Abstract
CRISPR-associated proteins 1 and 2 (Cas1–2) are necessary and sufficient for new spacer acquisition in some CRISPR-Cas systems (e.g., type I-E), but adaptation in other systems (e.g., type II-A) involves the crRNA-guided surveillance complex. Here we show that the type I-F Cas1–2/3 proteins are necessary and sufficient to produce low levels of spacer acquisition, but the presence of the type I-F crRNA-guided surveillance complex (Csy) improves the efficiency of adaptation and significantly increases the fidelity of protospacer adjacent motif selection. Sequences selected for integration are preferentially derived from specific regions of extrachromosomal DNA, and patterns of spacer selection are highly reproducible between independent biological replicates. This work helps define the role of the Csy complex in I-F adaptation and reveals that actively replicating mobile genetic elements have antigenic signatures that facilitate their integration during CRISPR adaptation.
Introduction
CRISPR loci and their associated cas genes are conserved components of adaptive immune systems that protect bacteria and archaea from foreign genetic elements.1,2 These immune systems are phylogenetically and functionally diverse, but all acquire immunity by preferentially integrating short fragments of foreign DNA at one end of a CRISPR locus.3–6 Conserved CRISPR-associated proteins 1 and 2 (i.e., Cas1 and Cas2) have been shown to be necessary and sufficient for integrating foreign DNA into the CRISPR loci in some system subtypes (e.g., I-E),7 while in vivo adaption in other CRISPR subtypes requires additional proteins (e.g., Cas4, Csn2, Cas9).8–11
The type II-A crRNA-guided surveillance complex (i.e., Cas9) facilitates integration of sequences flanked by a protospacer adjacent motif (PAM),11,12 and chimeric Cas9 proteins have been used to demonstrate that the PAM interacting domain is directly involved in this process.10 Similarly, the type I-F crRNA-guided surveillance (Csy) complex has been implicated in naive sequence adaptation,13 but the role played by this complex is yet to be determined.
Based on the previous work done in type II systems showing that Cas9 plays a necessary role in selecting sequences for integration (i.e., prespacers), we hypothesized that the type I-F crRNA-guided surveillance complex (i.e., Csy or I-F Cascade complex) has a similar role in I-F CRISPR adaptation. Here, we show that the Cas1 and Cas2/3 fusion proteins (Cas1–2/3), which form an integration complex in I-F systems,14–16 are capable of selecting PAM containing prespacers for inefficient adaptation independent of the Csy complex. However, the presence of the Csy complex increases the fidelity of PAM selection and enhances the efficiency of naive CRISPR adaptation. Moreover, our work reveals that protospacers are not selected at random; instead specific protospacers are consistently selected from specific regions of extrachromosomal DNA. Persistent protospacer preferences indicate that some DNA sequences may be recognized as antigenic and this work helps explain how CRISPR systems avoid autoimmunity.
Materials and Methods
Adaptation assays in Escherichia coli
Spacer acquisition assays were performed using an engineered strain of Escherichia coli BL21-AI that contains a type I-F CRISPR array (KD740) in place of the I-E CRISPR locus. These cells are identical to those described by Vorontsova et al.,13 but an orphan type I-F CRISPR locus has been removed using Red recombinase.17,18 The I-F CRISPR contains a single-spacer sequence (5′-ACGCAGTTGCTGAGTGTGATCGATGCCATCAG-3′) that targets the J protein of phage lambda and should not trigger priming, since this spacer has no homology to the chromosome or expression plasmids used in these studies.13,19,20 The I-F CRISPR of KD740 is flanked by a 134 bp leader sequence. The CRISPR is transcribed from LacI-repressed T7 RNA polymerase promoter.
The cas1–2/3 genes from Pseudomonas aeruginosa PA14 were cloned into a spectinomycin resistance (2S) LIC vector with a ColE1 origin of replication. The cas1–2/3 genes are cloned downstream of a T7 promoter. The construction of this plasmid (pCas1–2/3) has been previously described,15 and the sequence is available in Supplementary Table S1. pCsy was constructed by cloning genes coding for the Csy complex from PA14 (i.e., cas8f, cas5f, cas7f, cas6f) downstream of a T7 promoter in pRSF-1. Q5 mutagenesis was used to introduce a mutation in cas8f (N250A) that prevents Cas8f-mediated PAM-recognition (primers P-CsyPAMmut-F and P-CsyPAMmut-R in Supplementary Table S2),21 resulting in the pCsyPAM expression vector. Plasmid sequences can be found in Supplementary Table S1.
KD740 cells were transformed by electroporation with pCas1–2/3, and either the WT Csy expression vector (pCsy) or the PAM-recognition-deficient Csy mutant (pCsyPAM). Transformants were selected on Luria-Bertani (LB) agar plates containing 50 μg/mL spectinomycin (pCas1–2/3) and 50 μg/mL kanamycin (pCsy/pCsyPAM), but no antibiotics were used in downstream adaptation experiments. Cells were grown at 32°C in LB media supplemented with 1 mM l-arabinose and 1 mM IPTG for a total of 72 h in a shaking incubator at 225 RPMs. Cultures were diluted (1:100) in fresh media every 24 h.
Spacer acquisition was detected using “internal” and “degenerate” primers with Q5 polymerase (NEB) via the recently described CAPTURE method.22 Briefly, 1 μL of liquid culture was used as template for polymerase chain reactions (PCRs) with “internal” primers (i.e., P1 and P2) (Supplementary Table S2), and products were visualized on 1.5% agarose gels. PCR products were extracted from gels and purified using Zymoclean Gel DNA Recovery Kits (Zymogen) or the BluePippin size selection system (Sage Science) with a “Range” setting of 220–300 bp on a 2% agarose cassette. These size selected oligos were then used as templates for a subsequent round of PCR with “degenerate” primers (i.e., P3 and P2) (Supplementary Table S2), and products were subsequently visualized on 1.5% agarose gels. These experiments did not require approval by an IRB.
High-throughput sequencing
Products from the second PCR amplification were extracted from agarose gels and were purified using a Zymoclean Gel DNA Recovery Kit (Zymogen). These products were used as templates for PCR with Q5 polymerase (NEB) using barcoded primers P-HTS-F1–5 and P-HTS-R (Supplementary Table S2). Samples were sequenced with 1 × 150 bp single-end runs on an Illumina NextSeq and included two independent biological replicates of KD740 with pCsy and pCas1–2/3, and three independent biological replicates of KD740 with pCas1–2/3.
Spacer sequence analysis and statistics
High-throughput sequencing (HTS) data were processed with the ShortRead package from Bioconductor.23 Sequencing reads contained one, two, or three CRISPR, representing unexpanded arrays (i.e., no adaptation), arrays with one newly acquired spacer, and arrays with two newly acquired spacers, respectively. Only reads corresponding to one acquisition event were examined for downstream analyses, to prevent the analysis of spacers acquired through priming. Sequences between CRISPR (i.e., spacers) were extracted. Spacer sequences corresponding to the original WT spacer present in KD740 were removed.
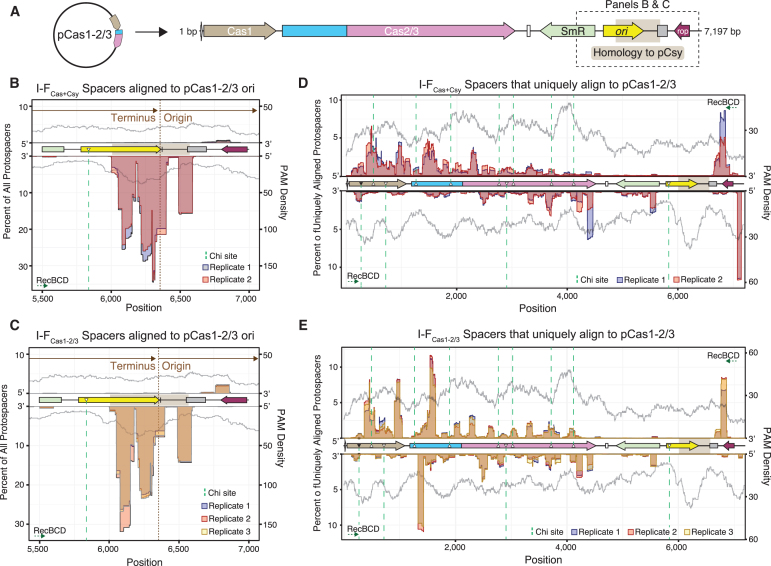
Extracted spacers were aligned to the E. coli genome (NC_012947.1) and cas gene expression plasmids present in each sample. Sequences were aligned using the Biostrings package, allowing for one mismatch.24 Spacer sequences that align with pCas1–2/3 were used for the analysis shown in Figure 3B and C. To assess spacers that uniquely align to pCas1–2/3 (Figs. 2 and 3D, E), we discarded sequences that produced more than one alignment or that aligned to a 565 bp region shared by the ori sequences of pCas1–2/3 and pCsy (94% nucleotide identity).
FIG. 3.
Protospacer hotspots are reproducible across multiple biological replicates. (A) Linearized genetic map of Cas1–2/3 expression plasmid. Regions shown in (B, C) are marked with dotted outline. (B, C) Positions and quantities of all protospacers that align to ori of pCas1–2/3, including spacers that produce alignments to additional sequences (e.g., pCsy). Protospacer locations are mapped using a 50 nt sliding window. Data from each biological replicate are represented as a different color. PAM densities (gray) mapped with a 150 nt sliding window. Direction and start/end positions of plasmid replication shown in brown. Consensus Chi sites are shown as triangles filled in with black, and Chi sites with a single base mismatch are shown as triangles with a white fill. Dotted green lines indicate the boundaries of Chi sites, and RecBCD directions of cleavage are shown for each strand of DNA. (D, E) Positions and quantities of protospacers that produce a single unique alignment to pCas1–2/3 are shown as in (B, C).
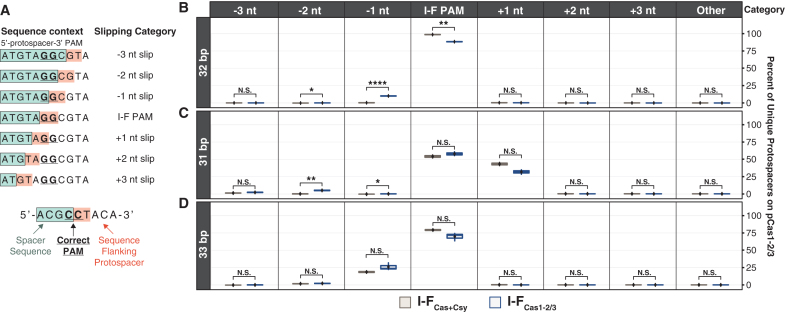
FIG. 2.
Cas1–2/3 acquire mostly PAM-flanked spacers, but Csy increases the fidelity of PAM selection. (A) Sequence context of protospacers for spacers acquired through Cas1–2/3 slipping. (B–D) Percentages of 31, 32, and 33 bp protospacers that uniquely map to pCas1–2/3 and were acquired through different categories of Cas1-/3 slipping. Spacers that produced more than one alignment to the genome or cas gene expression plasmids were not included in this analysis. Significances of T-tests performed for biological replicates between each condition are denoted as asterisks (N.S.: p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001).
Pearson correlation coefficients were calculated by comparing the quantities of uniquely aligned protospacers that were present at each position of pCas1–2/3 using the ggpubr package,25 and graphs of protospacer positions and quantities were rendered with the ggplot2 package.26 A summary of the HTS results can be found in Supplementary Table S3 and spacer sequences extracted from sequencing reads are included in Supplementary Table S4.
Results
Csy-PAM interactions increase I-F CRISPR adaptation efficiency
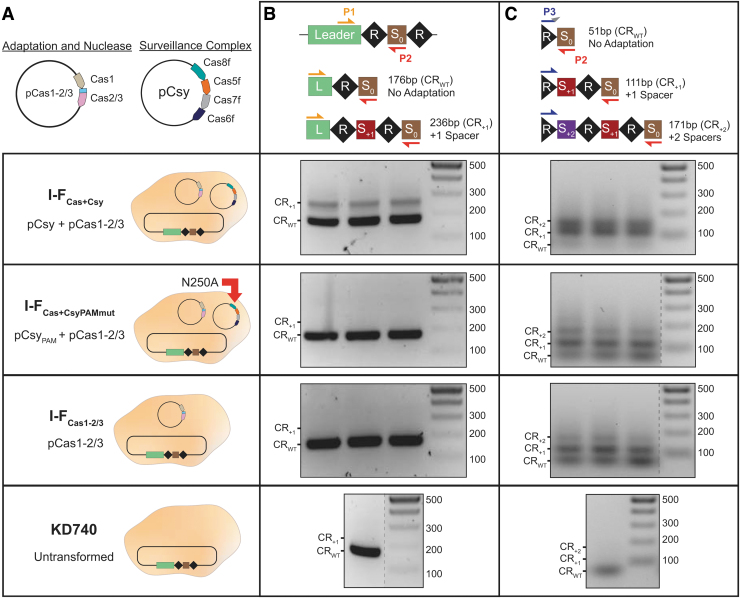
To determine if Csy-PAM interactions are required for I-F spacer acquisition, we screened for adaptation using E. coli KD740 cells overexpressing Cas1–2/3 (I-FCas1–2/3) and either the wild-type Csy complex (I-FCas+Csy) or a Csy mutant (I-FCas+CsyPAMmut) with reduced PAM recognition (Fig. 1A).21 Adaptation assays were performed by PCR amplification of the leader end of the I-F CRISPR array (Fig. 1B). PCR performed on cells expressing both Cas1–2/3 and the Csy complex (I-FCas+Csy) results in two clearly visible PCR products. The smaller product corresponds to the expected size of the wild-type, unexpanded CRISPR (CRWT), and the larger product corresponds to the addition of one spacer-repeat unit. In contrast, PCR performed on cells lacking the Csy complex (I-FCas1–2/3) or expressing the PAM sensing mutant (I-FCas+CsyPAMmut) resulted in only the CRWT product, in agreement with earlier data.13 These results suggest that Csy and more specifically Csy-PAM interactions are required for efficient I-F adaptation.
FIG. 1.
Cas1–2/3 proteins are necessary and sufficient for new spacer acquisition, but Csy accelerates adaptation. (A) Schematics of the Cas1–2/3 and Csy subunit (Cas8f, Cas5f, Cas7f, and Cas6f) expression vectors are shown above schematics for each of the conditions tested. Escherichia coli KD740 cells are shown as tan ovals. (B) PCR amplification with “internal” (P1 and P2) primers to detect adaptation events. (S0 is initial spacer that targets phage λ.) Expected sizes of PCR products for CRISPR with acquired spacers (CR+1 and CR+2) or no acquisition (CRWT) are labeled. Adaptation events detected after PCR with primers P1 and P2. Each lane represents a biological replicate. (C) PCR products from (B) were enriched for higher molecular weight species, and then used as templates for a subsequent round of CAPTURE PCR with primers P3 and P2. Products corresponding to expanded arrays are visible in all biological replicates for I-FCas+Csy, I-FCas+CsyPAMmut, and I-FCas1–2/3. Cas, CRISPR-associated protein; PAM, protospacer adjacent motif; PCR, polymerase chain reaction.
Cas1–2/3 are necessary and sufficient for inefficient I-F spacer acquisition
The observation that wild-type Csy is required for efficient spacer acquisition in vivo is consistent with previous results,13 but in vitro experiments have shown that Cas1–2/3 are sufficient for spacer processing and integration.16 We wondered if in vivo adaptation occurs without the Csy complex, but at levels below the limit of detection of the standard PCR-based assay.
To detect rare adaptation events in I-FCas+CsyPAMmut and I-FCas1–2/3 samples, we cut out regions of the gel that would contain higher molecular weight PCR products corresponding to expanded arrays and repeated the PCR according to a recently published CAPTURE PCR protocol.22 Size-enriched PCR products were amplified using a forward primer with a degenerate 3′ nucleotide that anneals in the CRISPR (P3) and a reverse primer that is complementary to the phage lambda (λ)-targeting spacer (P2) (Fig. 1C). This approach enables detection of rare (i.e., 1 in 105 cells) adaptation events, and is two orders of magnitude more sensitive than the standard PCR protocol.22
Using the CAPTURE primer set (P2 and P3), PCR products corresponding to expanded arrays (111 and 171 bp) were detected in samples from all three conditions (i.e., I-FCas+Csy, I-FCas+CsyPAMmut, and I-FCas1–2/3 cultures) (Fig. 1C). Notably, strong bands corresponding to one (111 bp) and two (171 bp) new spacer-repeat units were visible in cells expressing both Cas1–2/3 and Csy (I-FCas+Csy), suggesting that size selection of the PCR template successfully enriches expanded arrays (Fig. 1C). These results indicate that Cas1–2/3 proteins are necessary and sufficient for I-F spacer acquisition, but adaptation in this system is inefficient in the absence of the wild-type Csy complex.
The Csy complex increases the percentage of PAM-proximal spacers
Cas9 plays a critical role in prespacer selection during in vivo adaptation of type II CRISPR systems.10,11 While the mechanistic details of this process remain obscure, we do know that Cas9 has to be loaded with a tracrRNA and crRNA-guide and that the PAM sensing residues play a critical role in determining the sequence of newly acquired spacers.10,11 To determine if Csy plays a similar role in type I-F adaptation, we used Illumina NextSeq to sequence higher molecular weight CAPTURE PCR products from cells expressing Cas1–2/3 alone (I-FCas1–2/3) or Cas1–2/3 and Csy (I-FCas+Csy), then protospacer flanking sequences were analyzed for the presence of PAMs. We focused this analysis on 32 bp spacers that uniquely aligned to a single protospacer target on the pCas1–2/3 plasmid, since this represented the largest pool of uniquely aligned spacers in both I-FCas+Csy and I-FCas1–2/3 samples.
In total, we analyzed 227,834 and 205,354 sequences flanking I-FCas+Csy and I-FCas1–2/3 protospacers, respectively (Fig. 2A). In cells expressing Csy and Cas1–2/3, the majority (98.3–98.9%) of protospacers were flanked by a PAM (Fig. 2B). This trend was consistent across biological replicates, and these numbers agree with previous work showing high-fidelity PAM selection (97.8%) in this system.13 While the majority of protospacers acquired in cells expressing Cas1–2/3 alone (I-FCas1–2/3) were most often flanked by a PAM (87.9–88.6%), the occurrence of a PAM in these samples (i.e., Cas1–2/3 alone) was significantly less than what was measured for cells expressing Csy and Cas1–2/3 (T-test; p = 1.2 × 10−3) (Fig. 2B). These differences indicate that Cas1–2/3 are capable of acquiring spacers flanked by a PAM, but Csy significantly increases the fidelity of PAM selection.
Previous work has shown that newly acquired spacers are often flanked by incorrect PAMs due to slipping of Cas1–2/3 during prespacer capture or processing.20,27,28 To determine if uniquely aligned protospacers flanked by an incorrect PAM could have been produced by slipping, we searched for a PAM three nucleotides upstream and downstream of each protospacer (Fig. 2A).
For the three I-FCas1–2/3 biological replicates we sequenced, 10.6–11.4% of the 32 bp protospacers that were not directly flanked by a PAM were within three nucleotides of a PAM, suggesting that Cas1–2/3 slipping events and subsequent incorrect processing at the PAM-spacer boundary may be responsible for the integration of most spacers without a PAM (Fig. 2B). In particular, I-FCas1–2/3 spacers that were acquired with a slip of −1 nt accounted for most (92.2–93.5%) of the 32 bp protospacers that were not flanked by a PAM and represented a large portion (9.9–10.7%) of the uniquely aligned I-FCas1–2/3 spacer population. This represents a significant increase in −1 nt slipped spacers for I-FCas1–2/3 samples over I-FCas+Csy (0.2–0.6%) samples (p = 9.9 × 10−5; T-test) (Fig. 2C).
Collectively, these data indicate that Cas1–2/3 mediate prespacer trimming at the PAM-spacer boundary with about 90% accuracy in vivo, but the fidelity of this processing is increased by the presence of the Csy complex.
Since previous work has shown that aberrantly sized spacers are often associated with Cas1–2/3 slipping,20 we examined the PAMs of 31 and 33 bp spacers that uniquely align to pCas1–2/3. I-FCas1–2/3 samples had significantly more 31 bp spacers that were acquired with a slip of −1 or −2 nt compared with I-FCas+Csy samples (p = 1.6 × 10−2 and p = 3.1 × 10−3, respectively; T-test) (Fig. 2C). As previously reported,20 slipping of +1 nt was elevated in 31 bp spacers acquired in both conditions compared with +1 nt slipping for 32 bp spacers (Fig. 2B, C). Thirty-three base pair spacers had more −1 nt slipping overall, and although this trend was more pronounced in I-FCas1–2/3 spacers than in I-FCas+Csy spacers, the difference between these two conditions was not significant (p = 0.123; T-test) (Fig. 2D). Considered together, these data are consistent with the observation that slipping occurs more frequently when the Csy complex is not present.
Spacers are preferentially acquired from the terminus of plasmid replication
The origin of plasmid replication initiation (ori) has previously been shown to be a “hotspot” for spacer acquisition,6,13,20 however, it is unclear if the surveillance complex is involved in the spacer selection process. To determine if the Csy complex effects the frequency or location of spacer selection, we calculated the number of newly acquired spacers derived from the E. coli genome and the cas gene expression plasmids. The majority of spacers (44.7–45.8%) acquired in I-FCas+Csy cells were homologous to sequences that are identical in both expression plasmids (i.e., pCas1–2/3 and pCsy), and most of these spacers (97.3–98.3%) mapped to discrete locations near the origin of replication initiation (ori) (Fig. 3A, B). This preference for spacers derived from the ori occurred independent of Csy expression, since 69.6–77.1% of spacers acquired in I-FCas1–2/3 samples also mapped to the ori (Fig. 3A, C). These patterns suggest that the ori is a hotspot for acquisition, regardless of whether or not Csy is present.
The ColE1 ori of pCas1–2/3 shares ∼565 bp of homology with the RSF1030 ori of pCsy (94% nucleotide identity), and spans both the start and end of replication for these unidirectionally replicating plasmids.29 Most spacers acquired from this region are derived from sequences near the terminus of replication, rather than from replication initiation (Fig. 3B, C). Replication termini of bidirectionally replicating genomes and plasmids have previously been shown to be hotspots of spacer acquisition,6,13 and these data suggest that termini of unidirectionally replicating plasmids present similar antigenic signals that increase the number of spacers acquired from these regions.
Preference for terminus-centered acquisition have previously been explained by the action of RecBCD, which produces prespacer substrates during repair of dsDNA breaks (DSBs).6 DSBs occur more often near the terminus of replication where replication forks stall,6 and RecBCD nucleases degrade both strands of linear dsDNA until RecBCD reaches an asymmetric, octameric Chi site (5′-GCTGGTGG-3′).30 pCas1–2/3 and pCsy both contain a single imperfect Chi site (5′-GCTGGTAG-3′; mutation underlined) that is present near the ori. However, previous work has shown that this mutated Chi sequence is seldom recognized by RecBCD (<2% of consensus Chi-site activity),31,32 and this mutated Chi site is improperly oriented to stop RecBCD degradation, with respect to the terminus of replication (Fig. 3B, C).30 These data suggest that Chi-site distributions alone do not explain the boundaries of protospacer peaks near the pCas1–2/3 terminus.
Acquisition of spacers from the E. coli genome was rare (0.004–0.032%) and percentages of genomic protospacers were not significantly different between biological replicates of I-FCas+Csy and I-FCas1–2/3 (p = 0.15; T-test) (Supplementary Table S3). These results were expected for cells expressing the Csy complex, since the acquisition of “self” spacers would target the Cas2/3 nuclease to the E. coli chromosome, resulting in autoimmunity. However, the strong preference for plasmid-derived spacers in cells expressing Cas1–2/3 alone (I-FCas1–2/3) suggests that either RecBCD preferentially generates prespacers from replicating plasmids or that the Cas1–2/3 integration complex is capable of distinguishing self from nonself during I-F adaptation.
Some spacers are reproducibly acquired
To determine if there are hotspots for protospacer selection beyond the ori, we mapped the positions and quantities of spacers outside this region. Specifically, we examined 32 bp spacers extracted from I-FCas+Csy and I-FCas1–2/3 reads that uniquely align to pCas1–2/3. Protospacers outside the ori mapped primarily to the 5′ end of transcriptionally active genes, including cas1 and cas2/3, for both I-FCas+Csy and I-FCas1–2/3 samples (Fig. 3D, E). In contrast, very few spacers were acquired from the intergenic region between cas2/3 and the spectinomycin resistance gene (smR) in either I-FCas+Csy (0.2–0.4%) or I-FCas1–2/3 (0.0–0.1%) samples (Fig. 3D, E). These data are consistent with previous results suggesting that in addition to replication termini, protospacers are preferentially acquired from highly transcribed regions of foreign DNA.20
To determine if specific sequences are preferentially targeted for integration across biological replicates, as previously reported,6,13,20 we calculated Pearson correlation coefficients for positions and quantities of protospacers unique to pCas1–2/3 (Fig. 3D, E). This analysis revealed that I-FCas+Csy protospacer profiles from two biological replicates were positively correlated (R = 0.83, p < 2.2 × 10−16) (Supplementary Fig. S1A). In addition, protospacer profiles from three I-FCas1–2/3 replicates were nearly perfectly correlated (R = 0.98–0.99, p < 2.2 × 10−16) (Supplementary Fig. S1B). To determine how related the I-FCas+Csy and I-FCas1–2/3 protospacer profiles were, we calculated mean protospacer quantities from each biological replicate of each condition. This analysis reveals that the two data sets are positively correlated (R = 0.78, p < 2.2 × 10−16) (Supplementary Fig. S1C).
Taken together, these data indicate that hotspots of naive spacer acquisition are remarkably consistent with and without the Csy complex. Cas1–2/3, therefore, appears to be the primary determinant of spacer selection during naive I-F sequence adaptation. While we were unable to find similarities in the primary structure of highly acquired spacer sequences, the preponderance of protospacer hotspots in actively transcribed areas of the plasmid suggests that a higher level DNA structure may be recognized as antigenic by Cas1–2/3.13,20
Alternatively, DSBs that form as a result of transcription may lead to increased acquisition from these regions due to the increased availability of RecBCD degradation products.20 One complete Chi site and nine sites with a single mismatch to the consensus Chi sequence are present on pCas1–2/3. Previous work has shown that Chi sites with a single base mutation are sufficient, although less efficient (<2–38% of consensus Chi activity) for impeding RecBCD degradation of dsDNA.30–32 While some protospacer peaks occur near the boundaries of these RecBCD-halt signals, not all of the protospacer peaks are constrained by Chi sites as previous experiments in a I-E system have shown (Fig. 3B–E).6 These data suggest that the I-F integration complex may provide an additional level of prespacer selection specificity during CRISPR adaptation.
Discussion
We have shown that the Cas1–2/3 proteins are necessary and sufficient for low levels of PAM-proximal sequence acquisition in the type I-F CRISPR-Cas system of P. aeruginosa. However, it is probably unlikely that spacers are often acquired independent of the surveillance complex, since adaptation is difficult to detect even when Cas1–2/3 are overexpressed. Our data reveal that the Csy complex plays a role in increasing the efficiency of naive adaptation, and that this process is dependent on Csy-PAM interactions (Fig. 1). These findings differentiate Csy-facilitated adaptation from Cas9-dependent acquisition. In type II systems, PAM-sensing by Cas9 is required for the acquisition of spacers flanked by PAMs, but PAM-sensing mutants of Cas9 still support efficient adaptation of spacers that are not flanked by PAMs.10
While the mechanistic details of Csy-mediated enhancement of adaptation remain murky, our results suggest that the Csy complex not only increases the frequency of protospacers flanked by a PAM, but also helps define the PAM-spacer boundary. However, these data and their interpretations may be affected by additional selective pressures. Cells expressing both Csy and Cas1–2/3 can eliminate the cas gene expression plasmids via CRISPR-mediated interference. Cells replicating without the metabolic burden associated with supporting plasmid replication may have a growth advantage,33,34 and this advantage could account for some of the spacer acquisition enhancements that we detect in cells expressing both Cas1–2/3 and Csy. Additional experiments are necessary to clarify the influence of newly acquired spacers on cells during adaptation.
The Cas3 lobe of the genetic Cas2–3 fusion protein may be involved in spacer maturation,20 since the insertion of a stop codon between Cas2 and Cas3 or inactivating mutations of the SF2 helicase or HD nuclease active sites all prevent efficient spacer acquisition in vivo.13 However, these same mutations do not prevent spacer processing and integration in vitro.16 While our work does not address the role of Cas3 activity in naive spacer acquisition, our results do demonstrate that cells expressing Cas1–2/3 alone are sufficient for new spacer integration.
Previous work has shown that Cas1–2/3 are capable of processing prespacers at the PAM-boundary with ∼50% accuracy in vitro,16 but our results indicate that this process happens with greater accuracy (∼90%) in vivo (Fig. 2). These differences may arise from the availability of other cellular nucleases (e.g., DNA PolIII, ExoT, ExoI, or ExoIII) involved in trimming prespacer substrates in the I-E and II-A systems.35,36 However, the participation of cellular nucleases in I-F spacer maturation remains to be demonstrated, and additional studies are needed to determine if and how Csy-mediated spacer processing occurs.
Strikingly, we demonstrate that spacer sequences acquired in one experiment were frequently acquired in other biological replicates, reinforcing previous findings and suggesting that plasmids present antigenic signatures that prompt preferential adaptation from these elements.6,13 The vast majority of reproducible protospacers were confined to the origin and terminus of plasmid replication. Higher copy numbers of DNA at origins of replicating genomes have previously been used to explain high levels of spacer acquisition at these loci.6 In fact, when spacer counts are normalized to total cellular DNA content, protospacers derived from the origin become less pronounced.6 Similar dynamics may be at play in our data, since regions near the origin of plasmids will be replicated first and will thus have a higher copy number in the cell.
However, the high frequency of spacers derived from plasmid termini is more difficult to explain. Acquisition from the termini occurs reproducibly and independent of the Csy complex (Fig. 3). While we cannot rule out an increase in acquisition at this locus due to prespacer substrate generation by RecBCD DNA repair machinery, our data indicate that Chi-site distributions alone do not explain the reproducible patterns of protospacer acquisition. These data argue for an additional level of antigen-specific detection by the type I-F Cas1–2/3 integration complex, although the mechanistic details of this process remain to be determined.
Spacers acquired from the genome were rare in our data (0.004–0.032% of total spacers), regardless of whether or not cells expressed the Csy complex. A similar dearth (<1%) of genome-derived spacers has been detected in other I-F overexpression systems.13 In contrast, the I-F system of Pectobacterium atrosepticum has been reported to acquire more genomic spacers (∼16%) when cas genes are expressed at wild-type levels.20
Direct comparison between results of different experiments is, however, difficult due to differing experimental conditions (e.g., endogenous vs. heterologous expression, plasmid copy numbers, and different bacterial species). For example, naive adaptation experiments with I-E CRISPR systems result in the opposite dynamic. Cas1–2 overexpression increases I-E acquisition from the genome, while lower cas expression levels produce less genomic spacers.6 In contrast to both I-E and I-F adaptation, naive spacers acquired in the I-B system of Pyrococcus furiosus nearly all come from the genome.37 These subtype-specific differences may reflect differing mechanisms of Cas-mediated antigen recognition or may simply be the result of different experimental conditions.
While the specific antigenic signatures of plasmid DNA remain nebulous, cellular localization may play a role in recognition of these foreign genetic elements. Cas2 proteins from the I-E system of E. coli have previously been shown to localize to the poles of bacterial cells,38 and similar polar localization has been reported for plasmids with ColE1 origins of replication.39 Many bacteriophages also target cellular poles for entry and λ phages replicate at cellular poles.40 In contrast, the bacterial chromosome typically occupies the cellular midpoint.41 Collectively, these data suggest that CRISPR-associated adaptation proteins may rely on DNA location as one of the signatures that help distinguish self from nonself DNA. Future work is necessary to test this hypothesis, but localization could help explain why actively replicating foreign genetic elements are preferentially selected over the chromosome during CRISPR adaptation.
Supplementary Material
Author Confirmation Statement
The data presented in this article have not been published, nor are they in press or submitted elsewhere.
Authors' Contributions
B.W., T.W., E.S., and K.S. designed the research. K.D. engineered bacterial strain for experiments. T.W., E.S., and I.F. performed the research. A.S. designed tools for data analysis. T.W. analyzed data. B.W. and T.W. wrote the article with input from all authors.
Author Disclosure Statement
B.W. is the founder of SurGene LLC, VIRIS Detection Systems, Inc., and is an inventor on patent applications related to CRISPR-Cas systems and applications thereof.
Funding Information
Work in the Wiedenheft laboratory is supported by the National Institutes of Health (1R35GM134867 to Blake Wiedenheft), an Amgen Young Investigator award, the M.J. Murdock Charitable Trust, and the Montana State University Agricultural Experimental Station.
Supplementary Material
References
- 1. Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. DOI: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2. Cady KC, Bondy-Denomy J, Heussler GE, et al. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–5738. DOI: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Erdmann S, Le Moine Bauer S, Garrett RA. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol Microbiol. 2014;91:900–917. DOI: 10.1111/mmi.12503. [DOI] [PubMed] [Google Scholar]
- 4. Jackson SA, McKenzie RE, Fagerlund RD, et al. CRISPR-Cas: adapting to change. Science. 2017;356:eaal5056 DOI: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 5. Klompe SE, Sternberg SH. Harnessing “A Billion Years of Experimentation”: the ongoing exploration and exploitation of CRISPR-Cas immune systems. CRISPR J. 2018;1:141–158. DOI: 10.1089/crispr.2018.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levy A, Goren MG, Yosef I, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. DOI: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. DOI: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li M, Wang R, Zhao D, et al. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. DOI: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu T, Liu Z, Ye Q, et al. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 2017;45:8978–8992. DOI: 10.1093/nar/gkx612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heler R, Samai P, Modell JW, et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. DOI: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. DOI: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. DOI: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 13. Vorontsova D, Datsenko KA, Medvedeva S, et al. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 2015;43:10848–10860. DOI: 10.1093/nar/gkv1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richter C, Gristwood T, Clulow JS, et al. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype IF CRISPR/Cas system. PLoS One. 2012;7:e49549 DOI: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rollins MF, Chowdhury S, Carter J, et al. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci U S A. 2017;114:E5113–E5121. DOI: 10.1073/pnas.1616395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fagerlund RD, Wilkinson ME, Klykov O, et al. Spacer capture and integration by a type I-F Cas1-Cas2–Cas3 CRISPR adaptation complex. Proc Natl Acad Sci U S A. 2017;114:E5122–E5128. DOI: 10.1073/pnas.1618421114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Almendros C, Guzman NM, Garcia-Martinez J, et al. Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR-Cas I-F systems. Nat Microbiol. 2016;1:16081 DOI: 10.1038/nmicrobiol.2016.81. [DOI] [PubMed] [Google Scholar]
- 18. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. DOI: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richter C, Dy RL, McKenzie RE, et al. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. DOI: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Staals RH, Jackson SA, Biswas A, et al. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat Commun. 2016;7:12853 DOI: 10.1038/ncomms12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rollins MF, Chowdhury S, Carter J, et al. Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol Cell. 2019;74:132–142.e135. DOI: 10.1016/j.molcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKenzie RE, Almendros C, Vink JNA, et al. Using CAPTURE to detect spacer acquisition in native CRISPR arrays. Nat Protoc. 2019;14:976–990. DOI: 10.1038/s41596-018-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morgan M, Anders S, Lawrence M, et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. DOI: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pagès H AP, Gentleman R, DebRoy S Biostrings: efficient manipulation of biological strings. R package version 2520. Bioconductor. 2019. DOI: 10.18129/B9.bioc.Biostrings (last accessed October13, 2020)
- 25. Kassambara A. ggpubr: ‘ggplot2’ based publication ready plots. R package version 023. CRAN. 2019. https://CRAN.R-project.org/package=ggpubr (last accessed October13, 2020)
- 26. Wickham H. ggplot2: elegant graphics for data analysis. R package version 321. Springer-Verlag New York 2016. ISBN 978-3-319-24277-4. https://CRAN.R-project.org/package=ggplot2 (last accessed October13, 2020)
- 27. Jackson SA, Birkholz N, Malone LM, et al. Imprecise spacer acquisition generates CRISPR-Cas immune diversity through primed adaptation. Cell Host Microbe. 2019;25:250–260.e254. DOI: 10.1016/j.chom.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 28. Shmakov S, Savitskaya E, Semenova E, et al. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014;42:5907–5916. DOI: 10.1093/nar/gku226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lilly J, Camps M. Mechanisms of theta plasmid replication. Microbiol Spectr. 2015;3:PLAS-0029-2014. DOI: 10.1128/microbiolspec.PLAS-0029-2014. [DOI] [PubMed] [Google Scholar]
- 30. Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. DOI: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng KC, Smith GR. Recombinational hotspot activity of Chi-like sequences. J Mol Biol. 1984;180:371–377. DOI: 10.1016/s0022-2836(84)80009-1. [DOI] [PubMed] [Google Scholar]
- 32. Cheng KC, Smith GR. Cutting of chi-like sequences by the RecBCD enzyme of Escherichia coli. J Mol Biol. 1987;194:747–750. DOI: 10.1016/0022-2836(87)90252-x. [DOI] [PubMed] [Google Scholar]
- 33. Rozkov A, Avignone-Rossa CA, Ertl PF, et al. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol Bioeng. 2004;88:909–915. DOI: 10.1002/bit.20327. [DOI] [PubMed] [Google Scholar]
- 34. Ow DSW, Lee DY, Tung HH, et al. Plasmid regulation and systems-level effects on Escherichia coli metabolism. Syst Biol Biotechnol Escherichia Coli. 2009:273–294. DOI: 10.1007/978-1-4020-9394-4_14 [Google Scholar]
- 35. Kim S, Loeff L, Colombo S, et al. Selective loading and processing of prespacers for precise CRISPR adaptation. Nature. 2020;579:141–145. DOI: 10.1038/s41586-020-2018-1. [DOI] [PubMed] [Google Scholar]
- 36. Budhathoki JB, Xiao Y, Schuler G, et al. Real-time observation of CRISPR spacer acquisition by Cas1-Cas2 integrase. Nat Struct Mol Biol. 2020;27:489–499. DOI: 10.1038/s41594-020-0415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garrett S, Shiimori M, Watts EA, et al. Primed CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res. 2020;48:6120–6135. DOI: 10.1093/nar/gkaa381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang J, Akerboom J, Vaziri A, et al. Near-isotropic 3D optical nanoscopy with photon-limited chromophores. Proc Natl Acad Sci U S A. 2010;107:10068–10073. DOI: 10.1073/pnas.1004899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yao S, Helinski DR, Toukdarian A. Localization of the naturally occurring plasmid ColE1 at the cell pole. J Bacteriol. 2007;189:1946–1953. DOI: 10.1128/JB.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Edgar R, Rokney A, Feeney M et al. Bacteriophage infection is targeted to cellular poles. Mol Microbiol. 2008;68:1107–1116. DOI: 10.1111/j.1365-2958.2008.06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joyeux M. Preferential localization of the bacterial nucleoid. Microorganisms. 2019;7:204 DOI: 10.3390/microorganisms7070204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.