Abstract
Background/Aims:
The role of two polymorphisms rs1800591 and rs3816873 of the microsomal triglyceride transfer protein (MTTP) gene in the development of nonalcoholic fatty liver disease (NAFLD) remains controversial. A meta-analysis was conducted to determine the correlation between these MTTP polymorphisms and NAFLD.
Materials and Methods:
A systematic search was carried out using PubMed, Embase, and Cochrane Library to retrieve English studies that reported the relationship between MTTP polymorphisms (rs1800591 and rs3816873) and NAFLD published before February 18, 2020. Odds ratio (OR) and 95% confidence interval (CI) were used to appraise the risk of MTTP polymorphism in NAFLD.
Results:
A total of 10 case-control studies, including 1388 cases and 1690 healthy subjects, were included. No significant correlation between the rs1800591 (G vs. T: OR = 1.08, 95% CI = 0.68–1.70, P = 0.76) and rs3816873 (CT + CC vs. TT: OR = 1.23, 95% CI = 0.76–2.01, P = 0.398) polymorphisms of MTTP and NAFLD was found in any of the models. However, when NASH patients confirmed by liver biopsy were extracted alone for rs1800591 polymorphism analysis, it was found that the G allele significantly increased the risk of NASH under the heterozygote model (GT vs. TT: OR = 3.16, 95% CI = 1.13–8.83, P = 0.028) and dominant model (GT + GG vs. TT: OR = 3.03, 95% CI = 1.13–8.09, P = 0.027).
Conclusion:
The present meta-analysis revealed that the rs1800591 and rs3816873 polymorphisms of the MTTP gene are uncommon in NAFLD. However, the G allele of rs1800591 was more likely to be correlated to NASH susceptibility.
Keywords: Meta-analysis, MTTP, NAFLD, polymorphism
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is one of the major causes of liver disease.[1] The spectrum of NAFLD encompasses two subtypes: nonalcoholic fatty liver (NAFL) and nonalcoholic steatohepatitis (NASH).[2] In the later stages, NAFLD can progress to fibrosis, cirrhosis, and hepatocellular carcinoma (HCC).[3] However, the exact mechanism of its development has not been elucidated. Many pathogenetic factors, including genetic factors, environmental factors, and metabolic factors, have been found to be associated with NAFLD.[4] The risk of NAFLD development and its related complications vary among individuals.[5] The development of genome-wide association studies (GWASs) and high-throughput technologies have allowed for the in-depth research of the genetic factors of NAFLD.[4] Single nucleotide polymorphisms (SNPs) may be used as genetic biomarkers to screen individuals with genetic predisposition to NAFLD.[4] At present, TM6SF2 rs58542926[6] and PNPLA3 rs738409[7] are the widely studied genetic risk variants in NAFLD. These findings can help in developing remedies in the future.[5]
The microsomal triglyceride transfer protein (MTTP) gene is a key protein for lipid excretion from the liver[8] and one of the potential candidate genes found to be associated with NAFLD susceptibility. MTTP plays an important role in the assembly and secretion of very low-density lipoprotein (VLDL) in hepatocytes.[9] More precisely, MTTP catalyzes the transfer of triglycerides to nascent apolipoproteins B (ApoB) in the early stage of lipoprotein assembly. VLDL is produced to remove triglycerides from hepatocytes.[8,10] Abetalipoproteinemia (ABL) is a rare recessive monogenic disease, which is characterized by the lack of Apo B-containing lipoproteins in plasma and hepatic steatosis due to the inhibition of its transcriptional activity through mutations in the coding region of MTTP.[11,12]
Many studies have explored the association between the -493 G/T (rs1800591) and Ile128Thr (rs3816873) polymorphisms of MTTP genes and NAFLD. However, the effect of these polymorphisms on NAFLD remains uncertain due to the inconsistent results of different studies. A Japanese study on rs1800591 polymorphism revealed that the incidence of the G allele and G/G genotype was high in NASH patients. Furthermore, patients with the G/G genotype had an advanced stage of NASH, when compared to those with the G/T genotype.[13] Another study conducted in Iran revealed a significant difference in rs3816873 polymorphism between NAFLD and the control group, and patients with the CT genotype had increased susceptibility for NAFLD.[14] However, a case-control study that involved an Italian population reveals that there was no significant association between the rs1800591 polymorphism and NAFLD, its clinical, or histological characteristics.[15] Similarly, a Chinese study failed to demonstrate any significant association between the rs3816873 polymorphism and NAFLD.[16] Therefore, a meta-analysis that included all eligible studies was conducted to comprehensively determine the relationship between MTTP gene polymorphism and NAFLD.
MATERIALS AND METHODS
Retrieval strategy
Two authors independently retrieved relevant studies from PubMed, Embase, and Cochrane Library databases published before February 18, 2020. Any divergences were resolved by discussion. The keywords used were as follows: (“Non-alcoholic Fatty Liver Disease” or “Non alcoholic Fatty Liver Disease or NAFLD” or “Nonalcoholic Fatty Liver Disease” or “Fatty Liver, Nonalcoholic” or “Fatty Livers, Nonalcoholic” or “Liver, Nonalcoholic Fatty” or “Livers, Nonalcoholic Fatty” or “Nonalcoholic Fatty Liver” or “Nonalcoholic Fatty Livers” or “Nonalcoholic Steatohepatitis” or “Nonalcoholic Steatohepatitides” or “Steatohepatitides, Nonalcoholic” or “Steatohepatitis, Nonalcoholic”) AND (”microsomal triglyceride transfer protein” or “Microsomal triacylglycerol transfer protein” or “MTTP” or “MTP”). In addition, the cited references in the selected articles were also manually searched to obtain more relevant studies.
Selection criteria
The criteria to include studies for the present analysis were as follows: (1) case-control studies that involved adult human subjects (≥18 years old), and were designed to assess the relationship between MTTP gene rs1800591 and rs3816873 polymorphisms and the susceptibility to NAFLD; (2) the criteria for the diagnosis of NAFLD were clearly defined; (3) sufficient data were available to calculate the odds ratio (OR) and 95% confidence interval (CI); and (4) studies published in the English language. Studies were excluded based on the following: (1) the absence of a healthy control study group; (2) duplication of data; and (3) reviews, letters, conference abstracts, and conference papers.
Data extraction and quality assessment of the included studies
Two researchers independently extracted the data and assessed the quality of the studies based on the specified selection criteria, in order to validate the precision of the extracted data. Discrepancies were settled through discussion. The extracted data included the following: the first author of the study, year of publication, ethnicity of the participants, disease types, the number of subjects in the case and control group, the number of each genotype, source of control, Hardy–Weinberg equilibrium (HWE) data, genotype detection methods, and the characteristics of participants.
The quality of studies was assessed using the Newcastle-Ottawa Scale (NOS),[17] which has three aspects: selection of study groups (four items, 0–4 stars); comparability of the groups (one item, 0–2 stars); and ascertain of exposure or outcome (three items, 0–3 stars). According to the evaluation items, the total NOS score ranged from 0 to 9. The higher the score, the better the quality. A research with a score of 6 or more was considered to be of high methodological quality.
Statistical analysis
OR and 95% CI were used to measure the association between the rs1800591 and rs3816873 polymorphisms of the MTTP gene and NAFLD. The statistical significance of the pooled ORs was determined by Z-test. The heterogeneity of the studies was measured using the Cochran's Q and I2 index. Studies with a P value of <0.05 and I2> 50% were considered to have significant heterogeneity, and the fixed-effect model (FEM) was chosen for the analysis. Otherwise, the random-effect model (REM) was used. In order to determine the source of serious heterogeneity, subgroup analyses were implemented. In addition, in order to assess the impact of each study on the overall results and prevent the existence of separate studies leading to the reversal of the pooled results, a sensitivity analysis was performed. Furthermore, Funnel plots, Begg's rank correlation test and Egger's linear regression test were used to detect publication bias in the existing studies. A P value of <0.05 was considered statistically significant. All statistical analyses were performed using the STATA 12.0 (STATA Corp, College Station, TX, USA).
RESULTS
Study selection
A total of 428 studies were retrieved during the literature search from the PubMed (n = 128), Embase (n = 298), and Cochrane Library (n = 2) databases. After the elimination of 105 duplicate publications, 370 additional publications were removed by screening the title and abstract. Among these, 148 articles were reviews, letters, conference abstracts, meta-analysis, editorials, conference papers, short surveys, and notes, while 117 articles focused on animal or in vitro studies. Subsequently, the full texts of 58 studies were read, and 48 studies were further excluded due to the following reasons: other genes or other SNPs of MTTP were studied (n = 10), the study was not relevant to the gene (n = 18), other liver diseases were involved (n = 16), and the study had no available data or involved children (n = 4). Finally, 10 studies were retained for the present meta-analysis.[13,14,15,16,18,19,20,21,22,23] The flowchart for the literature search and selection process is illustrated in Figure 1.
Figure 1.
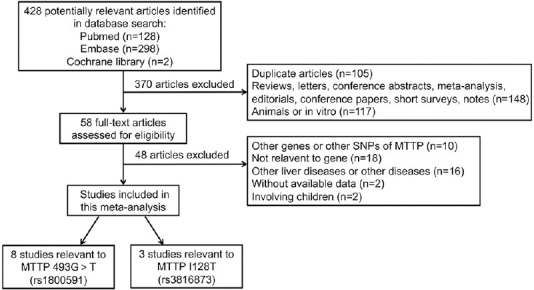
Flow diagram of the eligible study selection process
Characteristics of the included studies
A total of 1388 cases and 1690 healthy controls were included in the present meta-analysis. Among these 10 studies, four studies were conducted in Europe,[15,18,20,21] four studies were conducted in Asia,[13,14,16,23] one study was conducted in Africa,[19] and one study was conducted in South America.[22] In addition, all patients in these four studies underwent liver biopsy,[13,18,20,22] while in the remaining six studies, patients were partially biopsied or not biopsied.[14,15,16,19,21,23] In three studies, the control groups were hospital-based,[16,19,23] while in seven studies, the control groups were population-based.[13,14,15,18,20,21,22] For the detection of gene polymorphism, six studies used the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method,[13,15,18,19,20,21] while the remaining four studies used other detection methods.[14,16,22,23] Except for one of these studies,[14] the genotype distribution obeyed the HWE. The NOS score of eligible studies ranged within 5–9. The characteristics of each of the included studies are listed in Table 1.
Table 1.
Main characteristics of the studies included in the meta-analysis
| Authors | Year | Site | Ethnicity | Disease | NAFLD diagnosis | Source of Control | Gender (M/F) |
Genotyping Method | Sample size (Case/Control) | HWE (P) | NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||||
| Namikawa et al.[13] | 2004 | rs1800591 | Asia | NASH | LB | PB | 33/30 | 83/67 | PCR - RFLP | 63/150 | >0.05 | 7 |
| Gambino et al.[18] | 2007 | rs1800591 | Europe | NASH | LB | PB | 24/5 | 23/4 | PCR - RFLP | 29/27 | >0.05 | 9 |
| Musso et al.[21] | 2007 | rs1800591 | Europe | NAFLD | LB; US | PB | 50/14 | 60/14 | PCR - RFLP | 64/74 | >0.05 | 6 |
| Carulli et al.[15] | 2009 | rs1800591 | Europe | NAFLD | LB; US | PB | 74/40 | 28/51 | PCR - RFLP | 114/79 | >0.05 | 8 |
| Oliveir et al.[22] | 2010 | rs1800591 | South America | SSNASH | LB | PB | NA | NA | DNAsequencing | 129/113 | >0.05 | 5 |
| Gouda et al.[19] | 2017 | rs1800591 | Africa | NAFLD | US | HB | 60/114 | 81/60 | PCR - RFLP | 174/141 | >0.05 | 6 |
| Musso et al.[20] | 2010 | rs1800591 | Europe | NASH | LB | PB | 27/13 | 28/12 | PCR - RFLP | 40/40 | - | 8 |
| Peng et al.[16] | 2014 | rs1800591 rs3816873 | Asia | NAFLD | US | HB | 420/160 | 420/160 | MALDI - TOF MS | 580/580 | >0.05 | 6 |
| Jun et al.[23] | 2009 | rs3816873 | Asia | NAFLD | US | HB | NA | NA | TaqMan PCR | 113/393 | >0.05 | 6 |
| Hashemi et al.[14] | 2011 | rs3816873 | Asia | NAFLD | US | PB | 50/33 | 42/51 | ARMS - PCR | 83/93 | <0.05 | 7 |
SS = Simple steatosis, LB = Liver biopsy, US = Liver ultrasonographic, PB = Population-based, HB = Hospital-based, NA = Not available, PCR-RFLP = Polymerase chain reaction-restriction fragment length polymorphism, ARMS-PCR = Amplification refractory mutation system-polymerase chain reaction, MALDI-TOF MS = Allele-specific MALDI-TOF mass spectrometry assay, HWE = HardyWeinberg equilibrium
The rs1800591 in the MTTP gene and NAFLD
The present analysis revealed that there was substantial heterogeneity (I2> 50%, P(heterogeneity)<0.05) in the estimated effect sizes of each model. Hence, the random effect model was applied. Although the frequencies of the G allele in the control group (78.2%) and case group (74.3%) were different, there was no significant correlation between the G allele and NAFLD susceptibility (G vs. T: OR = 1.08, 95% CI = 0.68–1.70, P = 0.76; Figure 2a). In addition, the analyses of various models did not have any significant correlation between the MTTP gene rs1800591 polymorphism and NAFLD (heterozygous [GT vs. TT: OR = 1.38, 95% CI = 0.58–3.26; P = 0.46], homozygous [GG vs. TT: OR = 1.34, 95% CI = 0.45–3.95; P = 0.60], dominant [GT + GG vs. TT: OR = 1.37, 95% CI = 0.53–3.55; P = 0.51], and recessive (GG vs. GT + TT: OR = 0.98, 95% CI = 0.64–1.49; P = 0.91) [Table 2].
Figure 2.
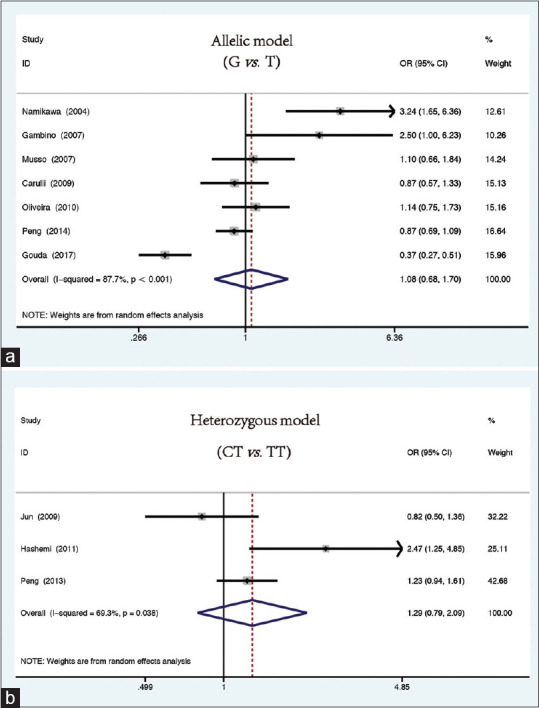
Forest plots for the association between MTTP rs1800591 and rs3816873 polymorphisms and susceptibility to NAFLD. (a) Allelic model (rs1800591 polymorphism): G vs. T. (b) Heterozygous model (rs3816873 polymorphism): CT vs. TT
Table 2.
The correlation between MTTP rs1800591 polymorphism with NAFLD and NASH under five genetic models
| Genetic model | NAFLD | NASH | |
|---|---|---|---|
| G vs. T | OR (95% CI) | 1.08 (0.681.70) | 1.88 (0.774.55) |
| P | 0.76 | 0.164 | |
| I-squared (%) | 87.7 | 81.4 | |
| PH | 0.000 | 0.005 | |
| GT vs. TT | OR (95% CI) | 1.38 (0.583.26) | 3.16 (1.13-8.83) |
| P | 0.46 | 0.028 | |
| I-squared (%) | 74.8 | 0 | |
| PH | 0.001 | 0.892 | |
| GG vs. TT | OR (95% CI) | 1.34 (0.453.95) | 3.24 (0.9810.68) |
| P | 0.60 | 0.053 | |
| I-squared (%) | 84.5 | 17.2 | |
| PH | 0.000 | 0.299 | |
| GT+GG vs. TT | OR (95% CI) | 1.37 (0.533.55) | 3.03 (1.138.09) |
| P | 0.51 | 0.027 | |
| I-squared (%) | 81.5 | 0 | |
| PH | 0.000 | 0.499 | |
| GG vs. GT+TT | OR (95% CI) | 0.98 (0.641.49) | 1.48 (0.69 3.18) |
| P | 0.91 | 0.319 | |
| I-squared (%) | 76.8 | 73.3 | |
| PH | 0.000 | 0.010 |
P: P-value of Z-test for statistical significance, PH: P-value of Q-test for heterogeneity test
Subgroup analyses were performed based on the ethnicity. There was no heterogeneity in Asian studies (heterozygous: GT vs. TT: OR = 2.09, 95% CI = 0.91–4.81, P = 0.082, I2= 0%, P(heterogeneity)= 0.444; dominant: GT + GG vs. TT: OR = 2.77, 95% CI = 0.42–18.08, P = 0.288, I2= 49%, P(heterogeneity)= 0.161) and European studies (heterozygous: GT vs. TT: OR = 1.17, 95% CI = 0.60–2.28, P = 0.639, I2= 0%, P(heterogeneity)= 0.576; homozygous: GG vs. TT: OR = 1.22, 95% CI = 0.53–2.81, P = 0.633, I2= 28.6%, P(heterogeneity)= 0.246; dominant: GT + GG vs. TT: OR = 1.17, 95% CI = 0.60–2.28, P = 0.635, I2= 8.1%, P(heterogeneity)= 0.337; recessive: GG vs. GT + TT: OR = 1.04, 95% CI = 0.72–1.51, P = 0.841, I2= 0%, P(heterogeneity)= 0.394).
Interestingly, when patients with NASH were analyzed alone, a significant association between the G allele of rs1800591 and NASH patients (heterozygous: GT vs. TT: OR = 3.16, 95% CI = 1.13–8.83, P = 0.028, Figure 3a; dominant: GT + GG vs. TT: OR = 3.03, 95% CI = 1.13–8.09, P = 0.027, Figure 3b) was detected [Table 2].
Figure 3.
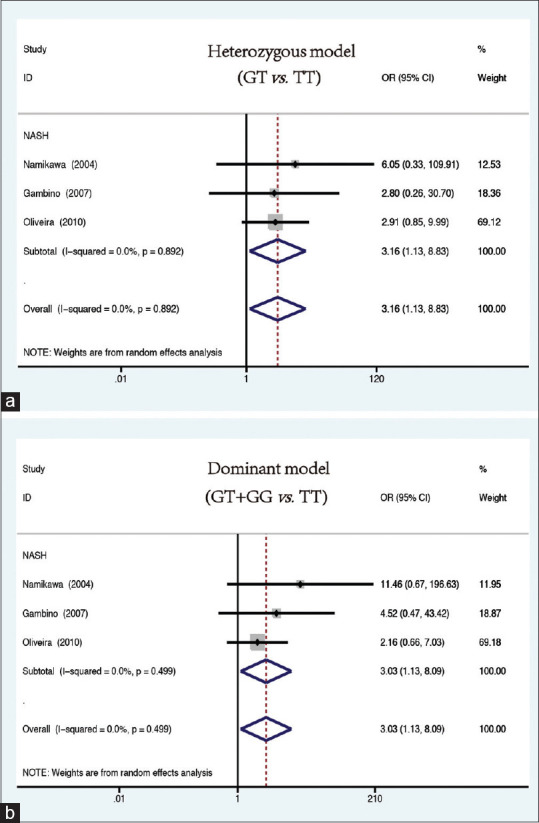
Forest plots for the association between MTTP rs1800591 xspolymorphism and NASH risk under the (a) heterozygous model (GT vs. TT) and (b) dominant model (GT + GG vs. TT)
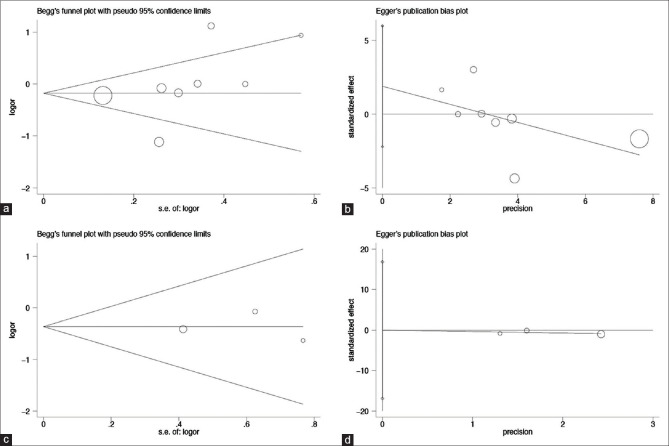
Through the sensitivity analysis, a significant change was found in the results after one study[19] was eliminated from the heterozygous model (GT vs. TT: OR = 1.71, 95% CI = 1.06–2.77). The visual observation of the symmetry of the funnel plot could not accurately determine the publication bias. Hence, this was further tested by Begg's rank correlation test [Figure 4a] and Egger's linear regression test [Figure 4b and Table 3].
Figure 4.
Begg's funnel plot and Egger's linear regression plot for detecting the publication bias through the recessive model. (a) Begg's funnel plot for the rs1800591 polymorphism; (b) Egger's linear regression plot for the rs1800591 polymorphism; (c) Begg's funnel plot for the rs3816873 polymorphism; (d) Egger's linear regression plot for the rs3816873 polymorphism
Table 3.
Publication bias of the five genetic models for rs1800591 and rs3816873 polymorphisms
| Genetic model | rs1800591 |
rs3816873 |
||
|---|---|---|---|---|
| PB | PE | PB | PE | |
| Allelic | 0.133 | 0.172 | 1.0 | 0.951 |
| Heterozygous | 0.368 | 0.091 | 1.0 | 0.803 |
| Homozygous | 0.133 | 0.067 | 1.0 | 0.851 |
| Dominant | 0.072 | 0.042* | 1.0 | 0.798 |
| Recessive | 0.035* | 0.302 | 1.0 | 0.988 |
PB: P-value of Begg’s rank correlation test. *P<0.05.PE: P-value of Egger’s linear regression test. *P<0.05.
In brief, the meta-analysis indicated that the MTTP gene rs1800591 polymorphism and NAFLD are not correlated, but the G allele of the rs1800591 polymorphism was more likely to be associated with NASH susceptibility.
The rs3816873 in the MTTP gene and NAFLD
A total of three studies were included in this SNP site for analysis. The fixed effect model (homozygous: CC vs. TT: I2= 0%, P = 0.417; recessive: CC vs. CT + TT: I2= 0%, P = 0.839) and random effect model (allelic: C vs. T: I2= 50.5%, P = 0.133; heterozygous: CT vs. TT: I2= 69.3%, P = 0.038; dominant: CT + CC vs. TT: I2= 71.2%, P = 0.031) were utilized to merge the results. After the analysis, it was found that the rs3816873 polymorphism and NAFLD had no association (allelic [C vs. T: OR = 1.08, 95% CI = 0.81–1.45, P = 0.602]; heterozygous [CT vs. TT: OR = 1.29, 95% CI = 0.79–2.09, P = 0.038, Figure 2b); homozygous [CC vs. TT: OR = 0.79, 95% CI = 0.43–1.47, P = 0.462); dominant (CT + CC vs. TT: OR = 1.23, 95% CI = 0.76–2.01, P = 0.398); recessive (CC vs. CT + TT: OR = 0.69, 95% CI = 0.37–1.27, P = 0.233]).
After removing the study which did not obey the HWE,[14] the heterogeneity decreased (allelic: C vs. T: I2= 46.8%, P(heterogeneity)= 0.17; heterozygous: CT vs. TT: I2= 47.8%, P(heterogeneity)= 0.166; homozygous: CC vs. TT: I2= 0%, P(heterogeneity)= 0.712; recessive: CC vs. CT + TT: I2= 0%, P(heterogeneity)= 0.799). The sensitivity analysis did not reveal any change in the original results. In addition, the funnel plot was symmetrical, and no publication bias was found on the analysis performed using the Begg's rank correlation test [Figure 4c] and Egger's linear regression test [Figure 4d].
DISCUSSION
The MTTP gene plays an important role in lipid metabolism. Its participation in the assembly of VLDL is the most important step for the liver to secrete triglycerides.[15] Therefore, the decrease or absence in activity of MTTP would result in the less secretion of triglycerides from the hepatocytes, thereby allowing the accumulation of lipids in the liver.[24] The resulting lipid export damage would render patients to be more prone to hepatic steatosis,[13] which is also correlated to the first hit phenomenon observed in the pathogenesis of NASH.[25]
MTTP gene rs1800591 polymorphism affects its transcriptional activity in HepG2 cells in vitro. Studies have shown that the transcriptional activity of the promoter structure containing the -493T site is almost twice of that of the -493G site.[26] However, the results of studies that analyzed the relationship between the MTTP gene rs1800591 polymorphism and NAFLD have been conflicting.[13,16] In a meta-analysis by Zheng et al. published in 2014, the authors reported that the rs1800591 polymorphism of MTTP was associated with an increased risk of NAFLD.[27] However, in the present meta-analysis, according to stricter selection criteria, after incorporating two newly published articles,[16,19] and excluding four studies on hepatitis C[28,29,30,31] and one pediatric study,[32] no correlation was found between NAFLD and the MTTP gene rs1800591 polymorphism in five gene models. However, considering that NAFLD patients have milder symptoms compared with NASH patients, we separately analyzed rs1800591 polymorphism in patients with (biopsy proven) NASH. This revealed that the G allele significantly increased the risk of NASH in both the heterozygous and dominant models. Hence, it was hypothesized that the G allele may play a role in the pathogenesis of NASH through the reduction in MTTP expression. It was also mentioned in one review that hepatic MTTP expression was found to be significantly lower in patients with NASH, when compared to NAFL.[33]
Although there was significant heterogeneity in each model, the subgroup analysis of these studies by ethnicity eliminated the heterogeneity, suggesting the potential impact of ethnicity on the results. During the sensitivity analysis, it was found that the original results were reversed after the elimination of one African study from the heterozygous model. The African study found that the T allele and TT genotype in the MTTP gene rs1800591 polymorphism were significantly elevated in NAFLD patients, which are contrary to the findings of other studies, and it also reflected the impact of ethnicity on the outcomes. Furthermore, the publication bias was found in the recessive model and the dominant model by Begg's rank correlation test and Egger's linear regression test, respectively (P < 0.05). Hence, further studies are needed to verify these findings.
MTTP gene rs3816873 polymorphism refers to the substitution of isoleucine by threonine at amino acid position 128, which reduces thermal stability.[34] So far, two studies have reported that rs3816873 polymorphism is not associated with NAFLD,[16,23] while a study conducted by Hashemi et al. revealed a significant correlation.[14] In the present study, the polymorphism was not found to be correlated to NAFLD susceptibility in a pooled analysis of the heterozygous model and dominant model. Although a study[14] did not meet the HWE (P < 0.05), the results of each model did not change after the removal of this study, and funnel plot was also symmetrical. Furthermore, the Begg's rank correlation test and Egger's linear regression test did not reveal any publication bias.
There were several limitations in the present meta-analysis. First, merely studies published in the English language with complete original data were included. Thus, the present study may have selection bias, which may affect the reliability of the present results. Second, the number of studies included was limited, and the sample size was small. Lastly, due to insufficient data, the results could not be adjusted according to other risk factors. Fortunately, despite these shortcomings, the present meta-analysis has some advantages. First, compared to the previously published meta-analysis in 2014, the total number of patients included in the present study was almost double, which significantly improves the statistical capacity of the analysis. Second, it was found that the quality of the included studies was better based on the NOS score.
CONCLUSION
In summary, the present meta-analysis revealed that there is no significant association between the rs1800591 and rs3816873 polymorphisms of the MTTP gene and NAFLD. However, there appeared to be a significant correlation between the G allele of rs1800591 and NASH. Therefore, MTTP gene rs1800591 polymorphism may be useful in the individualized management of NASH. Further large-scale studies in different ethnicity groups are needed to clarify the exact role of MTTP mutation in the susceptibility of NASH.
Financial support and sponsorship
This project was supported by the National Natural Science Foundation of China (Grant No. 31770837).
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Krawczyk M, Liebe R, Lammert F. Towards genetic prediction of non-alcoholic fatty liver disease trajectories: PNPLA3 and beyond. Gastroenterology. 2020;158:1865–80.e1. doi: 10.1053/j.gastro.2020.01.053. [DOI] [PubMed] [Google Scholar]
- 2.Lindenmeyer CC, McCullough AJ. The natural history of nonalcoholic fatty liver disease-an evolving view. Clin Liver Dis. 2018;22:11–21. doi: 10.1016/j.cld.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Yu Y, Cai J, She Z, Li H. Insights into the epidemiology, pathogenesis, and therapeutics of nonalcoholic fatty liver diseases. Adv Sci (Weinh) 2019;6:1801585. doi: 10.1002/advs.201801585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trepo E, Valenti L. Update on NAFLD genetics: From new variants to the clinic. J Hepatol. 2020;72:1196–209. doi: 10.1016/j.jhep.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Xia Y, Huang CX, Li GY, Chen KH, Han L, Tang L, et al. Meta-analysis of the association between MBOAT7 rs641738, TM6SF2 rs58542926 and nonalcoholic fatty liver disease susceptibility. Clin Res Hepatol Gastroenterol. 2019;43:533–541. doi: 10.1016/j.clinre.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Xu R, Tao A, Zhang S, Deng Y, Chen G. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: A HuGE review and meta-analysis. Sci Rep. 2015;5:9284. doi: 10.1038/srep09284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Zhang T, Lee S, Calabuig-Navarro V, Yamauchi J, Piccirillo A, et al. FoxO6 integrates insulin signaling with MTP for regulating VLDL production in the liver. Endocrinology. 2014;155:1255–67. doi: 10.1210/en.2013-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raabe M, Veniant MM, Sullivan MA, Zlot CH, Bjorkegren J, Nielsen LB, et al. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J Clin Invest. 1999;103:1287–98. doi: 10.1172/JCI6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamel R, Khan R, Pollex RL, Hegele RA. Abetalipoproteinemia: Two case reports and literature review. Orphanet J Rare Dis. 2008;3:19. doi: 10.1186/1750-1172-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetterau JR, Aggerbeck LP, Bouma ME, Eisenberg C, Munck A, Hermier M, et al. Absence of microsomal triglyceride transfer protein in individuals with abetalipoproteinemia. Science. 1992;258:999–1001. doi: 10.1126/science.1439810. [DOI] [PubMed] [Google Scholar]
- 13.Namikawa C, Shu-Ping Z, Vyselaar JR, Nozaki Y, Nemoto Y, Ono M, et al. Polymorphisms of microsomal triglyceride transfer protein gene and manganese superoxide dismutase gene in non-alcoholic steatohepatitis. J Hepatol. 2004;40:781–6. doi: 10.1016/j.jhep.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Hashemi M, Hoseini H, Yaghmaei P, Moazeni-Roodi A, Bahari A, Hashemzehi N, et al. Association of polymorphisms in glutamate-cysteine ligase catalytic subunit and microsomal triglyceride transfer protein genes with nonalcoholic fatty liver disease. DNA Cell Biol. 2011;30:569–75. doi: 10.1089/dna.2010.1162. [DOI] [PubMed] [Google Scholar]
- 15.Carulli L, Canedi I, Rondinella S, Lombardini S, Ganazzi D, Fargion S, et al. Genetic polymorphisms in non-alcoholic fatty liver disease: Interleukin-6-174G/C polymorphism is associated with non-alcoholic steatohepatitis. Dig Liver Dis. 2009;41:823–8. doi: 10.1016/j.dld.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Peng XE, Wu YL, Lu QQ, Hu ZJ, Lin X. MTTP polymorphisms and susceptibility to non-alcoholic fatty liver disease in a Han Chinese population. Liver Int. 2014;34:118–28. doi: 10.1111/liv.12220. [DOI] [PubMed] [Google Scholar]
- 17.Wells GA, Shea B, O′Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta analyses. [Last accessed on 2020 Apr 26]. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp .
- 18.Gambino R, Cassader M, Pagano G, Durazzo M, Musso G, et al. Polymorphism in microsomal triglyceride transfer protein: A link between liver disease and atherogenic postprandial lipid profile in NASH? Hepatology. 2007;45:1097–107. doi: 10.1002/hep.21631. [DOI] [PubMed] [Google Scholar]
- 19.Gouda W, Ashour E, Shaker Y, Ezzat W. MTP genetic variants associated with non-alcoholic fatty liver in metabolic syndrome patients. Genes Dis. 2017;4:222–8. doi: 10.1016/j.gendis.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musso G, Gambino R, Cassader M. Lipoprotein metabolism mediates the association of MTP polymorphism with beta-cell dysfunction in healthy subjects and in nondiabetic normolipidemic patients with nonalcoholic steatohepatitis. J Nutr Biochem. 2010;21:834–40. doi: 10.1016/j.jnutbio.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Musso G, Gambino R, De Michieli F, Biroli G, Premoli A, Pagano G, et al. Nitrosative stress predicts the presence and severity of nonalcoholic fatty liver at different stages of the development of insulin resistance and metabolic syndrome: Possible role of vitamin A intake. Am J Clin Nutr. 2007;86:661–71. doi: 10.1093/ajcn/86.3.661. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira CP, Stefano JT, Cavaleiro AM, Zanella Fortes MA, Vieira SM, Rodrigues Lima VM, et al. Association of polymorphisms of glutamate-cystein ligase and microsomal triglyceride transfer protein genes in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2010;25:357–61. doi: 10.1111/j.1440-1746.2009.06001.x. [DOI] [PubMed] [Google Scholar]
- 23.Jun DW, Han JH, Jang EC, Kim SH, Kim SH, Jo YJ, et al. Polymorphisms of microsomal triglyceride transfer protein gene and phosphatidylethanolamine N-methyltransferase gene in alcoholic and nonalcoholic fatty liver disease in Koreans. Eur J Gastroenterol Hepatol. 2009;21:667–72. doi: 10.1097/MEG.0b013e3283196adc. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Hegele RA. Abetalipoproteinemia and homozygous hypobetalipoproteinemia: A framework for diagnosis and management. J Inherit Metab Dis. 2014;37:333–9. doi: 10.1007/s10545-013-9665-4. [DOI] [PubMed] [Google Scholar]
- 25.Fernando DH, Forbes JM, Angus PW, Herath CB. Development and progression of non-alcoholic fatty liver disease: The role of advanced glycation end products. Int J Mol Sci. 2019;20:5037. doi: 10.3390/ijms20205037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpe F, Lundahl B, Ehrenborg E, Eriksson P, Hamsten A. A common functional polymorphism in the promoter region of the microsomal triglyceride transfer protein gene influences plasma LDL levels. Arterioscler Thromb Vasc Biol. 1998;18:756–61. doi: 10.1161/01.atv.18.5.756. [DOI] [PubMed] [Google Scholar]
- 27.Zheng W, Wang L, Su X, Hu XF. MTP -493G>T polymorphism and susceptibility to nonalcoholic fatty liver disease: A meta-analysis. DNA Cell Biol. 2014;33:361–9. doi: 10.1089/dna.2013.2238. [DOI] [PubMed] [Google Scholar]
- 28.Mirandola S, Osterreicher CH, Marcolongo M, Datz C, Aigner E, Schlabrakowski A, et al. Microsomal triglyceride transfer protein polymorphism (-493G/T) is associated with hepatic steatosis in patients with chronic hepatitis C. Liver Int. 2009;29:557–65. doi: 10.1111/j.1478-3231.2008.01892.x. [DOI] [PubMed] [Google Scholar]
- 29.Petit JM, Masson D, Minello A, Duvillard L, Galland F, Verges B, et al. Lack of association between microsomal triglyceride transfer protein gene polymorphism and liver steatosis in HCV-infected patients. Mol Genet Metab. 2006;88:196–8. doi: 10.1016/j.ymgme.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Siqueira ER, Oliveira CP, Correa-Giannella ML, Stefano JT, Cavaleiro AM, Fortes MA, et al. MTP -493G/T gene polymorphism is associated with steatosis in hepatitis C-infected patients. Braz J Med Biol Res. 2012;45:72–7. doi: 10.1590/S0100-879X2011007500160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zampino R, Ingrosso D, Durante-Mangoni E, Capasso R, Tripodi MF, Restivo L, et al. Microsomal triglyceride transfer protein (MTP) -493G/T gene polymorphism contributes to fat liver accumulation in HCV genotype 3 infected patients. J Viral Hepat. 2008;15:740–6. doi: 10.1111/j.1365-2893.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- 32.El-Koofy NM, El-Karaksy HM, Mandour IM, Anwar GM, El-Raziky MS, El-Hennawy AM. Genetic polymorphisms in non-alcoholic fatty liver disease in obese Egyptian children. Saudi J Gastroenterol. 2011;17:265–70. doi: 10.4103/1319-3767.82582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita K, Imajo K, Shinohara Y, Nozaki Y, Wada K, Yoneda M, et al. Novel findings for the development of drug therapy for various liver diseases: Liver microsomal triglyceride transfer protein activator may be a possible therapeutic agent in non-alcoholic steatohepatitis. J Pharmacol Sci. 2011;115:270–3. doi: 10.1254/jphs.10r14fm. [DOI] [PubMed] [Google Scholar]
- 34.Ledmyr H, Ottosson L, Sunnerhagen M, Ehrenborg E. The Ile128Thr polymorphism influences stability and ligand binding properties of the microsomal triglyceride transfer protein. J Lipid Res. 2006;47:1378–85. doi: 10.1194/jlr.M600072-JLR200. [DOI] [PubMed] [Google Scholar]