Abstract
The development of high-throughput, data-intensive biomedical research assays and technologies, such as DNA sequencing, imaging protocols, and wireless health monitoring devices, has created a need for researchers to develop strategies for analyzing, integrating and interpreting the massive amounts of data they generate. Although a wide variety of statistical methods have been designed to accommodate the ‘big data’ produced by these assays, experiences with the use of artificial intelligence (AI) techniques suggest that they might be particularly appropriate. In addition, the application of data-intensive biomedical technologies in research studies has revealed that humans vary widely at the genetic, biochemical, physiological, exposure and behavioral levels, especially with respect to disease processes and treatment responsiveness. This suggests that there is often a need to tailor, or ‘personalize,’ medicines to the nuanced and often unique features possessed by individual patients. Given how important data-intensive assays are to revealing appropriate intervention targets and strategies for personalizing medicines, AI can play an important role in the development of personalized medicines at all relevant phases of the clinical development and implementation of new personalized health products, from finding appropriate intervention targets to testing them for their utility. We describe many areas where AI can play a role in the development of personalized medicines, and argue that AI’s ability to advance personalized medicine will depend critically on the refinement of relevant assays and ways of storing, aggregating, accessing and ultimately integrating the data they produce. We also point out the limitations of many AI techniques, as well as consider areas for further research.
INTRODUCTION: EMERGING THEMES IN BIOMEDICAL SCIENCE
Modern biomedical science is guided, if not dominated, by many interrelated themes. Four of the most prominent and important of these themes are (see Figure 1): 1. Personalized medicine, or the belief that health interventions need to be tailored to the nuanced and often unique genetic, biochemical, physiological, exposure and behavioral features individuals possess; 2. The exploitation of emerging data-intensive assays, such as DNA sequencing, proteomics, imaging protocols, and wireless health monitoring devices; 3. ‘Big data’ research paradigms in which massive amounts of data, say of the type generated from emerging data-intensive biomedical assays, are aggregated from different sources, harmonized, and made available for analysis in order to identify patterns that would normally not be identified if the different data points were analyzed independently; and 4. Artificial Intelligence (AI; which we consider here to include algorithms based machine learning, deep learning, neural network constructs and a wide variety of related techniques[1]), which can be used to find relevant patterns in massive data sets.
Figure 1.
Four emerging complementary themes in biomedical science: personalized medicine, emerging data-intensive technologies, big data and information technologies (IT) infrastructure and artificial intelligence (AI). These technologies can be fuel, and be fueled by, AI in all phases of the development (T0-T4) of personalized medicines (see Figure 2).
These four themes are highly interrelated in that, e.g., personalizing a medicine or tailoring an intervention to a patient requires a very deep understanding of that patient’s condition and circumstances, and this requires the extensive use of sophisticated assays that generate massive amounts of data, such as DNA sequencing or an imaging protocol. Essentially, the data produced by these assays needs to be organized so that analyses can be pursued to identify features that the patient possesses that may indicate the optimal intervention. Research associated with each of these themes is often pursued independently of the others because of the very specialized expertise required. For example, there are scientific journals devoted to Personal Medicine (e.g., ‘Personalized Medicine,’ ‘Journal of Personalized Medicine’), emerging assays (e.g., ‘Nature Biotechnology,’ ‘Nature Digital Medicine’), big data (e.g., ‘Big Data,’ ‘Journal of Big Data,’ ‘Gigascience’), and artificial intelligence (e.g., ‘IEEE Transactions on Neural Networks and Learning Systems,’ ‘IEEE Transactions on Pattern Analysis and Machine Intelligence’) that publish very focused studies. However, the integrated use of the insights, information and strategies obtained from research associated with each of these themes is necessary for creating personalized health products, such as drugs and interventions.
Bringing together research activities associated with these four emerging themes is not trivial, as it will require communication and participation from researchers and practitioners with a wide variety of skills and expertise, including molecular biology, genetics, pathology, informatics, computer science, statistics, clinical science, and medicine. AI will have a special role to play in this integration process if the goal is to advance personalized medicine, since it is unclear how relevant clinically-meaningful insights can be drawn from big data-generating assays that would complement or build off the insights from experts in different domains. In this light, there are a number of phases in the development of medicines, general interventions, and other products, such as diagnostics, prognostics, decision support tools, etc., where AI could have a significant impact. These different phases are emphasized in subsections of this chapter that describe and comment on recent studies leveraging AI. This chapter does not provide an exhaustive literature review of AI in medicine, however, as there are some excellent reviews for this[2–4], but rather considers the potential that AI has in developing new medicines, health devices and products. In particular, a focus on the need for greater integration across the various phases of the development of health interventions and products could result in very radical yet positive changes in the way medicine is practiced. In this sense, this chapter is as much a summary of the ways in which AI can be exploited in modern medicine as it is a vision of the future.
THE TRANSLATIONAL WORKFLOW
As noted, the development of interventions and health products, as with the diagnosis and treatment of a patients, proceeds in different phases. There are various ways of defining and referring to these phases, however, and all of them point to opportunities for AI to have a substantial impact if leveraged appropriately. For example, in the context of clinical trials to vet a new drug or intervention for treating a disease like cancer, a common progression or workflow runs from Phase 0 trials, which involve characterizing the pharmacokinetic properties of a drug using what may turn out to be non-physiologic (i.e., does not have an appreciable effect on the body) and physiologic (i.e., does have an appreciable effect on the body) doses of a drug in a very small number of individuals, to Phase I trials, which involve establishing safe and effective doses of a drug in small number of individuals, to Phase II trials, which seek to establish whether a drug is likely to be efficacious in a moderately sized group of individuals, to Phase III trials, which attempt to establish the utility of a drug in the population at large by studying a very large number of individuals, to Phase IV trials, which evaluate the adoption, uptake and acceptance, as well as any evidence for adverse consequences, associated with the use of the drug in the population at large.
We take a broader view of the workflows behind developing new products than that reflected in the traditional clinical trial progression. This broader view is consistent with the scheme used by, for example, the National Institutes of Health (NIH)’s Clinical and Translational Science Award (CTSA) program.[5] The CTSA program focuses on all aspects of biomedical science associated with attempts to translate very basic biomedical insights concerning, e.g., pathogenic processes contributing to diseases, into clinically-useful products like drugs or interventions for those diseases. The CTSA scheme does, however, incorporate elements of the Phase 0-Phase IV clinical trials workflow or transition scheme for developing drugs or health products to treat or manage diseases in the population at large. Thus, in accordance with the CTSA scheme (left panel of Figure 2), T0 science involves very basic research focusing on that could lead to the identification of a drug or intervention target and then crafting an appropriate drug or intervention that modulates that target; T1 science focuses on testing an intervention or health device in a small clinical studies to determine if it is safe and at what dosages it should be used; T2 science involves vetting the drug or device in a large number of individuals in a well-designed study to assess its efficacy in the population at large; T3 science focuses on the implementation of the drug or device for use in the population, including adapting existing workflows (e.g., custody chains for tests or physician-patient interaction points); and T4 science involves a re-evaluation or assessment of the utility of the drug or device post-deployment and implementation. Each of these phases can leverage AI techniques if the right data and motivation is present.
Figure 2.
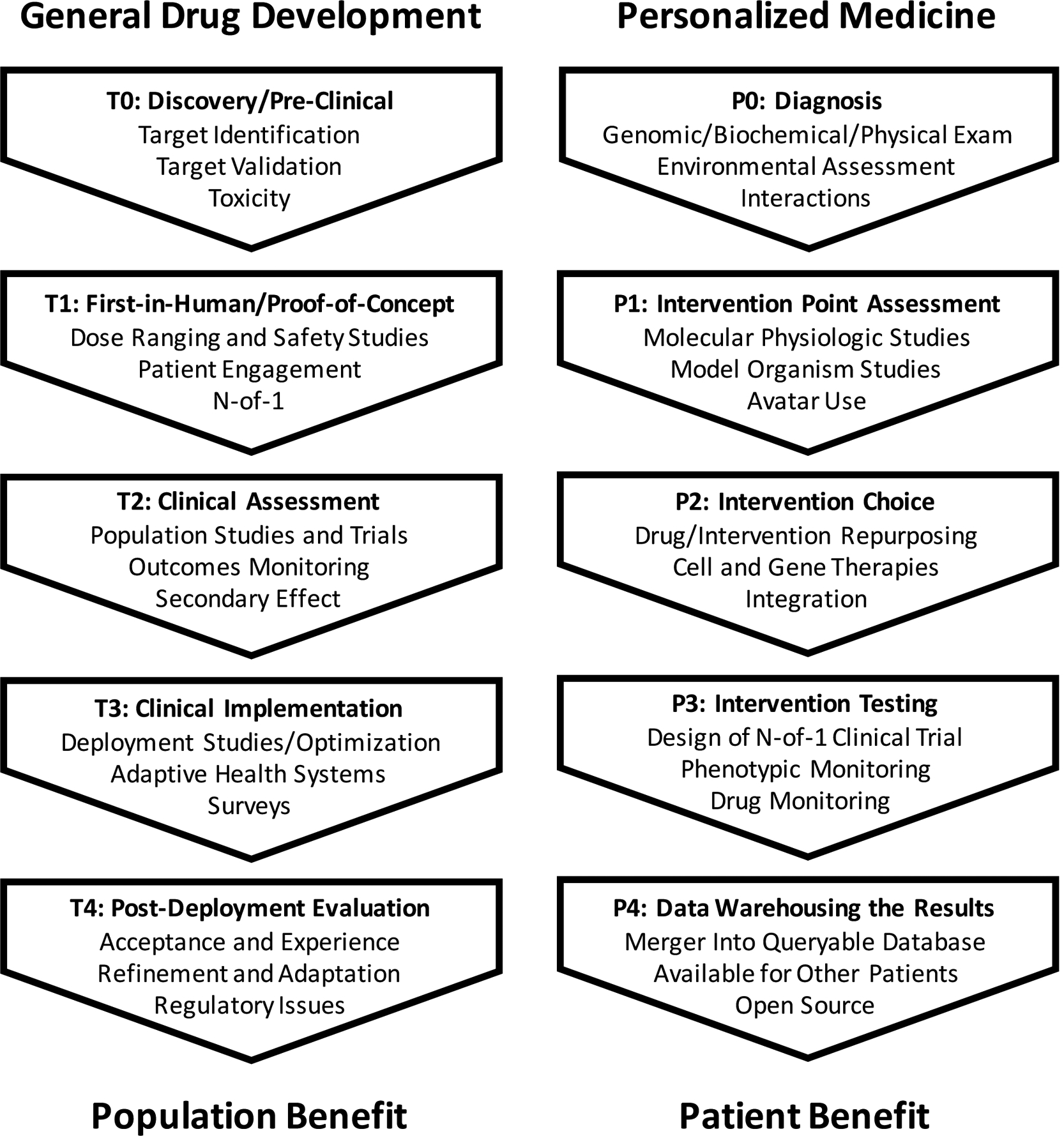
A representation of the stages in the ‘translation’ of basic insights into clinical useful products considered in initiatives such as the Clinical and Translational Science Award (CTSA) initiative overseen by the National Center on Advancing Translational Science of the United States National Institutes of Health (NCATS; left panel;[5]). An analogous representation of the stages in the diagnosis and treatment of an individual patient are provided in the right hand panel.
The actual practice of personalized medicine can be seen as involving an analogous process to the develop of drugs and health devices (right panel of Figure 2; see also Schork and Nazor [6]). Thus, P0 activity involves making a diagnosis or determining an individual’s risk of diseases; P1 activity involves identifying the key pathophysiologic processes, if not known, that are causing (or likely to cause) a disease that might be amenable to modulation by an appropriate intervention; P2 considers the identification of an appropriate intervention given what was identified in the P0 and P1 stages; P3 involves testing the intervention on the relevant individual undergoing the diagnosis and pathobiology assessment; and P4 involves warehousing the result in appropriate databases so that the insights and information obtained on a patient can be exploited in examinations of further patients or used in broader data mining initiatives to find further clinically-meaningful patterns.
A couple of items about these workflows are worth noting. First, as noted, although the science associated with each component involves unique insight and expertise and provide a fertile ground for collaborations with AI tools and scientists independently of the other components, the transmission of information from one component to another – or the transitions from one component to another – is of crucial importance (e.g., consider that a diagnostic would not be particularly useful if it did not help a physician choose an appropriate course of action). Second, a goal of personalized medicine, and the improvement of health care generally, is to make workflows like those in Figure 2 more efficient and reliable and AI can have a substantial role to play in this broader goal. These two notes are emphasized in the subsection on ‘Integration and the Personalized Medicine Workflow‘ below.
PRECLINICAL (T0) AND DIAGNOSTIC (P0) RESEARCH
The identification of targets for therapies (T0 research from the left hand side of Figure 2) can be greatly aided by AI in quite a few contexts. For example, many assays used to uncover potential therapeutic targets generate massive amounts of data and as such often require sophisticated statistical methods to identify meaningful patterns. AI has been used to identify such patterns from, e.g., DNA sequence data and molecular pathology imaging protocols.[7–10] For individual patients whose comprehensive diagnosis may lay the foundation for crafting a personalized treatment or intervention plan and exploit data-intensive assays (i.e., P0 in the right hand figure of Figure 2), many AI-based tools can be leveraged. For example, mining DNA sequence information obtained from an individual in order to make a genetic diagnosis of a disease can be greatly aided by AI-based analyses.[7] In addition, facilitating, e.g., cancer diagnoses with AI-based analyses of blood analytes has been shown to have great potential.[11]
If a particular therapeutic target has been identified that is consistent with the molecular pathology underlying an individual patient’s disease, then AI-based strategies for analyzing drug screening data collected to determine if any of a large number of extant drugs and compounds have activity against that target have been shown to be very reliable.[12] In addition, AI-based studies have been shown to reveal a great many insights into how drugs and compounds may impact various structural and functional features of a cell.[13] Finally, web sites with large databases like DeepChem (https://deepchem.io/about.html), which leverage AI in chemistry settings, can be used specifically to identify properties of drugs and compounds.
An interesting research area of relevance to the identification of pathologies underlying diseases that might be amenable to pharmacological modulation is the design of appropriate studies. For example, if the relationships between different potential drug targets and their effect on a molecular system or pathway is not known, researchers may have to systematically perturb each element in such a system and examine what the effect on the system these perturbations have.[14] As one can imagine, such studies can be tedious and laborious. However, recent studies suggest that one can use robotics and AI to conduct such experiments and, in fact, anticipate further experiments that might be called for based on the results of initial experiments.[15] Experimental infrastructure for pursuing AI-based experiments has also been developed, but only for a few select settings.[16, 17]
If an extant drug or compound is not found that could appropriately modulate a target, then creating a novel pharmacotherapeutic (i.e., drug) often requires insights obtained from materials science in order to make sure the molecular structure of the drug produced has favorable properties inside the body. Very recent work suggests that AI can be leveraged to identify materials that may not be easily identified with traditional brute-force approaches.[18, 19] In addition, AI has been used to design new structures, which may be relevant to crafting better interventions, whether a drug or mechanical device, as well as aid in the selection of appropriate chemical syntheses.[20–22] AI has also been used in studies of very basic phenomena, such as particle physics, to probe how materials interact.[23] Interestingly, the design of new drugs, for example in the context of refining the structure of a therapeutic protein or molecule, could also be greatly facilitated by AI. It has been shown that in many design contexts in which optimization of materials and the way they are put together, the use of AI can identify superior designs to those based on legacy strategies.[20] This theme of harnessing AI to identify ways of optimizing the assembly of materials, or the manner in which an objective function of whatever sort is optimized given some starting materials and appropriate yet basic principles for assembling them, was on display in the recent description of the system for playing the age-old game of GO developed by Google’s DeepMind group.[24] Essentially, the system developed at DeepMind was not only able to easily beat all human experts as well as other GO-playing systems, but was able to do this by identifying strategies and moves that were completely beyond those which humans had used to play and try to master the game for centuries.
If a therapeutic target has been identified, then one could potentially pursue compound screening studies to identify compounds that modulate the chosen target. Such studies often require a particular output or phenotype (e.g., the expression level of a target gene). In the absence of such a phenotype, high-content screens, in which many different phenotypes are evaluated to see if any of the compounds, often numbering in the thousands or tens of thousands, affects any one or some subset of these phenotypes as a sign of its activity. AI techniques have been used to identify potential compounds impacting a target in this setting.[25–27]
Once a therapeutic concept has been defined, relevant drugs or an intervention apparatus embodying the concept must be manufactured at scale for distribution. The manufacture of drugs and interventions of all sorts have been greatly facilitated by robotics and AI.[28] In the context of personalized medicines, it may be that the manufacture of drugs and interventions will require nuanced features based on patients’ profiles and therefore have to be designed and crafted in real-time as opposed to be created at scale, stored and distributed when needed – a topic to be discussed in a later section focusing on ‘Integration and the Personalized Medicine Workflow.[29, 30]’
FIRST-IN-HUMAN (T1) AND PATHOBIOLOGY (P1) STUDIES
Once a drug had been created, it must be shown to safe through Phase I clinical trials, or studies that are often referred to as ‘first-in-human-studies’ (T1 in the left panel of Figure 2). Such studies focus on safety and are typically pursued on a small number of individuals in case there are side-effects. To minimize the risk of exposing individuals to a new drug that might cause them harm, insights into the likelihood that individuals with a certain profile will have an adverse response are required. Studies that consider genetic factors that predispose to responses to drugs (good or bad) have revealed many compelling and clinically-useful connections between genetic variants and drug responses. Discovering such ‘pharmacogenetic’ insights have been greatly enhanced through the use of AI tools applied to very large databases with relevant genetic and drug response information.[31, 32]
The design of phase I clinical trials is an ongoing area of research. The fact that only a few individuals are enrolled in such trials, and a great sensitivity to the detection of the effects of the proposed drug or intervention are of focus, suggests that careful monitoring of the subjects enrolled in the trial is required. Extensions of N-of-1 and aggregated N-of-1 trial designs could be appropriate for Phase I trials.[33] Although discussed in greater detail in the context of vetting the efficacy of a personalized medicine in the section on the ‘Implementation (T3) and Clinical Assessment (P3) Studies‘ below, such studies can leverage massive amounts of data and AI techniques to identify patterns in a patient’s data that might be indicative of response to the intervention (see, e.g., Serhani et al.[34]). Table 1 lists examples of studies that have focused on monitoring a single individual over time to explore how they responded to a particular intervention, or how their health status may have changed over that time, using various data collection schemes.
Table 1.
Single patient-oriented studies leveraging intensive monitoring to identify either health status changes or the effects of an intervention.
| Authors | Reference | Study Elements | Comments |
|---|---|---|---|
| David et al. (2014)[93] | PMID: 25146375 | Diet and microbiome | 2 individuals fecal microbiome tracked for a year |
| Chen et al. (2012)[94] | PMID: 22424236 | Multiomics profiling | Individual tracked over a year (the ‘Snyderome’ study) |
| Magnuson et al. (2016)[95] | PMID: 28781744 | Sleep treatment study | Patient with multiple treatments for compromised sleep |
| Zeevi et al. (2015)[96] | PMID: 26590418 | Glycemic responses to diet | 800 people studied to develop personalized diets |
| Smarr et al. (2017)[97] | PMID: 29582916 | Colonoscopy effects | Gut microbiome changes post colonoscopy |
| Tremmell et al. (2016)[98] | PMID: 27721479 | NAD dose response | Single individual dosing study + study of others |
| Forsdyke (2015)[99] | PMID: 26055103 | Response to antihypertensives | Decade of seasonal variation in response to Losartan |
| O’Rawe et al. (2013)[100] | PMID: 24109560 | Deep brain stimulation (DBS) | 2 year study of a man with OCD treated with DBS |
| Li et al. (2016)[101] | PMID: 27140603 | Parkinson cell replacement | Single individual traced for 24 years post cell transplant |
| Bloss et a. (2016)[102] | PMID: 25790160 | Idiopathic neurologic disease | Severely disabled individual treated for sleep tremors |
| Piening et al. (2018)[103] | PMID: 29361466 | Multiomics weight loss study | 23 individuals observed over time |
| Zalusky et al. (1961)[104] | PMID: 14009735 | Folate supplementation | Study of an individual with severe dietary constraints |
| V. Herbert (1962)[105] | PMID: 13953904 | Folate supplementation | Study on the researcher himself; dietary restriction |
| P. H. Golding (2014)[106] | PMID: 25332850 | Folate supplementation | Study on the researcher himself; dietary restriction |
In the context of personalized medicine studies, once an individual is found to possess a certain pathology, a need to identify how that pathology can be corrected arises (P1 in the right hand panel of Figure 2). For many common chronic diseases this is obvious (e.g., for someone diagnosed with high blood pressure, providing blood pressure lowering medications makes sense). However, nuanced features of the patient, and the optimal way to correct the pathology given those nuances, are not often clear. Similar AI-based strategies for making diagnoses can be exploited to identify potentially correctable pathologies (P0 in the right panel of Figure 2). For example, the company Arterys recently announced the first FDA-approved AI-based application to be used in facilitating clinical diagnoses. The Arterys system used deep-learning applied to a medical imaging platform to help diagnose heart problems.[35] Other systems have been developed that consider more comprehensive approaches to understanding a patient’s profiles in a way that could facilitate the choice of an intervention.[36]
There is growing sophistication in the way that biopsied material or patient biosamples can be studied in a laboratory to identify targets for intervention. For example, emerging induced pluripotent stem cell (iPSC) and organoid technologies have shown great promise in yielding insights into patient-specific pathologies that could be overcome with specific interventions (see Table 2 of Schork and Nazor [6] as well as Rossi et al.[37]). When recently-developed single cell assays are combined with the use of organoids even greater resolution concerning pathologies and drug targets can be revealed and AI-based analyses have been shown to have great promise in this area.[38]
LATE PHASE HUMAN (T2) AND INTERVENTION CHOICE (P2) STUDIES
If a drug or health product has been shown to be safe and likely to be efficacious in early phase trials, then it must be tested for its general utility in the population at large. AI can be exploited in relevant large scale population trials that seek to minimize the deployment and use of inappropriate drugs or interventions studies for each participant in the trial, as described recently by Yauney and Shah.[39] Of great interest in this context are the design and conduct of bucket (or variations termed ‘basket’), umbrella and adaptive trial designs.[40] Although each has unique features to them, a description of basket trials provides the general strategy behind each and also points out where AI can be exploited. Essentially bucket trials enroll eligible patients, profile them to identify nuanced pathophysiologic profiles they possess (e.g., sequencing their tumor DNA in the context of a cancer clinical trial), and assign them one of possibly many treatments based on the mechanisms of action of the treatments (i.e., put them into one of many treatment ‘buckets’). If the patients provided with treatments dictated by their pathophysiologic profiles (i.e., assigned to the different baskets) have better outcomes than those provided treatments without recourse to the profiling and treatment matching scheme, then one could infer that the strategy or ‘algorithm’ for matching the treatments to the patients has merit. AI could be of great use in not only identifying treatment targets in the patient profiles, but also aid in determining the strategy for matching the treatments to the patient profiles. This would especially be the case if one could envision the use of many different treatment baskets (for example due to considering many treatment combinations or complicated temporal treatment schemes).
In the context of choosing an intervention for a particular individual via the personalized medicine paradigm (P2 in the right panel of Figure 1), if available drugs and interventions exist then the choice could be based on simply matching the patient’s pathophysiologic profile to the mechanisms of action of the drugs, consistent with the underlying theme governing basket trials. If the choice is not obvious, then one could leverage personalized drug screening strategies using biopsies or biomaterials obtained from the patient, as suggested by Kodack et al. in the context of cancer.[41] These, and more general, personalized drug screening strategies have been developed and could benefit from AI techniques to find patterns of relevance in the data that could indicate which drug or intervention is the most optimal.[42, 43]
IMPLEMENTATION (T3) AND CLINICAL ASSESSMENT (P3) STUDIES
If a drug or health product has been shown to benefit individuals in the general population, then considerations about the routine implementation and/or use of the product arise (T3 in the left side of Figure 2). Implementation can come in many forms. For example, if a drug shows sufficient proof to be safe and efficacious it can be approved for use by regulatory agencies such as the FDA and become embedded in clinical practice. Of greater relevance to AI is the implementation of insights that might benefit physicians with respect to intervention choices when confronted with patients with unique profiles (e.g., implementing a treatment strategy of the type tested in a basket trial). Implementing such insights require codifying and providing them to physicians through electronic medical record (EMR) systems typically used to convey patient information to physicians.[44] Implementation of AI-based insights is a major topic of discussion among pathologists since they are typically responsible for evaluating evidence that a patient has particular condition, as well as pointing out nuances associated with that condition that may require special attention when intervention decisions are made.[45] The provision of ‘decision support’ information to physicians and health care workers – especially that derived from AI-based analyses – opens up a number of thorny ethical issues, however, such as who to blame if the use of the decision support leads to poorer outcomes (i.e., the algorithm and its developers or the users who may be using it inappropriately).[46]
One important element of the implementation of AI-based decision support tools in EMR systems is that as new patient data are collected, the prediction models they are based on can be improved. Thus, ‘Learning Systems’ can be created that continually evolve and improve based on the accrual of more patient information and outcomes associated with the patients provided interventions based on the algorithms behind the learning systems’ recommendations.[47–49] Sophisticated AI-techniques can be used to enhance this learning, including aggregating data from multiple EMRs or sources.[50]
To vet the utility of an intervention for an individual patient (P3 in the right side of Figure 2), N-of-1 trials can be pursued (and could, as noted previously, be exploited in drug development phase I clinical trials)[33, 51]. AI techniques can be used to identify patterns in data collected on the patient – say through wireless sensors – that might be indicative of that patient’s response (or lack thereof) to the intervention.[34] The studies listed in Table 1 provide example published N-of-1 studies focusing on an individual’s response to a treatment or an individual undergoing monitoring for health status changes.
POST-DEPLOYMENT EVALUATION (T4) STUDIES AND WAREHOUSING (P4)
After the implementation and adoption of a new drug, treatment intervention or health product, continuous monitoring of that product must occur in order to determine if either unanticipated side effects are occurring or the product can be improved or replaced for various reasons (T4 in the left side of Figure 2). AI-based learning systems of the type mentioned in the previous section provide an excellent foundation for such monitoring, and early experiences with such systems bear this out.[52–54] In addition to the creation of learning systems, there are many initiatives to aggregate data on patients and patient materials to enable data mining and AI-based analyses, for example in cancer contexts,[55] but also for more general settings as well.[56, 57]
The implementation of AI-based products, such as EMR decision support tools and learning systems, will also affect doctor/patient relationships in profound ways. This is especially likely with respect to discussions around the justification of intervention choices,[58] but also with respect to discussions about predictions concerning future health care issues.[59] Large, government-sponsored national initiatives are being pursued to identify patterns among individuals tracked for health care-related phenomena that might be useful in clinical and public health practices in the future, such as the UK Biobank initiative in the United Kingdom and the ‘All-of-Us’ study in the United States.[60, 61] These studies raise important questions about the ethics, legal and social implications of aggregating data on individual patients for the purposes of benefitting individuals who may need focused care going forward.[61]
INTEGRATION AND THE PERSONALIZED MEDICINE WORKFLOW
Implicit in the P0-P4 personalized medicine translational workflow that has common elements with the T0-T4 drug and health product development workflow (Figure 2), is the suggestion that what is important for these workflows to function properly is to make sure the transitions from each stage of the workflow are eased and enabled. This can be difficult since the expertise and technologies needed at each stage are very different, often leading to their independent pursuit. However, emerging strategies and concepts in the way personalized medicine is practiced, coupled with the use of AI techniques, could lead to more holistic and efficient ways of treating individual patients that run through the entire P0-P4 workflow.[6]
Thus, the ideal setting for personalized medicine and health care is one in which the diagnoses, treatment and follow-up monitoring of individual patients is streamlined into a single process with very smooth and coordinated transitions from one relevant activity or sub-process to another. A good paradigm for this involves the creation of cell replacement therapies for a wide variety of conditions.[62, 63] Thus, for example, in certain immunotherapeutic-oriented cell replacement therapies for cancer, a patient’s tumor is profiled for the existence of unique ‘neo-antigens’ or mutations that might attract the host’s own immune system to attack cells harboring those mutations. If such neo-antigens are found, then cells from either a donor (allogeneic transplantation) or from the patient him or herself (autologous transplantation) are harvested and sensitized to recognize the neo-antigens. The basic idea is that these cells will attract the host’s immune cells towards the tumor cells harboring the neo-antigens when introduced into the patient’s body.[62, 63] Since the creation and manufacture of the cells cannot be pursued in advance of knowing what neo-antigens are present in the patient’s tumor, they must be created in near real time. The production of treatments in real-time based on the patient’s unique and immediate needs is termed the ‘magistral’ production of treatments, as opposed to the traditional or ‘officinal’ production of treatments.[29, 30] Magistral production of drugs is likely to be a reality for personalized medicine in many settings, even beyond cancer, since it would be too difficult to anticipate all the treatments (e.g., cells sensitive to every neo-antigen profile) and stockpile them for use in the future, as is assumed in the case with the officinal production of treatments.
To advance and generalize this concept of the magistral production of personalized medicine treatments, one could imagine leveraging AI-powered robotics technologies to enable the efficient and precise manufacture of relevant treatments.[64] 3D printing of treatments also has the potential to facilitate the real-time production of treatments in near real time, as the first US FDA-approval for a 3D printed drug was made in 2015.[65] One could also envision the immediate conduct of N-of-1 trials involving AI-based pattern discovery with sophisticated treatment outcome monitoring devices after a treatment has been crafted to assess its impact on the patient.[33, 51, 66] Further, one could potentially exploit AI-based simulation studies to anticipate directions that a treatment strategy might take.[67]
LIMITATIONS OF AI IN ADVANCING PERSONALIZED MEDICINE
There are many limitations to the use of AI in the development of personalized medicines. We briefly discuss some of the more salient issues. First, there is an argument that many big data analyses that combine information on many individuals to identify patterns that reflect population-level relationships between data points do not get at important individual-level relationships.[68] This potential lack of ‘ergodicity’ could result in models that are not useful for making individual treatment decisions. For example, in terms of identifying trends in a target individual’s health data that could indicate a health status change for that target individual based on data collected on a large number of individuals, as more data points are collected on each individual, any predictions of the target individual’s heath trajectory should rely more on the legacy data points on that target individual and less on the population level data.[69] Sensitizing AI techniques to this fact is crucial for advancing personalized medicine.
Second, there is a need to vet or test the utility of health care products rooted in AI. This is motivated by the inconsistent results observed with the use of some AI or big data based health care products, such as IBM’s Watson treatment decision support system. [70, 71] Testing such systems via traditional randomized clinical trials has been discussed in the literature, and some AI-based decision support tools have in fact been shown to pass muster in bona fide clinical trials.[72] A potential need for vetting, for example, AI-based decision support products, like IBM’s Watson, is that if the underlying system’s decision making capability is trained on an incomplete or biased data set, then the recommendations or predictions it provides are likely to be unreliable. A rather infamous case of this involves Google’s system for prediction flu outbreaks.[73] In addition, in the context of basket trials, in which the underlying scheme for matching drugs to patient profiles is being tested, if the scheme is shown not work better than standard of care or an alternative way of matching drugs to patient profiles, then a couple of questions could be raised. It could be that the drugs are ineffective, or some subset are ineffective, essentially negatively impacting the overall performance of the matching scheme. Alternatively, it could be that the drugs work, but simply are not matched properly to the patient profiles; i.e., the matching algorithm or scheme is simply wrong. These questions were raised in the context of the SHIVA trial – a bucket trail in which the drug matching scheme was shown not to benefit the patients any more than legacy ways of treating patients.[74]
Third, it may be that the best way to vet at least decision support tools, is not to test them in randomized clinical trials, but rather to implement them in learning systems in which the decision support rules or algorithms are continuously updated.[66, 75–78] However, this would not only require a lack of bias in the initial data sets used to seed the learning system in order to ensure generalizable results, but could also take a lengthy time period for the system to evolve into a system with accurate and reliable decision making.
Fourth, many AI-based decision support products leverage deep learning and neural network-based algorithms. Such algorithms can produce very reliable predictions if a large enough training set is used, but the connections between the inputs (i.e., data) and the outputs (predictions) can be very hard to understand. Thus, the ‘Black Box’ problem associated with many AI-based tools can be problematic and lead to a lack of confidence or sense of trepidation about relying on the predictions in the real world where real lives are at stake.[79] In addition, not all AI techniques are designed to identify causal relationships between various input and outputs, but rather mere associations or predictions (i.e., focus on correlation and not causation).[80] This may suffice if the goal is to develop accurate predictions, but if the goal is to, e.g., identify a drug target that, when modulated, leads to a desired effect, then identifying causal relationships is crucial.
FUTURE DIRECTIONS AND CONCLUDING REMARKS
The future contributions of AI in advancing personalized medicine are likely to be very pronounced, as this chapter makes clear. Not only will there be greater adoption of AI-based health products but such products could be developed and exploit emerging computing capabilities such as quantum computing[81, 82] to achieve increased speed and an ability to handle larger and larger data sets. These larger data sets are likely to derive from better and more sophisticated monitoring health monitoring devices which can be used to gather data to seed and key off for the development of more reliable predictions.[83]
In addition to exploiting greater speed and computational efficiency, AI-based health products and tools will likely incorporate greater understanding of biology in their formulations in the future. Thus, the discovery of simple input/output relationships among data points that has been focus of a great deal of AI, machine learning and statistical analysis research, could be pursued with constraints that are known to govern phenomena of relevance (e.g., known biophysical constraints involving the production of metabolites in a biochemical pathway, first principles having to do with Watson-Crick base pairing, etc.).[84, 85]
Finally, as a closing note, much of the use of AI in the development of personalized medicines is focused on the treatment of individuals with overt disease: identifying the underlying pathology, determining which interventions might make most sense to provide given what is known about that pathology and the mechanism of action of the intervention, and testing to see if the intervention works. Thus, the vast majority of AI-based products and tools used in advancing personalized medicine focus on the diagnosis, prognosis and treatment of individuals. This makes sense as there is a great need for advances and efficiency gains in treating patients given the costs of current treatments, especially in the context of cancer. However, the application of AI to disease prevention is gaining a great deal of attention and traction. For example, AI and machine learning techniques have been shown to be useful in the development of ‘polygenic risk scores’ that can be used to identify individuals with an elevated genetic risk for disease that could be monitored more closely.[86–88] In addition, by combining insights into genetic predisposition to disease with continuous monitoring to identify early signs of disease, one could potentially stop diseases in their tracks before complicated treatments are needed for fulminant forms of the disease manifest.[89, 90] Such monitoring could be greatly enhanced by applying AI techniques to novel sensors.[91, 92]
Ultimately, enthusiasm for leveraging AI techniques is not likely to slow down any time soon. AI is likely to impact virtually every industry, from manufacturing, to sales and marketing, to banking, to transportation. All of these industries can obviously be improved with AI playing an important role in the needed innovations. The health care industry is no less likely to benefit from AI, as this chapter has made clear, as long as appropriate integration and vetting occurs.
REFERENCES
- 1.Russell S and Norvig P, Artificial Intelligence: A Modern Approach (3rd Edition). 3rd ed 2009, Carmel, Indiana: Pearson. [Google Scholar]
- 2.Mahmud M, et al. , Applications of Deep Learning and Reinforcement Learning to Biological Data. IEEE Trans Neural Netw Learn Syst, 2018. 29(6): p. 2063–2079. [DOI] [PubMed] [Google Scholar]
- 3.Webb S, Deep learning for biology. Nature, 2018. 554(7693): p. 555–557. [DOI] [PubMed] [Google Scholar]
- 4.Fleming N, How artificial intelligence is changing drug discovery. Nature, 2018. 557(7707): p. S55–S57. [DOI] [PubMed] [Google Scholar]
- 5.Committee to Review the Clinical and Translational Science Awards Program at the National Center for Advancing Translational Sciences; Board on Health Sciences Policy; Institute of Medicine; Leshner AI TS, ed. The CTSA Program at NIH: Opportunities for Advancing Clinical and Translational Research. The National Academies Collection: Reports funded by National Institutes of Health. 2013, National Academies Press: Washington, DC. [PubMed] [Google Scholar]
- 6.Schork NJ and Nazor K, Integrated Genomic Medicine: A Paradigm for Rare Diseases and Beyond. Adv Genet, 2017. 97: p. 81–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telenti A, et al. , Deep learning of genomic variation and regulatory network data. Hum Mol Genet, 2018. 27(R1): p. R63–R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteva A, et al. , Dermatologist-level classification of skin cancer with deep neural networks. Nature, 2017. 542(7639): p. 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerstung M, et al. , Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat Genet, 2017. 49(3): p. 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulshan V, et al. , Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA, 2016. 316(22): p. 2402–2410. [DOI] [PubMed] [Google Scholar]
- 11.Cohen JD, et al. , Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science, 2018. 359(6378): p. 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray MA, et al. , A dataset of images and morphological profiles of 30 000 small-molecule treatments using the Cell Painting assay. Gigascience, 2017. 6(12): p. 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J, et al. , Using deep learning to model the hierarchical structure and function of a cell. Nat Methods, 2018. 15(4): p. 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ideker T, et al. , Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science, 2001. 292(5518): p. 929–34. [DOI] [PubMed] [Google Scholar]
- 15.Bohannon J, The cyberscientist. Science, 2017. 357(6346): p. 18–21. [DOI] [PubMed] [Google Scholar]
- 16.King RD, et al. , The automation of science. Science, 2009. 324(5923): p. 85–9. [DOI] [PubMed] [Google Scholar]
- 17.Sparkes A and Clare A, AutoLabDB: a substantial open source database schema to support a high-throughput automated laboratory. Bioinformatics, 2012. 28(10): p. 1390–7. [DOI] [PubMed] [Google Scholar]
- 18.Butler KT, et al. , Machine learning for molecular and materials science. Nature, 2018. 559(7715): p. 547–555. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Lengeling B and Aspuru-Guzik A, Inverse molecular design using machine learning: Generative models for matter engineering. Science, 2018. 361(6400): p. 360–365. [DOI] [PubMed] [Google Scholar]
- 20.Aage N, et al. , Giga-voxel computational morphogenesis for structural design. Nature, 2017. 550(7674): p. 84–86. [DOI] [PubMed] [Google Scholar]
- 21.Segler MHS, Preuss M, and Waller MP, Planning chemical syntheses with deep neural networks and symbolic AI. Nature, 2018. 555(7698): p. 604–610. [DOI] [PubMed] [Google Scholar]
- 22.Ahneman DT, et al. , Predicting reaction performance in C-N cross-coupling using machine learning. Science, 2018. 360(6385): p. 186–190. [DOI] [PubMed] [Google Scholar]
- 23.Radovic A, et al. , Machine learning at the energy and intensity frontiers of particle physics. Nature, 2018. 560(7716): p. 41–48. [DOI] [PubMed] [Google Scholar]
- 24.Silver D, et al. , Mastering the game of Go without human knowledge. Nature, 2017. 550(7676): p. 354–359. [DOI] [PubMed] [Google Scholar]
- 25.Madhukar NS, et al. , A New Big-Data Paradigm For Target Identification And Drug Discovery. BioRxiv ( 10.1101/134973), 2018. [DOI] [Google Scholar]
- 26.Chen B and Butte AJ, Leveraging big data to transform target selection and drug discovery. Clin Pharmacol Ther, 2016. 99(3): p. 285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y and Bajorath J, Entering the ‘big data’ era in medicinal chemistry: molecular promiscuity analysis revisited. Future Sci OA, 2017. 3(2): p. FSO179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez D, How Robots Are Making Better Drugs, Faster, in Wall Street Journal. 2018, Dow Jones & Company New York, New York. [Google Scholar]
- 29.Patient-centered drug manufacture. Nat Biotechnol, 2017. 35(6): p. 485. [DOI] [PubMed] [Google Scholar]
- 30.Schellekens H, et al. , Making individualized drugs a reality. Nat Biotechnol, 2017. 35(6): p. 507–513. [DOI] [PubMed] [Google Scholar]
- 31.Lavertu A, et al. , Pharmacogenomics and big genomic data: from lab to clinic and back again. Hum Mol Genet, 2018. 27(R1): p. R72–R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalinin AA, et al. , Deep learning in pharmacogenomics: from gene regulation to patient stratification. Pharmacogenomics, 2018. 19(7): p. 629–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schork NJ, Personalized medicine: Time for one-person trials. Nature, 2015. 520(7549): p. 609–11. [DOI] [PubMed] [Google Scholar]
- 34.Serhani MA, et al. , New algorithms for processing time-series big EEG data within mobile health monitoring systems. Comput Methods Programs Biomed, 2017. 149: p. 79–94. [DOI] [PubMed] [Google Scholar]
- 35.Marr B, First FDA Approval For Clinical Cloud-Based Deep Learning In Healthcare, in Forbes. 2017, Forbes Publishing Company: New York City. [Google Scholar]
- 36.Miotto R, et al. , Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci Rep, 2016. 6: p. 26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi G, Manfrin A, and Lutolf MP, Progress and potential in organoid research. Nat Rev Genet, 2018. [DOI] [PubMed] [Google Scholar]
- 38.Deng Y, et al. , Massive single-cell RNA-seq analysis and imputation via deep learning. BioRxiv 2018. https://t.co/EGBwlYFLLK. [Google Scholar]
- 39.Yauney G and Shah P, Reinforcement Learning with Action-Derived Rewards for Chemotherapy and Clinical Trial Dosing Regimen Selection. Proceedings of Machine Learning Research, 2018. 85. [Google Scholar]
- 40.Biankin AV, Piantadosi S, and Hollingsworth SJ, Patient-centric trials for therapeutic development in precision oncology. Nature, 2015. 526(7573): p. 361–70. [DOI] [PubMed] [Google Scholar]
- 41.Kodack DP, et al. , Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep, 2017. 21(11): p. 3298–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorshkov K, et al. , Advancing precision medicine with personalized drug screening. Drug Discov Today, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miranda CC, et al. , Towards Multi-Organoid Systems for Drug Screening Applications. Bioengineering (Basel), 2018. 5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott IA, et al. , Using EMR-enabled computerized decision support systems to reduce prescribing of potentially inappropriate medications: a narrative review. Ther Adv Drug Saf, 2018. 9(9): p. 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dreyer KJ and Geis JR, When Machines Think: Radiology’s Next Frontier. Radiology, 2017. 285(3): p. 713–718. [DOI] [PubMed] [Google Scholar]
- 46.Nabi J, How Bioethics Can Shape Artificial Intelligence and Machine Learning. Hastings Cent Rep, 2018. 48(5): p. 10–13. [DOI] [PubMed] [Google Scholar]
- 47.Etheredge LM, A rapid-learning health system. Health Aff (Millwood), 2007. 26(2): p. w107–18. [DOI] [PubMed] [Google Scholar]
- 48.Shrager J and Tenenbaum JM, Rapid learning for precision oncology. Nat Rev Clin Oncol, 2014. 11(2): p. 109–18. [DOI] [PubMed] [Google Scholar]
- 49.Shah A, et al. , Building a Rapid Learning Health Care System for Oncology: Why CancerLinQ Collects Identifiable Health Information to Achieve Its Vision. J Clin Oncol, 2016. 34(7): p. 756–63. [DOI] [PubMed] [Google Scholar]
- 50.Hastie T, Tibshirani R, and Friedman J, The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed 2009, New York, New York: Springer. [Google Scholar]
- 51.Schork NJ and Goetz LH, Single-Subject Studies in Translational Nutrition Research. Annu Rev Nutr, 2017. 37: p. 395–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwala V, et al. , Real-World Evidence In Support Of Precision Medicine: Clinico-Genomic Cancer Data As A Case Study. Health Aff (Millwood), 2018. 37(5): p. 765–772. [DOI] [PubMed] [Google Scholar]
- 53.Williams MS, et al. , Patient-Centered Precision Health In A Learning Health Care System: Geisinger’s Genomic Medicine Experience. Health Aff (Millwood), 2018. 37(5): p. 757–764. [DOI] [PubMed] [Google Scholar]
- 54.Rajkomar A, et al. , Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine, 2018. 1(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali M and Aittokallio T, Machine learning and feature selection for drug response prediction in precision oncology applications. Biophys Rev, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathe E, et al. , The Omics Revolution Continues: The Maturation of High-Throughput Biological Data Sources. Yearb Med Inform, 2018. 27(1): p. 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varghese J, et al. , CDEGenerator: an online platform to learn from existing data models to build model registries. Clin Epidemiol, 2018. 10: p. 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lerner I, et al. , Revolution in Health Care: How Will Data Science Impact Doctor-Patient Relationships? Front Public Health, 2018. 6: p. 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savage N, Machine learning: Calculating disease. Nature, 2017. 550(7676): p. S115–S117. [DOI] [PubMed] [Google Scholar]
- 60.Bycroft C, et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature, 2018. 562: p. 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sankar PL and Parker LS, The Precision Medicine Initiative’s All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet Med, 2017. 19(7): p. 743–750. [DOI] [PubMed] [Google Scholar]
- 62.Li C, et al. , Application of induced pluripotent stem cell transplants: Autologous or allogeneic? Life Sci, 2018. [DOI] [PubMed] [Google Scholar]
- 63.Graham C, et al. , Allogeneic CAR-T Cells: More than Ease of Access? Cells, 2018. 7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan R, Yang X, and Shen Y, Robot-aided electrospinning toward intelligent biomedical engineering. Robotics Biomim, 2017. 4(1): p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Osouli-Bostanabad K and Adibkia K, Made-on-demand, complex and personalized 3D-printed drug products. Bioimpacts, 2018. 8(2): p. 77–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schork NJ, Randomized clinical trials and personalized medicine: A commentary on deaton and cartwright. Soc Sci Med, 2018. 210: p. 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shamsuddin R, et al. Virtual patient model: An approach for generating synthetic healthcare time series data in IEEE Interantional COnference on Healthcare Informatics. 2018. IEEE Computer Society. [Google Scholar]
- 68.Fisher AJ, Medaglia JD, and Jeronimus BF, Lack of group-to-individual generalizability is a threat to human subjects research. Proc Natl Acad Sci U S A, 2018. 115(27): p. E6106–E6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drescher CW, et al. , Longitudinal screening algorithm that incorporates change over time in CA125 levels identifies ovarian cancer earlier than a single-threshold rule. J Clin Oncol, 2013. 31(3): p. 387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou N, et al. , Concordance Study Between IBM Watson for Oncology and Clinical Practice for Patients with Cancer in China. Oncologist, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt C, Anderson MD Breaks With IBM Watson, Raising Questions About Artificial Intelligence in Oncology. J Natl Cancer Inst, 2017. 109(5): p. 4–5. [DOI] [PubMed] [Google Scholar]
- 72.Abramoff MD, et al. , Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digital Medicine, 2018. 1(39). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazer D, et al. , Big data. The parable of Google Flu: traps in big data analysis. Science, 2014. 343(6176): p. 1203–5. [DOI] [PubMed] [Google Scholar]
- 74.Le Tourneau C, et al. , Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol, 2015. 16(13): p. 1324–34. [DOI] [PubMed] [Google Scholar]
- 75.Ioannidis JPA and Khoury MJ, Evidence-based medicine and big genomic data. Hum Mol Genet, 2018. 27(R1): p. R2–R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.AI diagnostics need attention. Nature, 2018. 555(7696): p. 285. [DOI] [PubMed] [Google Scholar]
- 77.Frieden TR, Evidence for Health Decision Making - Beyond Randomized, Controlled Trials. N Engl J Med, 2017. 377(5): p. 465–475. [DOI] [PubMed] [Google Scholar]
- 78.Abernethy A and Khozin S, Clinical Drug Trials May Be Coming to Your Doctor’s Office, in Wall Street Journal. 2017, Dow Jones & Company: New York, New York. [Google Scholar]
- 79.Voosen P, The AI detectives. Science, 2017. 357(6346): p. 22–27. [DOI] [PubMed] [Google Scholar]
- 80.Marwala T, Causality, Correlation and Artificial Intelligence for Rational Decision Making. 2015, New Jersey: World Scientific. [Google Scholar]
- 81.Ciliberto C, et al. , Quantum machine learning: a classical perspective. Proc Math Phys Eng Sci, 2018. 474(2209): p. 20170551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li RY, et al. , Quantum annealing versus classical machine learning applied to a simplified computational biology problem. npj Quantum Inf, 2018. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vashistha R, et al. , Futuristic biosensors for cardiac health care: an artificial intelligence approach. 3 Biotech, 2018. 8(8): p. 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palsson B, Systems Biology: Constraint-based Reconstruction and Analysis. 2nd ed 2015, Boston, MA: Cambridge University Press. [Google Scholar]
- 85.Ideker T, Dutkowski J, and Hood L, Boosting signal-to-noise in complex biology: prior knowledge is power. Cell, 2011. 144(6): p. 860–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khera AV, et al. , Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet, 2018. 50(9): p. 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warren M, The approach to predictive medicine that is taking genomics research by storm. Nature, 2018. 562(7726): p. 181–183. [DOI] [PubMed] [Google Scholar]
- 88.Schork AJ, Schork MA, and Schork NJ, Genetic risks and clinical rewards. Nat Genet, 2018. 50(9): p. 1210–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patel CJ, et al. , Whole genome sequencing in support of wellness and health maintenance. Genome Med, 2013. 5(6): p. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schork NJ, Genetic parts to a preventive medicine whole. Genome Med, 2013. 5(6): p. 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mapara SS and Patravale VB, Medical capsule robots: A renaissance for diagnostics, drug delivery and surgical treatment. J Control Release, 2017. 261: p. 337–351. [DOI] [PubMed] [Google Scholar]
- 92.Topol EJ, Deep Medicine: How Artificial Intelligence Can Make Healthcare Human Again. 2019, New York: Basic Books. [Google Scholar]
- 93.David LA, et al. , Host lifestyle affects human microbiota on daily timescales. Genome Biol, 2014. 15(7): p. R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen R, et al. , Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell, 2012. 148(6): p. 1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Magnuson V, Wang Y, and Schork N, Normalizing sleep quality disturbed by psychiatric polypharmacy: a single patient open trial (SPOT). F1000Res, 2016. 5: p. 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeevi D, et al. , Personalized Nutrition by Prediction of Glycemic Responses. Cell, 2015. 163(5): p. 1079–1094. [DOI] [PubMed] [Google Scholar]
- 97.Smarr L, et al. , Tracking Human Gut Microbiome Changes Resulting from a Colonoscopy. Methods Inf Med, 2017. 56(6): p. 442–447. [DOI] [PubMed] [Google Scholar]
- 98.Trammell SA, et al. , Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun, 2016. 7: p. 12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Forsdyke DR, Summertime dosage-dependent hypersensitivity to an angiotensin II receptor blocker. BMC Res Notes, 2015. 8: p. 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Rawe JA, et al. , Integrating precision medicine in the study and clinical treatment of a severely mentally ill person. PeerJ, 2013. 1: p. e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li W, et al. , Extensive graft-derived dopaminergic innervation is maintained 24 years after transplantation in the degenerating parkinsonian brain. Proc Natl Acad Sci U S A, 2016. 113(23): p. 6544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bloss CS, et al. , A genome sequencing program for novel undiagnosed diseases. Genet Med, 2015. 17(12): p. 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Piening BD, et al. , Integrative Personal Omics Profiles during Periods of Weight Gain and Loss. Cell Syst, 2018. 6(2): p. 157–170 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zalusky R and Herbert V, Megaloblastic anemia in scurvy with response to 50 microgm. of folic acid daily. N Engl J Med, 1961. 265: p. 1033–8. [DOI] [PubMed] [Google Scholar]
- 105.Herbert V, Experimental nutritional folate deficiency in man. Trans Assoc Am Physicians, 1962. 75: p. 307–20. [PubMed] [Google Scholar]
- 106.Golding PH, Severe experimental folate deficiency in a human subject - a longitudinal study of biochemical and haematological responses as megaloblastic anaemia develops. Springerplus, 2014. 3: p. 442. [DOI] [PMC free article] [PubMed] [Google Scholar]


