Abstract
Vaccinology is shifting toward synthetic RNA platforms which allow for rapid, scalable, and cell-free manufacturing of prophylactic and therapeutic vaccines. The simple development pipeline is based on in vitro transcription of antigen-encoding sequences or immunotherapies as synthetic RNA transcripts, which are then formulated for delivery. This approach may enable a quicker response to emerging disease outbreaks, as is evident from the swift pursuit of RNA vaccine candidates for the global SARS-CoV-2 pandemic. Both conventional and self-amplifying RNAs have shown protective immunization in preclinical studies against multiple infectious diseases including influenza, RSV, Rabies, Ebola, and HIV-1. Self-amplifying RNAs have shown enhanced antigen expression at lower doses compared to conventional mRNA, suggesting this technology may improve immunization. This review will explore how self-amplifying RNAs are emerging as important vaccine candidates for infectious diseases, the advantages of synthetic manufacturing approaches, and their potential for preventing and treating chronic infections.
Subject terms: Infectious diseases, RNA vaccines
Introduction
Since the 18th century vaccines have been developed to prevent and eliminate the spread of infectious diseases [1]. They form the foundation of global public health programs and have major socioeconomic benefits [2]. Prophylactic immunization offers pre-exposure protection and supports development of herd immunity. Therapeutic vaccination, a form of immunomodulation, assists in the treatment of infectious diseases and cancers. The longstanding clinical value of currently licensed vaccines continues to encourage further research on novel vaccination approaches. These are aimed at improving prophylactic and therapeutic efficacy, developing new technologies, streamlining manufacturing processes, and enabling a rapid response to emerging infectious diseases. Nucleic acid vaccines represent one such approach where synthetic sequences are used to express antigenic peptides or proteins in situ [3]. Genetic immunization may promote superior adaptive immunity by activating both humoral and cell-mediated responses, and has manufacturing advantages over traditional vaccines. Initial studies focused heavily on developing DNA rather than RNA candidates [4], because there were concerns about stability and large-scale production of RNA-based therapeutics. However DNA vaccines have generally performed poorly in human clinical trials [5], which has led to a renewed interest in RNA vaccinology for infectious diseases. To some extent this shift has been attributed to successes of cancer immunotherapy research. The transient nature and cytosolic location of RNA improves the safety profile of these nucleic acid vaccines. This is in contrast to their more stable DNA counterparts, which require nuclear delivery, promoter-driven expression, and risk integration within the host genome. These properties have led to major investments in RNA-based therapeutics over the past few years [6].
There are currently two different types of synthetic RNA vaccines: conventional mRNA and self-amplifying RNA (saRNA) (Fig. 1). Use of conventional mRNA strategies (also referred to as nonreplicating or non-amplifying mRNA) against infectious diseases and cancers has been investigated in several preclinical and clinical trials [7]. In vitro transcribed mRNAs encoding viral antigens have been explored as vaccines, while those encoding therapeutic proteins, such as antibodies or immune modulators, have been considered for immunotherapy. The incorporation of chemically modified nucleotides, sequence optimization, and different purification strategies improve efficiency of mRNA translation and reduce intrinsic immunogenic properties [7]. However, antigen expression is proportional to the number of conventional mRNA transcripts successfully delivered during vaccination. Achieving adequate expression for protection or immunomodulation may thus require large doses or repeat administrations. saRNA vaccines, which are genetically engineered replicons derived from self-replicating single-stranded RNA viruses [8, 9], address this limitation. They can be delivered as viral replicon particles (VRPs) with the saRNA packaged into the viral particle, or as a completely synthetic saRNA produced after in vitro transcription. To generate replication-defective VRPs, envelope proteins are provided in trans as defective helper constructs during production. Resulting VRPs therefore lack the ability to form infectious viral particles following a first infection, and only the RNA is capable of further amplification. VRPs may be derived from both positive-sense and negative-sense RNA viruses, however the latter are more complex and require reverse genetics to rescue the VRPs [10]. As with gene therapy, there are several issues associated with the use of viral vectors for vaccine development. These include immunogenicity of the vector itself, which can elicit an undesirable immune response and prevent subsequent booster administrations using the same vector [11]. Preexisting immunity to the viral vector can also render a vaccine ineffective. As with live-attenuated vaccines, replication-competent alphavirus vectors also pose the threat of viral reactivation [9]. To circumvent this, saRNA vaccines can be produced and delivered in a similar manner to conventional mRNA vaccines. Positive-sense alphavirus genomes that have been commonly used for saRNA vaccine design include the Venezuelan equine encephalitis virus (VEE), Sindbis virus (SINV), and Semliki forest virus (SFV) (Table 1). The alphavirus replicase genes encode an RNA-dependent RNA polymerase (RdRP) complex which amplifies synthetic transcripts in situ. The antigenic or therapeutic sequence is expressed at high levels as a separate entity and further proteolytic processing of the immunogen is not required. As a result of their self-replicative activity, saRNAs can be delivered at lower concentrations than conventional mRNA vaccines to achieve comparable antigen expression [12]. This review will explore the recent preclinical progress of prophylactic and therapeutic saRNA vaccine candidates, particularly for chronic infectious diseases such as the human immunodeficiency virus 1 (HIV-1), and emphasize their potential for innovative vaccine development.
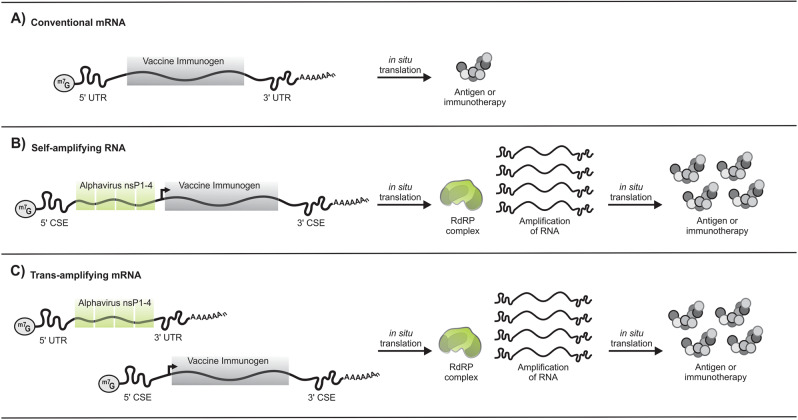
Fig. 1. Conventional, self-amplifying, and trans-amplifying RNA vaccine designs.
A 5′ cap (m7G) and poly A tail are common to all RNA transcripts. A Conventional mRNAs encode the vaccine immunogen and flanking 5′ and 3′ UTRs. An antigen or immunotherapy is translated from the nonreplicating transcript. B Self-amplifying RNA encodes 5′ and 3′ CSE sequences, the nsP1-4 genes, a subgenomic promoter, and the vaccine immunogen. Following in situ translation, the nsP1-4 proteins form an RdRP complex which recognizes flanking CSE sequences and amplifies vaccine-encoding transcripts. This results in an accumulation of the antigen or immunotherapy within the cell. C Trans-amplifying mRNAs use two different transcripts to achieve a similar effect to self-amplifying RNAs. A conventional mRNA encoding the nsP1-4 genes flanked by 5′ and 3′ UTRs is co-delivered with a separate transcript that encodes the viral CSE sequences, the subgenomic promoter, and the vaccine immunogen. In situ translation of the conventional mRNA results in the formation of the RdRP complex, which subsequently amplifies the vaccine-encoding transcript to result in the accumulation of the antigen or immunotherapy. UTR untranslated region, CSE conserved sequence elements, nsP1–4 nonstructural proteins 1–4, RdRP RNA-dependent RNA polymerase.
Table 1.
Clinical and preclinical synthetic saRNA vaccine studies for infectious diseases.
| Infectious disease | Replicon | Immunogen | Delivery | Animal | Year (reference) |
|---|---|---|---|---|---|
| Clinical studies | |||||
| Rabies | – | Glycoprotein G | CNE | Human | 2019 (NCT04062669) |
| COVID-19 | VEE | Spike protein | LNP | Human | 2020 (ISRCTN17072692) |
| Preclinical studies | |||||
| RSV | SFV | F glycoprotein | Naked | Miceb | 2001 [80] |
| VEE–SINV | F glycoprotein | LNP | Mice, ratsb | 2012 [81] | |
| VEE–SINV | F glycoprotein | CNE | Mice | 2014 [68] | |
| Influenza | SFV | NP | Naked | Mice | 1994 [79] |
| SFV | HA | Naked | Miceb | 2001 [80] | |
| VEE–SINV | HA | LNP | Mice | 2013 [14] | |
| CSFV | HA/NP | Chitosan NGA | Mice, rabbit | 2014 [71] | |
| VEE–SINV | HA | CNE | Miceb, ferretb | 2015 [125] | |
| VEE–SINV | NP | LNP | Mice | 2015 [126] | |
| VEE–SINV | M1/NP | LNP | Miceb | 2016 [85] | |
| VEE | HA | MDNP | Miceb | 2016 [127] | |
| CSFV | HA/NP | CPP PEI | Pigs | 2017 [128] | |
| CSFV | NP | Cationic lipid | Mice | 2018 [129] | |
| – | HA | PEI | Miceb | 2018 [12] | |
| VEE | HA | Neutral LPP | Mice | 2019 [55] | |
| – | HA | MLNP | Mice | 2019 [54] | |
| Trans-amplifying | HA | Naked | Miceb | 2020 [62] | |
| VEE | HA | pABOL | Miceb | 2020 [50] | |
| Coronavirus | VEE | Spike protein | LNP | Mice | 2020 [86] |
| LIV | SFV | prM-E | Naked | Miceb | 2001 [80] |
| TBEV | TBEV | Δ TBEV capsid | Gene gun | Miceb | 2004 [130] |
| TBEV | Δ TBEV capsid | Gene gun | Miceb | 2005 [131] | |
| HIV | VEE–SINV | Env | LNP | Mice | 2012 [81] |
| VEE–SINV | Env | Electroporation | Mice | 2013 [132] | |
| VEE–SINV | Env | CNE | Rabbit | 2014 [68] | |
| VEE–SINV | Env | CNE | NHP | 2015 [121] | |
| SFV | Gag/Pol mosaic | PEI | Mice | 2019 [123] | |
| VEE | eOD-GT8 | LNP | Mice | 2019 [120] | |
| VEE | Env | Exterior LNP | Mice | 2019 [58] | |
| CMV | VEE–SINV | gB/pp65-IE1 | CNE | NHP | 2014 [68] |
| Ebola | VEE | Glycoprotein | MDNP | Miceb | 2016 [127] |
| Toxoplasma gondii | VEE | Multimera | MDNP | Miceb | 2016 [127] |
| SFV | NTPase-II | LNP | Miceb | 2017 [133] | |
| GAS | VEE–SINV | SLOdm | CNE | Miceb | 2017 [134] |
| GBS | VEE–SINV | BP-2a | CNE | Miceb | 2017 [134] |
| Zika | VEE | prM-E | MDNP | Mice | 2017 [91] |
| VEE | prM-E | NLC | Miceb, guinea pigs | 2018 [90] | |
| VEE | prM-E | Naked | Miceb | 2019 [89] | |
| VEE | VEE | Attenuated VEE | CNE | Miceb | 2019 [88] |
| Rabies | VEE–SINV | Glycoprotein G | CNE | Rats | 2020 [92] |
| VEE–SINV | Glycoprotein G | Liposome, nanoparticle, CNE | Mice | 2020 [59] | |
BP-2a GBS pilus 2a backbone protein, CMV cytomegalovirus, CSFV classical swine fever virus, CNE cationic nanoemulsion, Env envelope, GAS group A streptococci, GBS group B streptococci, gB glycoprotein B, HA haemagglutinin, HIV human immunodeficiency virus, LIV louping ill virus, LNP lipid nanoparticle, LPP lipopolyplexes, M1 matrix protein 1, MLNP manosylated LNP, MDNP modified dendrimer nanoparticle, NGA nanogel alginate, NHP nonhuman primate, NLC nanostructured lipid carrier, NP nucleoprotein, pABOL poly(CBA-co-4-amino-1-butanol (ABOL)), PEI polyethylenimine, Pol polymerase, prM-E premembrane and envelope glycoproteins, RSV respiratory syncytial virus, SFV Semliki forest virus, SINV Sindbis virus, SLOdm double-mutated GAS Streptolysin-O, TBEV tick-borne encephalitis virus, VEE Venezuelan equine encephalitis virus, VEE–SINV alphavirus chimera based on the VEE and SINV replicons.
aMultimer comprised of granule protein 6 (GRA6), rhoptry protein 2A (ROP2A), rhoptry protein 18 (ROP18), surface antigen 1 (SAG1), surface antigen 2A (SAG2A), and apical membrane antigen 1 (AMA1).
bVaccination conferred protection.
Producing RNA vaccines
The need for rapid vaccine development in response to emerging pathogens has become devastatingly clear during the SARS-CoV-2 pandemic. A major caveat of live-attenuated, inactivated, toxin, or subunit vaccine manufacturing is the requirement for intricate cell culture technologies. These need dedicated facilities to produce individual vaccines as well as lengthy safety assessments to exclude risks posed by biological contaminants. In comparison RNA vaccine production is simple, can be easily adapted to accommodate new candidates within an established manufacturing pipeline, and is cost effective [13]. The in vitro transcription reaction used to produce both conventional mRNA and saRNA vaccines is cell-free and Good Manufacturing Practice-compliant reagents are available, facilitating quick turnaround times. This has been illustrated by Hekele et al. who produced a lipid nanoparticle (LNP) formulated saRNA vaccine for H7N9 influenza in 8 days [14]. Prompt RNA therapeutic manufacturing capabilities have more recently been revealed amidst the COVID-19 pandemic. The first SARS-CoV-2 vaccine to enter phase 1 clinical trials is the LNP-encapsulated mRNA-1273 developed by Moderna and the Vaccine Research Center at the National Institute of Health (ClinicalTrials.gov—NCT04283461) [15, 16]. Impressively it took only 25 days to manufacture the first clinical batch which commenced testing on the 16th of March 2020. With LNP mRNA-1273 receiving fast-track designation to phase 3 (NCT04470427), the efficiency of the vaccine as well as the capacity of the manufacturing pipeline will be tested.
Conventional and synthetic saRNA vaccines are essentially produced in the same manner [13, 17, 18]. Briefly, an mRNA expression plasmid (pDNA) encoding a DNA-dependent RNA polymerase promoter (typically derived from the T7, T3, or SP6 bacteriophages) and the RNA vaccine candidate is designed as a template for in vitro transcription. The flexibility of gene synthesis platforms is a key advantage. For conventional mRNA vaccines the antigenic or immunomodulatory sequence is flanked by 5′ and 3′ untranslated regions (UTRs). A poly(A) tail can either be incorporated from the 3′ end of the pDNA template, or added enzymatically after in vitro transcription [19]. saRNA vaccine pDNA templates contain additional alphavirus replicon genes and conserved sequence elements (Fig. 1). The nonstructural proteins 1, 2, 3, and 4 (nsP1-4) are essential for replicon activity as they form the RdRP complex [20]. In vitro transcription is performed on the linear pDNA template, typically with a T7 DNA-dependent RNA polymerase, resulting in multiple copies of the RNA transcript. After the RNA is capped at the 5′ end and purified, it is ready for formulation and delivery.
Refining saRNA pharmacokinetics
Substantial effort has gone into understanding and improving RNA production, stability, translation, and pharmacokinetics. Revising the 5′ cap structure, controlling the length of the poly(A) tail, including modified nucleotides, codon or sequence optimization, as well as altering the 5′ and 3′ UTRs are just some of the factors under consideration (recently reviewed in [21]). Balancing the intrinsic and extrinsic immunogenic properties of the synthetic RNA, the vaccine antigen, and delivery formulation are equally important for longer saRNA transcripts. As the field of synthetic RNA vaccinology is still relatively new it is difficult to decipher which technologies are indispensable. Some studies show that incorporating various pseudouridine-modified nucleotides during transcription enhanced translation and reduced RNA-associated immunogenicity [22, 23], whilst others show no discernible advantage of such modifications [24, 25]. As saRNAs use host-cell factors for mRNA replication, the addition of modified nucleotides may prove less valuable as they would be lost during amplification [26]. One practical approach to improving translation of saRNA vaccines is through optimization of 5′ and 3′ UTRs which is based on the evolution of naturally occurring alphaviruses [27]. The single-stranded RNA genome forms a variety of secondary structures to allow alphaviruses to bypass requirements of normal host-cell translation processes [28, 29] and evade immune responses [30–32]. Revising the sequence encoding the nsP1-4 replicon genes may also prove beneficial. An in vitro evolution strategy using interferon- (IFN-) competent cells was adopted by Li et al. to identify mutations within the VEE nonstructural proteins that improve in situ expression of the subgenomic RNA [33].
Five-prime Caps
A well-researched approach to protecting in vitro transcribed RNAs from nuclease digestion and augmenting translation is modification of the synthetic 5′ cap structure. The anti-reverse cap analog [34] and recent phosphorothioate derivatives [35] have been shown to improve in situ translation of RNAs by only capping transcripts in the forward orientation. Post-transcription capping enzymes derived from the vaccinia virus [36, 37] have high capping efficiencies, and when combined with 2′-O-methyltransferases, generate Cap 1 structures that mimic natural eukaryotic mRNAs [38]. To further improve co-transcriptional capping, TriLink BioTechnologies has recently developed the CleanCapTM system [39]. This technology can achieve Cap 1, Cap 2, and Cap 1 epitranscriptomic derivatives such as m6Am, which may further enhance RNA stability by evading de-capping metalloenzymes encoded by DCP2 [40, 41]. Interestingly the CleanCap Reagent AU has specifically been designed to generate Cap 1 structures on saRNA transcripts derived from positive-sense alphaviruses. Cap analogs play an important role in evading cellsʼ innate immune response as uncapped and Cap 0 RNAs may stimulate IFN responses. However, certain alphaviruses bypass the necessity for 2′-O-methylated caps [30], suggesting that careful design of the 5′ UTRs of may negate Cap 0 immunity.
Synthetic RNA immunogenicity
Stimulation of the innate immune system by in vitro transcribed saRNA vaccines is a complex topic which still requires empirical study. Immunization should ideally elicit an antigen-specific immune response, however it is currently unclear whether peripheral innate immune activation from in vitro transcribed RNAs enhances or compromises this response. Improving the in vitro transcription reaction [42, 43] and liquid chromatography purification [44] may reduce unintended immune stimulation by spurious by-products, including double-stranded RNAs (dsRNAs). For saRNA vaccines, the immunogenic benefits of purification may be less significant than for conventional mRNAs. Although purification may remove nonspecific dsRNAs, the formation of new dsRNA intermediates is inevitable during self-amplification. The co-delivery of immune modulating transcripts, particularly pattern recognition receptors (PRRs) antagonists, may help mitigate these effects. Conventional mRNAs encoding the E3, K3, and B18/B18R vaccinia virus immune evasion proteins, or the influenza A virus nonstructural protein 1 (NS1) have shown potential in this regard [45–47]. Beissert et al. augmented saRNA reporter gene expression in vitro and in vivo by combining conventional mRNAs encoding E3, K3, and B18, to reduce protein kinase R and IFN signaling in a synergistic manner [45].
Delivering large saRNA transcripts
Choice of delivery formulations, the inoculation site, and adjuvants are additional immunological considerations [7]. Nonviral formulations including cationic lipids, LNPs, polymers, and protamine sulfate, as well as physical methods such as electroporation may be used to deliver RNA therapeutics and vaccines [48, 49]. The inclusion of the nsP1-4 replicon sequence in saRNAs makes them much longer than their conventional counterparts which is important for formulation. Over the past year several new approaches aimed at improving the delivery of saRNAs have been described. Novel bioreducible polymer formulations using high molecular weight poly(CBA-co-4-amino-1-butanol) (pABOL) [50], ornithine dendrimers [51], mannosylated polyethyleneimine [52], or multiple linear peptides [53], may help facilitate intracellular trafficking and endosomal release. Blakney et al. demonstrated the preclinical efficacy of their 8 kDa pABOL polymer formulation to deliver haemagglutinin- (HA-) encoding saRNAs in mice [50]. Modifications to LNP formulations have also been described, where incorporating a mannose-cholesterol amine conjugate into LNPs improved intradermal (ID) immunization [54]. Manosylation of polyethyleneimine improved delivery of saRNA reporter constructs to human skin explants [52]. Intramuscular (IM) delivery of saRNAs as neutral lipopolyplexes (LPPs) resulted in an increase in antigen-specific T cells with a concurrent loss of antigen-expressing cells [55]. These LPPs were generated by creating core RNA/polyethylenimine polyplexes before encapsulation in PEG-anionic liposome formulations with mannosylated lipids. Inclusion of mannose is aimed at improving vaccine delivery to antigen presenting cells [56, 57]. Interestingly, saRNAs may not require conventional encapsulation to protect them from RNAse degradation. Complexing saRNAs on the exterior of positively charged LNPs formulated with dimethyldioctadecylammonium (DAA) cationic lipids conferred complete protection from direct RNaseA treatment [58]. Mice that were immunized with an HIV-1 Envelope (Env) saRNA complexed to the surface of DDA LNPs developed peak antibody titers following a single injection. When delivered as an encapsulated LNP, a boost injection was required to achieve equivalent antibody titres [58].
With so many different nonviral formulations available, Anderluzzi et al. sought to compare liposomes, solid LNPs, polymeric nanoparticles, and emulsions for the delivery of an saRNA vaccine-encoding the rabies virus glycoprotein [59]. In their hands, low-dose saRNA nanoparticles complexed with the non-ionizable cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) initially resulted in similar antibody titres to Rabipur (RabAvert), a commercially licensed rabies vaccine. However, the proprietary cationic nanoemulsion (CNE) 56 from GlaxoSmithKline (GSK, Rockville, MD, USA) outperformed all DDA and DOTAP formulations [59]. Comparing delivery approaches in side-by-side studies such as this would assist in determining the usefulness of each formulation. Reducing the size of the saRNA vaccine transcript by employing a trans-amplifying approach may also address concerns about inefficient delivery. Here the saRNA is split into two transcripts, the first encoding the nsP1-4 replicon complex and the second encoding the gene of interest as a “transreplicon” (Fig. 1) [60–62]. This approach was recently adopted by Beissert et al. [62] who generated an influenza vaccine transreplicon based on a SFV saRNA design. The trans-amplifying strategy worked best when the replicon was delivered as a sequence-modified mRNA transcript and not as an saRNA. Mice that received ID injections were successfully immunized, produced functional virus-neutralizing antibodies, and were protected following influenza challenge [62].
Augmenting vaccine immunity
Vaccines traditionally incorporate adjuvants to boost adaptive immunity and shape T-cell responses [63]. Aluminum salts, emulsions, lipid analogs, and virosomes are a few examples of adjuvants that are currently included in licensed vaccines. The importance of such adjuvants in RNA vaccinology remains unclear and may be contingent on the types of modifications included during design and manufacturing. To date, clinical trials with conventional mRNA vaccines have shown limited humoral immunity. As saRNAs contain native alphavirus motifs and mimic viral translation in situ they have the propensity to enhance immunization through stimulation of PRRs [64]. Further efforts to augment or direct RNA vaccine-associated immunity have also been described. RNActive®, a protamine complexed mRNA [65, 66] and RNAdjuvant®, a proprietary immunostimulatory cationic peptide formulation [67] have been developed by CureVac AG to improve the efficacy of vaccines. saRNA CNEs incorporating the adjuvant MF59 (Novartis) have shown comparable immunological responses to a subunit vaccine comprising the same adjuvant [68]. Lipid-based delivery formulations are well established adjuvants [69] and have been adapted to promote RNA vaccine immuno-potentiation [70]. The inclusion of mannosylated conjugates [52, 54, 55] and chitosan-based nanogel alginate [71] may help shape the immune response by enhancing receptor-mediated endocytosis of saRNA vaccines by dendritic cells. Manara et al. recently established a chemokine-adjuvant strategy using a murine granulocyte-macrophage colony-stimulating factor saRNA in conjunction with their influenza saRNA vaccine [72]. Including cytokine or chemokine adjuvants that augment cytolytic T-cell responses would improve the therapeutic potential of RNA vaccines, particularly for chronic infectious diseases. Another study by Blakney et al. described cationic adjuvant formulations with PPR agonists for saRNA delivery [73]. Surprisingly, the incorporation of toll-like receptor (TLR) 7/8 agonists R848 and 3M-052 had little effect on innate immunity as the saRNA dominated the IFN response. Perche et al. [55] included the stimulator of interferon genes (STING) agonist, cyclic diadenylate monophosphate (c-di-AMP), to modulate their LPP formulated saRNA influenza vaccine. This approach has also been used to improve the immunogenicity of cancer mRNA vaccines, where STING-activatable cyclic lipids were incorporated into LNPs [74].
Whether these adjuvant formulations improve the clinical potential of saRNA vaccines remains to be assessed. Preclinical studies indicate that both conventional and saRNAs elicit strong IFN responses that may be detrimental to vaccine efficacy [75, 76]. In this regard modifications to the RNA sequence, cap structure, or the inclusion of PRR antagonists may prove beneficial [77]. When using the VEE replicon architecture, Huysmans et al. showed that expression kinetics of saRNAs delivered by ID electroporation were superior to those of LNP-formulated saRNAs [78]. A rapid and strong innate immune response to the LNP saRNA had a negative effect on the translation and amplification of the bioluminescence reporter sequence. Strong IFN stimulation from both the saRNA and delivery formulation may impede in situ translation of the vaccine, ultimately preventing successful immunization. Extensive comparative and clinical studies will no doubt influence optimization of saRNA vaccine pharmacokinetics and shed light on which technologies are crucial for translating preclinical approaches into clinically relevant vaccines.
Expanding the scope of vaccines for infectious diseases
Licensed vaccines continue to have a major impact on global health as they reduce mortality associated with illnesses such as measles, mumps, pertussis, smallpox, and polio [2]. But for many longstanding and emerging infectious diseases, there is currently no prophylactic or therapeutic immunization available. The notion of using synthetic saRNAs as a vaccine was first described by Zhou et al. when they modified an SFV replicon to express the influenza nucleoprotein (NP) [79]. A few years later the same group described a similar approach to immunize mice against influenza, respiratory syncytial virus (RSV), or louping ill virus (LIV) [80]. These seminal studies demonstrated the potential of in vitro transcribed saRNA vaccines to elicit protective immune responses in mice following IM injection of unformulated RNA. More than a decade later Geall et al. [81] described the first LNP-formulated saRNA vaccine for RSV and HIV-1, using an alphavirus chimera based on the VEE and SINV replicons [82]. An ionizable cationic lipid was used to encapsulate the saRNA, which conferred protection from RNase degradation, and following IM injection increased immunogenicity when compared to an unformulated counterpart. Prophylactic protective immunity was achieved, which was equivalent to that of a VRP, and comparable to neutralizing titres required to protect infants from infection [81]. Notably, this study preceded the vast array of NVV formulations now used to deliver saRNA vaccines in preclinical models (Table 1) (reviewed by [7, 26, 83]).
Contagious respiratory viruses
Influenza is a popular candidate for saRNA vaccine development and a diverse range of delivery approaches have been established to improve immunization (Table 1). As discussed earlier, recent formulations under preclinical evaluation include the cationic polymer-based pABOLs [50], Mannosylated LNPs (MLNPs) [54], and neutral LPPs [55]. Strong antibody responses were attained with low-dose (1 µg per primer injection) saRNAs complexed with pABOLs, and the addition of mannose improved IgG response times in LNP formulations [50, 54]. Interestingly the pABOLs conferred protection following IM but not ID injections. MLNPs achieved enhanced ID immunization resulting in superior T-cell responses when compared to a monovalent inactivated influenza vaccine. This may facilitate the delivery of saRNA vaccines using needle-free devices. An important consideration for seasonal influenza is how antigenic drift could render a vaccine ineffective [84]. saRNAs can accommodate multiple antigens in a flexible pDNA template [85] which could be modified to include new epitopes. With conventional mRNA vaccines entering phase 3 clinical trials for the highly contagious SARS-CoV-2, an saRNA vaccine candidate developed by researchers at Imperial College London and Acuitas Therapeutics has recently entered a UK-based phase 1/2 clinical trial (isrctn.com—ISRCTN17072692). Preclinical studies in mice showed that the LNP-nCoVsaRNA vaccine, which encodes the SARS-CoV-2 spike protein, achieved a dose-dependent immune response following IM injection [86]. Vaccination elicited antigen-specific IgG antibodies and T-helper type I cells, with the lowest dose (0.01 µg) achieving higher SARS-2 antibody titres than in recovered COVID-19 patients. Dose-dependent neutralization of pseudo- and wild-type SARS-CoV-2 was shown, again with the lowest dose attaining a higher neutralization efficiency than COVID-19 patient samples. According to a report from Imperial College London the vaccine took only 14 days to develop and will embark on larger clinical trials later this year [87].
Insect-transmitted viruses
Encouraging preclinical results for protection against mosquito-borne diseases have also recently been described. By creating live- and irreversibly attenuated (LAV and IAV) versions of the VEE replicon, Samsa et al. developed a CNE formulated saRNA vaccine for VEE [88]. The safer (reversion-deficient) IAV was less immunogenic than the LAV, but still protected 70% of mice following VEE aerosol challenge. Using the same replicon, Zhong et al. [89] endeavored to immunize mice against Zika virus using an unformulated saRNA, however not all immunocompetent mice developed neutralizing antibodies following ID injection. Protective cellular and humoral immune responses were only detected in type I IFN receptor double knockout mice (Ifnar−/−), suggesting the self-adjuvant activity of naked saRNA compromised antigen-specific immunity. Previous Zika virus saRNA vaccine studies achieved protective immunity following IM injection of immunocompetent mice, but only when the RNA was formulated in a nanostructured lipid carrier [90]. Similarly, modified dendrimer nanoparticles [91] elicited antigen-specific IgG response in mice, highlighting the importance of formulation and injection site for vaccine efficacy.
Animal-transmitted virus
Rabies, another infectious disease with a licensed vaccine, has recently been the focus of GSK’s vaccinology program (RG-SAM GSK3903133A) which boasts the only clinical trial for a synthetic saRNA to date (NCT04062669). Preclinical evaluations of CNE formulated saRNA encoding the glycoprotein G antigen, showed that the vaccine was well tolerated following multiple IM injections in rats [92]. The saRNA-derived rabies RNA was detected up to 2 months after a single injection, emphasizing the durability of the approach. A second study evaluated immunogenicity in mice using the licensed inactivated rabies virus vaccine Rabipur (RabAvert) as a benchmark for immunization [59]. Positive protective antibody responses similar to Rabipur were observed following a single IM injection of DOTAP nanoparticle-encapsulated or CNE56-absorbed saRNAs (0.5 and 1.5 µg). A second injection boosted antigen-specific IgG titres in all encapsulation formulations (DOTAP nanoparticles, DOTAP liposomes, and DDA liposomes) but only high-dose CNEs elicited responses comparable to the licensed vaccine [59]. The first clinical trial of GSKs CNE formulated RG-SAM vaccine (NCT04062669) will establish the safety of a 3-dose IM regimen and determine the immunogenicity of saRNAs in humans. It will be interesting to compare the results of this trial with those of the CureVac conventional mRNA Rabies vaccine currently under investigation. An initial phase 1 trial of the protamine-formulated conventional mRNA (CV7201) proved disappointing, but following modification and LNP formulation has re-entered clinical testing (CV7202; NCT03713086) [93]. These parallel investigations will help ascertain the clinical relevance of conventional and saRNA vaccines.
The promise of an HIV-1 vaccine
HIV-1 remains an incurable global health challenge and despite intensive studies an effective vaccine is not yet available. Since 1984 scientists have attempted to develop a prophylactic vaccine, however numerous clinical trials have been met with few successes [94]. Sequence diversity of the HIV-1 viral Env glycoprotein required for entry into cells has presented a challenge to vaccine design [95, 96]. It has proven difficult to elicit broadly neutralizing antibodies (bNAbs) that are able to neutralize several HIV-1 strains with conventional vaccines. Protein-based vaccines have been widely investigated, but with some disappointing results in clinical trials [97]. New strategies currently under investigation include novel antibody approaches [98–100], antibody-like inhibitors [101], polyvalent immunogens [102], and adeno-associated viral vectors [103]. VRPs are also popular HIV-1 vaccine candidates which have shown promise in nonhuman primates [104–106] and mice [107]. While these traditional platforms continue to strive toward clinical efficacy, renewed interest in RNA vaccinology has led to development of unique HIV-1 candidates which have the potential to provide both prophylactic and therapeutic efficacy. Conventional mRNAs have been investigated in preclinical and clinical studies [108–110] with saRNA approaches gaining interest over the past few years. Phase I and II clinical trials have demonstrated the safety and feasibility of mRNA vaccines [111–116], but inadequate immune responses have resulted in disappointing clinical efficacy. An RNA (AGS-004) encoding multiple HIV-1 proteins and the CD40 ligand (CD40L) was tested in phase I and IIB clinical trials [113–115], but the vaccine did not reduce viral load or prevent viral rebound. When combined with the latency-reversing agent vorinostat, no substantial impact on the frequency of resting T-cell infection was observed [117]. A naked mRNA (iHIVRNA) encoding HIV-1 immunogens and the dendritic cell activator TriMix (CD40L, CD70, and caTLR4) was safe in a phase I trial, but did not show sufficient immunogenicity in a phase II trial which was recently terminated (NCT02888756) [118, 119].
The disproportion between preclinical and clinical success reinforces the need for strategies that improve RNA vaccine immunogenicity or immunotherapy in humans. The self-adjuvant effect of saRNAs may augment immunity and novel HIV-1 candidates have shown potential in preclinical models of infection (Table 1). In 2012, to highlight the flexibility of their LNP-formulated replicon, Geall et al. [81] described the first HIV-1 Env gp140 saRNA vaccine in mice. Env-specific immune responses were detected following multiple delivery routes (IM, ID, and subcutaneous), with IM injections achieving superior CD8+ T-cell responses. More recently, an saRNA encoding the HIV-1 Env gp140 protein was formulated on the interior or exterior of LNPs and administered to mice by IM injection, resulting in an antigen-specific IgG response [58]. Peak antibody titres were achieved after a single injection of the exterior formulated LNPs or a prime-boost of the encapsulated saRNA. LNPs have also been used to deliver an saRNA encoding a germline-targeting gp120 immunogen (eOD-GT8) designed to self-assemble into a 60-mer protein nanoparticle and prime B-cells capable of producing VRC01-like bNAbs [120]. Administration of this saRNA to transgenic mice expressing human inferred-germline B-cell receptors led to primed B-cell responses and somatic hypermutation characteristic of bNAb development. The efficacy and safety of MF59 CNE formulations was demonstrated in nonhuman primates where an saRNA vaccine encoding the HIV-1 gp140 Env protein was administered intramuscularly to rhesus macaques [121]. Higher levels of T-cell response, neutralizing antibodies and anti-envelope antibodies including V1V2 antibodies, which correlated with reduced risk of infection in the RV144 vaccine trial [122], were observed when compared to delivery using a corresponding VRP. An effective immune response was also achieved using a relatively low-dose (50 µg per primer injection) of the saRNA in the CNE formulation, thus demonstrating clinical utility. A PEI-based polymer formulation was recently used to deliver a mosaic vaccine-encoding six conserved regions of the gag and pol proteins in mice [123]. This vaccine induced plurifunctional CD4+ and CD8+ T cells at relatively high levels that persisted for up to 22 weeks post administration. These results are very promising, but also highlight the need for additional strategies to promote neutralization breadth and antibody durability for long-term prophylaxis or treatment of chronic HIV-1 infection. saRNAs should theoretically reduce the frequency or necessity for booster administrations, however, extensive clinical evaluations are still required.
Conclusions
Successful immunization programs have transformed our heath care systems by providing prophylactic protection against deadly infectious diseases. Pandemics such as those caused by SARS-CoV-2 emphasize not only the health, but also the social and economic impacts that a highly contagious virus can have on our everyday lives. Vaccine development and manufacturing strategies that allow rapid responses to such threats are currently limited. With many advantages, saRNA vaccines have the potential to fill this gap (Table 2). Simple cell-free manufacturing pipelines can easily be adapted to develop new vaccine candidates, ultimately streamlining production. The transient in situ translation of antigenic or immunotherapeutic sequences elicits humoral and cell-mediated immune responses as proteins can be presented by both major histocompatibility complexes (MHC I and MHC II). Immunopotent saRNAs amplify antigenic sequences as subgenomic transcripts and the accumulation of these immunomodulatory proteins in the cytoplasm may improve genetic immunization strategies, particularly for chronic infectious diseases. Preclinical studies confirm that saRNA vaccines establish antigen-specific immune responses against various infectious diseases and confer protection.
Table 2.
Advantages and disadvantages of saRNA-based vaccines.
| Property | Advantage | Disadvantage |
|---|---|---|
| Efficacy | Comparable efficacy to traditional protein-based vaccines | Prime/boost administrations may be required |
| High level of RNA amplification in situ | Little information is available on the effects of sustained, high-level amplification and expression of saRNAs | |
| saRNA activity occurs in the cytoplasm so nuclear import is not required as for DNA vaccines | ||
| Humoral and cellular responses are elicited against the expressed antigen | ||
| Protection against infection has been demonstrated in preclinical studies | ||
| Safety | Viral genes for structural proteins are removed from the saRNA replicon to prevent viral assembly | Little information is available regarding immunogenicity of the RdRP complex |
| Cytoplasmic mode of action, no danger of integration | Limited clinical data to date | |
| Synthesis | Amenable to large-scale synthesis using GMP in vitro transcription | |
| New sequences for different antigens can be synthesized easily | ||
| Flexibility to incorporate polyvalent or multipathogen sequences | ||
| Delivery by NVVs | Can be delivered using nonviral vectors | Delivery is not typically tissue-specific |
| Formulations are amenable to large-scale synthesis | Balancing immunogenicity of NVVs and saRNA | |
| Expression at delivery site following intramuscular, intradermal, or subcutaneous injection |
The RNA vaccinology field is constantly evolving as new studies aim to improve in vitro transcription, optimize adjuvant and delivery formulations, and ultimately refine in vivo pharmacokinetics. Yet with such a large variation in vaccination approaches and very few parallel comparisons, it is difficult to decipher which strategies are best. Establishing clinical efficacy is the next important step for synthetic saRNA vaccines as this will guide future design and production endeavors. It will also determine whether the preclinical trends are accurate, and if saRNAs are able to confer immunity at lower doses. Eliciting a strong adaptive immune response could lead to single-dose regimens and reduce the number of nonresponders. Comprehensive studies from a diverse population will help improve vaccine designs as environmental and genetic influences may affect immunization [124]. Balancing the innate immune response to enhance and not avert antigen-specific immunity will be central to clinical development. Establishing exactly how saRNA vaccines should be administered will assist in defining storage, distribution, and handling procedures. With the imminent launch of the first-in-human trials, the future of saRNA vaccinology is certainly exciting.
Acknowledgements
The authors gratefully acknowledge funding from the South African Medial Research Council, the National Research Foundation (Unique Grant Numbers 118022 and 120383) and the Poliomyelitis Research Foundation.
Compliance with ethical standards
Conflict of interest
KB is an associate editor of Gene Therapy.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plotkin SA. Vaccines: past, present and future. Nat Med. 2005;11:S5–11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orenstein WA, Ahmed R. Simply put: vaccination saves lives. Proc Natl Acad Sci USA. 2017;114:4031–3. doi: 10.1073/pnas.1704507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Restifo NP, Ying H, Hwang L, Leitner WW. The promise of nucleic acid vaccines. Gene Ther. 2000;7:89–92. doi: 10.1038/sj.gt.3301117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776–88. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobernik D, Bros M. DNA vaccines-how far from clinical use? Int J Mol Sci. 2018;19:3605. doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Zuroske T, Watts JK. RNA therapeutics on the rise. Nat Rev Drug Discov. 2020;19:441–2. doi: 10.1038/d41573-020-00078-0. [DOI] [PubMed] [Google Scholar]
- 7.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–79. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundstrom K. Self-replicating RNA viruses for RNA therapeutics. Molecules. 2018;23:3310. doi: 10.3390/molecules23123310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tews BA, Meyers G. Self-replicating RNA. Methods Mol Biol. 2017;1499:15–35. doi: 10.1007/978-1-4939-6481-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, et al. Rescue of measles viruses from cloned DNA. Embo J. 1995;14:5773–84. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirley JL, de Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther. 2020;28:709–22. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel AB, Lambert L, Kinnear E, Busse D, Erbar S, Reuter KC, et al. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol Ther. 2018;26:446–55. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson NAC, Kester KE, Casimiro D, Gurunathan S, DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11.. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, et al. Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice. Emerg Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020. 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed]
- 16.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020. 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed]
- 17.Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. Methods Mol Biol. 2011;703:29–41. doi: 10.1007/978-1-59745-248-9_3. [DOI] [PubMed] [Google Scholar]
- 18.Pardi N, Muramatsu H, Weissman D, Kariko K. In vitro transcription of long RNA containing modified nucleosides. Methods Mol Biol. 2013;969:29–42. doi: 10.1007/978-1-62703-260-5_2. [DOI] [PubMed] [Google Scholar]
- 19.Cao GJ, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–4. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kääriäinen L, Ahola T. Functions of alphavirus nonstructural proteins in RNA replication. Prog Nucleic Acid Res Mol Biol. 2002;71:187–222. doi: 10.1016/S0079-6603(02)71044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wadhwa A, Aljabbari A, Lokras A, Foged C, Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–40. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada TN. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release. 2015;217:337–44. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 24.Kauffman KJ, Mir FF, Jhunjhunwala S, Kaczmarek JC, Hurtado JE, Yang JH, et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. 2015;23:1456–64. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maruggi G, Zhang C, Li J, Ulmer JB, Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol Ther. 2019;27:757–72. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyde JL, Chen R, Trobaugh DW, Diamond MS, Weaver SC, Klimstra WB, et al. The 5′ and 3′ ends of alphavirus RNAs—non-coding is not non-functional. Virus Res. 2015;206:99–107. doi: 10.1016/j.virusres.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toribio R, Diaz-Lopez I, Boskovic J, Ventoso I. An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res. 2016;44:4368–80. doi: 10.1093/nar/gkw172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frolov I, Schlesinger S. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol. 1996;70:1182–90. doi: 10.1128/jvi.70.2.1182-1190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, et al. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343:783–7. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynaud JM, Kim DY, Atasheva S, Rasalouskaya A, White JP, Diamond MS, et al. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog. 2015;11:e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, et al. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506:245–8. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Teague B, Zhang Y, Su Z, Porter E, Dobosh B, et al. In vitro evolution of enhanced RNA replicons for immunotherapy. Sci Rep. 2019;9:6932. doi: 10.1038/s41598-019-43422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jemielity J, Fowler T, Zuberek J, Stepinski J, Lewdorowicz M, Niedzwiecka A, et al. Novel “anti-reverse” cap analogs with superior translational properties. Rna. 2003;9:1108–22. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–71. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 36.Ensinger MJ, Martin SA, Paoletti E, Moss B. Modification of the 5′-terminus of mRNA by soluble guanylyl and methyl transferases from vaccinia virus. Proc Natl Acad Sci USA. 1975;72:2525–9. doi: 10.1073/pnas.72.7.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuman S. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J Biol Chem. 1990;265:11960–6. [PubMed] [Google Scholar]
- 38.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–6. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaidyanathan S, Azizian KT, Haque A, Henderson JM, Hendel A, Shore S, et al. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol Ther Nucleic Acids. 2018;12:530–42. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–5. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCaffrey AP. RNA epitranscriptome: role of the 5′ Cap. Genet Eng Biotechnol News. 2019;39:59–61. [Google Scholar]
- 42.Wu MZ, Asahara H, Tzertzinis G, Roy B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. Rna. 2020;26:345–60. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gholamalipour Y, Johnson WC, Martin CT. Efficient inhibition of RNA self-primed extension by addition of competing 3′-capture DNA-improved RNA synthesis by T7 RNA polymerase. Nucleic Acids Res. 2019;47:e118. doi: 10.1093/nar/gkz700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kariko K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beissert T, Koste L, Perkovic M, Walzer KC, Erbar S, Selmi A, et al. Improvement of in vivo expression of genes delivered by self-amplifying RNA using vaccinia virus immune evasion proteins. Hum Gene Ther. 2017;28:1138–46. doi: 10.1089/hum.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Chin JM, Choo EL, Phua KKL. Messenger RNA translation enhancement by immune evasion proteins: a comparative study between EKB (vaccinia virus) and NS1 (influenza A virus) Sci Rep. 2019;9:11972. doi: 10.1038/s41598-019-48559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshioka N, Gros E, Li HR, Kumar S, Deacon DC, Maron C, et al. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13:246–54. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan S, Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–43. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 49.Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019;27:710–28. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blakney AK, Zhu Y, McKay PF, Bouton CR, Yeow J, Tang J, et al. Big is beautiful: enhanced sarna delivery and immunogenicity by a higher molecular weight, bioreducible, cationic polymer. ACS Nano. 2020;14:5711–27.. doi: 10.1021/acsnano.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saviano F, Lovato T, Russo A, Russo G, Shattock RJ, Alexander C, et al. Ornithine-derived oligomers and dendrimers for in vitro delivery of DNA and ex vivo transfection of skin cells via saRNA. J Mater Chem B. 2020;8:4940–9. doi: 10.1039/d0tb00942c. [DOI] [PubMed] [Google Scholar]
- 52.Blakney AK, Abdouni Y, Yilmaz G, Liu R, McKay PF, Bouton CR, et al. Mannosylated poly(ethylene imine) copolymers enhance saRNA uptake and expression in human skin explants. Biomacromolecules. 2020;21:2482–92. doi: 10.1021/acs.biomac.0c00445. [DOI] [PubMed] [Google Scholar]
- 53.Yang DC, Eldredge AC, Hickey JC, Muradyan H, Guan Z. Multivalent peptide-functionalized bioreducible polymers for cellular delivery of various RNAs. Biomacromolecules. 2020;21:1613–24. doi: 10.1021/acs.biomac.0c00211. [DOI] [PubMed] [Google Scholar]
- 54.Goswami R, Chatzikleanthous D, Lou G, Giusti F, Bonci A, Taccone M, et al. Mannosylation of LNP results in improved potency for self-amplifying RNA (SAM) vaccines. ACS Infect Dis. 2019;5:1546–58. doi: 10.1021/acsinfecdis.9b00084. [DOI] [PubMed] [Google Scholar]
- 55.Perche F, Clemencon R, Schulze K, Ebensen T, Guzman CA, Pichon C. Neutral lipopolyplexes for in vivo delivery of conventional and replicative RNA vaccine. Mol Ther Nucleic Acids. 2019;17:767–75. doi: 10.1016/j.omtn.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferkol T, Perales JC, Mularo F, Hanson RW. Receptor-mediated gene transfer into macrophages. Proc Natl Acad Sci USA. 1996;93:101–5. doi: 10.1073/pnas.93.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erbacher P, Bousser MT, Raimond J, Monsigny M, Midoux P, Roche AC. Gene transfer by DNA/glycosylated polylysine complexes into human blood monocyte-derived macrophages. Hum Gene Ther. 1996;7:721–9. doi: 10.1089/hum.1996.7.6-721. [DOI] [PubMed] [Google Scholar]
- 58.Blakney AK, McKay PF, Yus BI, Aldon Y, Shattock RJ. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–72. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderluzzi G, Lou G, Gallorini S, Brazzoli M, Johnson R, O’Hagan DT, et al. Investigating the impact of delivery system design on the efficacy of self-amplifying RNA vaccines. Vaccines. 2020;8:212. doi: 10.3390/vaccines8020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spuul P, Balistreri G, Hellstrom K, Golubtsov AV, Jokitalo E, Ahola T. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J Virol. 2011;85:4739–51. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blakney AK, McKay PF, Shattock RJ. Structural components for amplification of positive and negative strand VEEV splitzicons. Front Mol Biosci. 2018;5:71. doi: 10.3389/fmolb.2018.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beissert T, Perkovic M, Vogel A, Erbar S, Walzer KC, Hempel T, et al. A trans-amplifying RNA Vaccine strategy for induction of potent protective immunity. Mol Ther. 2020;28:119–28.. doi: 10.1016/j.ymthe.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med. 2013;19:1597–608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 64.Carpentier KS, Morrison TE. Innate immune control of alphavirus infection. Curr Opin Virol. 2018;28:53–60. doi: 10.1016/j.coviro.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rauch S, Lutz J, Kowalczyk A, Schlake T, Heidenreich R. RNActive® technology: generation and testing of stable and immunogenic mRNA vaccines. Methods Mol Biol. 2017;1499:89–107. doi: 10.1007/978-1-4939-6481-9_5. [DOI] [PubMed] [Google Scholar]
- 66.Kallen KJ, Heidenreich R, Schnee M, Petsch B, Schlake T, Thess A, et al. A novel, disruptive vaccination technology: self-adjuvanted RNActive(®) vaccines. Hum Vaccines Immunother. 2013;9:2263–76. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heidenreich R, Jasny E, Kowalczyk A, Lutz J, Probst J, Baumhof P, et al. A novel RNA-based adjuvant combines strong immunostimulatory capacities with a favorable safety profile. Int J Cancer. 2015;137:372–84. doi: 10.1002/ijc.29402. [DOI] [PubMed] [Google Scholar]
- 68.Brito LA, Chan M, Shaw CA, Hekele A, Carsillo T, Schaefer M, et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol Ther. 2014;22:2118–29. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allison AG, Gregoriadis G. Liposomes as immunological adjuvants. Nature. 1974;252:252. doi: 10.1038/252252a0. [DOI] [PubMed] [Google Scholar]
- 70.Perrie Y, Crofts F, Devitt A, Griffiths HR, Kastner E, Nadella V. Designing liposomal adjuvants for the next generation of vaccines. Adv Drug Deliv Rev. 2016;99:85–96. doi: 10.1016/j.addr.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 71.McCullough KC, Bassi I, Milona P, Suter R, Thomann-Harwood L, Englezou P, et al. Self-replicating replicon-RNA delivery to dendritic cells by chitosan-nanoparticles for translation in vitro and in vivo. Mol Ther Nucleic Acids. 2014;3:e173. doi: 10.1038/mtna.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manara C, Brazzoli M, Piccioli D, Taccone M, D’Oro U, Maione D, et al. Co-administration of GM-CSF expressing RNA is a powerful tool to enhance potency of SAM-based vaccines. Vaccine. 2019;37:4204–13. doi: 10.1016/j.vaccine.2019.04.028. [DOI] [PubMed] [Google Scholar]
- 73.Blakney AK, McKay PF, Christensen D, Yus BI, Aldon Y, Follmann F, et al. Effects of cationic adjuvant formulation particle type, fluidity and immunomodulators on delivery and immunogenicity of saRNA. J Control Release. 2019;304:65–74. doi: 10.1016/j.jconrel.2019.04.043. [DOI] [PubMed] [Google Scholar]
- 74.Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol. 2019;37:1174–85. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 75.Pollard C, Rejman J, De Haes W, Verrier B, Van Gulck E, Naessens T, et al. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol Ther. 2013;21:251–9. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pepini T, Pulichino AM, Carsillo T, Carlson AL, Sari-Sarraf F, Ramsauer K, et al. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J Immunol. 2017;198:4012–24. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–9. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 78.Huysmans H, Zhong Z, De Temmerman J, Mui BL, Tam YK, Mc Cafferty S, et al. Expression kinetics and innate immune response after electroporation and LNP-mediated delivery of a self-amplifying mRNA in the skin. Mol Ther Nucleic Acids. 2019;17:867–78. doi: 10.1016/j.omtn.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou X, Berglund P, Rhodes G, Parker SE, Jondal M, Liljestrom P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine. 1994;12:1510–4. doi: 10.1016/0264-410x(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 80.Fleeton MN, Chen M, Berglund P, Rhodes G, Parker SE, Murphy M, et al. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J Infect Dis. 2001;183:1395–8. doi: 10.1086/319857. [DOI] [PubMed] [Google Scholar]
- 81.Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA. 2012;109:14604–9. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perri S, Greer CE, Thudium K, Doe B, Legg H, Liu H, et al. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J Virol. 2003;77:10394–403. doi: 10.1128/JVI.77.19.10394-10403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scorza FB, Pardi N. New kids on the block: RNA-based influenza virus vaccines. Vaccines. 2018;6:20. doi: 10.3390/vaccines6020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paules CI, Fauci AS. Influenza vaccines: good, but we can do better. J Infect Dis. 2019;219:S1–4. doi: 10.1093/infdis/jiy633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Magini D, Giovani C, Mangiavacchi S, Maccari S, Cecchi R, Ulmer JB, et al. Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS ONE. 2016;11:e0161193. doi: 10.1371/journal.pone.0161193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McKay PF, Hu K, Blakney AK, Samnuan K, Brown JC, Penn R, et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scheuber A. Imperial social enterprise to accelerate low-cost COVID-19 vaccine. Imperial College London Health News. 2020. https://www.imperial.ac.uk/news/198053/imperial-social-enterprise-accelerate-lowcost-covid19/.
- 88.Samsa MM, Dupuy LC, Beard CW, Six CM, Schmaljohn CS, Mason PW, et al. Self-amplifying RNA vaccines for venezuelan equine encephalitis virus induce robust protective immunogenicity in mice. Mol Ther. 2019;27:850–65. doi: 10.1016/j.ymthe.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhong Z, Portela Catani JP, Mc Cafferty S, Couck L, Van Den Broeck W, Gorle N, et al. Immunogenicity and protection efficacy of a naked self-replicating mRNA-based zika virus vaccine. Vaccines. 2019;7:96. [DOI] [PMC free article] [PubMed]
- 90.Erasmus JH, Khandhar AP, Guderian J, Granger B, Archer J, Archer M, et al. A nanostructured lipid carrier for delivery of a replicating viral RNA provides single, low-dose protection against Zika. Mol Ther. 2018;26:2507–22. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chahal JS, Fang T, Woodham AW, Khan OF, Ling J, Anderson DG, et al. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Sci Rep. 2017;7:252. doi: 10.1038/s41598-017-00193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stokes A, Pion J, Binazon O, Laffont B, Bigras M, Dubois G, et al. Nonclinical safety assessment of repeated administration and biodistribution of a novel rabies self-amplifying mRNA vaccine in rats. Regul Toxicol Pharmacol. 2020;113:104648. doi: 10.1016/j.yrtph.2020.104648. [DOI] [PubMed] [Google Scholar]
- 93.Armbruster N, Jasny E, Petsch B. Advances in RNA vaccines for preventive indications: a case study of a vaccine against rabies. Vaccines. 2019;7:132. doi: 10.3390/vaccines7040132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Esparza J. A brief history of the global effort to develop a preventive HIV vaccine. Vaccine. 2013;31:3502–18. doi: 10.1016/j.vaccine.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 95.Kwong PD, Mascola JR. HIV-1 vaccines based on antibody identification, b cell ontogeny, and epitope structure. Immunity. 2018;48:855–71. doi: 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 96.Moore PL. The neutralizing antibody response to the HIV-1 Env protein. Curr HIV Res. 2018;16:21–8. doi: 10.2174/1570162X15666171124122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones LD, Moody MA, Thompson AB. Innovations in HIV-1 vaccine design. Clin Ther. 2020;42:499–514. doi: 10.1016/j.clinthera.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sok D, Burton DR. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol. 2018;19:1179–88. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Padte NN, Yu J, Huang Y, Ho DD. Engineering multi-specific antibodies against HIV-1. Retrovirology. 2018;15:60. doi: 10.1186/s12977-018-0439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tremouillaux-Guiller J, Moustafa K, Hefferon K, Gaobotse G, Makhzoum A. Plant-made HIV vaccines and potential candidates. Curr Opin Biotechnol. 2020;61:209–16.. doi: 10.1016/j.copbio.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 101.Gardner MR, Farzan M. Engineering antibody-like inhibitors to prevent and treat HIV-1 infection. Curr Opin HIV AIDS. 2017;12:294–301. doi: 10.1097/COH.0000000000000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Korber B, Hraber P, Wagh K, Hahn BH. Polyvalent vaccine approaches to combat HIV-1 diversity. Immunol Rev. 2017;275:230–44. doi: 10.1111/imr.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin A, Balazs AB. Adeno-associated virus gene delivery of broadly neutralizing antibodies as prevention and therapy against HIV-1. Retrovirology. 2018;15:66. doi: 10.1186/s12977-018-0449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davis NL, Caley IJ, Brown KW, Betts MR, Irlbeck DM, McGrath KM, et al. Vaccination of macaques against pathogenic simian immunodeficiency virus with Venezuelan equine encephalitis virus replicon particles. J Virol. 2000;74:371–8. doi: 10.1128/jvi.74.1.371-378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnston RE, Johnson PR, Connell MJ, Montefiori DC, West A, Collier ML, et al. Vaccination of macaques with SIV immunogens delivered by Venezuelan equine encephalitis virus replicon particle vectors followed by a mucosal challenge with SIVsmE660. Vaccine. 2005;23:4969–79. doi: 10.1016/j.vaccine.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 106.Mossman SP, Bex F, Berglund P, Arthos J, O’Neil SP, Riley D, et al. Protection against lethal simian immunodeficiency virus SIVsmmPBj14 disease by a recombinant Semliki Forest virus gp160 vaccine and by a gp120 subunit vaccine. J Virol. 1996;70:1953–60. doi: 10.1128/jvi.70.3.1953-1960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perri S, Greer CE, Thudium K, Doe B, Legg H, Liu H, et al. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J Virol. 2003;77:10394–403. doi: 10.1128/JVI.77.19.10394-10403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pardi N, Secreto AJ, Shan X, Debonera F, Glover J, Yi Y, et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pardi N, Hogan MJ, Naradikian MS, Parkhouse K, Cain DW, Jones L, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215:1571–88. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pardi N, LaBranche CC, Ferrari G, Cain DW, Tombacz I, Parks RJ, et al. Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol Ther Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Allard SD, De Keersmaecker B, de Goede AL, Verschuren EJ, Koetsveld J, Reedijk ML, et al. A phase I/IIa immunotherapy trial of HIV-1-infected patients with Tat, Rev and Nef expressing dendritic cells followed by treatment interruption. Clin Immunol. 2012;142:252–68. doi: 10.1016/j.clim.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 112.Gandhi RT, Kwon DS, Macklin EA, Shopis JR, McLean AP, McBrine N, et al. Immunization of HIV-1-infected persons with autologous dendritic cells transfected with mRNA encoding HIV-1 Gag and Nef: results of a randomized, placebo-controlled clinical trial. J Acquir Immune Defic Syndr. 2016;71:246–53. doi: 10.1097/QAI.0000000000000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gay CL, DeBenedette MA, Tcherepanova IY, Gamble A, Lewis WE, Cope AB, et al. Immunogenicity of AGS-004 dendritic cell therapy in patients treated during acute HIV infection. AIDS Res Hum Retrovir. 2018;34:111–22. doi: 10.1089/aid.2017.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jacobson JM, Routy JP, Welles S, DeBenedette M, Tcherepanova I, Angel JB, et al. Dendritic cell immunotherapy for HIV-1 infection using autologous HIV-1 RNA: a randomized, double-blind, placebo-controlled clinical trial. J Acquir Immune Defic Syndr. 2016;72:31–8. doi: 10.1097/QAI.0000000000000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Routy JP, Boulassel MR, Yassine-Diab B, Nicolette C, Healey D, Jain R, et al. Immunologic activity and safety of autologous HIV RNA-electroporated dendritic cells in HIV-1 infected patients receiving antiretroviral therapy. Clin Immunol. 2010;134:140–7. doi: 10.1016/j.clim.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Gulck E, Vlieghe E, Vekemans M, Van Tendeloo VF, Van De Velde A, Smits E, et al. mRNA-based dendritic cell vaccination induces potent antiviral T-cell responses in HIV-1-infected patients. AIDS. 2012;26:F1–12. doi: 10.1097/QAD.0b013e32834f33e8. [DOI] [PubMed] [Google Scholar]
- 117.Gay CL, Kuruc JD, Falcinelli SD, Warren JA, Reifeis SA, Kirchherr JL, et al. Assessing the impact of AGS-004, a dendritic cell-based immunotherapy, and vorinostat on persistent HIV-1 Infection. Sci Rep. 2020;10:5134. doi: 10.1038/s41598-020-61878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.de Jong W, Leal L, Buyze J, Pannus P, Guardo A, Salgado M, et al. Therapeutic vaccine in chronically HIV-1-infected patients: a randomized, double-blind, placebo-controlled phase IIa trial with HTI-TriMix. Vaccines. 2019;7:209. doi: 10.3390/vaccines7040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leal L, Guardo AC, Moron-Lopez S, Salgado M, Mothe B, Heirman C, et al. Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS. 2018;32:2533–45. doi: 10.1097/QAD.0000000000002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Melo M, Porter E, Zhang Y, Silva M, Li N, Dobosh B, et al. Immunogenicity of RNA replicons encoding HIV Env immunogens designed for self-assembly into nanoparticles. Mol Ther. 2019;27:2080–90.. doi: 10.1016/j.ymthe.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bogers WM, Oostermeijer H, Mooij P, Koopman G, Verschoor EJ, Davis D, et al. Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. J Infect Dis. 2015;211:947–55.. doi: 10.1093/infdis/jiu522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–86.. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moyo N, Vogel AB, Buus S, Erbar S, Wee EG, Sahin U, et al. Efficient Induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol Ther Methods Clin Dev. 2019;12:32–46. doi: 10.1016/j.omtm.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kollmann TR. Variation between populations in the innate immune response to vaccine adjuvants. Front Immunol. 2013;4:81. doi: 10.3389/fimmu.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brazzoli M, Magini D, Bonci A, Buccato S, Giovani C, Kratzer R, et al. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J Virol. 2016;90:332–44.. doi: 10.1128/JVI.01786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lazzaro S, Giovani C, Mangiavacchi S, Magini D, Maione D, Baudner B, et al. CD8 T-cell priming upon mRNA vaccination is restricted to bone-marrow-derived antigen-presenting cells and may involve antigen transfer from myocytes. Immunology. 2015;146:312–26. doi: 10.1111/imm.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chahal JS, Khan OF, Cooper CL, McPartlan JS, Tsosie JK, Tilley LD, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci USA. 2016;113:E4133–42. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Demoulins T, Ebensen T, Schulze K, Englezou PC, Pelliccia M, Guzman CA, et al. Self-replicating RNA vaccine functionality modulated by fine-tuning of polyplex delivery vehicle structure. J Control Release. 2017;266:256–71.. doi: 10.1016/j.jconrel.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 129.Englezou PC, Sapet C, Demoulins T, Milona P, Ebensen T, Schulze K, et al. Self-amplifying replicon RNA delivery to dendritic cells by cationic lipids. Mol Ther Nucleic Acids. 2018;12:118–34. doi: 10.1016/j.omtn.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kofler RM, Aberle JH, Aberle SW, Allison SL, Heinz FX, Mandl CW. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc Natl Acad Sci USA. 2004;101:1951–6. doi: 10.1073/pnas.0307145101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aberle JH, Aberle SW, Kofler RM, Mandl CW. Humoral and cellular immune response to RNA immunization with flavivirus replicons derived from tick-borne encephalitis virus. J Virol. 2005;79:15107–13. doi: 10.1128/JVI.79.24.15107-15113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cu Y, Broderick KE, Banerjee K, Hickman J, Otten G, Barnett S, et al. Enhanced delivery and potency of self-amplifying mRNA vaccines by electroporation in situ. Vaccines. 2013;1:367–83. doi: 10.3390/vaccines1030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Luo F, Zheng L, Hu Y, Liu S, Wang Y, Xiong Z, et al. Induction of protective immunity against toxoplasma gondii in mice by nucleoside triphosphate hydrolase-II (NTPase-II) self-amplifying RNA vaccine encapsulated in lipid nanoparticle (LNP) Front Microbiol. 2017;8:605. doi: 10.3389/fmicb.2017.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maruggi G, Chiarot E, Giovani C, Buccato S, Bonacci S, Frigimelica E, et al. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. 2017;35:361–8. doi: 10.1016/j.vaccine.2016.11.040. [DOI] [PubMed] [Google Scholar]