Abstract
Natural language processing (NLP) plays a vital role in modern medical informatics. It converts narrative text or unstructured data into knowledge by analyzing and extracting concepts. A comprehensive lexical system is the foundation to the success of NLP applications and an essential component at the beginning of the NLP pipeline. The SPECIALIST Lexicon and Lexical Tools, distributed by the National Library of Medicine as one of the Unified Medical Language System Knowledge Sources, provides an underlying resource for many NLP applications. This article reports recent developments of 3 key components in the Lexicon. The core NLP operation of Unified Medical Language System concept mapping is used to illustrate the importance of these developments. Our objective is to provide generic, broad coverage and a robust lexical system for NLP applications. A novel multiword approach and other planned developments are proposed.
Keywords: unified medical language system, natural language processing, lexicon, lexical tools, NLP tools
INTRODUCTION
The SPECIALIST Lexicon (https://umlslex.nlm.nih.gov/lexicon) (hereafter, the Lexicon) and Lexical Tools (https://umlslex.nlm.nih.gov/lvg), distributed by the National Library of Medicine as one of the Unified Medical Language System (UMLS) Knowledge Sources,1–3 provides an underlying resource for popular natural language processing (NLP) tools and applications, such as MetaMap,4,5 cTAKES (clinical Text Analysis and Knowledge Extraction System),6 CSpell,7 STMT (Sub-Term Mapping Tools),8 deepBioWSD,9 Part-Of-Speech (POS) Tagger,10 UMLS Metathesaurus (https://www.nlm.nih.gov/research/umls/knowledge_sources/metathesaurus/index.html), and ClinicalTrials.gov.
The Lexicon is a large syntactic lexicon of biomedical and general English, which provides the lexical information needed for NLP systems. Each entry records the syntactic, morphological, and orthographic information for a term (single word or multiword).11 Other lexical information for lexical entries, such as derivation pairs (dPairs) and synonym pairs, are generated through systematic methodologies.12,13
The Lexical Tools utilize the Lexicon data to provide a comprehensive toolset and Java application programming interfaces for lexical variant generation and other NLP fundamental functions, including retrieving syntactic category, inflectional variations, spelling variations, abbreviations, acronyms, derivational variations, synonyms, normalization, Unicode-to-ASCII conversion, tokenization, and stop word removal.14–19
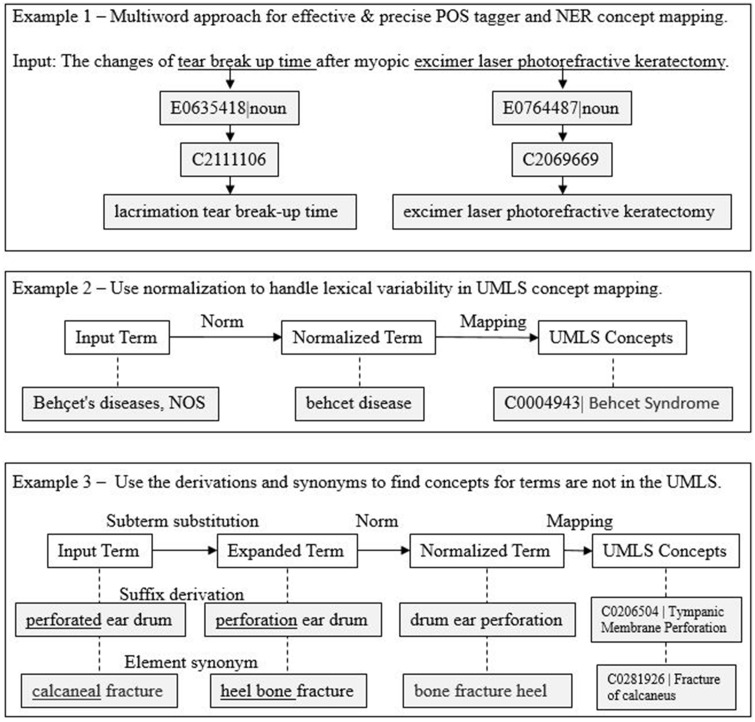
Since the first release in 1994, the Lexicon and Lexical Tools have been one of the richest and most robust NLP fundamental resources of the UMLS. The Lexicon provides the multiword thesaurus needed for a novel multiword approach in NLP, improving the precision of POS taggers and named entity recognition for UMLS concept mapping. In Figure 1, Example 1, “tear break up time” and “excimer laser photorefractive keratectomy” can be identified as units quickly (the longest multiwords in the Lexicon) for precise POS tagging and UMLS concept mapping, while they are very challenging for most popular POS parsers20,21 or require a laborious window shifting concept lookup algorithm.5,6,22
Figure 1.
Examples of applications of the Lexicon and Lexical Tools. NER: named entity recognition; POS: Part-of-Speech; UMLS: Unified Medical Language System.
Concept mapping is an important core operation in NLP applications. Normalization, UMLS synonyms, and query expansion are commonly used techniques to handle variability (number of terms for a concept) for better recall. In Figure 1, Example 2, the input term of “Behçet’s diseases, NOS” (one of many lexical variations of “Behçet Disease”) is normalized to “behcet disease” by the Lexical Tools norm program to abstract away from case, punctuation, possessive forms, inflections, stop words, and Unicode-to-ASCII conversion. The normalized term is then mapped to a UMLS concept. The mapping is a lookup operation either through UMLS-LUI (Lexical Unique Identifiers) or pre-indexed UMLS files, such as MRXNS_ENG.RRF or MRXNW_ENG.RRF. The Lexical Tools luiNorm and norm programs are used to handle lexical variations of terms (in the UMLS or not) for concept mapping.
Lexically dissimilar terms with the same concept are handled by UMLS synonymy to improve recall in concept mapping. For example, “behcet syndrome” and “behcet disease” are lexically dissimilar. They are UMLS synonyms because they carry the same concept. Query expansion can be used to find concepts for terms that are not lexically similar nor UMLS synonyms for further recall improvement. As shown in Figure 1, Example 3, by substituting the subterm “perforated” for its derivation “perforation,” the concept is found for “perforated ear drum.” Similarly, by substituting the subterm “calcaneal” for its synonym “heel bone,” the concept is found for “calcaneal fracture,” while no concept can be found otherwise.13 The success of query expansion relies on the broad coverage and quality of derivations and synonyms. A detailed description of applying the SPECIALIST tools to the examples in Figure 1 is available online (https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lvg/2020/docs/userDoc/examples/nlpApplicationExamples.pdf).
It is imperative to continuously develop a comprehensive lexical system with broad coverage and robust functions to ensure the success of NLP applications that use it. Here, we present recent developments and results of 3 key components in the Lexicon and Lexical Tools. They are multiword acquisition, derivation generation, and synonym generation.
DEVELOPMENT AND RESULTS
An element word (lowercase single words without punctuation) approach was used to build the Lexicon by National Library of Medicine linguists through a web-based computer-aided tool called LexBuild (http://umlslex. nlm.nih.gov/lexBuild) before 2014.23 Element words with high frequency (≥1500) from a corpus (MEDLINE titles and abstracts) were used to build the Lexicon. Multiword inclusion used element words to find new multiword candidates through the Essie search engine.24 This method is adequate for adding new single words but is not effective for adding multiwords. The Lexicon 2014 release covers 98.22% of single words in MEDLINE (2014). However, there were more single words than multiwords (47.74%) in the Lexicon, and the demand for multiwords for high-quality NLP applications has increased.25–27 Accordingly, we shifted our focus to multiword acquisition.
Lexical variation for a lexical record, such as inflection, spelling variations, abbreviations and acronyms, nominalizations, etc., are generated by computer programs with the growth of the Lexicon. However, the coverage of lexical variations among lexical records, such as derivations and synonyms, was low because they were generated by manual updates based on users’ requests. Accordingly, we developed systematic approaches to generate derivations and synonyms in the Lexicon. The development and results are described as follows.
Multiword acquisition
A new system was developed to effectively add multiwords to the Lexicon using an n-gram approach. MEDLINE was chosen as the corpus because it is the biggest and most commonly used resource in the biomedical domain. The MEDLINE n-gram set (n = 1-5) was generated to cover over 99.47% (estimated) of valid terms.28 Most of the n-grams (n ≥ 2) are not valid multiwords. Valid multiwords must meet 3 requirements: a single POS, corresponding inflections, and a lexical meaning. A set of high-precision filters (https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lexicon/current/docs/designDoc/UDF/multiwords/applyFilters/2019.html) were developed based on empirical rules to filter out n-grams that are invalid multiwords, with the result as the distilled MEDLINE n-gram set that contains only ∼38% of n-grams while preserving the recall rate (99.99%) of valid multiwords.29 Empirical models (https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lexicon/current/docs/designDoc/UDF/multiwords/candidates/index.html) based on matchers are applied to the distilled MEDLINE n-gram set to generate high-precision multiword candidates for linguists to build the Lexicon. Table 1 shows the accumulated precision of multiword candidates generated from developed models. The process has over 76.55% average precision on multiword candidates.
Table 1.
Results of multiword candidates from matcher models using the distilled MEDLINE n-gram set
| Models based on matchers | Candidates | Multiwords | Precision (%) |
|---|---|---|---|
| Abbreviation and acronyms expansion model | 1554 | 1452 | 93.44 |
| Parenthetic acronym matcher model | 8755 | 6224 | 71.09 |
| CUI-endword matcher model | 9134 | 8144 | 89.16 |
| Spelling variant matcher model | 7602 | 4882 | 64.22 |
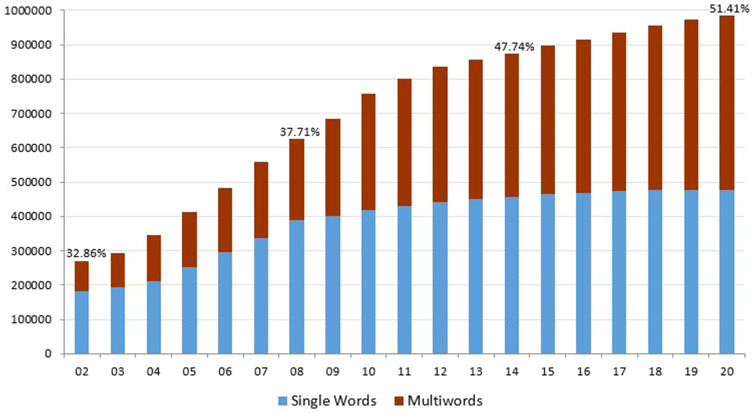
With this approach, the amount of multiwords in the Lexicon grew over single words to reach 51.41% (505 621 of 983 420) in the Lexicon 2020 release, as shown in Figure 2.
Figure 2.
Lexicon growth: single words vs multiwords.
Derivation generation
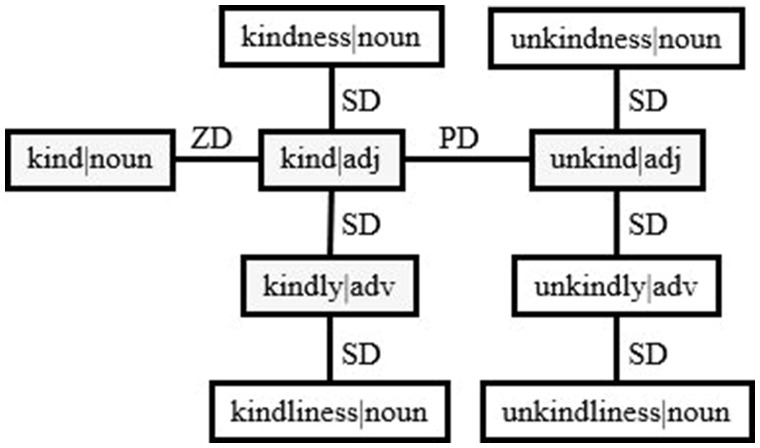
A systematic approach for generating derivational variants from the Lexicon was developed.12 This approach addressed prefix derivation (PD), zero derivation (ZD), and suffix derivation (SD) generation. Figure 3 shows an example of the derivation network for the “kind” family. Nodes of [unkind|adj], [kindly|adv], and [kind|noun] are all derived from the node of [kind|adj] through the derivational processes of prefixation (“un”), suffixation (“ly”), and ZD (category change without affixation), respectively. Nodes of [kindly|adv] and [kind|adj] are connected directly and thus form a dPair. Nodes of [kindly|adv] and [unkind|adj] are not a dPair in the Lexicon. They (and all nodes in this network) can be retrieved through the recursive derivation generation function in the Lexical Tools.
Figure 3.
Derivation network example for “kind” family. PD: prefix derivation; SD: suffix derivation; ZD: zero derivation
The processes of PD, ZD, and SD generation are similar. First, we retrieve all base forms (citations and spelling variants) from the Lexicon that match the patterns of PD, ZD, and SD. Second, expert systems are used to auto-tag valid and filter out invalid dPairs. Third, the remaining dPairs are manually tagged by linguists. The PD and SD are governed by prefix and suffix rules. We carefully review and add the most commonly used prefix and suffix rules. The number of prefix (https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lvg/current/docs/designDoc/UDF/derivations/prefixList.html) and suffix (https://lsg3.nlm.nih.gov/LexSysGroup/Projects/lvg/current/docs/designDoc/UDF/derivations/suffixDRules.html) rules have grown from 61 to 150 and 97 to 153, respectively, since the initial implementation in 2012. The use of a systematic data-mining approach, various expert systems and filter algorithms and expert tagging processes, increased derivations with the growth of both the Lexicon and prefix and suffix rules, and resulted in a dramatic increase in dPairs. As shown in Table 2, derivations have grown by 3200% (from 4559 in 2011 to 147 792 in 2020) since this system was implemented.
Table 2.
Growth of Lexicon derivations
| Year | suffixD | prefixD | zeroD | Total dPairs |
|---|---|---|---|---|
| 2020 | 57 573 | 74 369 | 15 850 | 147 792 |
| 2019 | 56 356 | 74 181 | 15 843 | 146 380 |
| 2018 | 56 339 | 74 153 | 15 831 | 146 323 |
| 2017 | 56 270 | 74 102 | 15 831 | 146 203 |
| 2016 | 55 528 | 74 001 | 15 810 | 145 339 |
| 2015 | 52 264 | 73 609 | 15 750 | 141 623 |
| 2014 | 51 337 | 73 167 | 15 699 | 140 203 |
| 2013 | 44 832 | 61 209 | 15 037 | 121 078 |
| 2012 | 18 509 | 56 694 | 14 747 | 89 950 |
| 2011 | 4202 | 4 | 353 | 4559 |
dPairs: derivation pairs.
New derivational features, such as negation, derivation types, and recursive derivations with options, are implemented in the Lexical Tools for better performance in NLP applications.15 Additionally, a methodology for optimizing the suffix rule set was developed for the Lexical Tools to generate SDs that are not in the Lexicon to reach 95% system precision.16
Synonyms generation
A new system was developed to generate element synonyms in subterm substitution. Element synonyms must be cognitive synonyms (have commutativity and transitivity) to increase recall and preserve precision in UMLS concept mapping. The sources of synonym pairs are Lexicon terms that are also UMLS synonyms, nominalizations in the Lexicon and synonyms in the lexical variant generation. Computer programs were developed to (1) remove abbreviations and acronyms to avoid overgeneration and preserve precision and (2) eliminate spelling variants and POS ambiguity. The produced synonym candidate list is then sent to linguists for manual tagging. Applying these high-quality and broad coverage element synonyms to test with the terminology of the UMLS core project, the result shows a 5% improvement of recall and F1, with similar precision for concept mapping.13,30 The number of synonyms has grown by 4300% (from 5198 in 2016 to 227 692 in 2020) since this system was implemented, as shown in Table 3.
Table 3.
Growth of Lexicon synonyms
| Year | UMLS | Lexicon | LVG | Total sPairs |
|---|---|---|---|---|
| 2020 | 155 488 | 67 432 | 4772 | 227 692 |
| 2019 | 140 726 | 67 664 | 4774 | 213 164 |
| 2018 | 124 508 | 67 830 | 4778 | 197 116 |
| 2017 | 118 468 | 67 584 | 4792 | 190 844 |
| 2016 | 0 | 0 | 5198 | 5198 |
LVG: lexical variant generation; sPairs: synonym pairs; UMLS: Unified Medical Language System.
New synonym features, such as POS, source information, and recursive synonyms with source options, were implemented in the Lexical Tools to provide better performance for downstream NLP applications.17
DISCUSSION AND FUTURE PLAN
The Lexicon and Lexical Tools provide comprehensive and precise lexical information to improve performance for NLP applications. For example, the Lexicon was used as the dictionary in CSpell for spelling error detection and collection and outperformed 3 other dictionaries by a large margin to reach an F1 score of 81.15.7,31
The multiword approach for NLP identifies the longest multiwords known to a lexicon as a unit in a sentence for faster and more precise performance. A sentence is more quickly understood by humans if readers already have the knowledge of multiwords. “Tradeoff,” “trade off,” and “trade-off” are spelling variants with the same concept and should be identified as a unit with or without spaces or hyphens. If we have enough multiwords, this preprocessed knowledge can be used for tokenization, POS tagging, and named entity recognition. A good collection of multiwords is the key to the success of the multiword approach. We plan to apply deep learning models for retrieving high-frequency multiwords to enrich the coverage of multiwords in the Lexicon.
High-quality and broad coverage of derivations and synonyms are used in subterm substitution for effective concept mapping.8 The results show improvements in recall and F1 and preserve precision.13 Derivations, synonyms, and other lexical variants are used in many NLP applications. For example, the fruitful variants, used in various NLP projects (Sophia, MetaMap Lite, and Custom Taxonomy Builder [https://metamap.nlm.nih.gov/CustomTaxonomyBuilder.shtml]) require the previously mentioned lexical information.4,22 The performance of these systems relies on the growth and quality of the Lexicon. Table 4 summarizes the usage and benefit of using some popular Lexicon and Lexical Tools components in NLP applications.
Table 4.
Usage, benefits, and applications of using lexical components
| Lexical Component | Descriptions |
|---|---|
| Norm |
|
| Derivations and synonyms |
|
| Fruitful variants |
|
| (The longest) Multiwords |
|
| The Lexicon |
|
cTAKES: clinical Text Analysis and Knowledge Extraction System; NER: named entity recognition; POS: Part-of-Speech; STMT: Sub-Term Mapping Tools; UMLS: Unified Medical Language System.
The Norm and LuiNorm programs in the Lexical Tools are used for UMLS concept mapping. We plan to further improve these 2 programs by optimizing both ambiguity (number of concepts for a term) and variability (number of terms for a concept) for better performance.
Consumer language often uses colloquial contexts and is not covered in the UMLS, such as “grandpa” for “grandfather.” A comprehensive consumer terminology collection is needed for consumer health informatics.7,32–35 A new classification type tag (class_type=informal) was added in the Lexicon for informal expressions.36 We also plan to expand corpus resources to consumer-oriented data, such as the WebMD community (http://exchanges.webmd.com), DoctorSpring (https://www.doctorspring.com/questions-home), and Genetic and Rare Diseases Information Center (http://rarediseases.info.nih.gov), for broader coverage in consumer terminology.
Negation detection and antonyms are essential for many NLP applications.37–42 Negation detection cue words can be derived from negative antonyms. For example, “without” and “fail,” from their antonyms of “with” and “succeed,” are negation cue words. We plan to develop a systematic approach to generate antonym pairs in the Lexicon and a comprehensive and generic negation detection tool.
CONCLUSION
Publicly available, open-source lexicon and fundamental NLP tools with broad coverage and robust functionality is important for the NLP community. The SPECIALIST Lexicon, Lexical Tools, and other fundamental NLP tools are available via an Open Source License (https://umlslex.nlm.nih.gov).
FUNDING
This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
AUTHOR CONTRIBUTIONS
All authors contributed sufficiently to the project to be included as authors.
ACKNOWLEDGMENTS
We would like to thank Dr. A.T. McCray, A. Browne, Dr. O. Bodenreider, Dr. D. Demner-Fushman, Dr. L. McCreedy, D. Tormey, G. Divita, and W.J. Rogers for their invaluable contribution.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Lindberg DAB, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med 1993; 32 (4): 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The unified medical language system: an informatics research collaboration. J Am Med Inform Assoc 1998; 5 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McCray AT, Aronson AR, Browne AC, Rindflesch TC, Razi A, Srinivasan S. UMLS knowledge for biomedical language processing. Bull Med Libr Assoc 1993; 81 (2): 184–94. [PMC free article] [PubMed] [Google Scholar]
- 4. Aronson AR. The effect of texture variation on concept based information retrieval. AMIA Annu Symp Proc 1996; 1996: 373–7. [PMC free article] [PubMed] [Google Scholar]
- 5. Demner-Fushman D, Wj R, Aronson AR. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. J Am Med Inform Assoc 2017; 24 (4): 841–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu CJ, Aronson AR, Shooshan SE, et al. Spell checker for consumer language (CSpell). J Am Med Inform Assoc 2019; 26 (3): 211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu CJ, Browne AC. Development of Sub-Term Mapping Tools (STMT). AMIA Annu Symp Proc 2012; 2012: 1845. [Google Scholar]
- 9. Pesaranghader A, Matwin S, Sokolova M, et al. deepBioWSD: effective deep neural word sense disambiguation of biomedical text data. J Am Med Inform Assoc 2019; 26 (5): 438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu K, Chapman W, Hwa R, et al. Heuristic sample selection to minimize reference standard training set for a part-of-speech tagger. J Am Med Inform Assoc 2007; 14 (5): 641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Browne AC, McCray AT, Srinivasan S. The SPECIALIST Lexicon (2018 Revision). Bethesda, MD: National Library of Medicine; 2018. [Google Scholar]
- 12. Lu CJ, Tormey D, McCreedy L, et al. A systematic approach for automatically generating derivational variants in lexical tools based on the SPECIALIST lexicon. IT Prof 2012; 14 (3): 36–42. [Google Scholar]
- 13. Lu CJ, Tormey D, McCreedy L, et al. Enhanced LexSynonym acquisition for effective UMLS Concept Mapping. MedInfo 2017; 245: 501–5. [PubMed] [Google Scholar]
- 14. McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care 1994; 1994: 235–9. [PMC free article] [PubMed] [Google Scholar]
- 15. Lu CJ, Tormey D, McCreedy L, et al. implementing comprehensive derivational features in lexical tools using a systematical approach. AMIA Annu Symp Proc 2013; 2013: 904. [Google Scholar]
- 16. Lu CJ, Tormey D, McCreedy L, et al. Generating SD-Rules in the SPECIALIST lexical tools-optimization for suffix derivation rule set. Biostec 2016; 5: 353–8. [Google Scholar]
- 17. Lu CJ, Tormey D, McCreedy L, et al. Enhancing LexSynonym Features in the Lexical Tools. AMIA Annu Symp Proc 2017; 2017: 2090. [Google Scholar]
- 18. Lu CJ, Browne AC. Converting unicode lexicon and lexical tools for ASCII NLP applications. AMIA Annu Symp Proc 2011; 2011: 1870. [Google Scholar]
- 19. Lu CJ, Browne AC, Divita G. Using lexical tools to convert Unicode characters to ASCII. AMIA Annu Symp Proc 2008; 2008: 1031. [PubMed] [Google Scholar]
- 20. Li X, Roth D. Exploring evidence for shallow parsing. Proc Comput Nat Lang 2001; 7: 107–10. [Google Scholar]
- 21. Manning C, Surdeanu M, Bauer J, et al. The Stanford CoreNLP Natural Language Processing Toolkit. In: 52nd Annual Meeting of the Association for Computational Linguistics; 2014: 5–60.
- 22. Divita G, Zeng QT, Gundlapalli AV, et al. Sophia: a expedient UMLS concept extraction annotator. AMIA Annu Symp Proc 2014; 2014: 467–76. [PMC free article] [PubMed] [Google Scholar]
- 23. Lu CJ, Tormey D, McCreedy L, et al. Using element words to generate (Multi)Words for the SPECIALIST Lexicon. AMIA Annu Symp Proc 2014; 2014: 1499. [Google Scholar]
- 24. Ide NC, Loane RF, Demner-Fushman D. Essie: a concept-based search engine for structured biomedical text. J Am Med Inform Assoc 2007; 14 (3): 253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rayson P, Piao S, Sharoff S, et al. Multiword expressions: hard going or plain sailing? Lang Resour Eval 2010; 44 (1–2): 1–5. Vol.Issue [Google Scholar]
- 26. Ramisch C. Multiword Expressions Acquisition: A Generic and Open Framework (Theory and Applications of Natural Language Processing). Berlin, Germany: Springer. [Google Scholar]
- 27. Constant M, Eryiğit G, Monti J, et al. Multiword expression processing: a survey. Comput Linguist 2017; 43 (4): 837–92. [Google Scholar]
- 28. Lu CJ, Tormey D, McCreedy L, et al. Generating the MEDLINE N-Gram Set. AMIA Annu Symp Proc 2015; 2015: 1569. [Google Scholar]
- 29. Lu CJ, Tormey D, McCreedy L, et al. Generating a distilled N-gram set: effective lexical multiword building in the SPECIALIST Lexicon. Biostec 2017; 5: 77–87. [Google Scholar]
- 30. Fung KW, Xu J. An exploration of the properties of the CORE problem list subset and how it facilitates the implementation of SNOMED CT. J Am Med Inform Assoc 2015; 22 (3): 649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu CJ, Demner-Fushman D. improving spelling correction with consumer health terminology. AMIA Annu Symp Proc 2018; 2018: 2053. [Google Scholar]
- 32. Zeng QT, Tse T. Exploring and developing consumer health vocabularies. J Am Med Inform Assoc 2006; 13 (1): 24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts K, Demner-Fushman D. Interactive use of online health resources: a comparison of consumer and professional questions. J Am Med Inform Assoc 2016; 23 (4): 802–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bakken S. The importance of consumer- and patient-oriented perspectives in biomedical and health informatics. J Am Med Inform Assoc 2019; 26 (7): 583–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Demner-Fushman D, Mrabet Y, Ben Abacha A. Consumer health information and question answering: helping consumers find answers to their health-related information needs. J Am Med Inform Assoc 2020; 27 (2): 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu CJ, Payne A, Demner-Fushman D. Classification types: a new feature in the SPECIALIST Lexicon. AMIA Annu Symp Proc 2019; 2019: 1661. [Google Scholar]
- 37. Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 34 (5): 301–10. [DOI] [PubMed] [Google Scholar]
- 38. Goryachev S, Sordo M, Zeng QT. Implementation and Evaluation of Four Different Methods of Negation Detection. DSG Technical Report. Boston, MA: Harvard Medical School; 2007.
- 39. Enger M, Velldal E, Øvrelid L. An open-source tool for negation detection: a maximum-margin approach. In: Workshop Computation SemBEaR Proceedings; 2017: 64–9.
- 40. Mohammad S, Dorr B, Hirst G. Computing word-pair antonymy. In: 2008 Conference on Empirical Methods in NLP Proceedings. 2008: 982–91.
- 41. Roth M, Walde SS. Combining word patterns and discourse markers for paradigmatic relation classification. In: 52nd Annual Meeting of the Association for Computational Linguistics; 2014: 524–30.
- 42. Santus E, Lu Q, Lenci A. Unsupervised antonym-synonym discrimination in vector space. In: first Italian Conference on Computational Linguistics CLiC-it Proceedings; 2014: 328–33.