ABSTRACT
Therapeutically intervening the function of RNA in vivo remains a big challenge. We here developed an exosome-based strategy to deliver engineered RNA-binding protein for the purpose of recruiting specific RNA to the lysosomes for degradation. As a proof-of-principle study, RNA-binding protein HuR was fused to the C-terminus of Lamp2b, a membrane protein localized in both exosome and lysosome. The fusion protein was able to be incorporated into the exosomes. Moreover, exosomes engineered with Lamp2b-HuR successfully decreased the abundance of RNA targets possibly via lysosome-mediated degradation, especially when the exosomes were acidified. The system was specifically effective in macrophages, which are lysosome enriched and resistant to routine transfection mediated RNAi strategy. In the CCl4-induced liver injury mouse model, we found that delivery of acidified exosomes engineered with Lamp2b-HuR significantly reduced liver fibrosis, together with decreased miR-155 and other inflammatory genes. In summary, the established exosome-based RNA-binding protein delivery strategy, namely “exosome-mediated lysosomal clearance”, takes the advantage of exosome in targeted delivery and holds great promise in regulating a set of genes in vivo.
KEYWORDS: HuR, Lamp2b, exosomes, lysosome, protein delivery
Introduction
RNA metabolism is a stepwise process, including transcription, pre-mRNA processing, mRNA export, subcellular localization, translation and degradation. Specific degradation of target mRNAs holds great importance for gene therapy. Up to now, there are few effective strategies to degrade the RNA in vivo, except the RNAi. However, sometimes RNAi is not effective as expected in the lysosome-enriched cells, especially when they are delivered via nanoparticles.
Exosomes are a kind of extracellular micro-vesicles with a diameter range of 30–150 nm, emerging as rational drug carriers. Recently, exosomes have been engineered towards better targeting specificity and larger loading capacity via modulating the exosome surface proteins [1,2]. Exosomes carry different types of cargos, such as RNAs, proteins and small molecule drugs, and release these cargos in the recipient cell. Together, these data indicate that exosomes could serve as a good carrier for protein delivery, either on the exosome surface or inside the exosome. Besides, there are also a great percentage of endocytosed exosomes destined to the lysosome and undergo degradation.
In this study, we repurposed the exosome for delivering engineered RNA-binding protein. By proof-of-principle experiments, we fused RNA-binding protein HuR to the Lamp2b protein, which is also known as CD107b (Cluster of Differentiation 107b), encodes one of the lysosome-associated membrane glycoproteins [3]. The Lamp2b-HuR fused protein functionalized exosomes were able to drive the RNA targets recognized by the RNA-binding motif to the lysosome for degradation in the recipient cells, acting as a dominant-negative analogue of the endogenous HuR. The system is specifically effective in lysosome-enriched macrophages and holds as an alternative strategy for RNA knockdown.
Materials and methods
Plasmid construction
Lamp2b alone or fusion protein was cloned into pcDNA 3.1(-) similarly as described before [2]. For Lamp2b-empty vector construction, the Lamp2b CDS was amplified using corresponding primers, followed by direct clone into pcDNA 3.1(-). For HuR-Lamp2b vector construction, Xho1 and BspE1 cloning sites were first introduced to the targeting region in the N terminus of Lamp2b by two-step cloning. HuR cDNA without stop codon was then amplified by PCR and inserted into the Lamp2b N terminus using Xho1 and BspE1. For Lamp2b-HuR vector construction, the HuR cDNA with stop codon was fused to the C-terminus of the Lamp2b CDS without stop codon. All the clones were confirmed by sequencing and the right clones were stored for following application. PCR primers used in this study are listed in Supplementary Table 1.
Cell culture
HEK293T cells were cultured in DMEM (Dulbecco's Modified Eagle Medium) medium (Logan, Utah, USA) containing 10% foetal bovine serum and 1% penicillin-streptomycin (Logan, Utah, USA). RAW 264.7 macrophages were cultured in RPMI medium (Logan, Utah, USA) with 10% FBS and 1% antibiotics. Cells were maintained at 37°C in a 5% CO2. Lysosomal function was inhibited by treatment of chloroquine (Sigma) at the dose of 50 µM.
To produce exosomes engineered with HuR-Lamp2b or Lamp2b-HuR, HEK293T cells were transfected with the corresponding vectors referred above with Lipofectamine 2000 (Invitrogen) as instructed. Six hours post transfection, cell culture medium were replaced with the growth medium supplemented with FBS without exosomes. Then, exosomes from the culture medium were isolated as described below.
For in vivo experiments, exosomes of mouse origin were used. Briefly, lentivirus was used to infect mouse mesenchymal stem cells to get fusion protein modified exosomes of mouse origin. For lentivirus packaging, lentivirus vectors expressing the fusion proteins were constructed by subcloning the insert from corresponding pcDNA3.1 plasmids referred above to pWPI vector. The expressing lentivirus vectors were transfected into HEK293T cells together with another two plasmids psPAX2 and pMD2G with Lipofectamine 2000 (Invitrogen) with the ratio of 10:5:1. Lentivirus particles were collected from the medium supernatant filtered through a 0.45-μm filter (Millipore) 48-h post transfection and stored at −80°C before use.
Exosome isolation and characterization
For the isolation of exosomes from cell culture, cells were maintained in the serum-free medium. Cell supernatants were centrifuged at 500 g for 10 min to remove cells and then at 10,000 g for 20 min to eliminate residual cellular debris. The resulting supernatants were then filtered through 0.22 μm filters, followed by exosome precipitation with Exoquick-TCTM as instructed. The isolated exosomes were re-suspended in PBS or DMEM and stored at −80°C and used after “Exosome RNA unloading”.
The isolated exosomes were uniformly diluted to 500 ng/ml and the size distribution was analysed by Nanosight (Malvern, UK). The morphology of isolated exosomes was analysed by electron microscopy. Briefly, the exosomes were added onto the grid. The exosomes were then stained (2% uranyl acetate) and imaged by the electron microscope (JEM-2000EX TEM, JEOL Ltd., Tokyo, Japan).
Exosome RNA unloading by acidification
To remove the encapsulated RNAs in the exosomes, 200 µg exosomes (protein concentration) were diluted in 400 µl PBS solution, with the pH value adjusted to 4. Then, the exosomes were electroporated in a 4 mm cuvette at 400 V and 125 µF for 4–5 pulses to remove the RNAs. Following electroporation, exosomes were further examined by TEM as described above.
Exosome capture with antibodies
To explore the orientation of HuR on the exosome surface, anti-HuR antibody was first incubated with protein A/G conjugated Magnetic Beads (#16-663, Sigma), and the isolated exosomes with indicated modifications in PBS were further incubated with the above antibody/beads complex for 2 h. The following immunoprecipitation process was performed as instructed. The pulled-down complex was then subjected to RIPA lysis buffer, followed by western blot analysis of the expression of Lamp2b.
To confirm the Lamp2b/HuR fusion expression in the exosomes, exosomes were first captured with anti-CD9 antibody similar as described above. The fusion protein expression was analysed by western blot analysis of the expression of HuR in the lysates of the exosomes.
RNA immunoprecipitation
For confirmation of the interaction between miR-155 and endogenous HuR, HEK293T cell lysates (Pierce™ IP Lysis Buffer, ThermoFisher) were used to incubate with IgG or anti-HuR, followed by precipitation with Protein A agarose beads. After washing out unbound miRNAs with PBS for three times, the precipitated RNA was isolated with TRIzol® reagent and miR-155 and negative control miRNAs were detected by qRT-PCR.
To explore whether different HuR/Lamp2b fusion proteins bind miR-155 similarly, HEK293T cells were forced to express Lamp2b-HuR or HuR-Lamp2b. The cell lysates were then incubated with anti-HuR, followed by precipitation with Protein A agarose beads. After washing out unbound RNAs with PBS for three times, the precipitated RNA was isolated with TRIzol® reagent and miR-155 abundance was detected by qRT-PCR.
Western blotting
Total protein of indicated cells or isolated exosomes was extracted using RIPA Lysis Buffer (Beijing, China) at 4°C for 30 min. Protein concentration was determined by PierceTM BCA Protein Assay Kit. Proteins were then concentrated on SDS-PAGE (6%) and separated by SDS-PAGE (12%), and transferred to nitrocellulose filter membranes. After blocking with 3% BSA, membranes were subsequently incubated with primary antibody anti-HuR (sc5261, Santa Cruz), anti-GM130 (11308-1-AP, Proteintech), anti-TSG101 (ab83, Abcam), anti-CD9 (ab92726, Abcam), anti-Lamp2b (ab18529, Abcam) and anti-GAPDH (D110016-0100, BBI Life Sciences). After washing three times in TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h.
qRT-PCR
Total RNA was extracted using TRIzol® reagent (Invitrogen, USA). miRNA and mRNA were reversely transcribed by miRcute Plus miRNA Synthesis Kit and Transcriptor First-strand cDNA Synthesis Kit (Roche), respectively, following the manufacturer’s protocol. qPCR reactions (20 μl) were performed by FastStart Essential DNA Green Master. MiR-155 and mRNA were normalized to U6 levels and β-actin, respectively. Relative expression was calculated by the 2−ddCt method. The sequences of PCR primers are provided in Supplementary Table 1.
Flow cytometry analysis
RAW264.7 macrophages incubated with 40 μg/ml (final concentration) indicated exosomes were additionally treated with Lipopolysaccharide (1 mg/ml) or IL-4 (20 ng/ml) for 24 h. Then, the cells were harvested and incubated with F4/80 antibody (Biolegend, 123120), anti-mouse CD11b (Biolegend, 101212), and anti-mouse CD86 (Biolegend, 105007) or CD206 (Biolegend, 141706) at 4°C for 30 min. Cells were then washed twice with flow-cytometry buffer, followed by analysis with BD FACSCalibur. Flow cytometry data were analysed by Flow Jo software.
Exosome labelling
Exosomes (1 µg/µL) were labelled with DiI, DiO or DiR by incubating with the dyes (1 mM) at the ratio of (500:1 in volume) for 30 min, followed by exosome isolation as described above. For visualization of the exosomes by confocal microscope, DiO or DiI-labelled exosomes were used. For ex vivo fluorescence tracing of exosomes, DiR-labelled exosomes were used.
Animal experiment
All animal experiments were carried out under protocols approved by the Animal Care and Use Committee of Fourth Military Medical University. Male C56BL/6 mice (8–10 weeks old, 22–25 g) were used to monitor the in vivo distribution and effects of modified exosomes.
For ex vivo fluorescence tracing of exosomes, mice were injected with 200 μg of DiR-labelled exosomes via the tail vein. The localization of the exosomes in different organs was detected by the IVIS® Lumina II system (PerkinElmer, Thermo Fisher, US).
CCL4 induced mouse liver fibrosis model was used to test the therapeutic effects of acidified exosomes. Briefly, male C57BL/6 mice were injected with CCL4 (0.6 ml/kg of body weight, intraperitoneally) diluted in corn oil every the fourth day. For exosome-mediated therapy, acidified control or Lamp2b-HuR exosomes were injected 24 h after the every CCL4 injection. At the end of the experiment, mice were sacrificed to harvest the tissues. Sections of formalin-fixed livers were stained with Sirius red staining for fibrosis analysis. RNA extracted from fresh liver tissues was reverse transcribed for qPCR analysis of the candidate genes.
Immunofluorescence analysis
Cells cultured in confocal dish were treated with DiO labelled exosomes for 6 h, followed by three times of PBS wash before further staining. Lysosomes were specially stained using LysoTracker Red (Thermo Fisher) according to the manufacturer’s instruction. The nuclei were counter-stained with Hoechst (1:1000 diluted in PBS). For tissue sections, tissues were fixed for 15 min by 4% paraformaldehyde and sections were stained with Hoechst (Invitrogen) to label the cell nuclei. Images were obtained by laser scanning confocal microscope (Nikon A1R, Tokyo, Japan).
Statistical analysis
Data are expressed as mean±SEM. Student’s t-test was used for two group comparison, and one-way ANOVA was used to compare the differences between groups while multiple comparisons were performed by Tukey’s post hoc test (Graphpad Prism 7.0). P values of <0.05 indicate statistical significance.
Results
Characterization of Lamp2b/HuR fusion protein modified exosomes
As a proof-of-principle study of RNA-binding protein delivery, HuR was selected due to its well-known function in multiple diseases [4]. Previous study has established that the tag would be exposed to the exosome surface when it is fused to Lamp2b N-terminus, and it would be localized inside of exosome when it is fused to the Lamp2b C-terminus [1]. To this end, it is reasonable to deduce that HuR would be exposed to the exosome surface when it is fused to the Lamp2b N terminus, and HuR would be inside the exosome when it is fused to the Lamp2b C terminus (Figure 1a,b). Thus, corresponding expression vectors (namely Lamp2b-empty, Lamp2b-HuR, and HuR-Lamp2b) were constructed and transfected into HEK293T cells. Western blot assay revealed that both fusion proteins were robustly expressed in the HEK293T cells and the derived exosomes (Figure 1c). Experiments from Corso et al. 2019 have shown that after transient transfections the proteins might contaminate the vesicle isolations [5]. To further confirm whether the exosomes carried HuR fusion protein, exosomes were captured by anti-CD9 antibody, and the exosome lysates were subjected to Western blot assay. The fusion protein HuR-Lamp2b and Lamp2b-HuR were detected in the exosomes derived from HuR-Lamp2b and Lamp2b-HuR-transfected cells (Supplementary Figure 1), suggesting that both Lamp2b fusion strategies help HuR fusion protein be loaded into exosomes.
Figure 1.

Exosome engineering with Lamp2b-HuR fusion protein.
(a) Schematic representation of membrane localization of the fusion protein, in which HuR is designed to fuse to N or C terminus of Lamp2b. (b) Plasmid structures of the corresponding fusion protein. The restriction endonuclease sites used also indicated. (c) Western blot analysis of the expression of fusion protein in both the parental cells and derived exosomes with indicated transfection. The fusion protein was identified with anti-HuR antibody with the band size of 147 kd. GAPDH served as the loading control. Representative data of three different experiments. (d) Size distribution of the exosomes from cells expressing indicated fusion protein or control Lamp2b were analysed by NanoSight. (e) Representative transmission electron microscope image of indicated functionalized exosomes. Scale bar = 100 nm. (f) Western blot analysis of the inclusive and exclusive exosomal markers. Representative image of three different experiments. (g) ExosLamp2b, ExosLamp2b-HuR, ExosHuR-Lamp2b were first captured by anti-HuR antibody. The exosomes could be only captured when HuR was exposed outside the exosomes. ExosHuR-Lamp2b were successfully captured, suggesting that HuR localized outside of the exosomes when fused at the N-terminus. Representative image of three different experiments.
Nanoparticle tracking analysis (NTA) (Figure 1d) and transmission electron microscopy (TEM) (Figure 1e) showed that Lamp2b, Lamp2b-HuR and HuR-Lamp2b modified exosomes were physically similar, with a size diameter ranging between 100 and 200 nm. Further analysis of the exosomal inclusive markers (CD9 and TSG101) and exclusive marker (GM130) by western blot additionally confirmed that Lamp2b/HuR fusion protein did not change the characteristics of the exosomes (Figure 1f). Moreover, pulldown assay with anti-HuR antibody revealed that the HuR was exposed to the exosomal surface only when it was fused to the N-terminus of Lamp2b, as expected (Figure 1g).
Lamp2b-HuR fusion protein modified exosomes represses M1 polarization in vitro
Consistent with previous finding that HuR plays an important role in regulating macrophage polarization [6], transfection of HuR promotes M1 marker gene expression in RAW264.7 (Supplementary Figure 2a). In addition, HuR transfection also results in increased expression of miR-155, a known promoter of M1 (Supplementary Figure 2b)[6]. Mechanistically, HuR directly interacted with miR-155, while had no obvious interaction with the control miR-27 and miR-186, as revealed by RNA-IP analysis (Supplementary Figure 2c). Moreover, fusion with Lamp2b did not change the binding ability of HuR with miR-155, as similar amount of miR-155 could be immunoprecipitated with Lamp2b-HuR or HuR-Lamp2b fusion protein (Supplementary Figure 2d).
In view of the above data that Lamp2b/HuR fusion protein was efficiently loaded into exosomes and they are effective in binding miR-155, we next asked whether the fusion protein could recruit the target RNA to the lysosome when endocytosed by the recipient cells. To reduce the influence of exosomal RNA from the parental cells, electroporation at acidified condition was employed (Supplementary Figure 3a). As expected, electroporation at acidified condition significantly reduced the RNA cargos in the exosomes as seen from both the abundance of total RNA and miR-155 (Supplementary Figure 3b, c), making delivery of exosomes with few RNA cargos possible. Notably, previous study has revealed that electroporation lead to RNA aggregate formation and siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles [7]. In contrast, we here found that electroporation in acid condition helps the RNA removal from the exosomes, which could be explained by the facts the RNAs were protected in the exosomes and the acid theoretically penetrated into EVs slowly. Both prevent the destruction of EVs and aggregation of RNAs. TEM and Nanoparticle tracking analysis further revealed that acidification plus electroporation did not change the morphology and size distribution of the exosomes (Supplementary Figure 3d, e).
In the following experiments, we examined the function of the acidified/fusion protein engineered exosomes. RAW264.7 cells were incubated with Lamp2b-empty, Lamp2b-HuR, and HuR-Lamp2b engineered exosomes, namely ExosLamp2b, ExosLamp2b-HuR, ExosHuR-Lamp2b, either unacidified or acidified (Figure 2a). Compared with the ExosLamp2b, ExosLamp2b-HuR efficiently reduced miR-155 expression (Figure 2b) and M1 polarization (Figure 2c), while ExosHuR-Lamp2b exosomes had no obvious effects on miR-155 expression and M1 polarization. The possible explanation should be that HuR exposure to the cytoplasm during endosomal sequestration only happened in the Lamp2b-HuR exosomes (Figure 2d). Acidification of the exosomes further increased the effects of the ExosLamp2b-HuR on reducing the expression of miR-155 and M1 marker Tnfα (Figure 2b,c). Notably, acidification of the exosomes did not change the endocytosis efficiency of the exosomes by RAW264.7 cells (Supplementary Figure 4). The explanation should be that a great proportion of HuR in the fusion protein Lamp2b-HuR has been already occupied by the endogenous RNA targets during the exosome biogenesis in the donor cells. Subsequently, the pre-occupied HuR has no additional capacity to bind RNA targets in the recipient cells when it is exposed to the cytoplasm during endosomal sequestration (Figure 2d). In contrast, acidification removes the endogenous cargos, making more empty HuR available to bind and thus reduce RNA targets (such as miR-155) in the recipient cells (Figure 2d). It is important to note that the possibility that acidification of the exosomes might promote the lysosome-mediated degradation could not be excluded.
Figure 2.
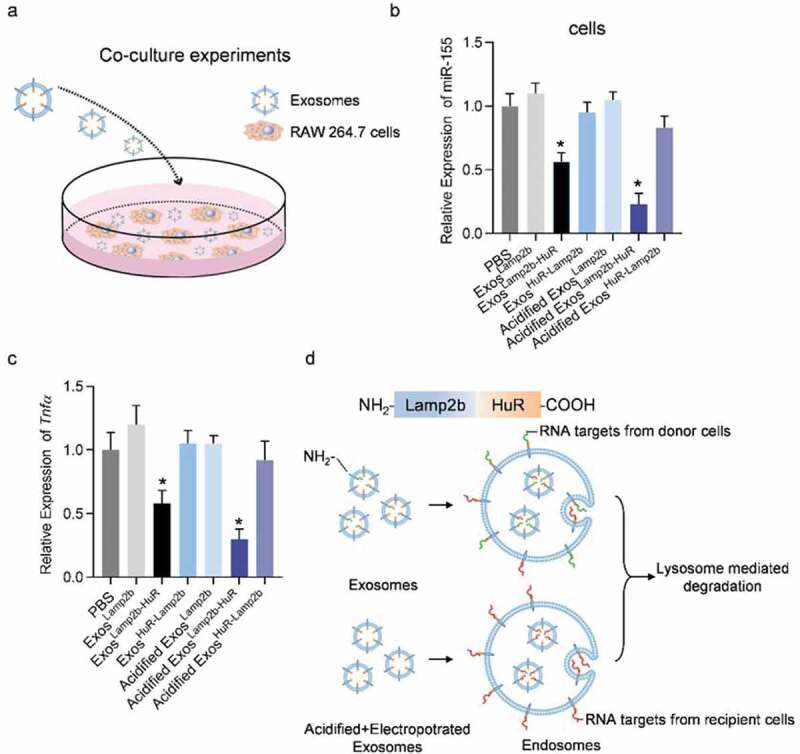
Acidified ExosLamp2b-HuR reduce endogenous miR-155.
(a) Schematic illustration of the procedure how engineered exosomes added into the RAW264.7 cells. (b) Expression of miR-155 in RAW264.7 cells receiving exosomes as indicated. U6b served as an internal control. Data are expressed as mean±SEM of three different experiments. *, p 0.05. (c) Expression of inflammatory genes in RAW264.7 cells receiving exosomes as indicated. β-actin served as an internal control. Data are expressed as mean±SEM of three biological replicates. *, p 0.05. (d) Schematic illustration of the conformational change of the fusion protein supposed to happen in the donor cells. A great proportion of the HuR has been already occupied by the RNA from the donor cells, limiting its capacity to bind additional RNA targets in the recipient cells. In contrast, acidification removes the endogenous cargos, making more free HuR available to bind and thus reduce RNA targets in the recipient cells.
The above data raise the possibility that exosome-mediated delivery of Lamp2b-HuR drives the endogenous RNA targets to the lysosome. To confirm the lysosome localization of the exosomes, exosomes were labelled with DiO, followed by incubation with RAW264.7 macrophages or HEK293T cells. As expected, colocalization of DiO-labelled (green) exosomes and lysosomes (red tracked by lysotracker) were visualized in RAW264.7 cells (Figure 4a). In contrast, there were few exosomes destinated into lysosomes in HEK293T cells, which had much fewer lysosomes (Supplementary Figure 5). Moreover, lysosome inhibitor chloroquine treatment significantly blocked the effects of ExosLamp2b-HuR on miR-155 (Figure 4b). Together, these data suggest that ExosLamp2b-HuR decreases HuR mRNA target abundance at least partially via lysosome-mediated degradation.
Figure 4.
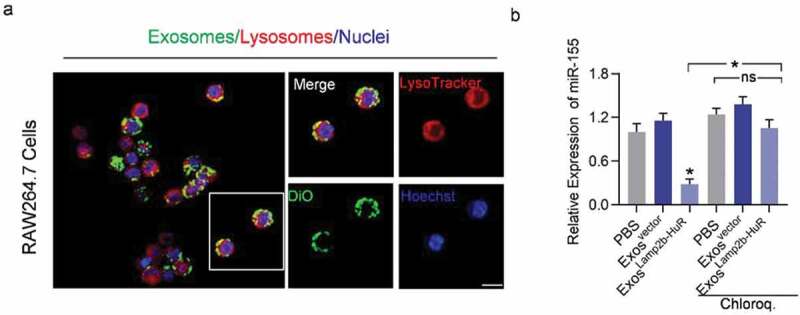
Acidified ExosLamp2b-HuR are destined to the lysosomes in macrophage RAW264.7.
(a) Exosomes were labelled with DiO and incubated with RAW264.7. Cells were stained with Lysotracker and counterstained with Hoechst. Images were taken under immunofluorescence microscope. (b) RAW264.7 cells were treated with control or chloroquine, followed by exosome treatments. Expression of miR-155 was examined 24 h later by qPCR. Data are expressed as mean±SEM of three different experiments. *, p< 0.05. Scale bar = 5 µm.
To benchmark the role of acidified ExosLamp2b-HuR in regulating M1/M2 polarization, the effects of the modified exosomes and miR-155 inhibitor were compared in LPS induced M1 polarization cell model. As expected, unacidified ExosLamp2b-HuR slightly reduced miR-155 expression and repressed M1 polarization, while acidified ExosLamp2b-HuR had a much stronger effect in miR-155 reduction and M1 polarization repression, to a similar extent as miR-155 antagonist (Figure 3a–d and Supplementary Figure 6a–c).
Figure 3.
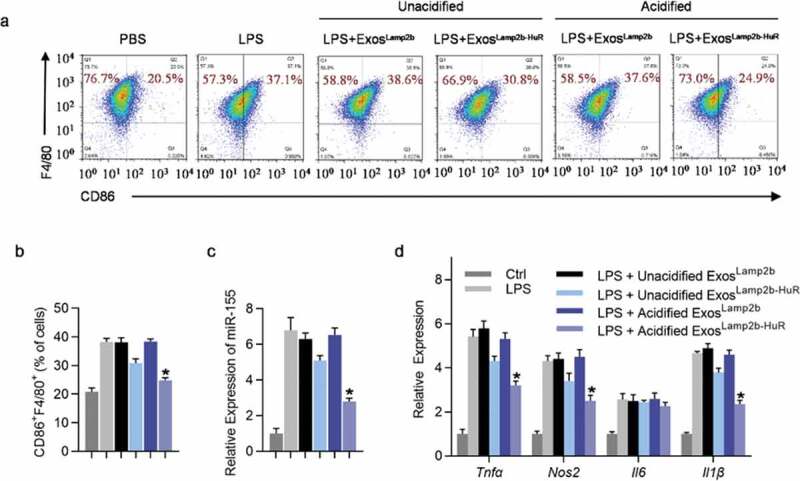
Acidified ExosLamp2b-HuR regulate M1/M2 polarization.
(a, b) Flow cytometry analysis (a) and quantification (b) of the effects of indicated exosomes on the RAW264.7 M1 polarization induced by LPS. Representative image of three biological replicates. (c-d) qPCR analysis of miR-155 (c) and M1 marker genes (d) in RAW264.7 cells treated as above. U6 and β-actin served as internal controls, respectively. Data are expressed as mean±SEM of three different experiments. *, p 0.05.
Acidified ExosLamp2b-HuR decrease target RNA expression in vivo
In the following experiments, we explored the ability of acidified ExosLamp2b-HuR to degrade RNA targets in vivo. Acidified ExosLamp2b-HuR were labelled with DiR or DiI before injection into wildtype mice via tail vein. Ex vivo imaging and immunofluorescent microscope analysis revealed that the engineered exosomes were mainly localized in the reticuloendothelial systems, such as liver, spleen, and lung (Figure 5a,b). Consistent with the role of ExosLamp2b-HuR in reducing miR-155 expression in vitro, in vivo injection of ExosLamp2b-HuR also reduced miR-155 expression in the liver and spleen significantly (Figure 5c). In contrast, no obvious change was found in the lung or kidney, where no obvious exosomes distributed (Figure 5c). These data suggest that the current strategy is ready for antagonizing HuR function in the liver and spleen.
Figure 5.
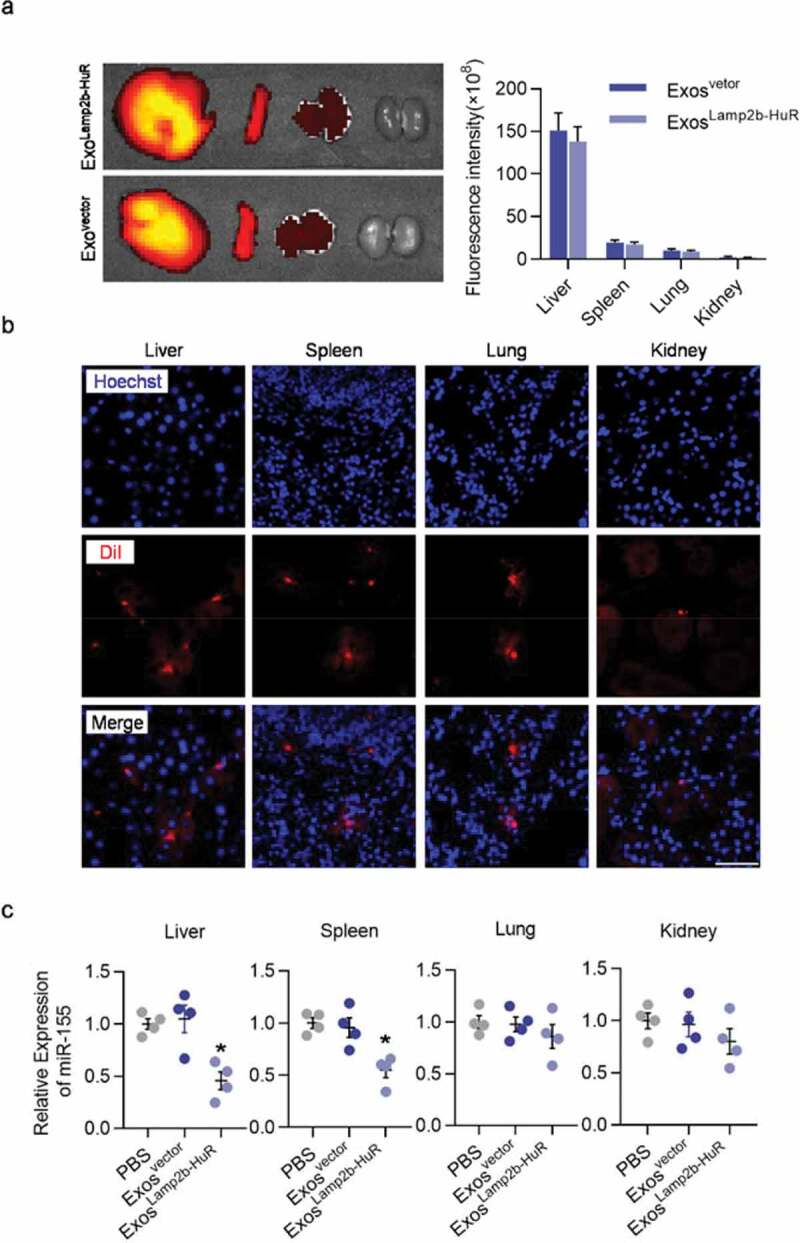
Distribution and function of acidified ExosLamp2b-HuR in vivo.
(a, b) Exosomes from empty vector or Lamp2b-HuR transfected mouse fibroblasts were acidified and labelled with DiI or DiR. About 100 µg (at protein level) DiI or DiR labelled exosomes were injected (in 100 µL) via tail vein. Liver and spleen dominant localization of the engineered exosomes as revealed by ex vivo bioluminescence imaging (a, DiR labelled exosomes) and immunofluorescence microscope of tissue sections (b, DiI labelled exosomes). Representative images of at least three mice. Scale bar = 50 µm. (c) Expression of miR-155 in different tissues from mice treated with control or ExosLamp2b-HuR as indicated. Data are presented as mean±SEM. n = 3. *, p 0.05.
Acidified ExosLamp2b-HuR alleviate CCl4-induced liver injury
Previous researches have confirmed that the pro-inflammatory miR-155 is a detrimental promoting factor of hepatitis and liver fibrosis, and therapeutically delivering miR-155 inhibitor would be beneficial for hepatitis prevention [8–10]. In the following experiments, we explored whether delivering acidified Exo-Lamp2b-HuR could be beneficial for liver fibrosis prevention by decreasing miR-155.
Liver injury was induced by injection of CCL4 every the fourth day. For exosome-mediated therapy, exosomes of mouse origin were included, which were derived from mouse mesenchymal stem cells infected with corresponding lentiviruses. Acidified control or Lamp2b-HuR modified exosomes were injected 24 h after the every CCL4 injection (Figure 6a). At the end of the experiment, mice were sacrificed to harvest the tissues (Figure 6a). RNA extracted from fresh liver tissues was reverse transcribed for qPCR analysis of the candidate genes. As expected, endogenous expression of miR-155 was significantly decreased by acidified ExosLamp2b-HuR treatment (Figure 6b). Consistently, expression level of inflammatory and fibrogenic genes, such as Tnfα, Mcp1, and Col1a1, were also significantly reduced in CCl4 mice additionally receiving acidified ExosLamp2b-HuR treatment (Figure 6c–e). Sirius red staining further revealed that acidified ExosLamp2b-HuR treatment alleviated the fibrosis in CCL4 mouse model efficiently (Figure 6f,g).
Figure 6.
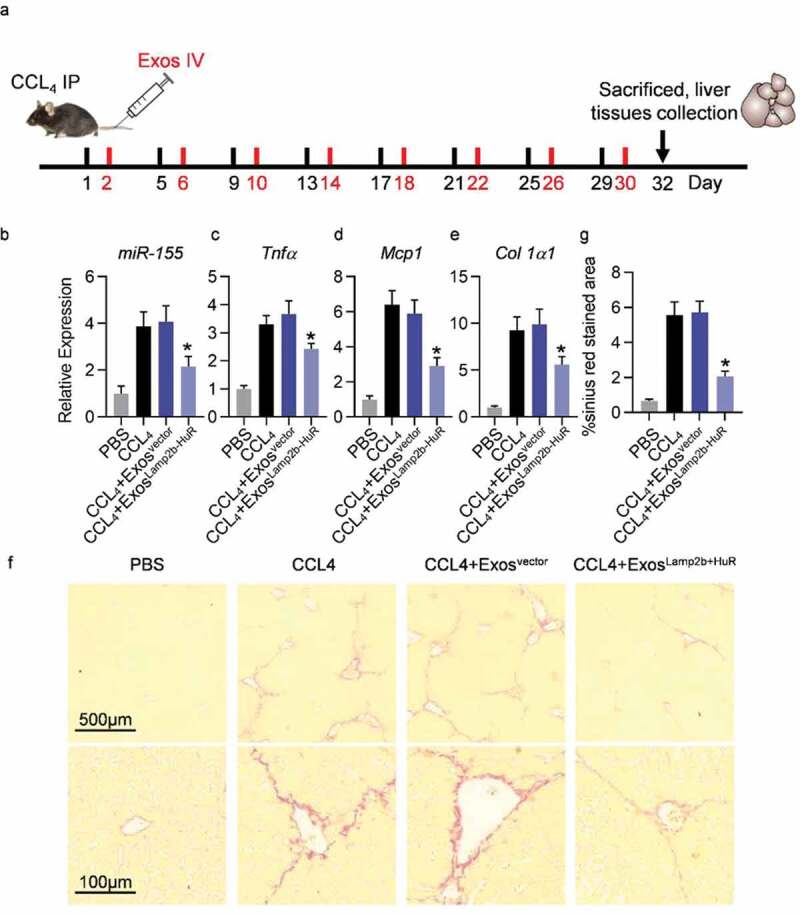
Acidified ExosLamp2b-HuR effectively alleviate CCL4-induced liver injury.
(a) Schematic representation of the experimental procedure. (b-d) Endogenous expression of miR-155 (b), Tnfα (c), Mcp1 (d), and Col1a1 (e) in livers from mice receiving indicated treatments. Livers were harvested 48 h after the last exosome injection. U6 or β-actin served as internal control, respectively. Data are expressed as mean±SEM of three different experiments. *, p 0.05. (f) Sirius red staining of the livers from mice receiving indicated treatments. Mice were treated with control or CCL4 for 4 weeks. About 200 µg indicated exosomes (at protein level) were injected 24 h after each CCL4 treatment. Livers were harvested 48 h after the last exosome injection for Sirius red staining. n = 5 mice in each group. (g) Quantification data of panel f.
Discussion
As a proof-of-principle study, we here developed an RNA-binding protein delivery strategy by fusing HuR to the N-terminus or the C-terminus of Lamp2b. The fusion protein was efficiently incorporated into the exosomes and the resultant exosomes successfully deliver the fusion protein to the recipient cells. Moreover, the fusion protein Lamp2b-HuR could drive the RNA targets towards the lysosome for degradation when the endosome fused with the lysosome (Figure 7). In vitro and in vivo experiments revealed that the system exerts a dominant-negative effect on endogenous HuR in macrophages, which are lysosome enriched and resistant to routine transfection mediated RNAi strategy.
Figure 7.
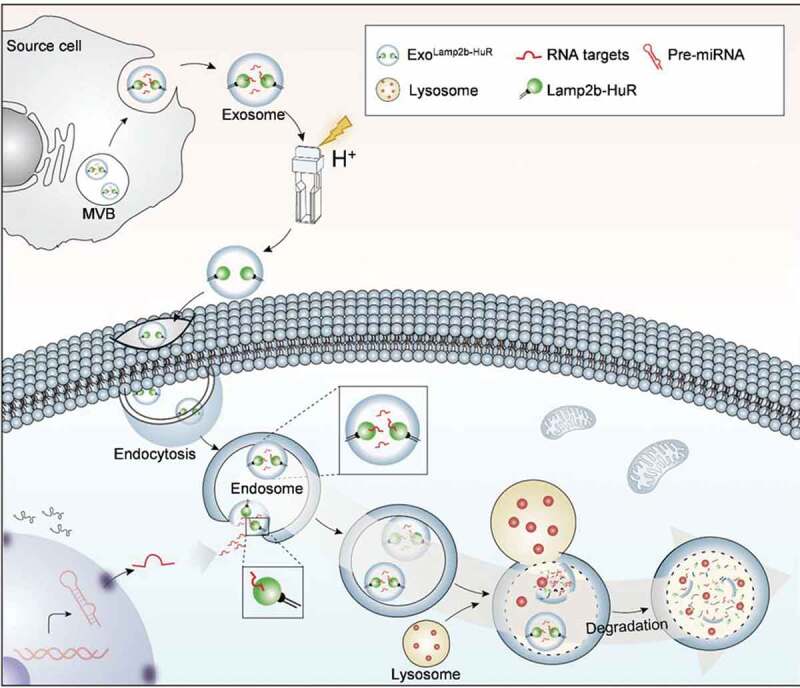
Working model of the engineered exosomes.
Exosomes are engineered with Lamp2b-HuR fusion protein in the donor cells. The exosomes are further acidified to remove the endogenous cargo. The exosomes are efficiently endocytosed by the macrophages in liver, spleen, etc. In the recipient cells, Lamp2b-HuR, as a surface protein on the endosome, drives the target RNAs of HuR to degradation when endosomes fused with the lysosomes.
Aberrant gene expression and regulation are the main causes of most diseases at the molecular level. Therapeutically targeting those aberrance holds promise for the disease treatment. Intracellular protein delivery is an important tool for both therapeutic and fundamental applications. Effective protein delivery faces two major challenges: efficient cellular uptake and avoiding endosomal sequestration [11]. We report here a general strategy for direct delivery of RNA-binding proteins bypassing the two challenges, via harnessing the advantages of exosomes and endosome sequestration. On one hand, exosomes, natural membrane vesicles, have been considered as potential vehicles to deliver miRNA, mRNA sand protein in vivo, due to their advantages in efficient taken up by different cells, especially the reticuloendothelial system (RES) [12]. On the other hand, different from other systems, we here harnessed the effects of endosomal sequestration and lysosome degradation on endocytosed nanoparticles. The strategy thus could be a generalized method for similar purposes.
Intracellular delivery of RNAi and its analogue miRNA mimics via nanoparticles or virus vectors serve as the main strategy to reduce the gene expression. Similar as the protein delivery, it is also widely established that escaping the lysosome-mediated degradation of the endocytosed nucleic acids are essential for optimum efficiency, which is especially true for macrophages and other lysosome-enriched cell types. Development of alternative strategies to knockdown genes overcoming the lysosome hurdle is urgently needed, as the macrophage is increasingly recognized as the cell target for multiple-complicated diseases. Numerous reports have shown that exosomes are highly efficient in delivering exogenous RNAs (siRNAs and miRNAs) to target tissues/cells in vivo [1,13,14]. Once endocytosed, the cargos inside the exosomes could be either released into the cytoplasm or degraded when the exosomes fused with the lysosomes [15,16]. We here identified that exosome-delivered Lamp2b-HuR might sequester on the endosome and thus recruit miRNA-155, followed by degradation when the endosome fused to lysosomes. It is interesting to note that HuR-Lamp2b fusion had much weaker effects. Besides the different orientation in the endosome, posttranslational modifications of HuR might be another reason, as HuR in HuR-Lamp2b locates around the signal peptide where additional modification, such as glycosylation, usually occurs [2]. As Lamp2b is a protein-enriched in lysosome membrane [17], it is thus reasonable to deduce that the free form of Lamp2b-HuR might be also recruit RNA targets to the lysosomes directly. Anyway, the strategy sheds light on an alternative strategy to reduce RNA expression independent of RNAi. RNA-binding proteins interact with target mRNA via the corresponding elements in the untranslated region (UTR) [18]. Combinatory fusion of different RNA-binding motifs would probably improve the specificity of the current system.
RNA-binding proteins (RBPs) are key components in RNA metabolism, regulating the temporal, spatial and functional dynamics of RNAs [19]. RNA-binding protein activities are highly regulated through protein levels, intracellular localization, and post-translation modifications. The impact of altered RNA-binding protein activities also affects human diseases, either due to gain-of-function or loss-of-function. Taking HuR for example, aberrant HuR activity has been found in many diseases, such as cancer, atherosclerosis, myocardial infarction, and inflammation [20–24]. Therapeutically targeting RNA-binding protein function and activity hold promise for aberrant RNA metabolism-related pathogenesis.
Uninterested RNA cargos in the exosome would produce unexpected off-target effects, and thus developing strategies to remove these RNAs is of great significance. We here found that electroporation at acidified condition could reduce the RNA cargos to some extent. Moreover, the strategy didn’t change the exosome morphology and endocytosis ability. Thus, the referred treatment would open the door for loading therapeutic RNAs. How to load the therapeutic RNAs, especially those long lncRNAs or mRNAs, into acidified exosomes, is of great interest and worth further study. It is also interesting to test whether the modification would affect other processes when endocytosed, such as the escape from the lysosomes.
In summary, we here have established an exosome-based RNA-binding protein delivery strategy, namely “exosome-mediated lysosomal clearance”, which delivers the RNA-binding protein destined to the lysosomes and thus functions with dominant-negative effects on the endogenous analogue. At least, the system could be an alternative for gene knockdown, especially for those cells or genes resistant to the RNAi. It would be promisingly expected that the method can be applied in the prevention of multiple macrophage associated diseases, such as inflammation, atherosclerosis, and even fibrosis. The strategy takes the advantage of exosome in targeted delivery and holds great promise in regulating a set of genes in vivo. In addition, the system could be easily modified by changing the targeting moiety and RNA-binding protein for other purposes.
Supplementary Material
Acknowledgments
This study was funded by NSFC81671690 and 81871357 to Yuan LJ, NSFC 31771507 and 81970737 to Yang GD, Provincial Scientific Foundation of Shaan’Xi (2020TD-038) and Major Clinical Renovation Project, Tangdu Hospital (NO.2013LCYJ003). We are grateful to the technical help to Jing Zhang from the Department of Ultrasound in Tangdu Hospital, Fourth Military Medical University.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–12. [DOI] [PubMed] [Google Scholar]
- [2].Hung ME, Leonard JN.. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290(13):8166–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Endo Y, Furuta A, Nishino I.. Danon disease: a phenotypic expression of LAMP-2 deficiency. Acta Neuropathol. 2015;129(3):391–398. [DOI] [PubMed] [Google Scholar]
- [4].Woodhoo A, Iruarrizaga-Lejarreta M, Beraza N, et al. Human antigen R contributes to hepatic stellate cell activation and liver fibrosis. Hepatology. 2012;56(5):1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Corso G, Heusermann W, Trojer D, et al. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule – single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J Extracell Vesicles. 2019;8(1):1663043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Squadrito ML, Etzrodt M, De Palma M, et al. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013;34(7):350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kooijmans SAA, Stremersch S, Braeckmans K, et al. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J Control Release. 2013;172(1):229–238. [DOI] [PubMed] [Google Scholar]
- [8].Blaya D, Aguilar-Bravo B, Hao F, et al. Expression of microRNA-155 in inflammatory cells modulates liver injury. Hepatology. 2018;68(2):691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bala S, Csak T, Saha B, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. 2016;64(6):1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Csak T, Bala S, Lippai D, et al. MicroRNA-155 deficiency attenuates liver steatosis and fibrosis without reducing inflammation in a mouse model of steatohepatitis. PLoS One. 2015;10(6):e0129251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tang R, Kim CS, Solfiell DJ, et al. Direct delivery of functional proteins and enzymes to the cytosol using nanoparticle-stabilized nanocapsules. ACS Nano. 2013;7(8):6667–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Conlan RS, Pisano S, Oliveira MI, et al. Exosomes as reconfigurable therapeutic systems. Trends Mol Med. 2017;23(7):636–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ohno S-I, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor MicroRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tian Y, Li S, Song J, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. [DOI] [PubMed] [Google Scholar]
- [15].Turturici G, Tinnirello R, Sconzo G, et al. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Physiol. 2014;306(7):C621–33. [DOI] [PubMed] [Google Scholar]
- [16].McKelvey KJ, Powell KL, Ashton AW, et al. Exosomes: mechanisms of uptake. J Circ Biomark. 2015;4(7). doi: 10.5772/61186. eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cheng X-T, Xie Y-X, Zhou B, et al. Revisiting LAMP1 as a marker for degradative autophagy-lysosomal organelles in the nervous system. Autophagy. 2018;14(8):1472–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zagrovic B. Of RNA-binding proteins and their targets: interaction determines expression. Genome Biol. 2014;15(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lukong KE, Chang K-W, Khandjian EW, et al. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–425. [DOI] [PubMed] [Google Scholar]
- [20].Stellos K, Gatsiou A, Stamatelopoulos K, et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med. 2016;22(10):1140–1150. [DOI] [PubMed] [Google Scholar]
- [21].Yashiro T, Nanmoku M, Shimizu M, et al. 5-Aminoimidazole-4-carboxamide ribonucleoside stabilizes low density lipoprotein receptor mRNA in hepatocytes via ERK-dependent HuR binding to an AU-rich element. Atherosclerosis. 2013;226(1):95–101. [DOI] [PubMed] [Google Scholar]
- [22].Gu L, Wang H, Wang J, et al. Reconstitution of HuR-inhibited CUGBP1 expression protects cardiomyocytes from acute myocardial infarction-induced injury. Antioxid Redox Signal. 2017;27(14):1013–1026. [DOI] [PubMed] [Google Scholar]
- [23].Shang J, Zhao Z. Emerging role of HuR in inflammatory response in kidney diseases. Acta Biochim Biophys Sin (Shanghai). 2017;49(9):753–763. [DOI] [PubMed] [Google Scholar]
- [24].Toyota K, Murakami Y, Kondo N, et al. Cytoplasmic Hu-Antigen R (HuR) expression is associated with poor survival in patients with surgically resected cholangiocarcinoma treated with adjuvant gemcitabine-based chemotherapy. Ann Surg Oncol. 2018;25(5):1202–1210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


