Abstract
The US National Library of Medicine regularly collects summary data on direct use of Unified Medical Language System (UMLS) resources. The summary data sources include UMLS user registration data, required annual reports submitted by registered users, and statistics on downloads and application programming interface calls. In 2019, the National Library of Medicine analyzed the summary data on 2018 UMLS use. The library also conducted a scoping review of the literature to provide additional intelligence about the research uses of UMLS as input to a planned 2020 review of UMLS production methods and priorities. 5043 direct users of UMLS data and tools downloaded 4402 copies of the UMLS resources and issued 66 130 951 UMLS application programming interface requests in 2018. The annual reports and the scoping review results agree that the primary UMLS uses are to process and interpret text and facilitate mapping or linking between terminologies. These uses align with the original stated purpose of the UMLS.
Keywords: Unified Medical Language System, terminology (as topic), National Library of Medicine (US), surveys and questionnaires, scoping review
INTRODUCTION
This perspective provides an overview of the growth and current dimensions of direct users and uses of the Unified Medical Language System (UMLS)1 and comments on how these align with the US National Library of Medicine’s (NLM) hopes and expectations when the first experimental edition of the UMLS Knowledge Sources (Metathesaurus and Semantic Network) was released in 1990.2–4 Expanded and updated versions of the UMLS resources have been issued at least annually since that time. The SPECIALIST lexicon and lexical tools first became part of the UMLS release in 1994. As was intended, the thousands of direct UMLS users are primarily informatics researchers and developers of databases and software. Some of them use the UMLS to build or enhance electronic resources and applications (eg, PubMed, ClinicalTrials.gov, clinical data warehouses, components of electronic health record (EHR) systems, natural language processing pipelines, test collections), which in total are used by millions of people worldwide. The amount and impacts of this indirect use of the UMLS resources are likely immense but literally incalculable.
NLM regularly reviews summary data on direct UMLS use as well as the few specific suggestions for UMLS improvements submitted to NLM customer service that are meant to inform decisions about changes to UMLS content, production, and distribution. The summary data sources include: UMLS user registration data, required annual reports submitted by registered users, and statistics on downloads and application programming interface (API) calls. From time to time, NLM also specially solicits feedback from heavy UMLS users to help set multiyear priorities for UMLS development and customer service improvements. Input from all these sources has influenced many UMLS enhancements including, for example, the addition of vocabularies to the UMLS Metathesaurus, improvements in API and download capabilities, and expansion of documentation and training materials.
In 2019, in preparation for a planned 2020 workshop to guide UMLS development priorities, we analyzed summary UMLS use data for 2018 and also conducted a scoping review of the literature to provide additional intelligence about the research uses of UMLS.
MATERIALS AND METHODS
Annual survey, user registration data, and usage statistics
NLM requires an annual user report from UMLS Metathesaurus Licensees. The licensees include users of UMLS, SNOMED CT, RxNorm, the Value Set Authority Center, and other NLM terminology products as well as people who execute a license for exploratory purposes. Failure to submit the annual report automatically cancels the license, thereby preventing access to new editions of NLM terminology products subject to the license. NLM requests the report from users via e-mail in January of each year. On January 11, 2019, 25 326 licensees were asked to complete the 2018 annual report. Of that number, 12 503 submitted responses, and 5043 reported using the UMLS. We joined the annual report data (hereafter referred to as the annual survey) for these respondents with user registration data and internal log files to produce an overall picture of UMLS users and uses.
Literature review
In 2019, NLM staff performed a scoping review and assessment of literature published from 2005 to Spring 2019 to obtain additional insight into current UMLS research uses and applications as input to future developments in UMLS production. This review focused on articles reporting original research that used the UMLS and its products (eg, MetaMap, a tool for identifying UMLS concepts in text). The search strategy was 2-fold: (1) broadly search for [Unified Medical Language System] in PubMed, Web of Science and Scopus (Supplementary Material 1) and (2) retrieve articles that cite key articles (see5) about the UMLS (Supplementary Material 2). After removing duplicates, 3510 articles remained. Three coauthors (SB, LA, AR) independently screened the titles, abstracts, materials/methods sections of a random 10% sample (n = 348) of the 3510 articles to identify English language papers that employed UMLS or related products as a methodological tool.
Research papers that discussed UMLS but did not report actual use of UMLS (or related products) were excluded. Articles with publication types such as perspectives, letters to the editor, and book chapters were also excluded. The 110 articles that remained were divided amongst pairs of coauthors (SB/LA and AR/BH) and independently reviewed to extract information about the UMLS products used, the corpus on which the research was conducted or the method/tool tested, and how the UMLS products were used. A Kappa interrater reliability of 0.6792 for the 2 pairs of coauthors was calculated. EndNote X96 was used for citation management. Colandr,7 an open-source tool for conducting collaborative reviews, was used for screening and data extraction.
RESULTS
Annual survey, user registration data, and usage statistics
In 2004, NLM began requiring users to fill out an annual survey in order to retain their UMLS accounts.8 The number of survey respondents indicating use of UMLS increased from 1427 in 2004 to 5043 in 2018 (see Table 1). This number represents users who maintain access to UMLS from 1 year to the next. The UMLS experienced a significant increase in overall use from 2013 to 2017 as measured by downloads, API requests, and survey responses. Use declined slightly in 2018.
Table 1.
Changes in UMLS use from 2013–2018
| 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | |
|---|---|---|---|---|---|---|
| Downloads | 2020 | 2248 | 2249 | 2691 | 4898 | 4402 |
| API Requests (millions) | 15.0 | 53.0 | 32.7 | 29.1 | 68.4 | 66.1 |
| Survey responses | 3366 | 4096 | 4169 | 4500 | 5145 | 5043 |
Comparable data prior to 2013 not available.
Based on registration data, 2018 UMLS users (n = 5043) self-identify as researchers (42%), software developers (28%), administrator/managers (20%), healthcare providers (7%), educators (5%), analysts (5%), and students (3%). UMLS users most commonly report affiliations with academic institutions (32%), for-profit entities (23%), not-for-profit entities (16%), and government (7%). In 2018, users representing 1339 different organizations downloaded the UMLS release files. Based on an analysis of e-mail domains, 49% of those organizations were for-profit. A majority of respondents reside in the United States (77%).
Most users report that they use UMLS products in their own research, but 18% of respondents reported providing “an application, tool, or system to external users that utilizes the UMLS.” These users provided free-text descriptions of their applications. The answers revealed a wide variety of implementations for use by researchers, the healthcare industry, and the general public.
In the annual survey, users indicate the purpose(s) for which they use the UMLS (see Table 2). In both research contexts and in application development, users primarily leverage the UMLS to identify meaning in text, make connections between terminologies, and improve information retrieval.
Table 2.
2018 Survey question: For what purpose(s) did you use the UMLS?
| For what purpose(s) did you use the UMLS? | Count (n = 5043) | % |
|---|---|---|
| Processing of texts to extract concepts, relationships or knowledge | 2553 | 51% |
| Facilitate mapping between terminologies | 2486 | 49% |
| Extract specific terminologies from the Metathesaurus (eg, MedDRA, MeSH, NDF-RT) | 1442 | 29% |
| Develop an information retrieval system | 949 | 19% |
| Creation and maintenance of local terminology | 943 | 19% |
| Research terminologies and ontologies beyond any of the above categories | 917 | 18% |
| Other | 405 | 8% |
| Support of a terminology server or service | 353 | 7% |
The most frequent free-text responses in the “Other” category involved education (teaching or learning), EHRs, and quality measures.
NLM and others have developed tools that leverage the UMLS to identify meaning in clinical text, biomedical abstracts, or other corpora. Fourteen percent (733) of respondents who chose UMLS from the picklist of terminology products used this year also chose NLM’s MetaMap.9 Four other tools were mentioned 10 or more times in free-text responses from declared UMLS users: Apache cTAKES10 164 times, CLAMP11 18, QuickUMLS12 17, and SemRep13 16.
The annual survey included several open-ended questions about enhancements users would suggest to improve various aspects of the UMLS, including coverage of terminologies and code systems in the UMLS Metathesaurus, the UMLS Terminology Services APIs and downloads, file formats, installation, site navigation, customer support, use case examples, training, documentation, and any other improvements UMLS users would like to propose. Most of these questions received responses from only a few percent of UMLS users, and many responses did not include suggestions for enhancements (eg, N/A, none, nothing, not sure, don’t know, OK). The substantive responses varied substantially (eg, documentation that some called difficult to use or inadequate was considered excellent by others). However, regarding use case examples, training, and documentation, most respondents asked for “more,” and many of them indicated that they wished the UMLS resources were simpler.
In 2018, the most frequently answered question about UMLS enhancements was “What terminology or code system would you like to see added to the UMLS?” Eighteen percent of respondents (924) provided a response, but 49% of those did not suggest additions. In most cases, their answers implied satisfaction with the current coverage. Some respondents stated it directly (eg, “I am fine with what exists – great service!”). Most specific terminologies and code systems suggested multiple times were already in the UMLS Metathesaurus. The most frequently mentioned systems not in the UMLS were: RadLex14 (15 mentions), the International Classification of Diseases, 11th Revision (ICD-11)15(15 mentions), and the International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3)16 (14 mentions).
Literature review
For the random 10% sample (n = 348) of the 3510 articles, the geographical distribution by first author was: North America (53%), including US 173, Canada 9; Europe, (25%); Asia and Australasia, (20%); Central and South America, (1%) and Africa, (1%).
Of the 348 110 English language research articles that employed UMLS or related products as a methodological tool were further analyzed for: the UMLS products used, the corpus on which the research was conducted or the method/tool tested, and how the UMLS products were used.
UMLS products
Seventy-five percent of articles indicated use of the Metathesaurus, although actual use may be closer to 100%. The second most commonly used UMLS product was MetaMap, which identifies concepts from the Metathesaurus in text (see Figure 1).
Figure 1.
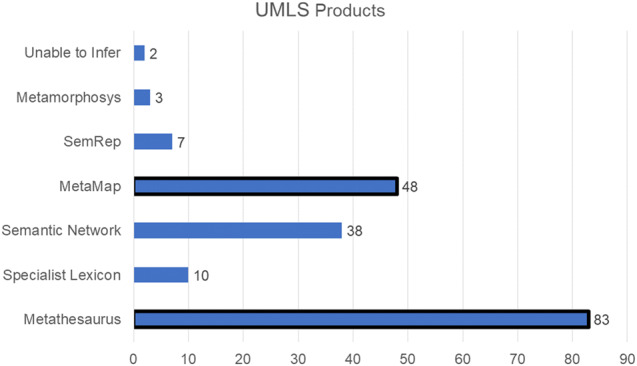
Counts of articles using various UMLS products.17
Corpora
The most common corpus processed in research that used the UMLS (see Figure 2) was the scientific literature (as represented by PubMed/MEDLINE citations and abstracts, full text articles, book chapters, etc.) and EHR data (medical records, physicians’ notes). Other corpora included: data from NCBI gene (GEO) and protein (Uniprot) databases; consumer questions and consumer-facing websites, user-generated data (patient-forums); tweets/Twitter, and Other (eg, ClinicalTrials.gov, clinical practice guidelines).
Figure 2.
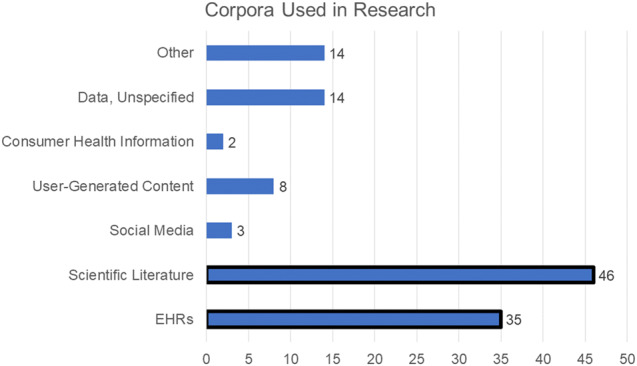
Counts of articles using different types of corpora.17
Applications of the UMLS in research
More than half of the 110 articles were tagged with more than 1 category of UMLS use (see Figure 3). Most (78% of articles) described using UMLS products to process text or to facilitate the mapping or linking of biomedical concepts. These articles often described NLP pipelines that process words and phrases, and then “assign UMLS CUIs” or create an index with UMLS CUIs. For example, Nawab et al18 used UMLS for query expansion to detect plagiarism in MEDLINE: “Input terms are mapped to UMLS CUIs using MetaMap. The UMLS Metathesaurus MRCONSO table is then consulted to identify synonymous terms for each CUI and these are used for query expansion.” Nawab’s information retrieval-based approach using UMLS outperformed the Kullback-Leibler distance approach on the plagiarism detection retrieval tasks.
Figure 3.
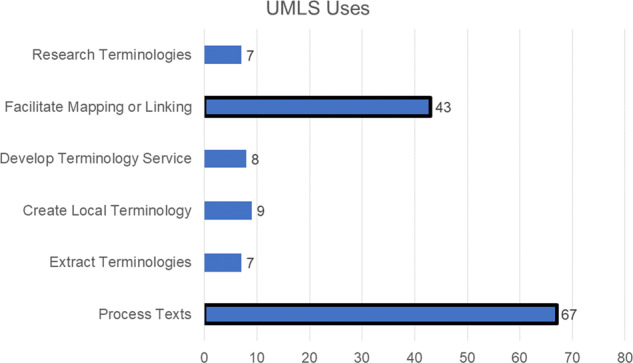
Counts of articles for different usages of UMLS.17
DISCUSSION
Millions of scientists, health professionals, and consumers use the UMLS indirectly through applications (eg, PubMed) that rely on it to some degree. The direct use of UMLS resources, the subject of this paper, is much smaller, but still substantial. After several years of rapid growth, the number of direct UMLS users (as measured by survey responses that indicated use of the UMLS), downloads, and API requests declined slightly in 2018. This pattern appears to be holding for users and downloads, but not for the more volatile API requests, which rose dramatically in 2019.
The significant increase in UMLS use over the past decade probably reflects broad environmental factors, including the designation of SNOMED CT, LOINC, and RxNorm as US national standards for clinical data in 2011, general developments in computing and communications that ease use of “big” UMLS data, and increasing research and development focus on natural language processing and data mining. There has also been an increase in the number of available tools that facilitate UMLS use in research and applications.
Most direct UMLS users are researchers or software developers who reside in the US. The percentage of US users (77%) has remained stable since 2004, although the number of users has increased more than 250%. In contrast, nearly half of the first authors in the random sample of research articles in our scoping review reside in other countries, suggesting a broader geographic distribution of research use. The UMLS Metathesaurus currently includes concept names in 25 different spoken languages, although coverage is sparse and uneven depending on the availability of electronic versions of translations of the English language terminologies and code sets used to build the Metathesaurus.
According to the results of both the user survey and the scoping review, the most heavily used UMLS resources are the Metathesaurus and MetaMap. Users of these products are also at least indirect users of the Semantic Network, SPECIALIST lexicon, and lexical tools, which are employed in building the Metathesaurus and in MetaMap functionality. Some authors who didn’t report use of the Semantic Network per se relied heavily on the semantic types assigned to Metathesaurus concepts in their research or applications. Some survey respondents who didn’t declare use of the UMLS itself reported that they use products, such as MetaMap, that depend on it.
Shifts in use from 2004 to 2018
In contrast to 2004,8 the first year that NLM used an annual Web-based survey, use of the UMLS has shifted from a focus on terminology research (53% of users in 2004 vs 18% of users in 2018) to addressing specific research and implementation problems related to text interpretation, use of now-mandated clinical terminologies and code systems, large scale analysis of EHR and administrative data, and development of applications for researchers, healthcare providers, and consumers. The decline in UMLS use in terminology research probably reflects the broader environmental factors that contributed to increased UMLS use overall.
In the 2018 survey, 51% of UMLS users indicated that they used UMLS for “processing of texts to extract concepts, relationships or knowledge.” In 2004,8 21% of users indicated using UMLS for “natural language processing.” (The categories in the survey have changed over time.) Processing of texts is done both in research contexts and in production applications. Research uses include: processing scientific literature,18,19 clinical notes,20,21 drug information,22,23 and social media.24,25 Text processing using the UMLS is generally performed to improve information retrieval, discover new knowledge, test hypotheses, and evaluate text processing methods to improve performance. In production applications, the UMLS is used to map local terms to standard terminologies, annotate records with standard terminology to improve information retrieval, process text for automated clinical coding, improve clinical documentation, and interpret patient questions.
In the 2018 survey, 49% of UMLS users indicated using UMLS to “facilitate mapping between terminologies,” an increase from 35% in the 2004 survey. Despite improving standardization in the clinical terminology space, mapping remains necessary. While mapping via UMLS synonymy and hierarchies cannot completely satisfy most use cases, it provides a critical starting point for cross-walking from codes in 1 vocabulary to codes in another. Users report mapping for a variety of purposes, but 1 common use case is mapping from local terminologies to standard terminologies for required reporting and health information exchange. Another common use case is mapping from SNOMED CT to ICD-10-CM for reimbursement and statistical purposes. NLM produces the SNOMED CT to ICD-10-CM Map for this purpose.26
Suggestions for UMLS enhancements
Respondents to the 2018 survey followed the pattern observed in the results of previous UMLS surveys by providing relatively few—but quite diverse—suggestions for enhancements. The responses reflect the wide range of disciplines, technical expertise, and interests within the UMLS user population, as well as differences in preferred distribution formats. A desire for less complexity and more training materials is evident in many of the responses received.
As was also the case in previous surveys, there were more recommendations to add terminologies and code systems that are already in the UMLS Metathesaurus than to add systems not yet in it. Possible reasons for this phenomenon include: use of UMLS-enabled products that do not include all the sources in the Metathesaurus, new users’ lack of familiarity with the UMLS, lack of attention to “new sources added” information distributed with UMLS releases, and the need for simpler documentation and lookup features to discover UMLS coverage. It is also highly likely that some respondents meant to recommend that an included source be updated more frequently in the UMLS or that it be freely available—not that it be added to the Metathesaurus.
Relationship of the literature review results to the survey results
The results of the scoping review of the literature confirm what the 2018 annual survey data convey: the most used UMLS products (Metathesaurus, Metamap) are used to process text and facilitate mapping or linking between terminologies. Scientific literature and EHR data are the most common corpora to which UMLS products are applied. These major current uses of the UMLS are directly aligned with the purpose stated at the outset of the UMLS project: to enable computers to behave as if they “understand” biomedical meaning,2 and its further elucidation in 1998: “Although the term is newer than the project, in essence the objective of the UMLS effort is to build ‘middle-ware’ that enables advanced capabilities in many different health information systems.”3
Current uses and original expectations
Along many dimensions that affect access and use of biomedical and health information, the world today is radically different from the pre-Web environment in which the UMLS was conceived. Nevertheless, key assumptions underlying the UMLS project have proven to be correct: “…the amount of useful biomedical information will continue both to increase and to be dispersed among many databases and systems… .many of the differences in the terminology used in databases and by users reflect important distinctions in purpose and perspective … Although current efforts to standardize the record structure, transmission formats, and terminology of specific types of biomedical information … may reduce the complexity of the UMLS task, they will not eliminate it.”2
The freely available and regularly maintained UMLS resources continue to provide value to researchers and developers in coping with this diversity. Critically, the UMLS Metathesaurus presents the names, concept information, hierarchies, relationships, and attributes from all its source terminologies in a common and explicitly tagged model. This substantially reduces or eliminates the need for time-consuming modeling of individual terminologies when a project requires the use of multiple terminologies with varying models and formats. Central to the UMLS model is concept orientation or the grouping of synonymous names into UMLS concepts, nonsemantic concept identifiers, and the assignment of semantic types to all those concepts. The UMLS Metathesaurus was the first biomedical terminology resource organized by concept, and its development had a significant impact on subsequent medical informatics theory and practice.27 The broad terminology coverage, synonymy, and semantic typing in the UMLS in combination with its lexical tools enable its primary use cases: identifying meaning in text, mapping between vocabularies, and improving information retrieval.
In addition, use of the UMLS Metathesaurus saves time and effort in obtaining access to different terminologies for evaluation, comparison, and research use. Annual survey answers and customer service queries tell us that users do not always understand all of the nuances of the intellectual property restrictions, but the UMLS Metathesaurus license effectively makes it possible to gain access to a broad set of biomedical terminologies for research and evaluation purposes without untangling the various restrictions associated with those terminologies. This can greatly simplify the process of determining which terminologies may also be needed in their native formats—as is usually the case for any vocabulary used in data creation.
CONCLUSION
Thirty years after the first experimental release of the UMLS Knowledge Sources, the UMLS resources are heavily used by researchers and system developers. They are used primarily as intended: to facilitate the interpretation of biomedical meaning in disparate electronic information and data in many different computer systems serving scientists, health professionals, and the public. Enhancements to UMLS structures and production methods that enable more efficient and effective determination of synonymy, assignment of semantic types, and incorporation of terminology updates are therefore likely to benefit the majority of UMLS users.
FUNDING
This research was supported by the Intramural Research Program of the US National Institutes of Health, National Library of Medicine, and, in part, by an appointment to the NLM Associate Fellowship Program sponsored by the National Library of Medicine and administered by the Oak Ridge Institute for Science and Education.
AUTHOR CONTRIBUTIONS
LA conceptualized overall project idea; DA was responsible for UMLS survey data extraction/analysis, writing; SB conducted literature review methodology; SB, LA, AR, BH conducted literature review analysis; SB performed literature review data extraction/analysis writing; LA, AR, DA synthesized the manuscript under BH guidance. LA and BH performed final edits. All authors reviewed, edited, and signed off on manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
ACKNOWLEDGMENTS
Thanks to Jim Mork and Olivier Bodenreider of NLM who kindly read a draft and provided comments. Stacy Brody was with the NLM/NIH during this work and is current at George Washington University, Himmelfarb Health Sciences Library.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/index.htmlAccessed February 7, 2020
- 2. Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med 1993; 32 (04): 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Humphreys BL, Lindberg DA, Schoolman HM, et al. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc 1998; 5 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc 1993; 81 (2): 170–7. [PMC free article] [PubMed] [Google Scholar]
- 5. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. EndNote. https://endnote.com/Accessed February 7, 2020
- 7. Colandr. https://www.colandrapp.com/Accessed February 7, 2020
- 8. Fung KW, Hole WT, Srinivasan S. Who is using the UMLS and how - insights from the UMLS user annual reports. AMIA Annu Symp Proc 2006; 2006: 274–8. [PMC free article] [PubMed] [Google Scholar]
- 9. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001; 2001: 17–21. [PMC free article] [PubMed] [Google Scholar]
- 10. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soysal E, Wang J, Jiang M, et al. CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25 (3): 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soldaini L, Goharian N. Quickumls: a fast, unsupervised approach for medical concept extraction. MedIR Workshop, Sigir 2016; 17: 1–4. [Google Scholar]
- 13. Kilicoglu H, Rosemblat G, Fiszman M, et al. Constructing a semantic predication gold standard from the biomedical literature. BMC Bioinformatics 2011; 12 (1): 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Radiological Society of North America (RSNA). Radlex. http://radlex.org Accessed March 19, 2020
- 15. World Health Organization (WHO). International Classification of Diseases. ICD-11. https://icd.who.int/browse11/l-m/en Accessed March 19, 2020
- 16. World Health Organization (WHO). International Classification of Diseases for Oncology. ICD-O. https://www.who.int/classifications/icd/adaptations/oncology/en/ Accessed March 19, 2020
- 17.Brody S. The Unifed Medical Language System: A Scoping Review of its Use in Research. https://www.nlm.nih.gov/about/training/associate/associate_projects/BrodyScopingReview2019.pdf Accessed May 11, 2020. [Google Scholar]
- 18. Nawab RMA, Stevenson M, Clough P. An IR-based approach utilising query expansion for plagiarism detection in MEDLINE. IEEE/ACM Trans Comput Biol Bioinform 2016; 99: 1. [DOI] [PubMed] [Google Scholar]
- 19. Choi S, Choi J, Yoo S, et al. Semantic concept-enriched dependence model for medical information retrieval. J Biomed Inform 2014; 47: 18–27. [DOI] [PubMed] [Google Scholar]
- 20. Divita G, Luo G, Tran LT, et al. General symptom extraction from VA electronic medical notes. Stud Health Technol Inform 2017; 245: 356–60. [PubMed] [Google Scholar]
- 21. Lee JH, Oh BJ, Ahn JY, et al. Effectiveness of automatic acute stroke alert system based on UMLS mapped local terminology codes at emergency department. AMIA Annu Symp Proc 2008; 2008: 1018. [PubMed] [Google Scholar]
- 22. Fiszman M, Rindflesch TC, Kilicoglu H. Summarizing drug information in Medline citations. AMIA Annu Symp Proc 2006; 2006: 254–8. [PMC free article] [PubMed] [Google Scholar]
- 23. Liu M, Wu Y, Chen Y, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc 2012; 19 (e1): e28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen D, Zhang R, Liu K, et al. Knowledge discovery from posts in online health communities using Unified Medical Language System. Int J Environ Res Public Health 2018; 15 (6): 1291. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jimeno-Yepes A, MacKinlay A, Han B, et al. Identifying diseases, drugs, and symptoms in Twitter. Stud Health Technol Inform 2015; 216: 643–7. [PubMed] [Google Scholar]
- 26. SNOMED CT to ICD-10-CM Map. https://www.nlm.nih.gov/research/umls/mapping_projects/snomedct_to_icd10cm.html Accessed February 7, 2020
- 27. Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998; 37 (04/05): 394–403. [PMC free article] [PubMed] [Google Scholar]


