ABSTRACT
Serology detection is recognized for its sensitivity in convalescent patients with COVID-19, in comparison with nucleic acid amplification tests (NAATs). This article aimed to evaluate the diagnostic accuracy of serologic methods for COVID-19 based on assay design and post-symptom-onset intervals. Two authors independently searched PubMed, Cochrane library, Ovid, EBSCO for case–control, longitudinal and cohort studies that determined the diagnostic accuracy of serology tests in comparison with NAATs in COVID-19 cases and used QUADAS-2 for quality assessment. Pooled accuracy was analysed using INLA method. A total of 27 studies were included in this meta-analysis, with 4 cohort, 16 case–control and 7 longitudinal studies and 4565 participants. Serology tests had the lowest sensitivity at 0–7 days after symptom onset and the highest at >14 days. TAB had a better sensitivity than IgG or IgM only. Using combined nucleocapsid (N) and spike(S) protein had a better sensitivity compared to N or S protein only. Lateral flow immunoassay (LFIA) had a lower sensitivity than enzyme-linked immunoassay (ELISA) and chemiluminescent immunoassay (CLIA). Serology tests will play an important role in the clinical diagnosis for later stage COVID-19 patients. ELISA tests, detecting TAB or targeting combined N and S proteins had a higher diagnostic sensitivity compared to other methods.
KEYWORDS: COVID-19, SARS-CoV-2, serology, immunoassays, metanalysis
Introduction
On 11 March 2020, the World Health Organization (WHO) described the global COVID-19 outbreak as a worldwide pandemic1. SARS-CoV-2 is the etiologic agent of COVID-19 and primarily attacks the human respiratory system and can cause respiratory infections, diarrohea, and even multiple organ failure in patients2. By 10 July 2020, there were 12,102,328 cases of COVID-19 diagnosed worldwide and 551,046 deaths had been reported3. At the time of writing, the pandemic was still severe and the likelihood of persistence of SARS-CoV-2 within the human population is increasing.
As no definitely effective drugs or vaccines are yet available, rapid diagnosis of SARS-CoV-2 infection and quick isolation of the patients and tracing of their close contacts are currently the most effective means of preventing transmission. At present, the definitive diagnosis of COVID-19 mainly depends on the detection of SARS-CoV-2 RNA by nucleic acid amplification tests (NAATs) such as RT-PCR4. Serological methods have also become an important auxiliary testing tool, and play an important role in the diagnosis and epidemiological investigation of COVID-19 cases5–10. At the time of writing, the United States Food and Drug Administration has granted Emergency Use Authorization for 31 serology test kits11. Serological test methods for the detection of anti-SARS-CoV-2 IgG and IgM antibody include enzyme-linked immunosorbent assay (ELISA), chemiluminescent immunoassay (CLIA), and lateral flow immunoassay (LFIA).
Compared with some NAATs, serological testing is relatively easier to perform and requires less technologically advanced equipment. In addition, the blood samples are less likely to contain infectious SARS-CoV-2 virus than respiratory specimens, decreasing the potential risk of infection to laboratory staff12. However, there are questions remaining to be answered concerning the serological diagnosis of COVID-19. First, studies have reported that the seroconversion happened at 3–14 days post symptom onsets13,14, which may not facilitate the early diagnosis of the disease. What’s more, the window periods of the different serological tests have not been directly assessed. Second, the specificity and sensitivity of serological methods can vary over the infection time course, and need to be further analysed15. Finally, the impact of assay design on the performance of serological tests has yet to be determined.
Meta-analysis is a quantitative evaluation method in evidence-based medicine and is widely accepted as one of the most reliable tools in clinical analysis. Our study evaluated all published case–control, longitudinal and cohort studies for the diagnostic efficacy and characteristics of the current serological tests for COVID-19.
Materials and methods
Selection criteria
The inclusion criteria for this meta-analysis were the following: (1) all cohort, case–control, and longitudinal studies published between 1 January 2020 and 30 June 2020; (2) all studies that evaluated the diagnostic performance of serological tests for COIVD-19 in comparison with a SARS-CoV-2 NAAT as a reference test; (3) studies from which we could directly or indirectly extract data on true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN); (4) participants were 18–85 years of age; (5) published articles as well as letters and corrected proofs; and (6) only articles in English were included.
The exclusion criteria were the following: (1) preprint articles which had not been peer reviewed; (2) studies that had crossed data with other published articles; (3) participants were immunocompromised (cancer, AIDS patients, etc.); and (4) studies published before 2020. (5) Studies with more than one “high risk of bias” in QUADAS-2 quality assessment domain 2–4 were excluded.
Search strategy
We searched the databases using the following Medical Subject Heading words and key words, or the combination: COVID-19, SARS-CoV-2, severe acute respiratory syndrome coronavirus 2, serology, serology test, antibody, antigen, diagnostic test. Main medical databases including PubMed, Cochrane library, EBSCO, and OVID were searched in this study (Full search strategy in supplementary material (1). We set a time limit published between 1 January 2020 and 30 June 2020 and a language limit of English only.
Study evaluation and data extraction
Two researchers (Wang and Ai) independently scrutinized abstracts and titles to include potentially eligible articles and acquire full texts online. Articles unavailable online were excluded. Then, the same two researchers examined the full texts individually using the preset inclusion and exclusion criteria.
As recommended by Cochrane Handbook for Systematic Reviews of Diagnostic Accuracy16, we adopted QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies -2) to evaluate the bias and quality of selected studies17. The following four domains were considered for risks of bias and application concerns as depicted in the assessment tool: (1) participant selection; (2) index test; (3) reference text; and (4) flowing and timing. Studies with more than one “high risk of bias” in the later 3 part were excluded (supplementary material 2).
The following information was extracted from final eligible studies: (1) details of the study: author, title, published date, countries where studies were conducted, study design, participant inclusion manner and criteria, number of enrolled participants and the grouping, number of participants whose results were available; (2) clinical characteristics of participants: age, gender, COVID-19 status; (3) target data: the results of serologic tests and NAATs for COVID-19 (TP, FP, FN, TN) and symptom onset-specimen collection interval (days). One sample per participant was included in the overall sensitivity and specificity, while the accuracy on different post-symptom interval directly used the respective data from the articles; and (4) test profile: methods for serology and SARS-CoV-2 RNA detection, profile of detected antibodies, and targeted antigen of serologic tests.
Statistical analysis
We assessed risks of bias and application concerns using QUADAS-2 tool on Review Manager 5.4 software18. Meta-analysis over selected studies was performed using R software (version 3.6.1) with the meta4diag package19. TAB was defined as combined IgG and IgM results, or directly described in the primary articles. Diagnostic performance of IgG, IgM, TAB (or combined IgG and IgM), were analysed. Sensitivity and specificity were calculated. Data synthesis was performed using Bayesian bivariate integrated nested Laplace approximation (INLA) method according to the protocol19. Forest plots of point estimates and 95% confidence intervals (95% CI) were provided. Summary receiver-operating characteristic (SROC) curves were plotted to evaluate the heterogeneity (threshold effect) between studies20.
Result
Search results
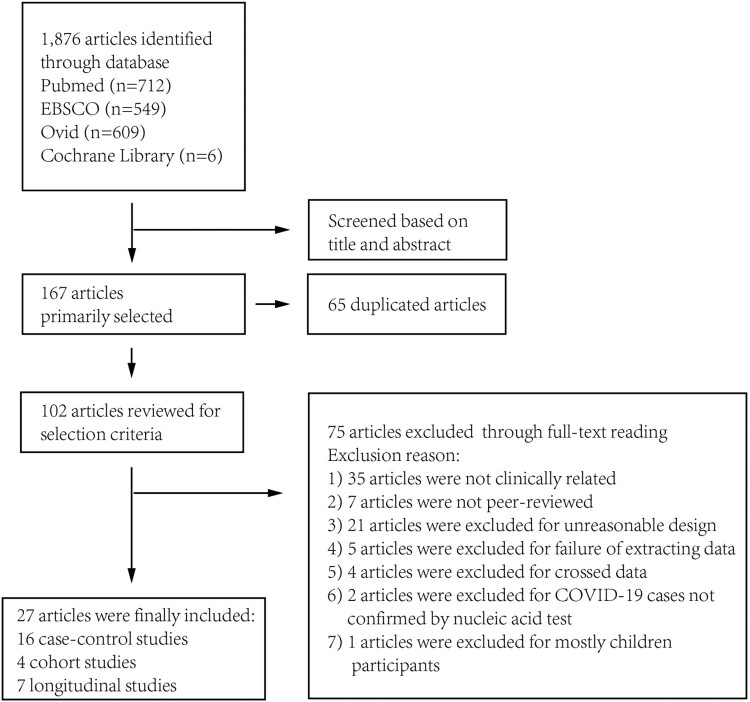
A total of 1876 articles were identified by systematic literature research as of30 June 2020. A total of 167 studies were selected through title and abstract, in which 65 were duplicated and 102 were selected for further review. Through full-text review, 75 articles were excluded as depicted in Figure 1 and supplementary Table 1. A total of 27 articles were finally included for analysis: 16 case–control studies; 7 longitudinal studies; and 4 cohort studies21–47.
Figure 1.
Search process of the meta-analysis.
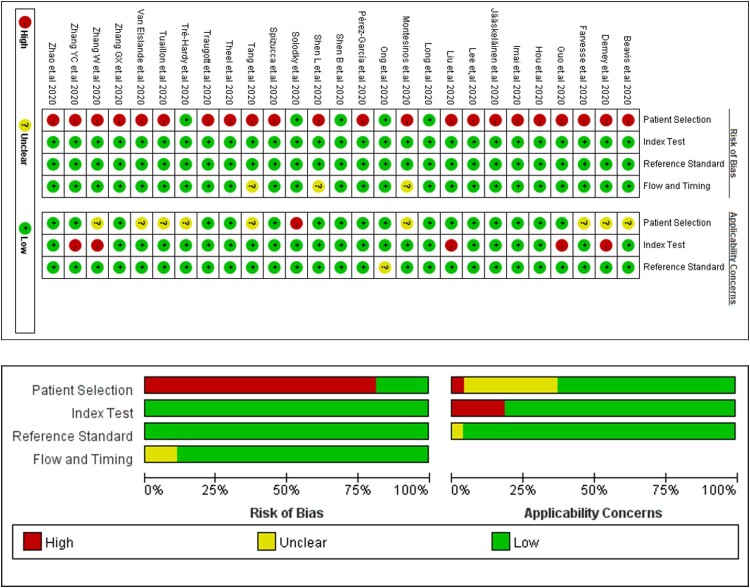
Assessment of risks of bias and application concerns are described in Figure 2. 85.1% (23/27) studies were present with a high risk of bias in patient selection, where these articles did not avoid case–control or longitudinal design. We involved these studies for later analysis and evaluated possible risks of bias in discussion.
Figure 2.
Risk of bias and application concerns of included studies assessed using QUADAS-2 tool. Red spots refer to high risk of bias or high concern, yellow refer to unclear and green refer to low.
Detailed characteristics of these 27 articles are shown in Table 1. A total of 4565 subjects were included for analysis. 37.0% (10/27) of the studies were conducted in China. 13, 6, 8, 9 studies performed ELISA, CLIA and LFIA for serology test, respectively. 77.8% (21/27) studies performed a serology test that targeted S protein/receptor binding domain (RBD) protein or N protein of COVID-19 virus.
Table 1.
Characteristics of studies included in the meta-analysis of serology test diagnostic performance.
| ID | Nation | Study Design | Included/Total Subjects (N) | Included Subject Distribution | Kit Company | Method | Item | Antigen | Age* | Male (%)* | Severe cases (%)* | Reason for not full inclusion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hou et al. 202021 | Wuhan, China | Longitudinal | 338/338 | 338 COVID-19 cases | YHLO Biotech Co. Ltd. Shenzhen, China | CLIA | IgG/IgM | nucleoprotein/spike protein | 63.3 | 50.6% | 22.2% | - |
| Long et al. 202022 | Chongqing, China | Part 1: Case-Control Part 2: Cohort Study |
478/501 | 262 COVID-19 cases 52 RT-PCR (-) suspected cases 164 close contacts |
Bioscience Co., Ltd, Chongqing, China | CLIA | IgG/IgM | nucleoprotein/spike protein | 47 | 55.4% | 13.7% | 23 cases with unclear records of symptom onset |
| Solodky et al. 202023 | France | Cohort Study | 244/329 | 244 health centre workers | TODA Pharma, Strasbourg, France | LFIA | IgG/IgM | unclear | - | - | - | 85 cases excluded were cancer patients, with abnormal immune response |
| Zhao et al. 202024 | Shenzhen, China | Longitudinal | 173/173 | 173 COVID-19 cases | Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China | ELISA | IgG/IgM | nucleoprotein | 48 | 48.6% | 18.5% | - |
| Guo et al. 202025 | Beijing, China | Case-Control | 425/425 | 82 COVID-19 cases 58 RT-PCT (-) suspected cases 135 cases before 2020 150 healthy blood donors before 2020 |
Self-produced | ELISA | IgG/IgM | nucleoprotein | - | - | 34.1% | - |
| Tang et al. 202026 | USA | Case-Control | 201/201 | 48 COVID-19 cases 80 RT-PCT (-) suspected cases 50 cases before 2020 23 other infection in 2020 |
EUROIMMUN, Lubeck, Germany et.al | ELISA | IgG | nucleoprotein/spike protein | - | - | - | - |
| Farvesse et al. 202027 | Belgium | Case-Control | 176/176 | 97 COVID-19 cases 79 cases before 2020 |
Hoffmann-La Roche Co., Ltd., Rotkreuz, Switzerland | CLIA | Tab | nucleoprotein | - | - | - | - |
| Van Elslande et al. 202028 | Belgium | Case-Control | 201/201 | 94 COVID-19 cases 103 cases before 2020 |
EUROIMMUN, Lubeck, Germany et.al | ELISA LFIA |
IgG/IgM | spike protein unclear |
67.5 | 70.2% | 30.9% | - |
| Ong et al. 202029 | Utrecht | Cohort Study | 228/228 | 99 COVID-19 cases 129 RT-PCR (-) suspected cases |
Orient Gene Biotech Co., Ltd., Zhejiang, China et.al | ELISA LFIA |
IgG/IgM | unclear spike protein |
61 | 52.0% | 9.0% | - |
| Liu et al. 202030 | Chongqing, China | Longitudinal | 32/32 | 32 COVID-19 cases | Xinsaiya Biotechnology Co., Ltd., Chongqing, China | Unclear | IgG/IgM | spike protein | 55 | 66.7% | 56.3% | - |
| Shen L et al. 202031 | Xiangyang, China | Case-Control | 188/188 | 103 COVID-19 cases 25 RT-PCR (-) suspected cases 10 other diseases in 2020 50 health donors before 2020 |
Outdo Biotech Co., Ltd., Shanghai, China | LFIA | IgM | nucleoprotein/spike protein | 25 | 45.0% | 13.6% | - |
| Zhang YC et al. 202032 | Nanjing, China | Longitudinal | 21/21 | 21 COVID-19 cases | Innovita Co., Ltd., Beijing, China | LFIA | IgG/IgM | nucleoprotein/spike protein | 37 | 61.9% | 23.8% | - |
| Demey et al. 202033 | France | Case-Control | 34/34 | 22 COVID-19 cases 12 cases before 2020 |
ISIA BIO-Technology Co., Ltd, Chongqing, China et.al | LFIA | IgG/IgM | unclear | - | - | - | - |
| Zhang W et al. 202034 | Wuhan, China | Longitudinal | 16/16 | 16 COVID-19 cases | Kyab Biotech Co., Ltd, Wuhan, China | ELISA | IgG/IgM | nucleoprotein | - | - | 18.8% | - |
| Tuaillon et al. 202035 | France | Case-Control | 58/58 | 38 COVID-19 cases 20 cases before 2020 |
EUROIMMUN, Lubeck, Germany et.al | ELISA LFIA |
IgG/IgM | nucleoprotein/spike protein | 67 | 57.9% | 68.4% | - |
| Spizucca et al. 202036 | Italy | Case-Control | 37/37 | 23 COVID-19 cases 7 RT-PCR (-) suspected cases 7 RT-PCR (-) asymptomatic controls |
Diagreat Biotechnologies Co., Ltd, Beijing, China | LFIA | IgG/IgM | unclear | 57 | - | 52.2% | - |
| Lee et al. 202037 | Taiwan, China | Longitudinal | 42/42 | 14 COVID-19 cases 28 RT-PCT (-) controls |
ALLTEST Biotech Co., Ltd. Hangzhou, China | LFIA | IgG/IgM | nucleoprotein | 51 | 50.0% | 42.9% | - |
| Theel et al. 202038 | USA | Case-Control | 205/310 | 56 COVID-19 cases 149 health donors before 2020 |
EUROIMMUN, Lubeck, Germany et.al | ELISA CLIA CMIA |
IgG | nucleoprotein/spike protein | 51 | 53.6% | - | 105 cases in early 2020 were not tested by RT-PCR for COVID-19 |
| Traugott et al. 202039 | Austria | Case-Control | 177/177 | 77 COVID-19 cases 60 RT-PCR (-) controls 40 cases before 2020 |
Euroimmun, Lübeck, Germany et.al | ELISA LFIA |
IgG/IgM | spike protein unclear |
63 | 62.3% | - | - |
| Shen B et al. 202040 | Taizhou, China | Cohort study | 150/150 | 150 suspected COVID-19 cases | Outdo Biotech Co. Ltd, Shanghai, China | LFIA | IgG/IgM | unclear | 40 | 59.3% | 21.6% | - |
| Beavis et al. 202041 | USA | Case-Control | 150/178 | 64 COVID-10 cases 70 RT-PCR (-) controls 16 cases before 2020 |
Euroimmun, Lübeck, Germany | ELISA | IgG | nucleoprotein | - | - | - | 28 cases in 2020 not tested by RT-PCR for COVID-19 |
| lmai et al. 202042 | Japan | Case-Control | 160/160 | 112 COVID-19 cases 48 cases before 2020 |
Artron, Burnaby, Canada | LFIA | IgG/IgM | unclear | 67 | 57.1% | - | - |
| Montesinos et al. 202043 | Belgium | Case-Control | 200/200 | 128 COVID-19 cases 62 cases before 2020 10 health donors in 2020 |
Euroimmun, Luebeck, Germany et.al | ELISA CLIA LFIA |
IgG/IgM | spike protein/ABEI | - | - | - | - |
| Tré-Hardy et al. 202044 | Belgium | Cohort Study | 125/125 | 125 clinically suspected COVID-19 cases | Euroimmun, Lübeck, Germany et.al | ELISA CLIA |
IgG | nucleoprotein/spike protein | - | - | - | - |
| Zhang GX et al. 202045 | Wuhan, China | Longitudinal | 112/112 | 112 COVID-19 cases | Yahuilong Biotechnology, Shenzhen, China | Unclear | IgG/IgM | nuceloprotein/envelop protein | 39 | 29.5% | - | - |
| Jääskeläinen et al. 202046 | Finland | Case-Control | 143/143 | 62 COVID-19 cases 81 cases before 2020 |
Abbott, Illinois, USA et.al | ELISA LFIA |
IgG/IgM | nucleoprotein/spike protein | 54 | 45.9% | 28.6% | - |
| Pérez-García et al. 202047 | Spain | Case-Control | 251/251 | 90 COVID-19 cases 61 PCR (-) cases 100 cases before 2020 |
AllTest Biotech, Hangzhou, China | LFIA | IgG/IgM | unclear | 64 | 57.8% | 28.9% | - |
*Age, male and severe cases were the mean value or percentage in RNA-confirmed COVID-19 cases.
Abbreviations: RT-PCR: real-time polymerase chain reaction; NAT: nucleic amplification test; ELISA: enzyme linked immune sorbent assay; CLIA: chemiluminescent immunoassay; LFIA: lateral flow (immune)assay.
Pooled diagnostic performance of IgG, IgM, TAB for COVID-19
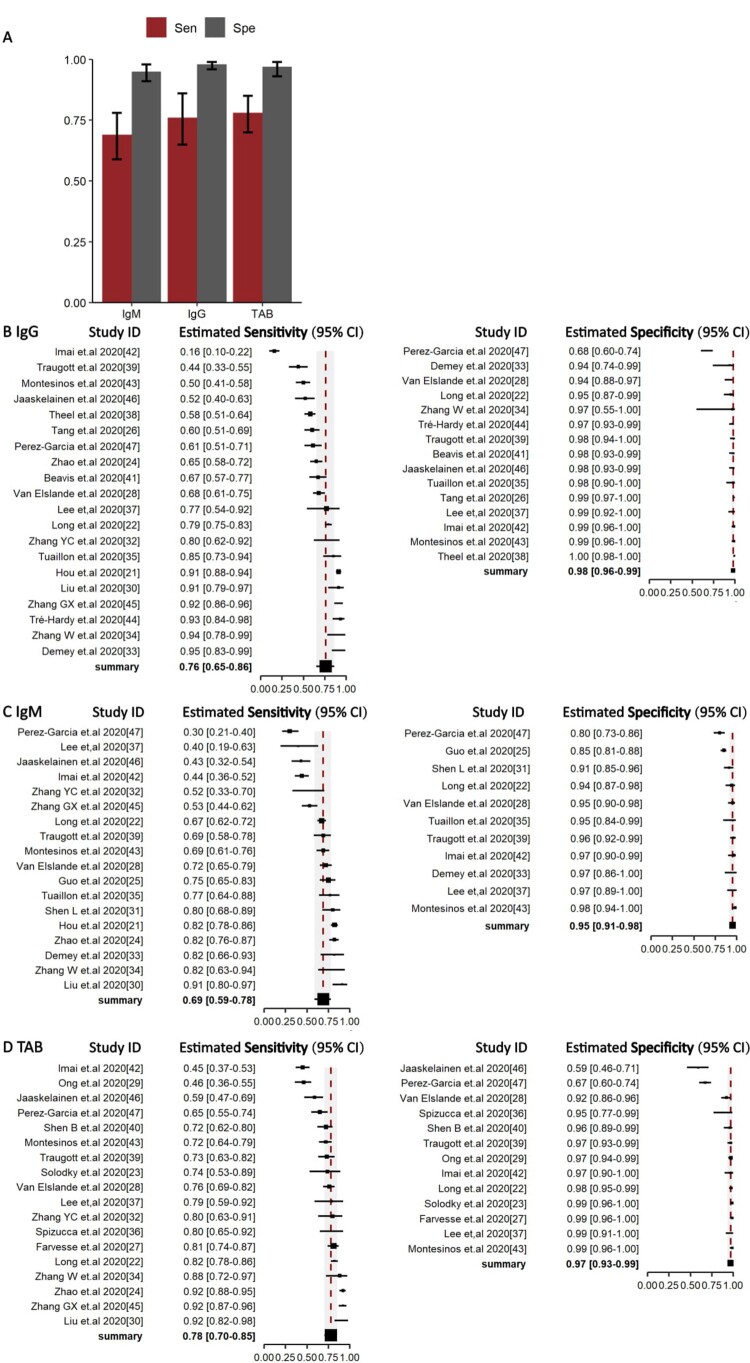
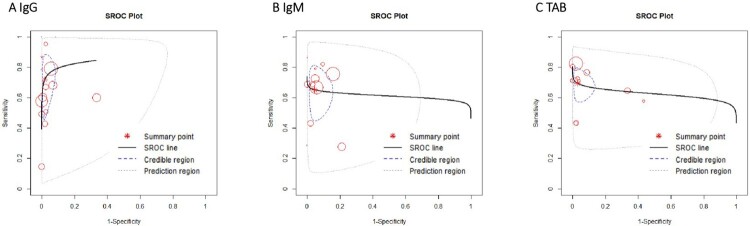
The pooled sensitivity of IgG, IgM, and TAB in RNA-positive COVID-19 cases was 0.76 (95%CI 0.65–0.86), 0.69 (95%CI 0.59–0.78), and 0.78 (95%CI 0.70–0.85) (Figure 3(B–D)), respectively. The specificity of IgG, IgM, and TAB was 0.98 (95%CI 0.96–0.99), 0.95 (95%CI 0.91–0.98), and 0.97 (95%CI 0.93–0.99), respectively (Figure 3(B–D)). There was no heterogeneity between studies (Figure 4).
Figure 3.
Overall Sensitivity and Specificity of Serology test in NAAT-confirmed COVID-19 cases. (A) Histogram of sensitivity and specificity in IgG, IgM, total antibody. Median (column) and 95% CI (error bar) were shown in the histogram. (B–D) forest plots of sensitivity (Right) and specificity (Left) in IgG, IgM, total antibody. Abbreviations: TAB: Total antibody.
Figure 4.
Summary receiver-operating characteristic of IgG (A), IgM (B), TAB (C).
Dynamic sensitivity of serologic tests after symptom onset
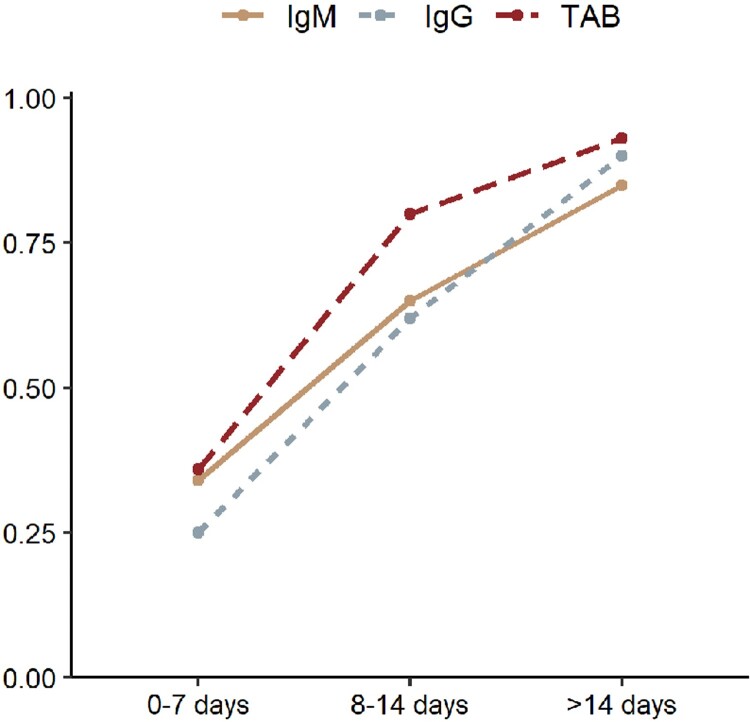
At 0–7 days, 12, 11, 10 articles were included for pooled analysis of IgG, IgM and TAB. At 7–14 days, 12, 10, 10 articles were included for pooled analysis of IgG, IgM and TAB. At over 14 days, 12, 11, 11 articles were included for pooled analysis of IgG, IgM and TAB. Sensitivity of IgG, IgM and TAB was 0.25 (95%CI 0.16–0.36), 0.34 (95%CI 0.25–0.42), and 0.36 (95%CI 0.28–0.43), respectively during the first 7 days after symptom onset, but increased to 0.62 (95%CI 0.52–0.71), 0.65 (95%CI 0.36–0.86), 0.80 (95%CI 0.69–0.99) at 8–14 days post symptom onset, and 0.90 (95%CI 0.86–0.93), 0.85 (95%CI 0.68–0.95), 0.93 (95%CI 0.80–0.98), respectively after 14 days post symptom onset in comparison with NAATs at diagnosis (Figure 5, supplementary figure).
Figure 5.
Dynamic change of the sensitivity of serology test at 0–7, 8–14, >14 days since symptom onset.
Diagnostic performance of different serologic test methods, and by targeted antigen
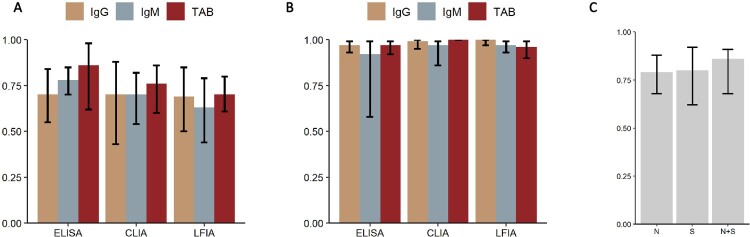
The sensitivity of different serologic methods is plotted in Figure 6(A). Seven studies provided direct comparison between different methods while 20 articles didn’t (supplementary Table 2). ELISA had the highest sensitivity in IgG, IgM and TAB with estimated sensitivity of 0.70 (95%CI 0.55–0.84), 0.78 (95%CI 0.70–0.85), 0.86 (95%CI 0.62–0.98), respectively. LFIA had the lowest sensitivity in IgG, IgM or TAB, with estimated sensitivity of 0.69 (95%CI 0.5–0.85), 0.63 (95%CI 0.44–0.79), 0.70 (95%CI 0.61–0.80), respectively. Pooled specificity of ELISA, CLIA, LFIA ranged from 92% to 100% (Figure 6(B)). The sensitivity of tests targeting N, S and both (combined) antigens was 0.79 (95%CI 0.68–0.88), 0.80 (95%CI 0.62–0.92), and 0.86 (85%CI 0.68–0.91), respectively (Figure 6(C)).
Figure 6.
Sensitivity of serology test in different method or targeted antigen. (A) Histogram of the sensitivity of serology test in ELISA, CLIA, and LFIA. (B) Histogram of the specificity of serology test in ELISA, CLIA, and LFIA. (C) Histogram of the sensitivity of serology test when targeted on spike protein (S), nucleoprotein (N) or both (N + S). Abbreviations: ELISA: Enzyme linked immune sorbent assay; CLIA: Chemiluminescent immunoassay; LFIA: Lateral flow (immuno)assay.
Discussion
Our meta-analysis included 27 articles, with 4 cohort studies, 16 case–control studies and 7 longitudinal studies to evaluate the overall diagnostic performance of serology tests for diagnosis of COVID-19, including the optimum time window and best performing methodology. Serology tests had a sensitivity of less than 40% at 0–7 days post symptom onset. Serology tests detecting TAB had a higher sensitivity than IgM or IgM alone. Targeting combined N and S proteins had a higher sensitivity than targeting N or S protein alone. LFIA tended to have a lower sensitivity than ELISA or CLIA.
The overall sensitivity of serology tests was poor, thus negative serological results alone cannot exclude the diagnosis of COVID-19. However, significant variation was observed in the forest plots of the sensitivity of serology tests (Figure 3(B–D)), with a range of 16%–93% in IgG, 42%–92% in IgM and 45%–92% in TAB. We attributed this mostly likely to different seroconversion times for different antibody classes, and further divided included articles according to symptom onset-specimen collection interval48. Our analysis suggested that serology tests had the lowest sensitivity at 0–7 days post symptom onset and the highest sensitivity at >14 days. Our findings and those of others suggest that 14 days post symptom onset is a point when the sensitivity serology tests is sufficiently high to replace NAATs for the optimal diagnosis of COVID-1913,49–52. During the early acute phase of infection, antibody detection might cause numerous false negatives cases. Nonetheless, there have been rare detectable antibody responses during the early phase of COVID-19 concurrent with high virus load and a high risk of transmission53. In the late phase of disease, on the contrary, seroconversion occurs when virus load begins to decline, and serological tests might play a more important role in the diagnosis of COVID-19. Overall, our pooled analysis suggests a preferred diagnostic algorithm based on days post symptom onset: NAAT alone at 0–14 days, NAAT combined with a serology test at over 14 days, when virus shedding might drop below the detection limit of most NAATs54.
As for the serology test methodology, our analysis suggested that serology tests detecting TAB (or combined IgG and IgM), targeting N and S combined may provide greater sensitivity than tests based on N or S alone. LFIA had a relatively low sensitivity than ELISA or CLIA but provided a fast turn-around time and convenience, and had been authorized by FDA for emergency use. The choice of serology test methodology should be based on testing environment and patient population. LFIA tests could prove useful in the emergency room, ambulatory and outpatient settings rather than simply abandoned for its relatively poor performance. We didn’t pool our analysis based on assays from different companies, but other head-to-head studies had shown a variable accordance between different assays within only a small group of participants28,38,46,55. A recent study showed a high accordance between Abbott Architect, DiaSorin Liaison, Ortho VITROS, and Euroimmun among 1200 serum samples56. Considering that the clinical performance of commercial assays was varied from laboratory condition, immune status of participants, time from symptoms onset to sample collection, etc., more head-to-head comparison was needed to figure out the accordance between commercial assays on a relatively larger scale.
In this study, most studies remained to had no risk of bias in the domain 2–4 or fewer application concern compared with other meta-analysis of diagnostic test accuracy. We attributed this phenomenon due to the following reasons. First, studies with high risk of bias in the domain 2–4 were excluded. The detailed exclusion reasons included no prespecified threshold for serology test, not using NAATs as reference tests, not all participants receiving the NAATs, etc. All of these problems were considered to bring high risk of concerns while the first domain, with a non-cohort study design or unclear consecutive enrolment were considered to bring less effect to the analysis. Second, COVID-19 was a global public health problem broke out within less than one year and thus studies on serology test accuracy of COVID-19 had some similar features: (1) Participant enrolment was confined to a short time and the criterion was usually not complex, with no clear exclusion criterion. (2) NAATs is the only method suggested by WHO to diagnose COVID-19. (3) Most case–control studies used preserved serum or blood before 2020 as the control group for determining the accuracy for serology test. These features also led to a high agreement between enrolled articles in the assessment of risk of bias and application concern using QUADAS-2 tool.
Previously, NAATs were the recommended gold standard for COVID-19 diagnosis by the WHO, while antigen tests were not recommended due to insufficient performance data57,58. Another concern raised by the WHO regarding serology tests was the relatively long antibody window, with seroconversion occurring during the second week after symptom onset52. At present, antibody detection was only suggested for epidemiological research or disease surveillance5,9,59,60. This is the first study that meta-analysed the sensitivity of serology tests across different time windows. It also provides a general review of different serology test methods. Combined IgG and IgM, as well as combined N and S protein-based tests had better performance than IgG/IgM alone, or N/S protein alone based tests, while among method formats, LFIA had lower sensitivity than ELISA or CLIA.
This study has some limitations. First, we did not analyse the cross-reactivity/specificity of serology tests for COVID-19. This was limited by data extraction, where most qualified articles did not provide specificity data. Previous studies had reported that the serological cross-reactivity between COVID-19 and other coronavirus disease like SARS-CoV seemed to be high, suggesting that serology tests might bring more false negativities and should only be applied as a supplementary tool for clinical diagnosis61. Second, 23/27 (85.2%) of enrolled articles were present with high risks of bias for case–control or longitudinal design. Specificity in our study might be overestimated because most of the control group used samples from healthy donors before 2020, which avoid possible cross-reactivities as mentioned above. Another limitation was that we did not analyse the combined diagnostic performance of NAATs and serology tests, because clinically confirmed COVID-19 cases without positive RNA or serology test results were not enrolled into this meta-analysis. According to our study, the combination of these two tests was preferred during the late phase of disease progression. However, the actual sensitivity remains to be evaluated in the future.
Our results highlight that serology tests could play an important role in the diagnosis of suspected COVID-19 infections during later stage of the disease. In clinical practice, COVID-19 serological tests could contribute to the understanding of the immunological state of the population.
Supplementary Material
Acknowledgements
We acknowledge all health-care workers involved in the diagnosis and treatment of patients and show the greatest appreciation to all health workers for their valuable input to the control of diseases.
Funding Statement
This study was supported in part by the National Natural Science Foundation of China (grant number 82041010), Shanghai Association for Science and Technology (grant number 20411950400), Shanghai Youth Science and Technology Talents Sailing Project (grant number 20YF1404300), and the Investigator Initiated Study grants (grant number Cepheid-IIS-2020-0001) to WHZ.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Contributors
HYW, JWA contributed equally to this article. YWT and WHZ conceptualized the paper. HYW collected and analysed the data. JWA, HYW, MJL wrote the initial draft, with all authors providing critical feedback and edits to subsequent revisions. All authors approved the final draft of the manuscript. YWT and WHZ is the guarantor. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Competing interests
MJL and YWT are employees of Cepheid, the commercial manufacturer of the Xpert Xpress SARS-CoV-2 test. HYW, JWA and WHZ declare no competing interests.
Data sharing
Additional data will be available on request.
Transparency
The lead authors and manuscript’s guarantor affirm that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained.
References
- 1.World Health Organizaiton . Coronavirus disease 2019 (COVID-19) Situation Report – 51 [Internet]. 2020 [cited 2020 Jul 10]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10.
- 2.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 Hospitalized patients With 2019 Novel coronavirus–infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Coronavirus diseases . (2019). (COVID-19) Situation Report-172 [Internet]. 2020 [cited 2020 Jul 10]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200710-covid-19-sitrep-172.pdf?sfvrsn=70724b90_2.
- 4.Office of the State Administration of Traditional Chinese Medicine, National Health Commission . Novel Coronavirus Pneumonia (trial version 7) [Internet]. 2020 [cited 2020 Jul 10]; Available from: http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml.
- 5.Stringhini S, Wisniak A, Piumatti G, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. The Lancet. 2020;396(10247):313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, Sun J, Nie S, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020;26(8):1193–1195. [DOI] [PubMed] [Google Scholar]
- 7.To KK-W, Cheng VC-C, Cai J-P, et al. Seroprevalence of SARS-CoV-2 in Hong Kong and in residents evacuated from Hubei province, China: a multicohort study. The Lancet Microbe. 2020;1(3):111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng D, Goldgof G, Shy B, et al. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient blood from the San Francisco Bay Area. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 9.Loeffelholz MJ, Tang Y-W.. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect. 2020;9:747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol. 2020;58(8):e00941–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food And Drug Administration . Independent Evaluations of COVID-19 Serological Tests [Internet]. openFDA. [cited 2020 Jul 1]. Available from: https://open.fda.gov/apis/device/covid19serology/.
- 12.Zainol Rashid Z, Othman SN, Abdul Samat MN, et al. Diagnostic performance of COVID-19 serology assays. Malays J Pathol. 2020;42:13–21. [PubMed] [Google Scholar]
- 13.Lou B, Li T-D, Zheng S-F, et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 2020;56(3):2000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung IF-N, Cheng VC-C, Li X, et al. SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: a case series. Lancet Infect Dis. 2020;20(9):1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espejo AP, Akgun Y, Al Mana AF, et al. Review of current Advances in serologic testing for COVID-19. Am J Clin Pathol. 2020;154(3):293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macaskill P, Gatsonis C, Deeks J, et al. Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy. Cochrane Training [Internet] 2020 [cited July 10]. Available from: https://training.cochrane.org/resource/cochrane-handbook-systematic-reviews-diagnostic-test-accuracy.
- 17.Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. [DOI] [PubMed] [Google Scholar]
- 18.Review Manager (RevMan) . [Internet]. The Cochrane Collaboration; 2020. Available from: https://training.cochrane.org/online-learning/core-software-cochrane-reviews/revman.
- 19.Guo J, Riebler A. meta4diag: Bayesian Bivariate Meta-analysis of Diagnostic Test Studies for Routine Practice. arXiv. 2016.
- 20.Kim KW, Lee J, Choi SH, et al. Systematic review and meta-analysis of studies Evaluating diagnostic test accuracy: A Practical review for clinical researchers-part I. general Guidance and Tips. Korean J Radiol. 2015;16(6):1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou H, Wang T, Zhang B, et al. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology. 2020;9(5):e01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. [DOI] [PubMed] [Google Scholar]
- 23.Solodky ML, Galvez C, Russias B, et al. Lower detection rates of SARS-COV2 antibodies in cancer patients versus health care workers after symptomatic COVID-19. Ann Oncol. 2020;31(8):1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Ren L, Yang S, et al. Profiling early Humoral response to diagnose Novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang MS, Hock KG, Logsdon NM, et al. Clinical performance of Two SARS-CoV-2 serologic assays. Clin Chem. 2020;66(8):1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Favresse J, Eucher C, Elsen M, et al. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies. Clin Chem. 2020;66(8):1104–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Elslande J, Houben E, Depypere M, et al. Diagnostic performance of 7 rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26(8):1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong DSY, de Man SJ, Lindeboom FA, et al. Comparison of diagnostic accuracies of rapid serological tests and ELISA to molecular diagnostics in patients with suspected COVID-19 presenting to the hospital. Clin Microbiol Infect. 2020;26(8):1094.e7–1094.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Wang J, Xu X, et al. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020;9:1269–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen L, Wang C, Zhao J, et al. Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg Microbes Infect. 2020;9:1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yongchen Z, Shen H, Wang X, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9:833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demey B, Daher N, François C, et al. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J Infect. 2020;81(2):e6–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Du R-H, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuaillon E, Bolloré K, Pisoni A, et al. Detection of SARS-CoV-2 antibodies using commercial assays and seroconversion patterns in hospitalized patients. J Infect. 2020;81(2):e39–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spicuzza L, Montineri A, Manuele R, et al. Reliability and usefulness of a rapid IgM-IgG antibody test for the diagnosis of SARS-CoV-2 infection: A preliminary report. J Infect. 2020;81(2):e53–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y-L, Liao C-H, Liu P-Y, et al. Dynamics of anti-SARS-Cov-2 IgM and IgG antibodies among COVID-19 patients. J Infect. 2020;81(2):e55–e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theel ES, Harring J, Hilgart H, et al. Performance characteristics of four high-Throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(8):e01243–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traugott M, Aberle SW, Aberle JH, et al. Performance of SARS-CoV-2 antibody assays in different stages of the infection: comparison of commercial ELISA and rapid tests. J Infect Dis. 2020;222(3):362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen B, Zheng Y, Zhang X, et al. Clinical evaluation of a rapid colloidal gold immunochromatography assay for SARS-Cov-2 IgM/IgG. Am J Transl Res. 2020;12:1348–1354. [PMC free article] [PubMed] [Google Scholar]
- 41.Beavis KG, Matushek SM, Abeleda APF, et al. Evaluation of the EUROIMMUN anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol. 2020;129:104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imai K, Tabata S, Ikeda M, et al. Clinical evaluation of an immunochromatographic IgM/IgG antibody assay and chest computed tomography for the diagnosis of COVID-19. J Clin Virol. 2020;129:104393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montesinos I, Gruson D, Kabamba B, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tré-Hardy M, Wilmet A, Beukinga I, et al. Validation of a chemiluminescent assay for specific SARS-CoV-2 antibody. Clin Chem Lab Med. 2020;58(8):1357–1364. [DOI] [PubMed] [Google Scholar]
- 45.Zhang G, Nie S, Zhang Z, et al. Longitudinal change of SARS-Cov2 antibodies in patients with COVID-19. J Infect Dis. 2020;222(2):183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jääskeläinen A, Kuivanen S, Kekäläinen E, et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol. 2020;129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-García F, Pérez-Tanoira R, Romanyk J, et al. Alltest rapid lateral flow immunoassays is reliable in diagnosing SARS-CoV-2 infection from 14 days after symptom onset: A prospective single-center study. J Clin Virol. 2020;129:104473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndrome-Related coronavirus 2: A Narrative review. Ann Intern Med. 2020;172:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamilton F, Muir P, Attwood M, et al. Kinetics and performance of the Abbott architect SARS-CoV-2 IgG antibody assay. J Infect. 2020;S0163-4453(20)30510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long Q, Deng H, Chen J, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020.
- 52.Suhandynata RT, Hoffman MA, Kelner MJ, et al. Longitudinal Monitoring of SARS-CoV-2 IgM and IgG Seropositivity to Detect COVID-19. J Appl Lab Med. 2020;5(5):908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral load in Upper respiratory specimens of Infected patients. N Engl J Med. 2020;382(12):1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohmer N, Westhaus S, Rühl C, et al. Clinical performance of different SARS-CoV-2 IgG antibody tests. J Med Virol. 2020;92(10):2243–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prince HE, Givens TS, Lapé-Nixon M, et al. Detection of SARS-CoV-2 IgG targeting nucleocapsid or spike protein by four high Throughput immunoassays authorized for emergency Use. J Clin Microbiol. 2020;JCM.01742-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organizaiton . Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance [Internet]. 2020 [cited 2020 July 10]. Available from: https://www.who.int/internal-publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected.
- 58.World Health Organizaiton . Advice on the use of point-of-care immunodiagnostic tests for COVID-19 [Internet]. 2020 [cited 2020 July 10]. Available from: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19.
- 59.World Health Organizaiton . Serology in the context of COVID-19 [Internet]. 2020 [cited 2020 Jun 25]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/serology-in-the-context-of-covid-19.
- 60.Tang Y-W, Schmitz JE, Persing DH, et al. Laboratory diagnosis of COVID-19: current Issues and Challenges. J Clin Microbiol. 2020;58(6):e00512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chia WN, Tan CW, Foo R, et al. Serological differentiation between COVID-19 and SARS infections. Emerg Microbes Infect. 2020;9(1):1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.