Abstract
Poly(β-L-malic acid) (PMLA) is a natural polyester produced by numerous microorganisms. Regarding its biosynthetic machinery, a nonribosomal peptide synthetase (NRPS) is proposed to direct polymerization of L-malic acid in vivo. Chemically versatile and biologically compatible, PMLA can be used as an ideal carrier for several molecules, including nucleotides, proteins, chemotherapeutic drugs, and imaging agents, and can deliver multimodal theranostics through biological barriers such as the blood–brain barrier. We focus on PMLA biosynthesis in microorganisms, summarize the physicochemical and physiochemical characteristics of PMLA as a naturally derived polymeric delivery platform at nanoscale, and highlight the attachment of functional groups to enhance cancer detection and treatment.
Keywords: polymalatase, synthetase, nanomedicine, cancer diagnosis, tumor therapy
Biopolymer: From Benchtop to Bedside
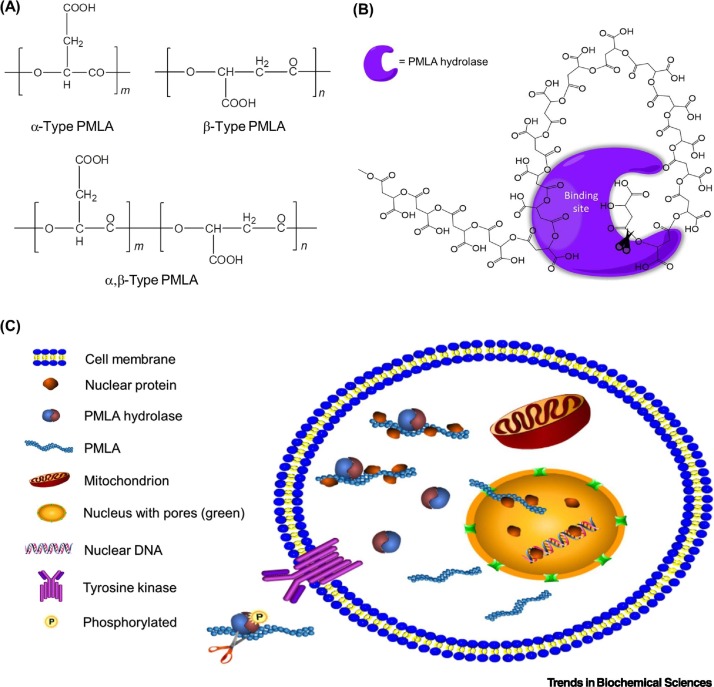
PMLAs are water-soluble polyesters with repeating malyl units (see Glossary) that were discovered in aqueous extracts of Penicillium cyclopium culture in 1969 [1]. To date, three major types of PMLAs have been identified that differ in their ester bonds between hydroxyl groups and either α- or β-carboxyl groups in the polymeric chain, namely α-, β-, and α,β-PMLAs (Figure 1A). However, naturally available PMLA, such as from Aureobasidium pullulan or Physarum polycephalum, has a β-type structure, whereas α- and α,β-PMLAs are obtained by chemical synthesis [2].
Figure 1.
Structures of Poly(β-L-Malic Acids) (PMLAs) and Their Degradation by PMLA Hydrolases In Vivo.
(A) Structures of α, β, and α,β types of polymalic acid. (B) PMLA hydrolases break down the first ester bond from OH-terminus while binding to the 12th malyl unit along the polymer chain towards the C-terminus [73]. (C) Intracellular trafficking of PMLA hydrolases and their extracellular activation. In typical eukaryotes, PMLA hydrolase binds to PMLA and remains inactive, carrying nuclear proteins into the nucleus while PMLA hydrolase remains at the surface of nuclear envelope. Cargo-discharged PMLA exits from the nucleus into the cytoplasm via nuclear pores; it binds to PMLA hydrolases that then translocate across the cell membrane where they are phosphorylated by membrane-bound tyrosine kinases, restoring their activity to degrade PMLAs extracellularly [74,75].
In addition to functions in carbon storage and as an energy reservoir for other polyhydroxyalkanoates (PHAs) in their host microorganisms [3], PMLA acts as a molecular transporter to bind to and carry nuclear proteins related to DNA replication, demonstrating its unique role in maintaining protein homeostasis and assisting DNA synthesis [4]. Unlike bioderived water-insoluble polyhydroxybutyrate (PHB), PMLA possesses a high capacity to attach hydrophobic molecules while remaining water-soluble owing to the exceptional hydrophilicity of abundant carboxylic acids that are present as pendant groups from its polymeric chain [5., 6., 7.]. This characteristic of PMLA diminishes its innate toxicity and immunogenicity, and makes it compatible with various biological systems [8., 9., 10.]. Moreover, hydrolyzable ester bonds in the backbone of the polymeric chain facilitate the complete decomposition of PMLA in biological milieux, where the sole end-product of malates is reused by the citric acid cycle in the mitochondrial matrix as a part of central cellular metabolism [11]. Only the L-form of malic acid is obtained naturally via microbiological production [12] (Figure 1A). Non-enzymatic hydrolysis of PMLA takes place via spontaneous random breakdown of ester bonds and concurrent formation of intermediate oligomers, whereas enzymatic hydrolysis in the presence of PMLA hydrolases (Box 1 ) uncaps one unit of malic acid after another from one end (the hydroxyl terminus) of the polymeric chain to the other [13]. The PMLAs discussed in this review focus on biogenic poly(β-L-malic acid), although synthetic PMLAs are discussed where relevant.
Box 1. PMLA Hydrolases as Opponents of Biosynthesis.
PMLA hydrolases (also called PMLA depolymerases or polymalatases) have been purified and characterized from both eukaryotic and prokaryotic microorganisms [73,76]. In eukaryotes, particularly PMLA-producing myxomycetes, soluble PMLA hydrolase serves as a molecular chaperone of PMLA and binds to its hydroxyl terminus at the penultimate malyl residue for catalytic cleavage, although another binding site 12 malyl residues down the polymeric chain was also proposed to govern stability and carriage function (Figure 1B) [73]. Intracellular PMLA hydrolase remains inactive and forms complexes with PMLA that may carry other nuclear proteins, assisting the translocation of PMLA into the nucleus while leaving PMLA hydrolase on the surface of nuclear envelope, possibly because of either its binding to envelope proteins and/or the outer nuclear membrane, or disruptive variation in local ionic strength (Figure 1C) [75]. After discharging its nuclear protein cargo, PMLA exits through nuclear pores and to join newly synthesized PMLA in the cytoplasm for the next deliveries [75]. PMLA thus shuttles between the nucleus and the cytoplasm to maintain homeostasis in different phases of the cell cycle. Excess PMLAs bound to hydrolases are excreted through the cell membrane, where a membrane-bound tyrosine kinase then phosphorylates PMLA hydrolases and activates their hydrolytic activity, leading to degradation of concomitantly released PMLAs in the culture medium (peak activity at pH 3–5) [74]. However, the hydrolases may differ across different eukaryotes such as PMLA-producing myxomycetes and fungi: PMLA isolated from P. polycephalum had higher molar mass and polydispersity than that from A. pullulan [38]. In prokaryotes such as bacteria that are devoid of PMLA, insoluble PMLA hydrolases are confined to the outer cell membrane where they hydrolyze outsourced PMLA into malic acid monomers that are further taken up for cellular metabolism [76]. Unlike PHB depolymerase, PMLA hydrolase does not exhibit serine esterase activity in which a nucleophilic serine in the active site initiates substrate hydrolysis, and is not inhibited by serine protease inhibitors [76].
Alt-text: Box 1
Given its great sustainability because it is produced from natural sources, PMLA serves as a platform for potential multimodal conjugates with unmatched bioavailability, biocompatibility, and biodegradability. For this reason, PMLA has been intensively and extensively used in biomedical and medicinal research, particularly in drug delivery and in bioimaging for cancer theranostics [14,15].
Biosynthesis of PMLA in Different Microorganisms
Following its discovery in P. cyclopium as an acidic substance containing no nitrogen (molar mass M = ~5000 g/mol), PMLA was also found in the myxomycete P. polycephalum (M = 10 000–12 000 g/mol) as an inhibitor to DNA polymerases, and in the yeast-like fungi Aureobasidium sp. (M = 6000–11 000 g/mol) and A. pullulan (M = 3000–5000 g/mol) as an extracellular secretion in culture broth [16,17]. Bacteria producing PMLA have not yet been identified. A study that examined PMLA bioproduction from 56 strains of A. pullulan of a diversity of phylogenetic clades indicated high productivity but low molar mass (5100–7900 g/mol), where PMLA was bound to polysaccharides of varying molar mass depending on the exact strain type [17]. By screening various Aureobasidium spp. and optimizing the culture conditions, efforts have been made to obtain efficient biosynthesis of PMLA with high molar mass (up to 20 000 g/mol) [18]. Using a highly productive Aureobasidium sp. strain isolated from mangrove systems, purified PMLA with M = 205 400 g/mol was reported [19]. By contrast, PMLA produced from the plasmodial stage of P. polycephalum sustains a much elongated linear chain in its pure form, with M = 30 000–300 000 g/mol [12,20].
Generic Biosynthetic Pathways of PMLA
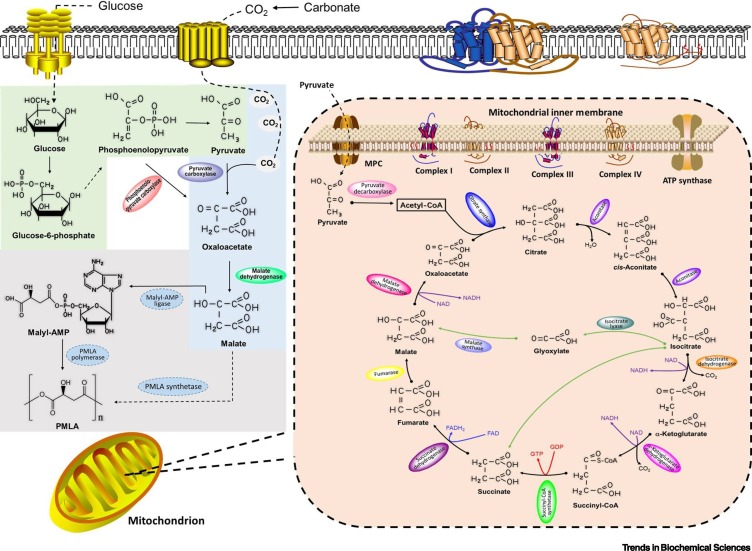
Using D-glucose as the most efficient carbon source, and relying on nutrients available in the extracellular culture medium, independent extramitochondrial and intramitochondrial metabolic routes for generating L-malate (the immediate precursor to PMLA) in vivo were unraveled [21] (Figure 2 ). On the one hand, in the presence of exogenous carbonates (e.g., CaCO3 or Na2CO3), L-malate is produced via a reductive pathway in the cytoplasm; because of the high level of CO2 in the cytoplasm, pyruvate is prioritized for carboxylation to oxaloacetate by pyruvate carboxylase, and this is further reduced by malate dehydrogenase to malate [22]. Alternatively, in a similar manner, phosphoenolpyruvic acid is directly converted by phosphoenolpyruvate carboxylase to oxaloacetate, forming malate [23]. Therefore, phosphoenolpyruvic acid and pyruvate from the glycolysis pathway are indispensable molecules for malate synthesis in vivo, whereas their carboxylases govern carbon flux for malate biosynthesis because regulated carboxylase activity influences the ultimate level of PMLA bioproduction [23,24].
Figure 2.
Generic Biosynthetic Pathways of Poly(β-L-Malic acids) (PMLAs) in Eukaryotic Cells.
Glucose is transported into the cytoplasm where it is metabolized by glycolysis to produce pyruvate (green). Excess CO2 activates the reductive extramitochondrial pathway to generate malate (blue). When exogenous carbonates are absent, intramitochondrial malate formation via the oxidative pathway takes place in the mitochondrial matrix through either the tricarboxylic acid (TCA) cycle or the glyoxylate shunt (orange). Through either pathway, malates are polymerized to form PMLA in the cytoplasm (gray). A PMLA synthetase has been proposed that possibly combines the activities of a malyl-AMP ligase and an unknown PMLA polymerase, in conjunction with an auxiliary peptide or enzyme (e.g., spherulin 3b in P. polycephalum) [30,39].
On the other hand, if exogenous carbonates are absent, malate formation via the oxidative pathway is mostly achieved in the mitochondrial matrix through either the tricarboxylic acid (TCA) cycle or the glyoxylate bypass [25]. Glucose or another sugar is transformed into pyruvate through glycolysis, followed by import into mitochondria by mitochondrial pyruvate carrier (MPC) proteins and subsequent decarboxylation by pyruvate decarboxylase complex to produce acetyl coenzyme A (acetyl-CoA), thus entering the TCA cycle and being converted to four-carbon oxaloacetate and subsequently to six-carbon citrate [26,27]. The TCA cycle continues as a series of biochemical transformations that take place in an enzyme-mediated cascade, generating products including cis-aconitate, isocitrate, α-ketoglutarate, succinate, fumarate, malate, and eventually oxaloacetate again. In parallel, taking a shortcut in the TCA cycle, a process known as the glyoxylate shunt, isocitrate is directly converted into glyoxylate and succinate; glyoxylate then reacts with acetyl-CoA to produce malate, and succinate generates malate catalyzed by succinate dehydrogenase and fumarase [28]. The glyoxylate pathway could thus be driven to produce malate and PMLA if key enzymes (e.g., fumarase, succinate dehydrogenase) in the TCA cycle are blocked [25], or if malate synthase is upregulated by exogenous ethanol stress [29].
Because the inner membrane of mitochondria remains a barrier to most molecules, malate and oxaloacetate from the oxidative TCA cycle or glyoxylate shunt interchange with their counterparts in the cytosolic reductive pathway via the malate/aspartate shuttle, where oxaloacetate is reduced to malate both extramitohchondrially and intramitochondrially to permit transportation and avoid oxaloacetate accumulation in the cytoplasm [21]. In cultures of P. polycephalum and A. pullulan, the addition of exogenous carbonates augments CO2 fixation and pyruvate carboxylation into oxaloacetate by pyruvate carboxylase in the cytoplasm, abolishing the intramitochondrial pathways for L-malate production and ensuing PMLA synthesis (Figure 2) [23,30]. Under these conditions, the addition of TCA cycle metabolites into the cell culture might not promote PMLA production [11]. In other words, environmental carbonate works as a switch between oxidative and reductive pathways to produce malate. Furthermore, carbonate adjusts the acidity of the culture medium, noting that low pH values accelerate PMLA hydrolysis. In contrast to P. polycephalum, that merely uses D-glucose as a carbon source in malate and PMLA bioproduction, A. pullulan takes up a broad spectrum of saccharides (e.g., sucrose, fructose, and maltose) because it expresses a remarkable diversity of polysaccharide lyases, glycoside hydrolases, carbohydrate esterases, glycosyltransferases, and sugar transporters, enabling its efficient utilization of diverse carbohydrates to produce malate [31., 32., 33.]. By depleting the nitrogen supply or augmenting the carbon/nitrogen ratio in the cell medium, PMLA bioproduction is significantly enhanced per unit of cell mass because nitrogen starvation upregulates the expression of key enzymes in PMLA biosynthetic pathways, uncoupling cell growth from PMLA production [34,35].
At the cost of ATP probably derived from glycolysis, malates are polymerized to form PMLA in the cytoplasm [36]. The acellular slime mold P. polycephalum only synthesizes PMLA in its multinucleated plasmodia, whereas the yeast-like fungus A. pullulan spends its entire life cycle (except hyphae form) producing PMLA [37,38]. Although the search for PMLA synthetase is still underway, a nonribosomal peptide synthetase (NRPS) machinery was proposed that first forms malyl-AMP via the action of malyl-AMP ligase, and this is then assembled into a polymeric chain by an unidentified polymerase [36]. In addition, a plasmodium-specific polypeptide spherulin 3b (i.e., NKA48) was found to assist PMLA synthesis, although similar enzymes have not been identified in fungi [39]. Alternatively, in A. pullulan, it was proposed that cytosolic malate may be polymerized into PMLA using malyl-CoA as the precursor, followed by the action of malate-CoA ligase and PMLA synthetase [40]. In one variety of A. pullulan, namely Aureobasidium melanogenum, a characteristic NRSP was recently reported to be a putative PMLA synthetase that contains an adenylation domain for ATP binding and malyl-AMP formation, an activatable thiolation domain for phosphopantetheine attachment and polymerization of malyl-AMP into PMLA, and a hexa-transmembrane region for transport of PMLA out of the cytoplasm [41]. Because the whole-genome sequences of several PMLA-producing fungal strains have been determined, a conserved PMLA synthetase across species is expected to be unmasked in the near future [22,42].
Non-Enzymatic Degradation of PMLA
Biosynthetic PMLA in aqueous solution (2% w/v) has a pH of 2.0 but a pK a of 3.4–3.6 (average M = 10 000–24 000 g/mol) [43]. At acidic pH less than the pK a (e.g., pH 2–3), PMLA remains protonated, promoting the formation of intramolecular double hydrogen bonding between side-chain carboxylic acids and the construction of dense, inflexible, double-stranded segments [44]. In phosphate buffer (pH 7.4) at 37°C, PMLA with fully ionized carboxylic groups retains an open-coil conformation owing to the negatively charged neighboring side-chains, and undergoes hydrolysis with a half-life of 10 h, initially following first-order kinetics, whereas elevated temperature and acidic pH dramatically accelerate its hydrolytic degradation [37,45,46]. Moreover, random hydrolysis prioritizes the breakdown of intrachain ester bonds over those at the ends of the molecule, producing oligomers instead of malates until hydrolysis proceeds to completion, whereas hydrophobic substitution (e.g., alkylation) of PMLA side-chains delays this hydrolytic degradation, possibly because of limited water access to ester bonds in the backbone as the polymer conformation alters [47,48]. In addition, a larger substituent group or a higher degree of substitution in the side-chain that increases hydrophobicity, leads to the slower degradation in aqueous solutions [47,48].
Physiochemical Properties of PMLA
In a pilot study, repeated intraperitoneal injection of synthetic PMLA into rabbits returned no detectable immune response, demonstrating non-immunogenicity, whereas the same injection into mice revealed nearly no acute toxicity (LD 50 = 3.3 g/kg body weight) [49]. Intravenous (i.v.) injection of PMLA sodium salt into mouse tail vein, with repetitive administration at low dosage, showed no mortality or adverse effects at doses up to 3 g/kg body weight; however, mice only tolerated a one-time i.v. high-dose injection (up to ~2 g/kg body weight or otherwise) toxicity was due to the hyperosmolarity of the concentrated polymer solution injected rather than to the polymer concentration per se [50]. The elimination of injected PMLA from blood was very fast, with a t 1/2 of 8 min or much less, and injection might not even be complete before the polymer is exported into urine [50,51]. Indeed, 70% of injected PMLA was excreted after 1 h, and 90% after 6 h, although there was low but persistent liver accumulation 24 h after injection, yet no substantial accumulation in other organs, including kidney, lung, intestine, spleen, heart, muscle, and brain [50,51]. Notably, these early studies on the pharmacokinetics and biodistribution of synthetic PMLA laid a solid foundation for recent preclinical research using natural PMLA as a pharmaceutical carrier [20]. Bioproduced PMLAs possess a similar or even superior biocompatibility profile to synthetic PMLAs, and their end-product is only L-malic acid. Intriguingly, L-malate administered i.v. into the tail vein in mice had a half-life of only 10 min, and one third of injected dose ended up in exhaled CO2, whereas the majority of the remainder accumulated in tissues via renal tubular reabsorption, participating in the TCA cycle [51].
Permeation of Cellular Membrane by PMLAs
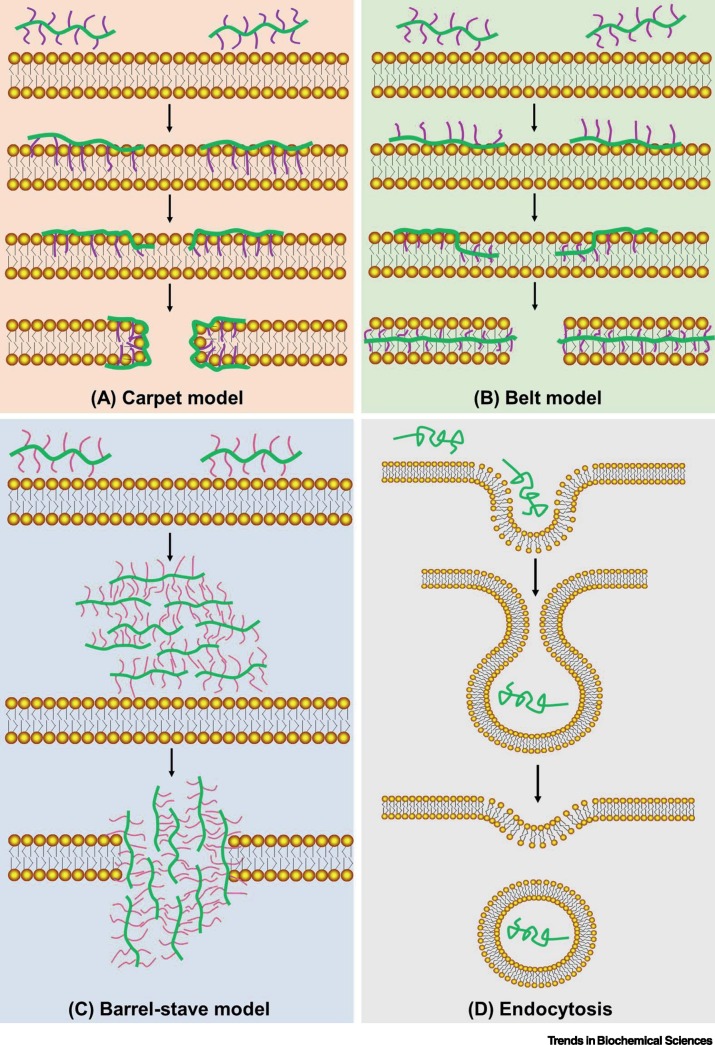
PMLA is negatively charged at pH 7.4, and hence poses no disruptive threat to phospholipid membranes because they have the same charge. Given that there is no specific cell-surface receptor of PMLA, water-soluble PMLA is transported into the cytoplasm through the invagination of cell membrane via a process of non-specific endocytosis, although the efficiency of transmembrane transport can be very low. In contrast to PMLA, its copolymers with lipophilic ligands undergo hydrophobic interactions in aqueous solution and assemble into lipophilic patches, and these can interact with the cell membrane, leading to anchoring of lipophilic patches in close proximity to lipid bilayers [52]. Depending on the attached hydrophobic ligands, three distinct mechanisms have been proposed by which different PMLA-conjugated copolymers can induce membrane permeation (Figure 3 ); these are discussed in the following text:
-
(i)
The carpet model is typified by PMLA leucine ethyl ester (PMLA-LOEtxH100−x) [53]. At physiological pH, esterification of carboxylic acid groups in PMLA side chains leads to permanent charge neutralization, excluding further protonation even when the environmental pH drops. Upon binding to the cell membrane, PMLA-LOEtxH100−x orients itself to insert the hydrophobic LOEt side chain into the phospholipid layer, leaving the outer membrane surface expansively covered by the hydrophilic backbone of the polymeric chain. The hydrophobic binding energy is sufficient to strongly bend the plasma membrane into a curved structure, creating a transient pore that enables membrane permeation [54,55]. This process is independent of pH and its membranolytic efficiency varies according to the ratio of hydrophobic/hydrophilic moieties (Box 2 ) in the polymer (i.e., ).
-
(ii)
The belt model is typified by PMLA tritryptophan (PMLA-WWWxH100−x) [56]. Tritryptophan contains three side-chain indoles and one terminal α-carboxylic acid, constituting a non-polar hydrophobic tripeptide. At pH 7.4, the terminal α-carboxylic acid in the side chain is deprotonated and ionized; this would be repelled from the cell membrane, but, because of strong hydrophobic interactions, indole in the side chain can attract and intercalate into phospholipids, generating PMLA tritryptophan–lipid complexes and releasing binding energy to stabilize the structure. In another scenario, pH reduction from neutral to acidic may protonate (and neutralize) the end-group carboxylate in the side chain, whereas protonation of the indole moieties is constrained because this would lead to loss of aromatic stabilization. Under both circumstances, the PMLA backbone is sandwiched between two layers of phospholipids inserted with outflanking tryptophans, forming a 'belt-like' or 'dental brace' configuration [56]. The pH-dependent charge neutralization of the carboxylic acid end-groups does not hamper strong hydrophobic interactions between indole and membrane lipids, thereby leading to pH-independent membrane permeation. This permeation thus resembles the 'boomerang model' that was proposed to mediate viral membrane fusion with the host cell [57]. A highly conserved tryptophan-rich domain has been found in many human viruses, including coronavirus, influenza virus, and HIV, and has been proposed to be a key determinant of viral entry through strong interactions between the aromatic rings of the tryptophan-rich domain and lipids in the target membrane lipid, thus perturbing the lipid bilayer and mediating membrane fusion [58].
-
(iii)
The barrel-stave model typified by PMLA trileucine (PMLA-LLLxH100−x) [53]. Trileucine in the side chain of PMLA-LLLxH100−x has three hydrophobic isobutyl groups and one α-carboxylic acid end-group that is subject to pH-dependent protonation. At neutral pH, ionized PMLA-LLLxH100−x is likely to generate a random-coil conformation, similarly to PMLA, and is largely unable to penetrate the cell membrane owing to its negative charge. As the pH was decreased below 6, PMLA trileucine was found to form aggregates via oligomerization that vertically pierce the membrane core and tentatively form a transmembrane pore to allow entry. In this manner, an increase in the fraction of hydrophobic substituents or the molar mass of the amphiphilic polymer could augment its membranolytic activity [59,60]. Importantly, acidic pH-triggered membranolysis would enable selective disruption of intracellular membranes by ionizable polymeric carriers, including endosomes that are of particular interest for drug delivery (pH ~5.5), thus escaping endosomal capture and lysosomal degradation, leading to release of drug payloads in the cytosol, thereby promoting intracellular drug trafficking and targeting.
Figure 3.
Structure-Related Membrane Permeation of Poly(β-L-Malic Acid) (PMLA) and Its Derivatives.
Mechanisms by which different types of PMLA-based polymers induce cell membrane permeation [53,56]. (A) The carpet model: PMLA polymers first bind to the cell membrane and generate a layer of polymers in which their hydrophobic side chains are inserted into the phospholipid; this is followed by induced curvature of the plasma membrane, forming a transient pore. (B) The belt model: anionized PMLA polymers are first repelled from the cell membrane, but strong interactions between indole moieties in the side chain and membrane phospholipids induces the PMLA backbone to be sandwiched between two layers of phospholipids, resulting in a 'belt-like' configuration. (C) The barrel-stave model: PMLA polymers first self-aggregate to form oligomers and then vertically penetrate the membrane core, generating a transmembrane pore. (D) Endocytosis: cellular engulfment of PMLA. Once PMLA comes into contact with the cell surface, the cell membrane forms vesicles that wrap around the random-coil polymers to generate early endosomes; these can mediate secretion from the cell or develop into late endosomes.
Box 2. Hydrophillic–Hydrophobic Balance of PMLAs.
In view of its hydrophilicity and negative charge at physiological pH, PMLA is a polyanion that has little affinity for negatively charged lipid bilayers and does not translocate through cell membranes. To increase the interaction between the biopolymer and the plasma membrane, methylation of carboxylic acid groups with different levels of diazomethane was used to generate a PMLA-MexH100−x copolymer (where x is the percentage of methyl units) [77,78]. As x increases, the hydrophobicity of the molecule increases in the order PMLA < PMLA-Me25H75 < PMLA-Me50H50 < PMLA-Me75H25 < PMLA-Me. Both PMLA-Me75H25 and PMLA-Me were completely insoluble in water, similarly to PMLA benzyl esterification. The same order was observed for hydrolysis in saline and plasma, rupture of liposome membranes, and cytotoxicity [77]. In addition, hydrophobic amino acids or peptides can be conjugated to PMLA side-chain carboxylic acids to modulate its hydrophilicity and net charge, thus tuning the interplay with cellular/subcellular membranes in a pH-responsive manner. Adjacent carboxylic acid pendants on the PMLA backbone are five atoms apart, equal to their distance in poly(aspartic acid) that has a similar membranolytic profile [79]. Importantly, this spacing dictates an optimized combination of physicochemical parameters for side-chain substituents, including their individual hydrophobicity, charge-neutralizing capacity, ligand length, and density, thus determining the optimal molecular geometry and charge distribution for effective membrane disruption [79,80].
Alt-text: Box 2
In addition to PMLA conjugation with hydrophobic ligands through covalent coupling, methods to regulate its membrane permeability have been developed by promoting non-covalent interactions between the pendant carboxylic acids of PMLA and attaching moieties to the polymer. Given that protonated PMLA can only form hydrogen bonds with functional groups containing electronegative atoms at acidic pH [44], and these are much weaker than covalent or ionic bonds, PMLA complexes generated by hydrogen bonding are unsuitable for membranolytic modification or pharmaceutical loading. Nonetheless, the generation of PMLA ionic complexes via electrostatic interactions offers an alternative strategy for enhanced cellular uptake. At neutral pH, PMLA is a polyanion in which negatively charged carboxylates can form stable complexes with positively charged compounds [61]. Depending on the stoichiometry and chemistry of the attached cations, the polyelectrolyte complexes generated can have a variety of sizes, surface charges, water solubilities, molecular structures, and morphologies, each modulating their membrane penetration. In a scenario where anionic PMLA segments are preferentially situated on the surface of polyelectrolyte complexes, they are internalized via non-specific endocytosis. Conversely, cationized PMLA complexes translocate into cells in a similar manner to polycations, which first adsorb onto the hydrophilic outer surface of the cell membrane and then induce the formation of aqueous pores or defects in the membrane hydrophobic core, and subsequently integrate into or permeabilize the cell membrane [62]. Once transported into the cytoplasm, pH reduction or, to a lesser extent an increase in ionic strength, would accelerate hydrolytic degradation of the PMLA backbone and dissociation of ionic bonds, liberating the complexed moiety for intracellular utility [61].
PMLA as a Nanosized Platform for Cancer Diagnosis and Therapy
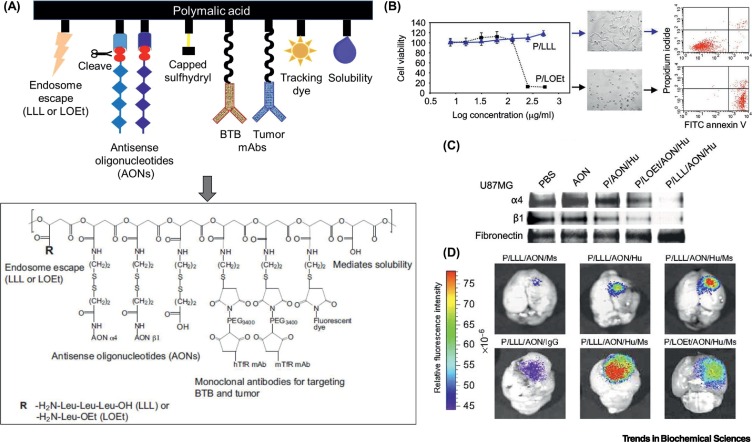
A novel type of PMLA-based nanoconjugate has been developed, termed Polycefin [63]. Through step-by-step chemical synthesis, PMLA was activated by N-hydroxysuccinimidyl ester to enable direct conjugation or further modification by linker molecules, permitting linkage with a variety of chemical and biological ligands, including polyethylene glycol (PEG), monoclonal antibody against transferrin receptor (TfR), an endosome escape unit (LLL or LOEt), and two antisense oligonucleotides (AONs) [64], to synergistically inhibit the α4 and β1 chains of laminin-8 that are overexpressed by human glioblastoma (glioblastoma multiforme, GBM); a fluorescent reporter was also covalently bonded with the PMLA backbone to visualize and localize the biodistribution of Polycefin (Figure 4 ) [63]. The attachment of pendant functionality groups established a new hydrophobic–hydrophilic balance (Box 2) in the macromolecular structure, as the sizes of pure PMLA and variant Polycefins were determined to be <10 and ~20 nm, respectively [65]. These PMLA-based nanoconjugates provide a multifunctional delivery system that can effectively pass through the blood–brain and blood–tumor barriers, leading to enhanced accumulation in brain tumors following i.v. injection into the mouse tail, thus allowing visualization of cancerous lesions and liberating medicinal agents to inhibit tumor angiogenesis or growth [15].
Figure 4.
Poly(β-L-Malic Acid) (PMLA)-Based Nanoconjugates for Brain Tumor Imaging and Therapy.
(A) Scheme and chemical formula of Polycefin, attached to (from left to right) an endosome escape unit (LLL or LOEt), AONs to laminin-411 α4 and β1 chains with disulfide linkages cleavable by glutathione, capped unused sulfhydryls, mAb (Ms) targeting the blood–tumor barrier (BTB) endothelium (mouse TfR), mAb (Hu) targeting tumor cells (human TfR), tracking dye Alexa fluor 680, pendant carboxylates for water solubility. (B) Viability of human U87MG glioma cells treated with PMLA conjugates containing pH-dependent (LLL, i.e., P/LLL) or pH-independent (LOEt, i.e., P/LOEt) endosome escape units for 24 h. Cells treated with P/LOEt (not P/LLL) showed low viability at high concentrations, and their early apoptosis could be visualized by microscopy (20× magnification) and examined by FACS analysis after double staining with propidium iodide and FITC-labeled annexin V. (C) Western blot analysis showing inhibition of laminin-411 α4 and β1 chain synthesis in human U87MG glioma cells after treatment with PBS, AON, P/AON/Hu, P/LOEt/AON/Hu, and P/LLL/AON/Hu, where P/LLL/AON/Hu was the most effective. (D) U87MG cells (105) were implanted intracranially in mice 21 days before they were treated with different PMLA nanoconjugates for 24 h after intravenous (i.v.) injection, namely P/LLL/AON/Ms (Ms, anti-mouse TfR), P/LLL/AON/Hu (Hu, anti-human TfR), P/LLL/AON/Hu/Ms, P/LOEt/AON/Hu/Ms, and P/LLL/AON/IgG (unrelated IgG), all labeled with Alexa fluor 680 to visualize tumor accumulation. The PMLA nanoconjugate with both Ms and Hu mAbs showed the highest tumor accumulation, whereas LLL ensured higher drug tumor accumulation than LOEt, and a control with an unrelated IgG showed low accumulation. Abbreviations: AON, antisense oligonucleotide; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; Hu, human (mAb); LLL, trileucine; LOEt, leucine ethyl ester; mAb, monoclonal antibody; Ms, mouse (mAb); P, PMLA moiety; PBS, phosphate-buffered saline; PEG, polyethylene glycol; TfR, transferrin receptor. Figure reproduced, with permission, from [65].
Since this invention, variant Polycefin biopolymers have been made by combining them with different therapeutic antibodies [66,67], penetrating peptides [5,68], and fluorescent or magnetic contrast agents [69., 70., 71.] that are linked to the PMLA chain through direct or indirect conjugation chemistry, and these have demonstrated improved targeting and accumulation in specific tumors such as breast and brain cancers. Notably, by targeting and inhibiting the laminin α4 and β1 subunits, treatment of mice bearing intracranial human GBM xenografts with PMLA nanoconjugates led to significantly prolonged survival and reduced tumor sizes relative to mice bearing GBMs in which laminins had been knocked out using CRISPR/Cas9 [10]. The minute physical size, potential for multifunctional conjugation, and outstanding water solubility of these PMLA conjugates give them enormous advantages over many other drug delivery systems in terms of finding their way through a labyrinth of cancers, highlighting their huge potential for clinical translation.
Concluding Remarks
Microbial production of PMLA currently remains a challenge because of its low yield, high cost, and the difficulty in defining the length of the biopolymer produced (see Outstanding Questions). Novel gene-editing techniques are urgently needed that would allow efficient delivery of exogenous gene sequences into Physarum plasmodia or filament fungal cells to upregulate PMLA production. CRISPR/Cas9-mediated gene modification was recently successfully applied in A. pullulan, leading to a mutation rate nearly ten-fold higher than that obtained by traditional homologous recombination [72]. It is likely that further genes involved in PMLA biopolymerization will soon be identified, with the prospect of producing PMLA in a controlled manner by using novel gene-editing tools that preselect high-yielding strains. Indeed, PMLA per se could be used as an effective transmembrane gene-delivery shuttle. Future genetic modifications will be necessary not only to increase PMLA yield but also to predetermine the molecular weight of the polymer. Furthermore, suppression of polymalatase to inhibit the decomposition of PMLA could also be achieved through gene editing, thus increasing control over both polymer yield and length. Therefore, to harness the potential of biopolymer production, further elucidation of the genes and molecular machinery that mediate the biosynthesis of PMLA and its decomposition will be essential.
Because PMLAs represent a versatile platform for both disease diagnosis and treatment, further research is required into the PMLA biosynthetic machinery, the biosafety of bioproduced PMLA, and clinical translation of PMLA for the detection and therapy of diseases (particularly at early stage) including cancers. There is also an urgent need to scale-up PMLA production from the laboratory to the industrial pilot plant. In addition, multiple new uses of biodegradable PMLA in daily life can be envisaged, and this would not only stimulate industrial interest in PMLA bioproduction and increase the popularity of PMLA as a natural biomaterial, but would also promote wider medical applications of these remarkable biopolymers.
Outstanding Questions.
Given the complications of scalability from bench to industrial pilot plant production, how best to scale up PMLA microbial production and further functionalization for medicinal purposes?
What is the best way to uncover the genes that govern PMLA synthesis in relevant microorganisms such that they can be engineered in industrial bacteria or yeasts for large-scale PMLA bioproduction?
How can we tune the length of biosynthesized PMLA?
What is the precise relationship between the molar mass of PMLA and its suitability as a delivery platform for cancer medicine?
What are the potential applications of PMLA-based biopolymers other than as delivery platforms?
Alt-text: Outstanding Questions
Acknowledgements
We thank Jiangsu University for financial support.
Glossary
- Antisense oligonucleotides (AONs)
synthetic, short, single-stranded nucleotides that target DNA or mRNA.
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats, DNA fragments originally derived from bacteriophages, whereas CRISPR-associated protein 9 (Cas9) is an endonuclease that specifically recognizes and cleaves double-stranded DNA at a sequence complementary to the CRISPR sequence.
- Endosome escape
release of entrapped molecules by endosomes to avoid further degradation.
- Ester bond
a chemical bond between an acid and an alcohol via the elimination of water.
- First-order kinetics
a reaction that proceeds at a rate proportional to the reactant concentration.
- Glioblastoma (GBM)
one of the most aggressive brain tumors; has a poor prognosis.
- Hyperosmolarity
fluid of abnormally high osmolarity.
- Immunogenicity
the ability of a molecule to provoke an immune response in the host.
- LD50
lethal dose 50%, a dose that kills a half the group of animals studied.
- Malyl unit
the repeating unit in polymalic acid.
- Mitochondrial pyruvate carrier (MPC)
a protein gatekeeper that governs the transport of pyruvate into mitochondria to provide oxidative fuel.
- Molecular chaperone
a class of proteins that assist in the folding and translocation of newly synthesized polypeptides.
- Myxomycetes
mostly known as slime molds, these are fungus-like microorganisms whose life cycle includes spore, plasmodium, and fruiting body stages.
- Nonribosomal peptide synthetase (NRPS)
microbial enzymes that synthesize peptides independently of ribosomes and mRNAs.
- Nucleophilic
a substance that can provide an electron pair to generate a chemical bond.
- Oligomers
intermediate products generated by polymer breakdown or by condensation that contain only a few repeating units.
- Phylogenetic clade
a group of lineages that share a common ancestor in their phylogenetic tree.
- Plasmodial
related to the plasmodium, a single cell that contains multiple nuclei but a single cytoplasm.
- Polyhydroxyalkanoates (PHAs)
a group of plastic polyesters produced by various microorganisms in nature.
- Polyhydroxybutyrate (PHB)
a bacteria-generated plastic that is biodegradable and water-insoluble.
- Protein homeostasis
biological pathways for balanced protein synthesis, modification, trafficking, and degradation in living organisms.
- Renal tubular reabsorption
a process in which functional units within the kidney remove water and solutes from pre-urine tubular fluid, and these are reabsorbed into the bloodstream for reuse.
- Serine protease
a proteolytic enzyme that contains a catalytic serine residue within its active site
- t1/2
half-life, the time needed to reduce the concentration of a drug administered to half its original level.
- Tyrosine kinase
an enzyme that covalently adds a phosphate group to tyrosine residues of proteins to modify their conformation and function.
References
- 1.Shimada K., et al. Poly-(L)-malic acid; a new protease inhibitor from Penicilliumcyclopium. Biochem. Biophys. Res. Commun. 1969;35:619–624. doi: 10.1016/0006-291x(69)90449-5. [DOI] [PubMed] [Google Scholar]
- 2.Chi Z., et al. Poly(beta-L-malic acid) (PMLA) from Aureobasidium spp. and its current proceedings. Appl. Microbiol. Biotechnol. 2016;100:3841–3851. doi: 10.1007/s00253-016-7404-0. [DOI] [PubMed] [Google Scholar]
- 3.Choi S.Y., et al. Microbial polyhydroxyalkanoates and nonnatural polyesters. Adv. Mater. 2020;32 doi: 10.1002/adma.201907138. [DOI] [PubMed] [Google Scholar]
- 4.Gottler T., Holler E. Screening for beta-poly(L-malate) binding proteins by affinity chromatography. Biochem. Biophys. Res. Commun. 2006;341:1119–1127. doi: 10.1016/j.bbrc.2006.01.064. [DOI] [PubMed] [Google Scholar]
- 5.Ding H., et al. HER2-positive breast cancer targeting and treatment by a peptide-conjugated mini nanodrug. Nanomedicine. 2017;13:631–639. doi: 10.1016/j.nano.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao Y., et al. Enhanced endocytic and pH-sensitive poly(malic acid) micelles for antitumor drug delivery. J. Biomed. Nanotechnol. 2019;15:28–41. doi: 10.1166/jbn.2019.2673. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q., et al. Dual-pH sensitive charge-reversal nanocomplex for tumor-targeted drug delivery with enhanced anticancer activity. Theranostics. 2017;7:1806–1819. doi: 10.7150/thno.18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ljubimova J.Y., et al. Toxicity and efficacy evaluation of multiple targeted polymalic acid conjugates for triple-negative breast cancer treatment. J. Drug Target. 2013;21:956–967. doi: 10.3109/1061186X.2013.837470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andorko J.I., et al. Intrinsic immunogenicity of rapidly-degradable polymers evolves during degradation. Acta Biomater. 2016;32:24–34. doi: 10.1016/j.actbio.2015.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun T., et al. Blockade of a laminin-411–Notch axis with CRISPR/Cas9 or a nanobioconjugate inhibits glioblastoma growth through tumor–microenvironment cross-talk. Cancer Res. 2019;79:1239–1251. doi: 10.1158/0008-5472.CAN-18-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee B.S., Holler E. Beta-poly(L-malate) production by non-growing microplasmodia of Physarum polycephalum. Effects of metabolic intermediates and inhibitors. FEMS Microbiol. Lett. 2000;193:69–74. doi: 10.1111/j.1574-6968.2000.tb09404.x. [DOI] [PubMed] [Google Scholar]
- 12.Holler, E., Production of long chain unbranched beta-poly (L-malic acid) by large scale Physarum cultivation and high-grade purification of the same, US Patent 8,377,652, 2013.
- 13.Gasslmaier B., Holler E. Specificity and direction of depolymerization of beta-poly(L-malate) catalysed by polymalatase from Physarum polycephalum – fluorescence labeling at the carboxy-terminus of beta-poly(L-malate) Eur. J. Biochem. 1997;250:308–314. doi: 10.1111/j.1432-1033.1997.0308a.x. [DOI] [PubMed] [Google Scholar]
- 14.Loyer P., Cammas-Marion S. Natural and synthetic poly(malic acid)-based derivates: a family of versatile biopolymers for the design of drug nanocarriers. J. Drug Target. 2014;22:556–575. doi: 10.3109/1061186X.2014.936871. [DOI] [PubMed] [Google Scholar]
- 15.Ljubimova J.Y., et al. Covalent nano delivery systems for selective imaging and treatment of brain tumors. Adv. Drug Deliv. Rev. 2017;113:177–200. doi: 10.1016/j.addr.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holler E., et al. Specific inhibition of Physarum polycephalum DNA-polymerase-alpha-primase by poly(L-malate) and related polyanions. Eur. J. Biochem. 1992;206:1–6. doi: 10.1111/j.1432-1033.1992.tb16894.x. [DOI] [PubMed] [Google Scholar]
- 17.Manitchotpisit P., et al. Poly(beta-L-malic acid) production by diverse phylogenetic clades of Aureobasidium pullulans. J. Ind. Microbiol. Biotechnol. 2012;39:125–132. doi: 10.1007/s10295-011-1007-7. [DOI] [PubMed] [Google Scholar]
- 18.Cao W., et al. High molecular weight beta-poly(L-malic acid) produced by A. pullulans with Ca2+ added repeated batch culture. Int. J. Biol. Macromol. 2016;85:192–199. doi: 10.1016/j.ijbiomac.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y.K., et al. Enhanced production of Ca2+-polymalate (PMA) with high molecular mass by Aureobasidium pullulans var. pullulans MCW. Microb. Cell Factories. 2015;14:115. doi: 10.1186/s12934-015-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black, K.L. et al. Cedars-Sinai Medical Center. Polymalic acid based nanoconjugates for imaging, US-20120258049-A1
- 21.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J., et al. Metabolome- and genome-scale model analyses for engineering of Aureobasidium pullulans to enhance polymalic acid and malic acid production from sugarcane molasses. Biotechnol. Biofuels. 2018;11:94. doi: 10.1186/s13068-018-1099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H., et al. Toward understanding the key enzymes involved in β-poly (L-malic acid) biosynthesis by Aureobasidium pullulans ipe-1. Eng. Life Sci. 2018;18:379–386. doi: 10.1002/elsc.201700209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J., et al. Reconstruction of a genome-scale metabolic model and in silico analysis of the polymalic acid producer Aureobasidium pullulans CCTCC M2012223. Gene. 2017;607:1–8. doi: 10.1016/j.gene.2016.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Zeng W., et al. Analysis of the L-malate biosynthesis pathway involved in poly(beta-L-malic acid) production in Aureobasidium melanogenum GXZ-6 by addition of metabolic intermediates and inhibitors. J. Microbiol. 2019;57:281–287. doi: 10.1007/s12275-019-8424-0. [DOI] [PubMed] [Google Scholar]
- 26.Tavoulari S., et al. The yeast mitochondrial pyruvate carrier is a hetero-dimer in its functional state. EMBO J. 2019;38 doi: 10.15252/embj.2018100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzig S., et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- 28.Ahn S., et al. Role of glyoxylate shunt in oxidative stress response. J. Biol. Chem. 2016;291:11928–11938. doi: 10.1074/jbc.M115.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., et al. Enhanced polymalic acid production from the glyoxylate shunt pathway under exogenous alcohol stress. J. Biotechnol. 2018;275:24–30. doi: 10.1016/j.jbiotec.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Lee B.-S., et al. β-Poly (L-malate) production by Physarum polycephalum. Appl. Microbiol. Biotechnol. 1999;52:415–420. [Google Scholar]
- 31.Feng J., et al. Efficient production of polymalic acid from xylose mother liquor, an environmental waste from the xylitol industry, by a T-DNA-based mutant of Aureobasidium pullulans. Appl. Microbiol. Biotechnol. 2019;103:6519–6527. doi: 10.1007/s00253-019-09974-x. [DOI] [PubMed] [Google Scholar]
- 32.Zeng W., et al. Poly(malic acid) production from liquefied corn starch by simultaneous saccharification and fermentation with a novel isolated Aureobasidium pullulans GXL-1 strain and its techno-economic analysis. Bioresour. Technol. 2020;304 doi: 10.1016/j.biortech.2020.122990. [DOI] [PubMed] [Google Scholar]
- 33.Gostinčar C., et al. Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics. 2014;15:549. doi: 10.1186/1471-2164-15-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., et al. Effects of nitrogen availability on polymalic acid biosynthesis in the yeast-like fungus Aureobasidium pullulans. Microb. Cell Factories. 2016;15:146. doi: 10.1186/s12934-016-0547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song X., et al. GATA-type transcriptional factor Gat1 regulates nitrogen uptake and polymalic acid biosynthesis in polyextremotolerant fungus Aureobasidium pullulans. Environ. Microbiol. 2020;22:229–242. doi: 10.1111/1462-2920.14841. [DOI] [PubMed] [Google Scholar]
- 36.Willibald B., et al. Is beta-poly(L-malate) synthesis catalysed by a combination of beta-L-malyl-AMP-ligase and beta-poly(L-malate) polymerase? Eur. J. Biochem. 1999;265:1085–1090. doi: 10.1046/j.1432-1327.1999.00834.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee B.S., et al. Water-soluble aliphatic polyesters: poly(malic acid)s. Biopolymers Online. 2005 doi: 10.1002/3527600035.bpol3a03. Published online January 15, 2005. [DOI] [Google Scholar]
- 38.Rathberger K., et al. Comparative synthesis and hydrolytic degradation of poly (L-malate) by myxomycetes and fungi. Mycol. Res. 1999;103:513–520. [Google Scholar]
- 39.Pinchai N., et al. Stage specific expression of poly(malic acid)-affiliated genes in the life cycle of Physarum polycephalum spherulin 3b and polymalatase. FEBS J. 2006;273:1046–1055. doi: 10.1111/j.1742-4658.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 40.Zou X., et al. Biosynthesis of polymalic acid in fermentation: advances and prospects for industrial application. Crit. Rev. Biotechnol. 2019;39:408–421. doi: 10.1080/07388551.2019.1571008. [DOI] [PubMed] [Google Scholar]
- 41.Wang K., et al. A novel PMA synthetase is the key enzyme for polymalate biosynthesis and its gene is regulated by a calcium signaling pathway in Aureobasidium melanogenum ATCC62921. Int. J. Biol. Macromol. 2019;156:1053–1063. doi: 10.1016/j.ijbiomac.2019.11.188. [DOI] [PubMed] [Google Scholar]
- 42.Jia S.-L., et al. Genome sequencing of a yeast-like fungal strain P6, a novel species of Aureobasidium spp.: insights into its taxonomy, evolution, and biotechnological potentials. Ann. Microbiol. 2019;69:1475–1488. [Google Scholar]
- 43.Holler E., Lee B. Analysis of poly (β-L-malic acid) in tissue and solution. Recent Res. Dev. Anal. Chem. 2002;2:177–192. [Google Scholar]
- 44.Francis B., et al. Double hydrogen bonding between side chain carboxyl groups in aqueous solutions of poly (β-L-malic acid): implication for the evolutionary origin of nucleic acids. Life. 2017;7:35. doi: 10.3390/life7030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larrañaga A., Lizundia E. A review on the thermomechanical properties and biodegradation behaviour of polyesters. Eur. Polym. J. 2019;121 [Google Scholar]
- 46.Zou X., et al. Production of polymalic acid and malic acid by Aureobasidium pullulans fermentation and acid hydrolysis. Biotechnol. Bioeng. 2013;110:2105–2113. doi: 10.1002/bit.24876. [DOI] [PubMed] [Google Scholar]
- 47.Lanz-Landazuri A., et al. Nanoparticles of esterified polymalic acid for controlled anticancer drug release. Macromol. Biosci. 2014;14:1325–1336. doi: 10.1002/mabi.201400124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanz-Landazuri A., et al. Modification of microbial polymalic acid with hydrophobic amino acids for drug-releasing nanoparticles. Macromol. Chem. Phys. 2012;213:1623–1631. doi: 10.1002/macp.201200134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fournie P., et al. In vivo fate of end-chain radiolabelled poly (β-malic acid), a water-soluble biodegradable drug carrier. J. Bioact. Compat. Polym. 1990;5:381–395. [Google Scholar]
- 50.Fournie P., et al. In vivo fate of repeat-unit-radiolabelled poly (β-malic acid), a potential drug carrier. J. Bioact. Compat. Polym. 1992;7:113–129. [Google Scholar]
- 51.Domurado D., et al. In vivo fates of degradable poly (β-malic acid) and of its precursor, malic acid. J. Bioact. Compat. Polym. 2003;18:23–32. [Google Scholar]
- 52.Marie E., et al. Amphiphilic macromolecules on cell membranes: from protective layers to controlled permeabilization. J. Membr. Biol. 2014;247:861–881. doi: 10.1007/s00232-014-9679-3. [DOI] [PubMed] [Google Scholar]
- 53.Ding H., et al. Distinct mechanisms of membrane permeation induced by two polymalic acid copolymers. Biomaterials. 2013;34:217–225. doi: 10.1016/j.biomaterials.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shinoda W. Permeability across lipid membranes. Biochim. Biophys. Acta Biomembr. 2016;1858:2254–2265. doi: 10.1016/j.bbamem.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 55.Goda T., et al. Translocation mechanisms of cell-penetrating polymers identified by induced proton dynamics. Langmuir. 2019;35:8167–8173. doi: 10.1021/acs.langmuir.9b00856. [DOI] [PubMed] [Google Scholar]
- 56.Ding H., et al. Polymalic acid tritryptophan copolymer interacts with lipid membrane resulting in membrane solubilization. J. Nanomater. 2017;2017 doi: 10.1155/2017/4238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pastore A., et al. Why does the Abeta peptide of Alzheimer share structural similarity with antimicrobial peptides? Commun. Biol. 2020;3:135. doi: 10.1038/s42003-020-0865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., et al. Tryptophan-dependent membrane interaction and heteromerization with the internal fusion peptide by the membrane proximal external region of SARS-CoV spike protein. Biochemistry. 2015;54:1819–1830. doi: 10.1021/bi501352u. [DOI] [PubMed] [Google Scholar]
- 59.Ramadurai S., et al. Dynamic studies of the interaction of a pH responsive, amphiphilic polymer with a DOPC lipid membrane. Soft Matter. 2017;13:3690–3700. doi: 10.1039/c6sm02645a. [DOI] [PubMed] [Google Scholar]
- 60.Checkervarty A., et al. Formation and stabilization of pores in bilayer membranes by peptide-like amphiphilic polymers. Soft Matter. 2018;14:2526–2534. doi: 10.1039/c7sm02404e. [DOI] [PubMed] [Google Scholar]
- 61.Lanz-Landázuri A., et al. Poly (β,L-malic acid)/doxorubicin ionic complex: a pH-dependent delivery system. React. Funct. Polym. 2014;81:45–53. [Google Scholar]
- 62.Awasthi N., et al. Molecular mechanism of polycation-induced pore formation in biomembranes. ACS Biomater. Sci. Eng. 2019;5:780–794. doi: 10.1021/acsbiomaterials.8b01495. [DOI] [PubMed] [Google Scholar]
- 63.Lee B.S., et al. Polycefin, a new prototype of a multifunctional nanoconjugate based on poly(beta-L-malic acid) for drug delivery. Bioconjug. Chem. 2006;17:317–326. doi: 10.1021/bc0502457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018;14:9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 65.Ding H., et al. Inhibition of brain tumor growth by intravenous poly(beta-L-malic acid) nanobioconjugate with pH-dependent drug release. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18143–18148. doi: 10.1073/pnas.1003919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galstyan A., et al. Blood–brain barrier permeable nano immunoconjugates induce local immune responses for glioma therapy. Nat. Commun. 2019;10:3850. doi: 10.1038/s41467-019-11719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chou S.T., et al. Simultaneous blockade of interacting CK2 and EGFR pathways by tumor-targeting nanobioconjugates increases therapeutic efficacy against glioblastoma multiforme. J. Control. Release. 2016;244:14–23. doi: 10.1016/j.jconrel.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Israel L.L., et al. A combination of tri-leucine and angiopep-2 drives a polyanionic polymalic acid nanodrug platform across the blood–brain barrier. ACS Nano. 2019;13:1253–1271. doi: 10.1021/acsnano.8b06437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patil R., et al. Polymalic acid chlorotoxin nanoconjugate for near-infrared fluorescence guided resection of glioblastoma multiforme. Biomaterials. 2019;206:146–159. doi: 10.1016/j.biomaterials.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patil R., et al. MRI virtual biopsy and treatment of brain metastatic tumors with targeted nanobioconjugates: nanoclinic in the brain. ACS Nano. 2015;9:5594–5608. doi: 10.1021/acsnano.5b01872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patil R., et al. Single- and multi-arm gadolinium MRI contrast agents for targeted imaging of glioblastoma. Int. J. Nanomedicine. 2020;15:3057–3070. doi: 10.2147/IJN.S238265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y., et al. CRISPR/Cas9-mediated efficient genome editing via protoplast-based transformation in yeast-like fungus Aureobasidium pullulans. Gene. 2019;709:8–16. doi: 10.1016/j.gene.2019.04.079. [DOI] [PubMed] [Google Scholar]
- 73.Gasslmaier B., et al. Synthetic substrates and inhibitors of beta-poly(L-malate)-hydrolase (polymalatase) Eur. J. Biochem. 2000;267:5101–5105. doi: 10.1046/j.1432-1327.2000.01573.x. [DOI] [PubMed] [Google Scholar]
- 74.Mueller W., et al. Physarum polymalic acid hydrolase: recombinant expression and enzyme activation. Biochem. Biophys. Res. Commun. 2008;377:735–740. doi: 10.1016/j.bbrc.2008.09.127. [DOI] [PubMed] [Google Scholar]
- 75.Karl M., et al. Localization of fluorescence-labeled poly(malic acid) to the nuclei of the plasmodium of Physarum polycephalum. Eur. J. Biochem. 2003;270:1536–1542. doi: 10.1046/j.1432-1033.2003.03521.x. [DOI] [PubMed] [Google Scholar]
- 76.Godde C., et al. Isolation of poly(beta-L-malic acid)-degrading bacteria and purification and characterization of the PMA hydrolase from Comamonas acidovorans strain 7789. FEMS Microbiol. Lett. 1999;173:365–372. doi: 10.1111/j.1574-6968.1999.tb13527.x. [DOI] [PubMed] [Google Scholar]
- 77.Portilla-Arias J., et al. Nanoconjugate platforms development based in poly(β,L-malic acid) methyl esters for tumor drug delivery. J. Nanotechnol. 2010;2010 doi: 10.1155/2010/825363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Portilla-Arias J.A., et al. Synthesis, degradability, and drug releasing properties of methyl esters of fungal poly(beta,L-malic acid) Macromol. Biosci. 2008;8:540–550. doi: 10.1002/mabi.200700248. [DOI] [PubMed] [Google Scholar]
- 79.Ding H., et al. The optimization of polymalic acid peptide copolymers for endosomolytic drug delivery. Biomaterials. 2011;32:5269–5278. doi: 10.1016/j.biomaterials.2011.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang S., et al. Physical principles of nanoparticle cellular endocytosis. ACS Nano. 2015;9:8655–8671. doi: 10.1021/acsnano.5b03184. [DOI] [PMC free article] [PubMed] [Google Scholar]