Abstract
CRISPR-Cas-based transcriptional activators allow genetic engineers to specifically induce expression of one or many target genes in trans. Here we review the many design variations of these versatile tools and compare their effectiveness in different eukaryotic systems. Lastly, we highlight several applications of programmable transcriptional activation to interrogate and engineer complex biological processes.
Introduction
The engineering of CRISPR-Cas (CRISPR-associated) systems has provided a means for simple, accurate, and efficient genome editing. The type II CRISPR-Cas9 system from Streptococcus pyogenes is the most commonly used for genome editing.1 This ribonucleoprotein complex consists of a DNA endonuclease (Cas9) and two RNAs, CRISPR RNA (crRNA) and transacting RNA (tracrRNA). Together, these components make a blunt cut in DNA upon binding to the target sequence.2–4
The crRNA component of the complex provides sequence specificity by base-pairing to the complementary 20 nucleotides of the target DNA (protospacer) upstream of an “NGG” protospacer adjacent motif (PAM).3,4 crRNA and tracrRNA can be combined into a small guide RNA (sgRNA) that is sufficient for function.4 Thus, the two-component sgRNA-Cas9 complex constitutes an RNA-guided platform for cleaving specific genomic regions. This tool has transformed the cost and throughput of genome editing in recent years.
CRISPR-Cas9 systems have been engineered to remove their DNA cutting activity for applications that exploit their sequence-programmable DNA-binding ability. Cas9 contains two endonuclease domains, HNH and RuvC, each involved in the cleavage of a single DNA strand.4 Thus, to create a catalytically inactive or “dead” Cas9 (dCas9), both domains were inactivated.4 Later, it was demonstrated that dCas9 proteins harboring mutations that inactivate the endonuclease catalytic sites retain their DNA binding abilities.5,6 Qi and colleagues used a dCas9 with D10A and H840A substitutions in the RuvC and HNH domains, respectively.4,5 A second dCas9 version was designed using four mutations in the nuclease domains, D839A and N863A in addition to the abovementioned D10A and H840A.6 Both dCas9 versions are devoid of nuclease activity but remain strong RNA-guided DNA binding proteins.5,6
dCas9 has been fused to many effector domains with the goal of recruiting different activities to locally modify the target DNA or its associated proteins. When transcription activation domains (ADs) are fused to dCas9, the resulting protein can induce expression of genes in the vicinity, thus becoming a programmable transcriptional activator (PTA), also known as a CRISPR activator (CRISPRa). This review focuses on PTAs created using CRISPR-Cas systems, describing the different designs, common features, and in vivo applications. We focus on PTAs developed and tested in eukaryotic organisms.
Activation Domains
ADs are defined as motifs capable of recruiting the transcription preinitiation complex (PIC) to a core promoter. RNA polymerase II and the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH comprise the PIC.7,8 The mediator, a large complex of reversibly-associating transcriptional regulatory subunits, is part of the PIC through interactions with RNA polymerase II.9 Strong ADs interact with components of the PIC, accelerating its assembly at a core promoter.10 The mechanism by which these interactions occur relates to the conserved architecture of many ADs.
ADs commonly used in conjunction with dCas9 programmable DNA binding domains are shown in Table 1. VP64, a common AD, is a tetrameric repeat derived from the VP16 protein of herpes simplex virus.11 Fusing four end-to-end repeats of the VP16 motif (VP64) enhanced its ability to activate transcription.12 Furthermore, fusion of the VP64 domain to additional ADs resulted in even greater transcription activation.13,14 The ADs from p65 (nuclear factor kappaB, 65 kDa subunit) and Rta (Epstein–Barr virus R transactivator) are two of the domains commonly used as a fusion to VP64.
Table 1.
Summary of CRISPR-Cas activation systems
| PTA system | CRISPR Cas systema | No. of elementsb | ADs/copy number recruitedc | Organisms | Addgene catalog number | References |
|---|---|---|---|---|---|---|
| First-generation | ||||||
| dCas9-AD | Class 2/Cas9, Class 2/Cpf1 (Cas12a) | 2 | VP64/1X; VP640/1X; p65/1X; EDLL/1X; TAL/1X; CBF1/1X | Mammals, fish, worms, insects, yeast, plants | 48214, 48218–48228, 48236–48240, 49013–49016, 46912–46923, 50918, 50920, 47753, 47754, 47314–47321, 47106–47108, 69303, 132334 | 6,13,20,26–32,40,80,81 |
| Second-generation | ||||||
| dCas9-VPR | Class 2/Cas9, Class 2/Cpf1 (Cas12a) | 2 | VP64-p65-Rta/1X | Mammals, insects, yeast, plants | 63798–63802, 64046, 104567 | 13,39,40,88 |
| dCas9-TV | Class 2/Cas9, Class 2/Cpf1 (Cas12a) | 2 | TAL/6X, VP64/2X | Plants, mammals | n.a. | 14 |
| SAM | Class 2/Cas9 | 3 | VP64/1X, p65-HSF1/4X; VP64 5X | Mammals, insects, plants | 78901, 78902, 78905, 61422, 61427, 73795, 99884–99897, 99905–99907, 100044, 122835–122839, 122856, 122857, 122860, 122861 | 33,56,83,84,88 |
| scRNA | Class 2/Cas9 | 3 | VP64/4X; EDLL/1X, VPR/4X | Mammals, yeast, plants | 47314–47321, 78906, 62277, 62279–62283, 62313–62322, 62325, 62327, 62328, 62330–62342, 62344, 66564, 66565 | 6,34,56,88 |
| SunTag | Class 2/Cas9, Class 2/Cpf1 (Cas12a) | 3 | VP64/10X; p65-HSF1/10X | Mammals, insects, plants | 60903, 60904, 78899, 78900, 119672, 120249–120252 | 35,41,56,60,72 |
| TREE | Class 2/Cas9 | 4 | VP64/1X, HSF1-p65/16-32X; VP64/1X, VP64-p65-Rta/16-32X | Mammals | na | 36 |
| Split ddCpf1 | Class 2/Cpf1 (Cas12a) | 3 | HSF1-p65/4X | Mammals | na | 42 |
| Sth Cascade | Class 1/CRISPR-Cascade | 6 | CBF1/3X | Plants | 132334–132340, 132342–132353 | 20 |
The number of elements takes into account the sgRNA or sgRNA2.0 component.
A semicolon (;) separates different activator sets used in the same system.
Copy number refers to the theoretical maximum of ADs recruited.
AD, activation domain; na, not available; PTA, programmable transcriptional activator; SAM, synergistic activation mediator; scRNA, scaffold RNA; TREE, Three-component repurposed technology for enhanced expression.
Transcriptional activation mediated by p65 is conferred by two distinct C-terminal transactivation domains, TA1 and TA2.15 Similarly, Rta contains two acidic C-terminal activation subdomains, 1 and 2, from which domain 2 confers the most potent transcriptional activity.16 Another domain used in the engineering of PTAs in animals is from human heat shock factor 1 (HSF1). HSF1 AD comprises regions B and C located at the C-terminus of the protein.17
ADs such as EDLL (APETALA2 family activation domain) and CBF1 (C-REPEAT/DRE BINDING FACTOR 1) are derived from plant species and activate target genes in both plant and mammalian cells.18–20 EDLL is characterized by a distinctive distribution of acidic residues and hydrophobic leucines located at the C-terminus of AP2/ERF family of plant transcription factors.19 The AD of CBF1 comprises acidic amino acid residues located at the C-terminal half of the protein.18
Transcription activator-like acidic activation domain (TAL) is a prokaryotic AD from the C-terminal region of the transcription activator-like effector (TALE) protein, a Xanthomonas transcription factor secreted into plant cells to regulate gene expression in the host.21 The TAL AD was demonstrated to been able to induce expression of target genes in yeast and plant cells.21
ADs commonly used in PTA systems (Table 1) tend to be intrinsically disordered motifs. ADs have previously been classified as “acidic blobs” or “negative noodles” enriched in acidic, proline, serine, threonine, and glutamine residues.22 This trend is described by a model in which disordered acidic ADs recruit coactivators by concurrent AD-coactivator phase separation at a promoter.10 This mechanism relies upon the propensity for intrinsically disordered regions to form scaffold-like structures by exposing short repeating peptide motifs along with conformational flexibility.23 ADs, coactivators, and RNA Pol II itself contain intrinsically disordered low-complexity regions.24
The following model has emerged for dynamic and reversible assembly of the PIC. Genomic targeting of an intrinsically disordered AD, which can interact with coactivators, forms a phase-separated condensate at a core promoter. This promotes clustering of other intrinsically disordered proteins at the droplet such as additional coactivators, general transcription factors, and RNA Pol II CTD, resulting in an active PIC.25
CRISPR-Cas Activation Systems
First-generation PTAs
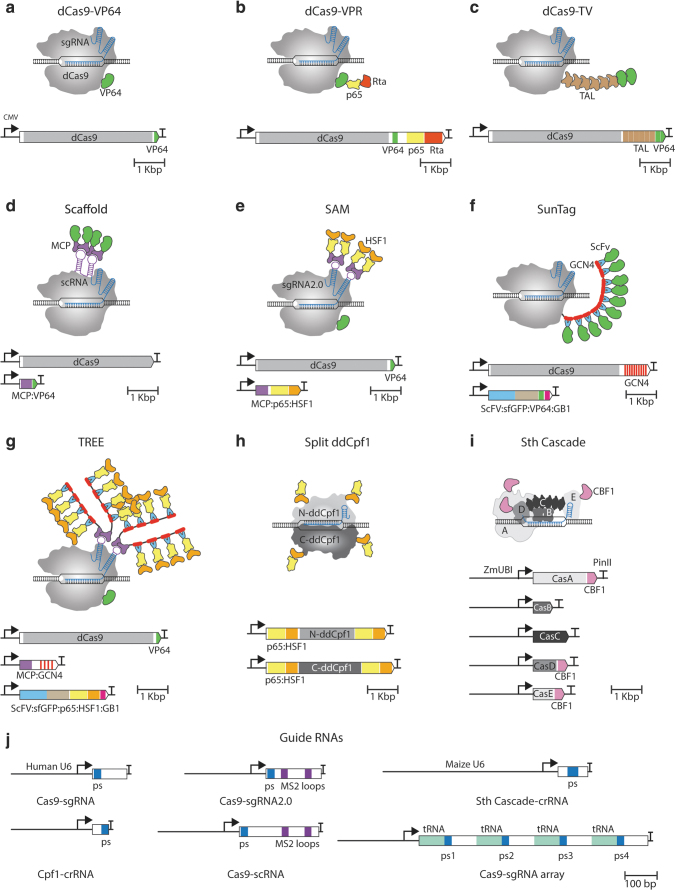
The first CRISPR-based transcriptional activators were created by fusing an AD to the C-terminus of dCas9 (Fig. 1 and Table 1). The AD of choice was VP16 in any of its multiple iterations (four tandem repeats, VP64 or 10 repeats, VP160) (Fig. 1a). The chimeric dCas9-VP64 is able to activate transcription of reporter and endogenous genes when targeted to promoter regions.6,26–31 Transcriptional activation of some genes led to the increased accumulation of endogenous protein.29–31 However, most targeted genes showed only modest to low levels of transcriptional activation.6,28,31 Even for dCas9 constructs with the stronger activator VP160, only 10-fold activation was observed.26
FIG. 1.
Diagram of different CRISPR-Cas transcriptional systems. For each PTA described, a schematic representation of the CRISPR-Cas protein in complex with its sgRNA, target DNA sequence, and additional protein modules, when present, is shown in the upper panels. Expression constructs encoding the protein components of the activator systems are shown in the lower panels. Each construct is driven by the human cytomegalovirus promoter and the HSV thymidine kinase polyadenylation signal sequence as a terminator, unless indicated otherwise. Genetic elements are drawn to scale. Cas proteins are shown in different shades of gray, and the sgRNA and crRNA are indicated by blue lines inside the Cas proteins. ADs are shown in different colored shapes. (a) dCas9-VP64. (b) dCas9-VPR. (c) dCas9-TV. (d) scRNA. The MS2 loops appended to the sgRNA to form the (scaffold sgRNA) scRNA are indicated by purple lines. (e) SAM. The MS2 loops in the sgRNA2.0 are indicated by purple lines. (f) TREE. (g) SunTag. (h) split ddCpf1. The N-terminal and C-terminal fragments of ddCpf1 are indicated as N-ddCpf1 and C-ddCpf1, respectively. (i) Sth Cascade. The different domains that form the DNA binding Cascade complex are CasA, A; CasB, B; CasC, C; CasD, D; and CasE, E. Cascade expression constructs are driven by the ZmUBI promoter and the PinII terminator. (j) Schematic representation of different sgRNA expression cassettes used in the CRISPR-Cas transcriptional activators shown. Unless indicated otherwise, the human U6 RNA polymerase III promoter is used in all constructs. The terminator in all sgRNA constructs is a poly T-stretch of 6-8 nucleotides. sgRNA scaffold is represented by an open box and the ps in blue. AD, activation domain; HSV, herpes simplex virus; PinII, potato proteinase inhibitor; ps, protospacer; PTA, programmable transcriptional activator; SAM, synergistic activation mediator; scRNA, scaffold RNA; sgRNA, small guide RNA; TREE, three-component repurposed technology for enhanced expression; ZmUBI, Zea mays ubiquitin.
Other direct fusion PTAs have been created. For instance, a dCas9-p65 fusion is capable of transcription activation although to lower levels than dCas9-VP64.28 In plants, dCas9 fused to EDLL, TAL, and CBF1 activator domains, all increase gene activation at significant, but still modest levels.20,32
Second-generation PTAs
The second generation of CRISPR-based activators leverage the synergistic effect that the recruitment of multiple transcription factors has in natural activating systems.13 Thus, the main premise is that recruiting multiple ADs to the promoter, as single repeated domains, heterogeneous combinations, or both, will enhance transcriptional activation. This has been achieved through diverse and creative mechanisms (Table 1).
dCas9-VPR
In the dCas9-VPR system, the efficiency of transcriptional activation was greatly improved by making tandem fusions of different ADs (Fig. 1b). First, to identify suitable ADs, Chavez et al. screened 22 single-AD dCas9 C-terminal fusions for their ability to activate transcription of a reporter.13 VP64, p65, and Rta were the strongest ADs.13 To increase the strength of the transcriptional activity, tripartite activators were created by sequential fusion of the ADs to the C-terminus of dCas9 in different orders.
The fusion providing the highest transcriptional activation was VP64-p65-Rta, VPR for short. dCas9-VPR performs significantly better than dCas9-VP64, any of the single ADs fused to dCas9, and constructs containing double AD fusions.13 dCas9-VPR can activate transcription of endogenous genes in animal cell lines, in some cases to levels comparable with those observed in native tissues. Furthermore, activation of neurogenin 2 or neurogenic differentiation factor 1 by sgRNA-guided dCas9-VPR was robust enough to induce differentiation of human-induced pluripotent stem cells into induced neurons, which was not possible using dCas9-VP64.13 Thus, dCas9-VPR constitutes a strong transcriptional activator capable of inducing gene expression to levels meaningful enough to exert phenotypic changes.
dCas9-TV
dCas9-TV was similarly developed by fusing tandem repeats of ADs to the C-terminus of dCas9 (Fig. 1c). Increasing the number of AD repeats led to an increase in transcriptional activation. However, too many repeats fused to dCas9 led to decreased protein accumulation, possibly due to instability triggered by the repetitive nature of the constructs.14 By testing different combinations of four ADs, Li et al. created an efficient PTA with a number of AD repeats that balances PTA stability and target gene overexpression.14 This optimized activator, named dCas9-TV, is a fusion of dCas9 to six tandem copies of TAL followed by eight copies of VP16 (Fig. 1c).14
dCas9-TV was tested in cells from eudicot and monocot plants and significantly activates transcription of endogenous genes. dCas9-TV is also capable of activating ASCL1 and OCT4 in HEK 293T human cells. Transgenic plants expressing dCas9-TV and an sgRNA targeting the promoter RLP23, a leucine-rich repeat receptor protein that mediates immune response, displayed enhanced immune response in the presence of the peptide elicitor, nlp20, of RLP23.14
Scaffold RNA and synergistic activation mediator
Scaffold RNA (scRNA) and synergistic activation mediator (SAM) are based on engineering hairpins in the sgRNA structure that allow the use of RNA-binding proteins to tether ADs to the dCas9 ribonucleoprotein complex (Fig. 1d, e). In the scRNA system, two MS2 RNA hairpin loops are covalently attached to the 3′ end of the sgRNA. MS2 hairpin loops are bound specifically and avidly by dimers of the MS2 bacteriophage coat protein (MCP). When a chimeric MCP-VP64 protein is coexpressed with the modified sgRNA and dCas9, the assembled ribonucleoprotein complex will contain up to four copies of the VP64 domain (Fig. 1d).6
A similar approach was used in the SAM system, however, in this case, the MS2 RNA hairpin loops are appended to the tetraloop and stem-loop 2 of the sgRNA to create the so-called sgRNA2.0.33 Since both the tetraloop and stem-loop 2 extend beyond the surface of the dCas9-sgRNA complex, the addition of the hairpins does not affect the DNA-binding of dCas9 and still allows recruitment of MCP-AD fusions.33 In addition, to increase the effectiveness of the SAM system, p65 and HSF1 ADs were fused in tandem to MCP and a single VP64 AD was directly linked to dCas9 (Fig. 1e).33
SAM is a potent transcriptional activator, consistently outperforming dCas9-VP64.33 Conversely, the initial iteration of scRNA was up to three times less effective than dCas9-VP64.6 The low performance of the first iteration of the scRNA system may be due to the instability of the modified sgRNA caused by the addition of multiple repeats of the MS2 loop.34 Indeed, redesign of the multihairpin to improve stability resulted in a more potent activator.34
scRNA and SAM can be engineered as platforms for broad control of gene expression. The use of orthogonal sets of RNA-hairpins:binding-proteins produces distinct regulons. sgRNAs that recruit different effector domains to the dCas9 protein, will confer unique effects at each sgRNA target locus.6,34 This flexibility allows for the creation of complex sgRNA-encoded programs using dCas9 as a master regulator.34
SunTag
In the SunTag system, multiple copies of an AD are targeted to the dCas9 ribonucleoprotein through an epitope/antibody interaction.35 The SunTag PTA is composed of an sgRNA and two protein modules: (1) dCas9 protein fused to a tandem array of GCN4 epitope motifs separated by flexible GS linkers, and (2) a single-chain variable fragment antibody (ScFv), with affinity for the GCN4 motif, fused to VP64. Upon expression of both protein components, the GCN4 epitope array on dCas9 recruits up to 10 copies of the ScFv-VP64 AD. This complex is targeted to the promoter-of-interest by the sgRNA (Fig. 1f).35
SunTag is a powerful activator of gene expression. When introduced into mammalian cells, the SunTag system increased expression of target genes and produced the expected phenotypes.35 In contrast to the results obtained with SunTag, activation of these same genes by dCas9-VP64 was very inefficient and did not yield the expected cell responses.35 Thus, transcriptional activation by SunTag is robust enough to produce the biological response predicted by the overexpression of a target gene.
Three-component repurposed technology for enhanced expression
The three-component repurposed technology for enhanced expression (TREE) system enhances the recruitment of multiple AD copies via a hierarchical multitag system. It combines SunTag with the RNA tethering system used by SAM in a tree-resembling architecture (Fig. 1g).36 The primary tag is the RNA hairpin loop bound by an MCP-GCN4-array fusion protein. Finally, the AD is recruited to the complex by fusion to an ScFv with affinity for GCN4. Both the p65-HSF1 and tripartite VPR ADs have been used in the TREE system (Fig. 1g).36
Like other second-generation PTAs, the TREE system gives strong transcriptional activation. TREE outperformed dCas9-VP4, the SAM system, and dCas9-VPR when using p65-HSF1 and VPR as ADs, respectively. A direct comparison with the canonical 10-copy SunTag-VP64 was not published, but TREE appears to outperform a SunTag version recruiting up to four and eight copies of VP64.36
Engineering PTAs using novel CRISPR-Cas systems
Most PTA designs currently available for RNA-guided activation use the S. pyogenes dCas9 ribonucleoprotein complex. However, characterization of alternative CRISPR systems has provided researchers with a broader set of CRISPR-Cas proteins for engineering of novel PTAs (Table 1).
One of these proteins is Cpf1 (CRISPR from Prevotella and Francisella 1), also known as Cas12a, an RNA-guided endonuclease producing staggered DNA double-stranded breaks.37 A DNase-dead Cpf1 (ddCpf1) that retains the sgRNA-guided DNA binding function was generated by inactivating the RuvC domain.38 ddCpf1 fused to VP64 or p65 ADs activates transcription of target genes, although not very efficiently.39,40 However, more robust gene activation was obtained when ddCpf1 was used in ddCpf1-VPR and ddCpf1-SunTag systems.39–41 Conversely, replacing dCas9 by ddCpf1 in the TV system yielded only a weak activator.14 Attempts to create the ddCpf1 equivalent of SAM or scRNA were unsuccessful because the stem loop region and the 3′ end of the crRNA do not protrude from the Cpf1 ribonucleoprotein complex.42
Recently, a design not first demonstrated with dCas9 was used to create a novel ddCpf1 activator. Nihongaki et al. generated a split form of ddCpf1 that spontaneously associates to yield a functional heterodimer.42 This split form duplicated the number of N- and C-terminal ends available for attaching ADs.42 The bipartite AD, p65-HSF1, was fused to each end of the two halves of ddCpf1 (Fig. 1h). This split ddCpf1 PTA was able to activate multiple endogenous genes in HEK293T cells. Compared with the dCas9-based SAM system, split ddCpf1 consistently reached higher activation levels.42
In addition to the effectiveness of ddCpf1 for the design of potent PTAs, this protein possesses additional features that make its use compelling: (1) binding specificity for Cpf1 to its DNA target is greater than for Cas943–45; (2) the Cpf1 ribonucleoprotein complex comprised a crRNA while lacking the tracrRNA, simplifying the design of the sgRNAs37; (3) the T-rich PAM used by Cpf1 enables the targeting of promoter regions not covered by the G-rich PAM of Cas937; and (4) the endogenous RNase activity of Cpf1 simplifies the generation of multiple crRNAs from the processing of a single transcript carrying a crRNA array.46
Both Cas9 and Cpf1 are Class 2 CRISPR systems that utilize a single-protein component for nuclease activity. This is in contrast to class 1 systems that feature multicomponent nucleases.47 Despite the added complexity, the diversity of class 1 CRISPR-Cas systems offers some advantages for the development of PTAs. Young et al. have reported a novel PTA based on a Class 1/type I-E complex from Streptococcus thermophilus DGCC7710 called Cascade (CRISPR-associated complex for antiviral defense).20 Type I-E Cascade comprised 6 subunits (Cas3, CasA, CasB, CasC, CasD, and CasE) from which CasE is involved in crRNA processing, CasABCD in target recognition, and Cas3 as a single-stranded exonuclease.20
Since Cas3 is recruited only after the complex is poised at its target sequence, a DNA binding complex could be obtained by simply excluding Cas3 from the system. To fashion a Cascade PTA, the CBF1 AD was fused to the C-terminal end of CasA, CasD, and CasE (Fig. 1i). The resulting PTA, Sth Cascade, activated transcription of a reporter and an endogenous target gene (r) when transiently expressed in maize embryos. Activation of the transcription factor R by Sth Cascade was robust enough to produce the expected anthocyanin phenotype. However, it was not significantly better than that obtained using dCas9-CBF1.20
Common Features of Gene Activation Mediated by CRISPR Activation Systems
Despite the diversity of PTA designs, target genes, and experimental organisms, some features common to all systems have started to emerge (Fig. 2).
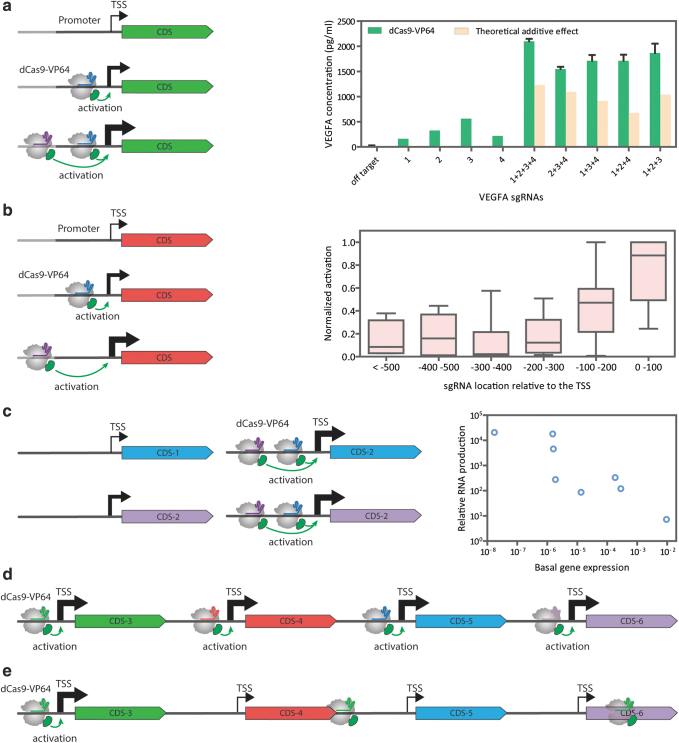
FIG. 2.
Features of transcriptional stimulation mediated by CRISPR transcriptional activators. (a) Synergy. When two dCas9-VP64 copies are recruited, they act concertedly to yield stronger transcriptional activation than a single copy (left panel). In the right panel, dCas9-VP64 can induce the production of VEGFA protein. However, VEGFA protein production is synergistically increased when three or four sgRNAs are coexpressed with the dCas9-VP64 activator.30 (b) Position effects. Binding of dCas9-VP64 to the region upstream to the TSS induces robust gene activation (right panel). Induction of expression in 12 human genes using the SAM activator is the strongest when using sgRNAs located between −200 and 0 bp of the TSS.33 (right panel) (c) Gene to gene variability. The relative change in gene expression induced by dCas9-VP64 will be larger for weakly induced genes than for genes with higher basal expression (left panel). There is a negative correlation between the basal expression of a gene and the fold-activation attained by dCas9-VPR.13 (right panel). (d) Multiplexing. dCas9-VP64 can be used for the targeted activation of multiple genes by using pooled sgRNAs binding to the promoter of different genes. (e) Specificity. Despite having more than one binding site, dCas9-VP64 specifically activates the expression of its target genes with little to no off-target effects. This is, in part, due to the position effect requiring dCas9-VP64 to target the region upstream of the TSS for maximum efficiency. The TSS is indicated by a bent arrow. The thickness of the arrow represents the strength of gene expression. The dCas9-VP64 activator consists of the dCas9 protein in gray, the VP64 activation domain in green, and the colored line inside the dCas9 protein is the sgRNA. VEGFA, vascular endothelial growth factor A; TSS, transcription start site.
Synergy
Before the discovery of CRISPR-Cas systems, it was established that many promoters contain multiple binding motifs for the same transcription factor. When multiple copies of a transcription factor are recruited to a promoter, they interact synergistically to enhance transcriptional activation.48–50 Thus, early in the inception of CRISPR-based PTAs, targeting multiple regions of a gene was explored as an approach to increase potency. Certainly, when multiple dCas9-VP64 complexes are recruited to a promoter, they act in concert to yield a stronger transcriptional activation (Fig. 2a).6,30,31 This same observation holds true for most second-generation PTAs and across multiple organisms.
This effect had been previously reported with TALE-based PTAs.51 However, an advantage of CRISPR-Cas-based PTAs is that DNA targeting is mediated by complementarity to a crRNA (or sgRNA). This facilitates targeting multiple sites upstream of a gene-of-interest. Doing the same with TALEs would require engineering and expressing multiple 4 kb transgenes in the same construct.
Position effects
sgRNA targeting of a PTA to different regions of a gene, in any orientation, may confer some degree of gene activation.30,52 However, the strength of overexpression is influenced by the proximity of PTA binding to the transcription start site (TSS). Systematic screens determined that binding in the upstream region close to the TSS induces the strongest gene activation, although to unpredictable levels (Fig. 2b).6,26
Targeting the dCas9-VP160 activator using sgRNAs binding the region 300 bp upstream of the TSS of IL1RN, SOX2, and OCT4 was most effective, whereas using sgRNAs binding the region downstream of the TSS had adverse effects on activation.26 Similarly, the most potent sgRNAs for dCas9-VP64 targeted a window 147–89 bp upstream of the TSS of the mouse OCT4 gene.53 For the SunTag activator, sgRNAs showing the highest activation bound 400–50 bp upstream of the TSS.52 For the targeting of the SAM activator, the strongest induction of expression was obtained with sgRNAs located within 200 bp upstream of the TSS of 12 human genes.33
In yeast cells, robust activation was obtained when dCas9-VPR was targeted within 400 bp upstream of the start codon, yet the efficiency decreased when the sgRNAs were located within 20 bp of the TATA box.54 In rice plants, binding of dCas9-TV to the region within 300 bp of the TSS of OsWOX11 and OsYUC1 yielded the strongest gene activation responses.55 Similar targeting windows were found for TALE-based PTAs.53 Consequently, hitting this “sweet spot,” a couple of hundred base pairs upstream of the TSS, should be a general consideration when designing optimal sgRNAs. It is unknown how optimal targeting sites will covary with PTA designs, an important point when considering studies that compare diverse PTA designs at a single binding site.
Gene to gene variability in relative efficiency
dCas9-VP64-mediated activation of gene expression in the human cell line HEK293T was observed using one sgRNA for some genes, while others required the synergistic activity of pooled sgRNAs.31 A similar gene-to-gene variability in the relative strength of transcriptional activation was observed with dCas9-VPR and SAM systems.13,33 The chromatin state around the promoter was thought to cause this variability, but open chromatin, as determined by the presence of DNase I hypersensitivity sites, is not a prerequisite for gene activation.31
A negative correlation exists between basal gene expression and relative fold-change in activation conferred by CRISPR-Cas activators (Fig. 2c).13,33,56 In animal cells, this correlation was observed in all dCas9-based activators tested, dCas9-VP64, dCas9-VPR, SAM, and SunTag.56 Likewise, in Drosophila, the ability of dCas9-VPR to activate transcription of a given gene depended on its basal expression level.57 In plants, a similar correlation exists for genes activated by dCas9-TV.14
Even though the relative change in gene expression induced by most PTAs will be larger for weakly induced genes, the absolute transcription rate that a gene may reach depends on the system used.56 Thus, when activating a gene, its steady-state rate should be taken into account and the use of multiple sgRNAs considered when designing PTAs to achieve maximum gene expression.
Multiplexing
One unique advantage of CRISPR-based PTAs is the ease of reprogramming the target site. This property allows for multiplexed gene activation by directing a single PTA to different targets via multiple coexpressed sgRNAs (Fig. 2d). Multiplexed regulation was first reported using dCas9-VP160. Transfection of dCas9-VP160 and sgRNAs targeting SOX2, IL1RN, and OCT4 led to the concurrent activation of each gene.26 Furthermore, the relative activation of the three genes could be modulated by changing the dosage of the individual sgRNAs.26
Multiplexed gene activation of up to 10 genes, using 10 sgRNAs, was achieved using the SAM system in mammalian cells.33 However, this led to a global reduction in the absolute activation levels of each gene.33 In addition, the relative activation efficiencies among genes change in single- versus multiplexed assays.33 A similar reduction in overall activation was observed in the multiplexed activation of three genes, twist, snail, and engrailed using dCas9-VPR in Drosophila cells.57
These studies suggest that the concentration of dCas9 sets the upper limit for transactivation, and this resource is allocated between the various sgRNAs expressed in the cell. In contrast, another study did not find differences in efficiency between single and multiplexed activation of six target genes with dCas9-VP64, dCas9-VPR, SunTag, or the SAM system.56 However, variability in the levels of basal expression of target genes makes direct comparisons of these experiments challenging.56 Multiplexing CRISPR-based PTAs can be exploited for the manipulation of metabolic pathways and for the rewiring of gene expression networks to yield complex phenotypes.34
Specificity
The specificity of CRISPR-Cas PTAs is a major concern for their use in living systems. Unintended gene expression changes caused by off-target binding may cause undesired effects and may even lead to reduced fitness or survival. Off-target effects have been documented with the Cas9 nuclease, which induced cleavage of up to five off-target sites in the human genome.58,59
In contrast, transcriptome-wide analysis by RNAseq demonstrated that CRISPR-Cas-based PTAs are very specific with little off-target effects. In human cells, gene activation mediated by the relatively weak activators dCas9-VP64 and dCas9-VP160 resulted in activation of only the target genes even when multiple sgRNAs were used.26,52,56
Transcriptomic analysis of animal cells expressing second-generation activators dCas9-VPR, SAM, and SunTag demonstrates the specificity of these PTAs even when higher target gene expression levels are achieved.33,56 In plants, direct activation of RPL23 by dCas9-TV was shown to be specific, but a few nontarget genes were indirectly induced because of RPL23 expression.14 Similarly, activation of FLOWERING WAGENINGEN (FWA) by the SunTag system in the model plant Arabidopsis resulted in specific activation of FWA with only a few other upregulated genes.60
In Drosophila, RNA sequencing revealed misregulation of many genes besides the target genes, twist and snail, activated by dCas9-VPR. Because twist and snail are transcription factors themselves, some of the misregulated genes may represent direct and indirect targets and not result from off-target PTA activation.57
The apparent specificity of CRISPR-Cas activators may be due to the fact that they need to be targeted to a couple of hundred base pairs upstream of the TSS for maximum efficiency (Fig. 2e) (see Position Effects section). Most of the possible off-target binding sites simply will not produce a measurable phenotype.
When bound to a target site, the influence of CRISPR-Cas activators in the transcription of the surrounding genomic regions is limited. Analysis of 112 genes using a SunTag screen combined with single-cell RNA-sequencing (Perturb-Seq) showed that activation of a target gene does not affect the expression of neighboring genes unless they share promoter regions.61 In addition, the dCas9-VP64 ribonucleoprotein complex appears to be sensitive to guide-target mismatches; it can tolerate only three mismatches at its binding positions, while off-target sites for Cas9 nucleases may contain up to five.6,58 Even with these considerations, the careful design of sgRNAs that provide strong transcriptional activation with minimal off-target sites will be the best way to provide a potent and specific induction of the desired genes.
Applications of CRISPR-Cas Activators
The advantages provided by dCas9-based PTAs have been leveraged in genetic screens and the creation of novel therapies. Most of the development of novel transcriptional activators, especially in mammals, has been carried out using transient transfection in cell lines. The ability to activate gene expression combined with the generation of genome-wide sgRNA libraries has allowed for the generation of novel gain-of-function (GOF) screens. In addition, in vivo applications using CRISPR-Cas have been developed in whole multicellular organisms.
Genome-wide screenings using CRISPR-Cas activators
Among the early applications of dCas9-based PTAs is the development of genome-wide activation screens using sgRNA libraries (CRISPRa libraries) to identify genes whose overexpression will confer a phenotype easily scored in a high-throughput manner.33,52 sgRNA enrichment in cells displaying the phenotype-of-interest is measured via high-throughput sequencing and used to identify the target GOF genes.52
Using the SAM system, Konermann et al. designed a screen to identify genes whose overexpression protects A375 malignant melanoma cells from cell cycle arrest and apoptosis induced by the BRAF inhibitor, PLX-4720.33 The lentiviral expression library designed for this screen, consisting of three sgRNA2.0s per every coding isoform (23,430 isoforms) of the human RefSeq database, was transformed into A375 cells expressing dCas9-VP64 and MCP-p65-HSF1. After selection in PLX-4720, enriched sgRNAs were sequenced and their corresponding target genes identified. These target genes, 13 of which were independently validated, were shown to correspond to known but also novel targets of PLX-4720.33
Another GOF genome-wide screen used a library composed of sgRNAs binding to 10 sites upstream of the TSS of 15,977 human genes.52 After this library was transformed into K562 human cells expressing the SunTag system, one screen was used to determine enrichment of sgRNAs before and after 10 days of growth, and another after exposure to a chimeric cholera/diphtheria fusion toxin (CTx-DTA).52 Thus, while one screen identified genes affecting cell growth, the other identified genes modulating the response to the CTx-DTA toxin.52 Among the overexpressed genes that cause growth suppression were tumor suppression genes, transcription factors involved in tissue development and differentiation, and mitotic genes.52
Similar screens using genome-wide CRISPRa libraries have been performed to identify overexpressed genes promoting neuronal differentiation, reprogramming of somatic cells to induced pluripotent stem cells, and conferring resistance to the anticancer drug imatinib.62–64
In contrast, other screens have made use of specific libraries to activate a specific subset of transcripts.65,66 For instance, a CRISPRa library designed to activate all putative cell surface proteins was used to find extracellular receptors recognizing ligands of interest.66 Another CRISPRa library targeting long-noncoding RNAs (lncRNAs) was used to find overexpressed lncRNAs conferring resistance to the PLX-4720 analogue, vemurafenib.65 Interestingly, when some of these GOF genome-wide screens were paired with loss-of-function CRISPR interference (CRISPRi) screens, both provided complementary and comprehensive insights toward the phenotypes interrogated.52
CRISPRa/i screens allow for the systematic identification of individual genes associated with the phenotypes of interest. However, a biological phenotype often results from synergistic interactions of combinations of genes rather than the summed activity of individual genes. Thus, a genetic interaction (GI) between two genes will result in a deviation of the expected phenotypes resulting from simply adding their phenotypic effects.67
To measure interactions between the genes identified in CRISPRa screens, additional GI libraries have been generated and tested. To find GIs among 19 genes identified in a screen for factors promoting neuronal differentiation, Liu and colleagues developed a combinatorial CRISPRa gene activation library. The lentiviral library consisted of a combination of paired sgRNAs, validated in the previous screen, each of which will activate a target gene to two levels (high or low). To allow comparisons between the effects of activating a single gene versus pairs, paired sgRNAs in which one will target a gene and the other will not were also included in the library.62 This screen not only identified positive and negative interactions between genes promoting neuron formation, but also sheds light on the role that expression levels play in the intensity of the GI.62 It also allowed for the discovery of gene pairs that can readily induce differentiation of fibroblasts into neurons.62
For a screen of GI modulating resistance to the drug imatinib, two orthogonal CRISPR-Cas9 systems, one producing gene knock outs (Staphylococcus aureus Cas9 nuclease) and the other activating gene expression (SunTag system using S. pyogenes dCas9), were used. The library design contained pairwise sgRNAs, one for SaCas9 nuclease and other for the SunTag system, so that one of the genes will be activated while the other knocked out. It also contained sgRNA pairs that will knock out or activate a single gene, while not perturbing the other.63
The sgRNAs in this library target 87 genes identified in a previous screen for imatinib resistance and 1327 genes involved in cancer-associated pathways combined in a total of 100,000 pairwise combinations.63 As expected, a set of positive and negative GIs were seen. However, by evaluating the range of interactions resulting from combining gene activation and knock outs, this screen was able to determine the direction of the GI between pairs of genes, thereby allowing the inference of more complex interaction maps than those obtained with pairwise CRISPRa libraries.63
While initial GOF library screens were successful in identifying genes involved in the cellular processes analyzed, improvements in sgRNA design may produce libraries with enhanced performance and result in more robust screens. In the first two CRISPRa libraries described, for each gene, multiple sgRNAs were designed to target a couple of hundred base pairs upstream of the TSS.33,52
However, to obtain libraries with improved sgRNA activity, a machine learning approach was used that created a predictive model incorporating nucleosome positioning, sequence features, and improved sgRNA design rules.68 This model allowed the design of more active sgRNAs that in turn were used to construct an improved version (CRISPRa v2) of the library previously used by Gilbert and colleagues (CRISPR v1).68 When tested, the CRISPRa v2 library was shown to identify 540 genes affecting cell growth, whereas a similar screen with CRISPR v1 identified only 283 genes.68 Furthermore, the sgRNAs in the CRISPRa v2 library were shown to be more active than in CRISPR v1.
Another optimized library was created by using two approaches to modify an SAM-like system.69 First, a novel guide, tracr-v14, was designed that includes two MS2 and two PP7 stem loops, thereby increasing the flexibility in the use of recruiting domains. Second, a narrow window (150–75 nucleotides upstream of the TSS) in which sgRNA activity was predicted to be highest was used for targeting.69
The resulting CRISPRa library, named Calabrese, was tested using a screen for resistance to a BRAF inhibitor, similar to the one used by Konermann and colleagues.69 The screen performed with the Calabrese library was able to identify previously known and also novel genes that when activated confer resistance to vemurafenib. In addition, the number of genes identified using the Calabrese library was substantially larger than those identified in a similar screen performed by Konermann and colleagues using the SAM library.69
Thus, as suggested by these studies, refinement of the sgRNA design rules and accurate gene annotation, especially TSS and nucleosome positioning, are crucial for the design and construction of CRISPRa libraries with significantly increased activation potential.
Use of CRISPR-Cas activators in in vivo systems
CRISPR-Cas activators and their cognate sgRNAs have also allowed for the development of novel gene and cellular therapies. Some of these in vivo studies involve either injecting transfected cells or viral constructs containing the activators and sgRNAs into adult organisms or making use of transgenic animals to express the components of the PTA system in the desired organs or tissues (or a combination of both).
For instance, dCas9-VP64 was used in mice to identify genes that affect the sensitivity to the DNA-damage inducing chemotherapy drug, temozolomide (TMZ). B cell lymphoblastic leukemia cells (B-ALL) transfected with dCas9-VP64 and sgRNAs targeting the Mgmt (O6-methylguanine–DNA methyltransferase) gene were shown to be more resistant than control cells to TMZ treatments after lymphoma transplantation in adult mice.70 Resistance of B-ALL cells to TMZ treatment required activation of Mgmt, a gene involved in the detoxification of TMZ-induced DNA damage. Using this system, a screen was designed to test for gene regulators of the DNA damage response that mediate sensitivity to TMZ.70
A GOF screen using the SunTag system was developed to study the effect of gene activation in a mouse liver injury and repopulation model. In this screen, the dCas9-GCN4 component of SunTag was expressed as a Cre-inducible transgene in mice. Then, to activate gene expression, the activator component (ScFv-VP64), Cre recombinase, and sgRNAs were delivered by injection before induction of liver injury. After liver repopulation, high-throughput sequencing determined which sgRNAs, and by extension which target genes, promote cell proliferation and the formation of hepatocellular carcinomas.71 A similar approach using SPH, a SunTag-like system that uses p65-HSF1 instead of VP64 as an AD, was used for the in vivo multiplexed activation of genes in the brain.72
Another GOF screen, this time for genes whose transcriptional activation induced heart failure in mice, was also recently developed. This system combined the expression of dCas9-VPR driven by the Myh6 (myosin heavy chain) promoter in the heart of transgenic mice with the injection of adeno-associated virus (AAV) vectors containing sgRNAs to induce gene expression of target genes specifically in cardiomyocytes.73
An antitumor therapy called multiplexed activation of endogenous genes as an immunotherapy (MAEGI) was developed using PTAs. In MAEGI, the SAM system was used to induce the expression of endogenous genes in tumors, some of which encode tumor antigens that enhance antitumor immune responses.74 To induce the multiplexed expression of tumor genes, a genome-scale or customized tumor-specific sgRNA2.0 library, including the MCP-p65-HSF1 module, was created in AAV vectors. Intratumoral injection of AAV libraries in dCas9-VP64 expressing mice bearing orthotopic tumors led to increased tumor remission. Both libraries elicit a potent and specific antitumor immune response that showcases the therapeutic efficacy of MAEGI.74
In another study, the SAM system was used in rats for multiplexed activation of two osteogenesis-promoting genes, Wnt10b and Forkhead c2 (Foxc2), in bone mesenchymal stem cells (BMSCs). These BMSCs, with activated Wnt10b and Foxc2, when implanted in gelatin scaffolds to calvarial bone defects were able to greatly enhance bone healing.75
Therapies to correct pathologies caused by haploinsufficiency, due to the loss-of-function of one gene copy, have been designed using CRISPR-Cas activators to increase expression of the remaining wild-type copy.76 The proof of concept of this therapy was applied to either Single-minded homologue 1 (Sim1) or Melanocortin 4 receptor (Mc4r) haploinsufficient mice (Sim1+/− and Mc4r+/−, respectively) that develop obesity phenotypes.
Expression of dCas9-VP64 and a single sgRNA targeting either the Sim1 promoter or the Sim1 candidate enhancer 2 in transgenic mice increased the expression of Sim1 to levels high enough to correct the obesity phenotype of Sim1+/−. Even though dCas9-VP64 was expressed constitutively, it only increased transcription in tissues in which Sim1 was already expressed, suggesting a need for a tissue-specific transcription factor for Sim1 expression.
As an alternative approach, dCas9-VP64 and sgRNAs, targeting Sim1 or Mc4r, were delivered using AAV vectors injected directly to the tissues in which the target genes are active, the hypothalamus in this case. AAV-mediated delivery of the PTA system led to increased expression of the target genes and mice with long-lasting reduced weight, suggesting the feasibility of this approach to treat haploinsufficiency.76
In Drosophila, the first study of CRISPR-Cas activators in transgenic flies was carried out using dCas9-VP64 and dCas9-VPR activated by the Gal4-UAS system and two sgRNAs (sgRNA-wg) to target the wingless (wg) gene. Constitutive expression of dCas9-VP64 or dCas9-VPR was not toxic in transgenic flies. Furthermore, when dCas9-VPR was expressed together with the two sgRNA-wg, ectopic Wg production was observed accompanied by morphological abnormalities reminiscent of wg overexpression phenotypes.57 In contrast, dCas9-VP64 was not able to activate wg expression, which is consistent with results obtained from cell lines.57
Ewen-Campen et al. also used dCas9-VPR and sgRNAs to activate several genes in transgenic Drosophila.77 Out of 36 target genes, they observed activation of 27 (75%) although to a consistently lower expression than that obtained by expressing cDNAs of the same genes driven by the Gal4-UAS system.77 Despite the lower activation levels, several of the predicted overexpression phenotypes were observed.77
Attempts to use the SAM system in Drosophila transgenic lines were motivated by previous results showing better performance than other activators in cell lines.56,77 Expression of the SAM system in transgenic flies resulted in lethality that was avoided when its AD was replaced by ADs previously used in Drosophila.77 However, none of these modified SAM systems performed better than dCas9-VPR in vivo. One of the reasons for the lethality of SAM was believed to be the high expression levels achieved in initial constructs, because even dCas9-VPR was shown to be toxic when expressed at higher levels.77
With these considerations, the SAM system was modified by expressing from a weaker promoter dCas9-VP64 in tandem with MCP-p65-HSF1, separated by a T2A self-cleaving peptide. This system, dubbed FlySAM, is not lethal or toxic when expressed in transgenic Drosophila and performs significantly better than dCas9-VPR at activating endogenous genes.78 Phenotypes induced by FlySAM, even with the use of a single sgRNA2.0, are similar in severity to those produced by the Gal4-UAS overexpression systems.78
Inspired by the success of dCas9-VPR and FlySAM, Zirin et al. developed The Transgenic RNAi Project-CRISPR overexpression (TRiP-OE) collection of Drosophila lines.79 Each TRiP-OE stock expresses either sgRNAs or sgRNA2.0s for FlySAM, binding to the region upstream of the TSS of a target gene. Gene activation of the target gene is initiated by crossing the sgRNAs or sgRNA2.0s containing stocks with another harboring the dCas9-VPR or FlySAM activators, respectively.79 There are also some stocks that express sgRNA2.0 together with FlySAM under the UAS control that are activated by crossing the lines with the Gal4-UAS activating system.79
In the nematode Caenorhabditis elegans, dCas9-VP160 was tested as an activator to induce expression of endogenous genes. Expression of dCas9-VP160 and single sgRNAs targeting dbl-1 (a transforming growth factor-β family gene) in transgenic nematodes did not provide any target gene activation. Only when expressing the PTAs together with six sgRNAs targeting the dbl-1 promoter was a modest increase in gene expression observed. The level of activation was, however, enough to induce the elongated body length phenotype expected from dbl-1 overexpression.80
In plants, a few PTAs and target genes have been tested for activation in vivo. Transgenic plants expressing dCas9-VP64 and three sgRNAs targeting either PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1) or the microRNA gene, miR319, were made in the model plant Arabidopsis thaliana.81 Although these plants showed modest activation of the target genes (up to sevenfold increase), no phenotype was observed in any of the plants suggesting that higher expression levels may be needed to induce an overexpression phenotype.81
SAM and SAM-like systems have also been used to induce gene expression in transgenic plants. The SAM system with two sgRNAs2.0s was used in Arabidopsis to individually activate the expression of two endogenous genes, PAP1 and ARABIDOPSIS VACUOLAR H+-PYROPHOSPHATASE (AVP1). SAM was capable of inducing only moderate gene expression in both target genes, but robust enough to display overexpression phenotypes.82 An SAM-like system, in which the p65-HSF1 activator was replaced by VP64, was designed independently by two groups and used to generate Arabidopsis transgenic plants. One of these SAM-like systems, namely CRISPR-Act2.0, was used for the targeted overexpression of PAP1 and Fertilization-Independent Seed2 (FIS2), whereas the other SAM-like system was used to target the flowering-promoting gene FLOWERING LOCUST T (FT).83,84
Both SAM-like systems were able to induce gene expression of their target genes by one to two orders of magnitude. However, only the expected early flowering phenotype resulting from the activation of FT using the SAM-like system was reported.83
Other CRISPR-Cas systems have been used to generate transgenic plants. As shown in the dCas9-TV section, dCas9-TV was used to increase the expression levels of receptor RLP23 leading to plants with enhanced immune response.14 The SunTag system was used in Arabidopsis to activate the expression of three genes, FWA, CLAVATA3 (CLV3), and APETALA3 (AP3). SunTag-mediated activation of these genes reached several hundred-fold transcript levels compared with wild-type, and phenotypes associated with the overexpression of FWA and CLV3 were observed in transgenic plants.60
Thus far, to our knowledge, there has not been any reports on the toxicity or lethality associated with the expression of CRISPR-Cas activators in plants, even when expressed from strong constitutive promoters. However, protein instability from the expression of dCas9-TV designs with an increased number of AD repeats, and toxic effects specifically associated with the activation of PAP1 in Arabidopsis have been observed.14,82 Thus, in comparison with Drosophila, plants appear to be more resilient to the constitutive expression of activators.
Future Remarks
The development of CRISPR-Cas-based PTAs capable of robust and specific gene induction and with the ease of multiplexing provides the means to design genetic screens to interrogate biological systems, especially transcriptional networks. Novel biotechnologies based on the rewiring of transcriptional programs have emerged from these developments, such as the metabolic engineering of organisms.34,54,85,86 PTAs may also lead to the development of therapies based on the corrective activation of genes or pathways that fail to express during disease. In insects, PTAs have been used to engineer synthetic speciation events, an approach that could improve the biocontainment of transgenes and lead to novel biocontrol approaches for pests and disease vectors.87
Continuous development of new activator architectures that allow even further activation levels will pave the way for new applications. Furthermore, combination of these activators with other CRISPR-Cas effectors such as repressors or chromatin modulators will exponentially increase the range of applications in which they could be used. However, more research is needed in the performance of these activators in in vivo systems, especially in vertebrates. In addition, deep knowledge of the transcriptional networks involved in the processes of interest is required to efficiently deploy PTAs to obtain the desired outcomes.
Authorship Confirmation Statement
All authors contributed to the writing and revision of the article. The final version of the article was approved by all authors.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
J.A.C.M. and M.J.S. are part of a team supported by the Advanced Plant Technologies program, DARPA Award HR001118C0146. M.H.Z. is supported by an NIH NIGMS Biotechnology Training grant NIHT32GM008347.
References
- 1. Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911 DOI: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. DOI: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 3. Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. DOI: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. DOI: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. DOI: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. DOI: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Green MR. Eukaryotic transcription activation: Right on target. Mol Cell. 2005;18:399–402. DOI: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 8. Petrenko N, Jin Y, Dong L, et al. Requirements for RNA polymerase II preinitiation complex formation in vivo. Elife. 2019;8:e43654 DOI: 10.7554/eLife.43654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allen BL, Taatjes DJ. The Mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. DOI: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boija A, Klein IA, Sabari BR, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855 e1816. DOI: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sadowski I, Ma J, Triezenberg S, et al. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. DOI: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 12. Beerli RR, Segal DJ, Dreier B, et al. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci U S A. 1998;95:14628–14633. DOI: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. DOI: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Z, Zhang D, Xiong X, et al. A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants. 2017;3:930–936. DOI: 10.1038/s41477-017-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmitz ML, Baeuerle PA. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 1991;10:3805–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hardwick JM, Tse L, Applegren N, et al. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Green M, Schuetz TJ, Sullivan EK, et al. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol Cell Biol. 1995;15:3354–3362. DOI: 10.1128/mcb.15.6.3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1997;94:1035–1040. DOI: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tiwari SB, Belachew A, Ma SF, et al. The EDLL motif: A potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 2012;70:855–865. DOI: 10.1111/j.1365-313X.2012.04935.x. [DOI] [PubMed] [Google Scholar]
- 20. Young JK, Gasior SL, Jones S, et al. The repurposing of type I-E CRISPR-Cascade for gene activation in plants. Commun Biol. 2019;2:383 DOI: 10.1038/s42003-019-0637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu W, Yang B, Chittoor JM, et al. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Mol Plant Microbe Interact. 1998;11:824–832. DOI: 10.1094/MPMI.1998.11.8.824. [DOI] [PubMed] [Google Scholar]
- 22. Sigler PB. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988;333:210–212. DOI: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 23. Babu MM. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44:1185–1200. DOI: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kwon I, Kato M, Xiang S, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. DOI: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boehning M, Dugast-Darzacq C, Rankovic M, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833–840. DOI: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- 26. Cheng AW, Wang H, Yang H, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. DOI: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013;2:604–613. DOI: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. DOI: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kearns NA, Genga RM, Enuameh MS, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. DOI: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maeder ML, Linder SJ, Cascio VM, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. DOI: 10.1038/nmeth.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. DOI: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piatek A, Ali Z, Baazim H, et al. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13:578–589. DOI: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- 33. Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. DOI: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zalatan JG, Lee ME, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. DOI: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tanenbaum ME, Gilbert LA, Qi LS, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. DOI: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kunii A, Hara Y, Takenaga M, et al. Three-component repurposed technology for enhanced expression: Highly accumulable transcriptional activators via branched tag arrays. CRISPR J. 2018;1:337–347. DOI: 10.1089/crispr.2018.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. DOI: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang X, Wang J, Cheng Q, et al. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3:17018 DOI: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tak YE, Kleinstiver BP, Nunez JK, et al. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods. 2017;14:1163–1166. DOI: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, Xu L, Fan R, et al. Genetic editing and interrogation with Cpf1 and caged truncated pre-tRNA-like crRNA in mammalian cells. Cell Discov. 2018;4:36 DOI: 10.1038/s41421-018-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang X, Wang W, Shan L, et al. Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell. 2018;9:380–383. DOI: 10.1007/s13238-017-0491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nihongaki Y, Otabe T, Ueda Y, et al. A split CRISPR-Cpf1 platform for inducible genome editing and gene activation. Nat Chem Biol. 2019;15:882–888. DOI: 10.1038/s41589-019-0338-y. [DOI] [PubMed] [Google Scholar]
- 43. Kim D, Kim J, Hur JK, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863–868. DOI: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 44. Kleinstiver BP, Tsai SQ, Prew MS, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. DOI: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh D, Mallon J, Poddar A, et al. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a). Proc Natl Acad Sci U S A. 2018;115:5444–5449. DOI: 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zetsche B, Heidenreich M, Mohanraju P, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35:31–34. DOI: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. DOI: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anderson GM, Freytag SO. Synergistic activation of a human promoter in vivo by transcription factor Sp1. Mol Cell Biol. 1991;11:1935–1943. DOI: 10.1128/mcb.11.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carey M, Lin YS, Green MR, et al. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. DOI: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 50. Pettersson M, Schaffner W. Synergistic activation of transcription by multiple binding sites for NF-kappa B even in absence of co-operative factor binding to DNA. J Mol Biol. 1990;214:373–380. DOI: 10.1016/0022-2836(90)90187-q. [DOI] [PubMed] [Google Scholar]
- 51. Perez-Pinera P, Ousterout DG, Brunger JM, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. DOI: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. DOI: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu J, Lei Y, Wong WK, et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res. 2014;42:4375–4390. DOI: 10.1093/nar/gku109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deaner M, Alper HS. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab Eng. 2017;40:14–22. DOI: 10.1016/j.ymben.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 55. Gong X, Zhang T, Xing J, et al. Positional effects on efficiency of CRISPR/Cas9-based transcriptional activation in rice plants. aBIOTECH. 2020;1:1–5. DOI: 10.1007/s42994-019-00007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chavez A, Tuttle M, Pruitt BW, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–567. DOI: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lin S, Ewen-Campen B, Ni X, et al. In vivo transcriptional activation using CRISPR/Cas9 in drosophila. Genetics. 2015;201:433–442. DOI: 10.1534/genetics.115.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. DOI: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pattanayak V, Lin S, Guilinger JP, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. DOI: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Papikian A, Liu W, Gallego-Bartolome J, et al. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun. 2019;10:729 DOI: 10.1038/s41467-019-08736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Norman TM, Horlbeck MA, Replogle JM, et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science. 2019;365:786–793. DOI: 10.1126/science.aax4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y, Yu C, Daley TP, et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell. 2018;23:758–771 e758. DOI: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boettcher M, Tian R, Blau JA, et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat Biotechnol. 2018;36:170–178. DOI: 10.1038/nbt.4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang J, Rajan SS, Friedrich MJ, et al. Genome-Scale CRISPRa Screen Identifies Novel Factors for Cellular Reprogramming. Stem Cell Reports. 2019;12:757–771. DOI: 10.1016/j.stemcr.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Joung J, Engreitz JM, Konermann S, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. DOI: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chong ZS, Ohnishi S, Yusa K, et al. Pooled extracellular receptor-ligand interaction screening using CRISPR activation. Genome Biol. 2018;19:205 DOI: 10.1186/s13059-018-1581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ramkumar P, Kampmann M. CRISPR-based genetic interaction maps inform therapeutic strategies in cancer. Transl Cancer Res. 2018;7:S61-S67. DOI: 10.21037/tcr.2018.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Horlbeck MA, Gilbert LA, Villalta JE, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016;5 DOI: 10.7554/eLife.19760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sanson KR, Hanna RE, Hegde M, et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9:5416 DOI: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Braun CJ, Bruno PM, Horlbeck MA, et al. Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc Natl Acad Sci U S A. 2016;113:E3892–E3900. DOI: 10.1073/pnas.1600582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wangensteen KJ, Wang YJ, Dou Z, et al. Combinatorial genetics in liver repopulation and carcinogenesis with a in vivo CRISPR activation platform. Hepatology. 2018;68:663–676. DOI: 10.1002/hep.29626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhou H, Liu J, Zhou C, et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci. 2018;21:440–446. DOI: 10.1038/s41593-017-0060-6. [DOI] [PubMed] [Google Scholar]
- 73. Schoger E, Carroll KJ, Iyer LM, et al. CRISPR-mediated activation of endogenous gene expression in the postnatal heart. Circ Res. 2020;126:6–24. DOI: 10.1161/CIRCRESAHA.118.314522. [DOI] [PubMed] [Google Scholar]
- 74. Wang G, Chow RD, Bai Z, et al. Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity. Nat Immunol. 2019;20:1494–1505. DOI: 10.1038/s41590-019-0500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hsu MN, Huang KL, Yu FJ, et al. Coactivation of endogenous Wnt10b and Foxc2 by CRISPR activation enhances BMSC osteogenesis and promotes calvarial bone regeneration. Mol Ther. 2020;28:441–451. DOI: 10.1016/j.ymthe.2019.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Matharu N, Rattanasopha S, Tamura S, et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science. 2019;363 DOI: 10.1126/science.aau0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ewen-Campen B, Yang-Zhou D, Fernandes VR, et al. Optimized strategy for in vivo Cas9-activation in Drosophila. Proc Natl Acad Sci U S A. 2017;114:9409–9414. DOI: 10.1073/pnas.1707635114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jia Y, Xu RG, Ren X, et al. Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc Natl Acad Sci U S A. 2018;115:4719–4724. DOI: 10.1073/pnas.1800677115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zirin J, Hu Y, Liu L, et al. Large-scale transgenic drosophila resource collections for loss- and gain-of-function studies. Genetics. 2020;214:755–767. DOI: 10.1534/genetics.119.302964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Long L, Guo H, Yao D, et al. Regulation of transcriptionally active genes via the catalytically inactive Cas9 in C. elegans and D. rerio. Cell Res. 2015;25:638–641. DOI: 10.1038/cr.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lowder LG, Zhang D, Baltes NJ, et al. A CRISPR/Cas9 Toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169:971–985. DOI: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park JJ, Dempewolf E, Zhang W, et al. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS One. 2017;12:e0179410 DOI: 10.1371/journal.pone.0179410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee JE, Neumann M, Duro DI, et al. CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS One. 2019;14:e0222778 DOI: 10.1371/journal.pone.0222778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lowder LG, Zhou J, Zhang Y, et al. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act Systems. Mol Plant. 2018;11:245–256. DOI: 10.1016/j.molp.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 85. Jensen ED, Ferreira R, Jakociunas T, et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb Cell Fact. 2017;16:46 DOI: 10.1186/s12934-017-0664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lian J, HamediRad M, Hu S, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat Commun. 2017;8:1688 DOI: 10.1038/s41467-017-01695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Maselko M, Feltman N, Upadhyay A, et al. Engineering multiple species-like genetic incompatibilities in insects. Nat Commun. 2020;11:4468 DOI: 10.1038/s41467-020-18348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Selma S, Bernabe-Orts JM, Vazquez-Vilar M, et al. Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol J. 2019;17:1703–1705. DOI: 10.1111/pbi.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]