] [PubMed] [Google Scholar]
- 33.Barberino MG, Cruvinel SA, Faria C, Salvino MA, Silva MO. Isolation of blaNDM-producing Enterobacteriaceae in a public in Salvador, Bahia, Brazil. Braz J Infect Dis. 2018;17:30–32. doi: 10.1016/j.bjid.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Da Silva IR, Aires CAM, Conceição-Neto OC, Santos ICO, Pereira NF, Senna JPM, et al. Distribution of Clinical NDM-1-Producing Gram-Negative Bacteria in Brazil. Microb Drug Resist. 2019;25(3):394–399. doi: 10.1089/mdr.2018.0240. [DOI] [PubMed] [Google Scholar]
- 35.Scavuzzi AM, Firmo EF, Oliveira ÉM, Lopes ACS. Emergence of blaNDM-1 associated with the aac(6?)-Ib-cr, acrB, cps, and mrkD genes in a clinical isolate of multi-drug resistant Klebsiella pneumoniae from Recife-PE, Brazil. Rev Soc Bras Med Trop. 2019;52:1–4. doi: 10.1590/0037-8682-0352-2018. [DOI] [PubMed] [Google Scholar]
- 36.Sharples GJ, Lloyd RG. A novel repeated sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18:6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira PS, Borghi M, Albano RM, Lopes JC, Silveira MC, Marques EA, et al. Coproduction of NDM-1 and KPC-2 in Enterobacter hormaechei from Brazil. Microb Drug Resist. 2015;21(2):234–236. doi: 10.1089/mdr.2014.0171. [DOI] [PubMed] [Google Scholar]
- 38.Matsumura Y, Peirano G, Bradford PA, Bradford PA, Motyl MR, DeVinney R, et al. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J Antimicrob Chemother. 2018;73(11):3034–3038. doi: 10.1093/jac/dky303. [DOI] [PubMed] [Google Scholar]
- 39.Aoki K, Harada S, Yahara K, Ishii Y, Motooka D, Nakamura S, et al. Molecular Characterization of IMIP-1-producing Enterobacter cloaceae complex isolates in Tokyo. Antimicrobi Agents Chemother. 2018;62(3):1–13. doi: 10.1128/AAC.02091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sana F, Mabrouka S, Elaa M, Sarra H, Hamzaoui Z, Hosni H, et al. Escherichia coli colonizing healthy children in Tunisia: High prevalence of extra-intestinal pathovar and occurrence of non-extended-spectrum-β-lactamase-producing ST131 clone. Internat J Antimicrob Agents. 2018;52:878–885. doi: 10.1016/j.ijantimicag.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 41.Nicoletti AG, Marcondes MFM, Martins WMBS, Almeida LGO, Nicolás MF, Vasconcelos ATR, et al. Characterization of BKC-1 class A carbapenemase from Klebsiella pneumoniae clinical isolates in Brazil. Antimicrob Agents Chemother. 2015;59(9):5159–5164. doi: 10.1128/AAC.00158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yigit H, Queenan AM, Anderson GJ, Sanchez AD, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae . Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordmann P, Naas T, Poirel L. Global Spread of Carbapenemase producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong F, Xu XW, Song WQ, Lu P, Yu SJ, Yang YH, et al. Characterization of multidrug-resistant and metallo-betalactamase-producing Pseudomonas aeruginosa isolates from a paediatric clinic in China. Chin Med J. 2008;121:1611–1616. [PubMed] [Google Scholar]