Abstract
Diversification of chain stereochemistry offers a tremendous increase in protein design space. We have designed a minimal fluorescent protein, pregnant with β-(1-azulenyl)-l-alanine in the hydrophobic core of a heterotactic protein scaffold, employing automated design tools such as automated repetitive simulated annealing molecular dynamics and IDeAS. The de novo designed heterochiral protein can be selectively excited at 342 nm, quite distant from the intrinsic fluorophore, and emits in the blue region. The structure and stability of the designed proteins were evaluated by established spectroscopic and calorimetric methods.
Introduction
Protein molecules found in nature are polymers of amino acids with definite handedness or chirality, with their spatial orientation being preferentially left-handed or l-chiral.1,2 Of late, this disproportionate or rather complete excess of l-amino acids is argued to be preordained and eventually turned out to be one of the most powerful arguments of creationists who believe that species on earth are not formed by evolution but by “intelligent design”.3
Tacticity is a relative term in stereochemistry, which deals with the arrangement of adjacent chiral centers within a given polymeric molecule. Based on the spatial arrangement, a polymer can be isotactic, syndiotactic, or heterotactic in nature. An isotactic polypeptide sequence is characterized by amino acids having l- or d-stereochemistry, whereas a syndiotactic sequence has a stereoregular arrangement with alternate l- and d-amino acids (or vice versa) in succession, and a heterotactic polymer is a random distribution of l- and d-amino acids in a sequence.4,5 If we can hypothetically add one more variable as “stereochemistry of amino acids” in the backbone, by adding d-amino acids at “n” sequence positions, the number of stereoisomeric diversities increases exponentially from 1n to 2n, but the entire protein universe is made of just only one of them, which implies that (2n – 1) is yet to be explored.2 The origin of this apparent stereochemical filtration in protein biogenesis remains unclear; however, its consequence on peptide assemblies can be investigated and explored for the future design of bioinspired materials.6 Our primary interest is to use the backbone stereochemistry as an additional variable, exploring novel architectures in the 2n – 1 design space available for novel peptide-based molecular constructs.
In one of the pioneering studies, Flory observed that isotactic polypeptides are far more “stiff” than could be explained on the grounds of his statistical coil model with effects of atomic excluded volumes included.7 The stiffness is measured in terms of characteristic ratio, which is unusually high in poly l-chiral peptides with a limiting value of 9.0.8 However, when the random polypeptide is modified from poly l (isotactic) to alternating l, d (syndiotactic), the characteristic ratio deflates from 9.0 to 0.9, which is a value lower than the theoretical minimum for the unperturbed random coil. The drastic drop in stiffness without making any difference in its residue-level inter-amide electrostatics remained a “puzzle” to him.9 In an apparent attempt to study this phenomenon, we simulated equilibrium ensembles of octa-alanine peptides in the poly l and alternating l, d-chiral stereochemical sequences in water at different temperatures. We observed that the short- and long-range electrostatic interactions are found to be in the opposite directions in the folded state and in harmony in the unfolded state.10 The yin-yang of these two opposing interactions enables a third variable such as amino acid side-chain sequence or solvent dielectric to be detrimental in selecting the conformational fold. It may be this effect that makes an isotactic polypeptide such as protein, so powerfully responsive to a stimulus, and eventually manifests itself as a sensor transducer apparatus in living systems.11 In this work, we introduce the next level of complexity in “polypeptide stereochemistry”, by adopting a heterotactic (random l and d) backbone, and examine the scope of this prototype, for the design of future biomaterials.
The de novo design of proteins and the determination of minimum requirements for the formation of protein-like structures are currently the subjects of active research.12−17 Significant advances have been established toward the design of stable, well-folded proteins with novel sequences.18−20 The design of the stable protein-like structure with a smaller number of residues requires both an exquisite optimization of hydrophobic packing and a strategy to decrease the backbone configurational entropy of the unfolded state.21 The discovery of a small and fast-folding protein, a 20 residue Trp-cage protein which folds in 4 μs to the native state, by Andersen and co-workers contributed much to the understanding of the protein structure and folding mechanisms.21,22 Owing to its small size and fast folding rate, tryptophan (Trp)-cage has been an extremely popular model for computational studies of protein folding dynamics.23
The aromatic amino acids responsible for fluorescence are relatively rare in proteins.24 Trp’s significant red shift, when compared to that of all other amino acids, high sensitivity to the local environment, and low abundance in proteins are particularly well suited for the studies such as protein structure, function, and dynamics.25,26 However, Trp needs to be excited by UV light with wavelengths less than 300 nm, which interferes with other chromophores often present in a biological sample.27 These factors make Trp rarely useful as a fluorophore for single-molecule measurements.
One approach to address this problem is to use the unnatural analogue of Trp which can be excited above ∼300 nm, where Trp absorption is negligible. A pseudoisosteric analogue of Trp, β-(1-azulenyl)-l-alanine (AzAla), which is an azulene moiety,28,29 fulfills the above criteria. This can be excited at 342 nm, independently of Trp, and the emission is in the visible region.
The limitations of such methods are low protein yields, unpredictability of efficiencies, and complicated working protocols.30 Peptides have been in use as reliable scaffolds for novel biomaterials.31−33 Many sensitive fluorophores can respond rapidly to environmental changes by way of a change in their spectral characteristics. We report the design, synthesis, and characterization of a heterochiral artificial protein of a length of just 30 amino acids, which was further examined to explore the possibility of converting it to a fully functional molecule. An unnatural amino acid AzAla is incorporated in a heterotactic protein scaffold which has a clear distinction in spectral characteristics compared to the native Trp signal.
Results and Discussion
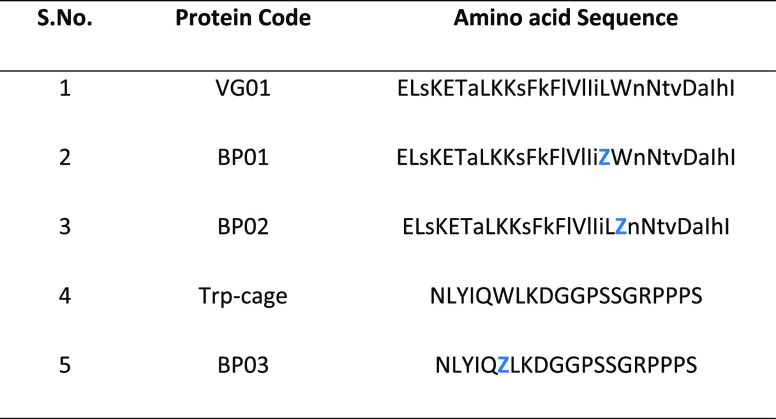
A series of four new “mini proteins” have been synthesized by solid-phase peptide synthesis (SPPS, Table 1). The structure and stability were examined by calorimetric methods to evaluate their characteristics to be qualified as a self-folding fluorescent mini protein. To facilitate the visible range fluorescence of the protein, we designed and synthesized three sequences, incorporating an unnatural fluorophore that has an emission wavelength in the visible region of the electromagnetic spectrum. Trp-cage and an AzAla-substituted variant of Trp-cage are used as the controls for objective analysis. The designs are implemented in three steps. First, the amino acids were folded by an already established automated repetitive simulated annealing molecular dynamics (AR-SAMD) procedure.11,34 At high temperature, the polypeptide chain can switch from l to d (or otherwise), and this is achieved by modifying the force function governing the improper dihedral angle in the GROMOS force field. As the folds evolve in the prescribed design space, they were assessed and scored on the basis of their potential utility to be sequence-engineered as functional foldamers, involving an automated inverse design scheme. The collection of 30-mer polyleucine folds generated as a result of AR-SAMD was subjected to molecular dynamics simulation at 298 K in water.
Table 1. Details of the Designed Proteins.
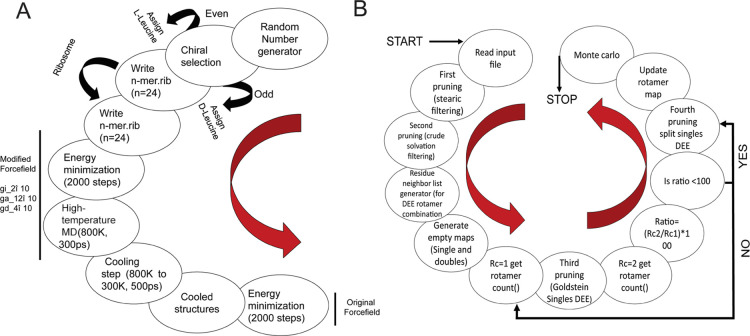
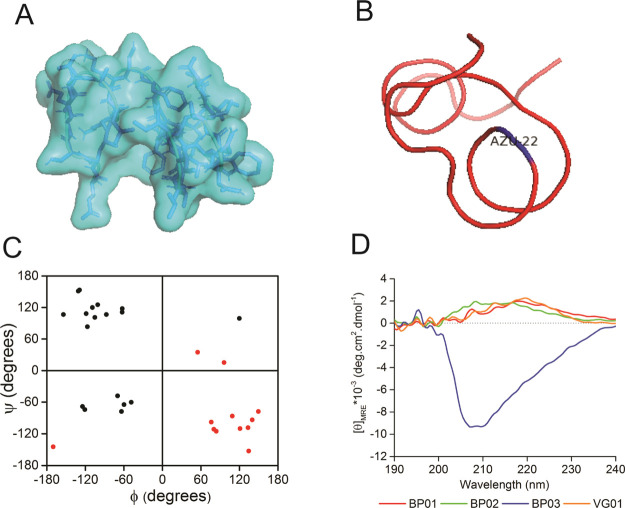
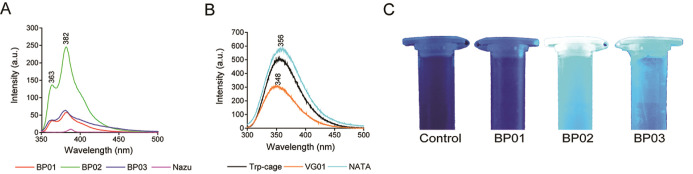
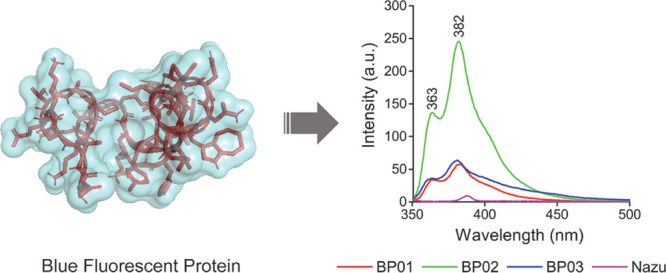
By doing so, we intend to evaluate the possibility of generating metastable folds that can further be sequence-designed as “artificial proteins”. This is achieved by performing sequence design of a few selected folds using an automated sequence design tool, IDeAS (Figure 1). Details of the computational procedure employed for the generation of an optimal sequence solution have been described elsewhere.34,35 In the third step, we selectively mutate one or two core residues with a blue fluorescent unnatural amino acid. To facilitate the visible range emission, we designed four artificial fluorescent proteins, incorporating AzAla (denoted as Z). The solvent exposure of the fluorophore has been verified using a spectrofluorimeter. The earlier-established de novo designed mini protein Trp-cage and its W6Z mutant were chosen as the controls. The first sequence (VG01) was a de novo designed heterochiral fold evolved as a result of systematic stereochemical and sequence selection routines. BP01 and BP02 are L20Z and W21Z mutants of VG01, respectively. BP03 is the W-to-Z mutant of our control protein, Trp-cage. The dihedral angle distribution of all amino acids in the designed sequences and C-alpha traces of the foldamer BP02 are shown in Figure 2. The predicted secondary structure of the designed proteins was mainly an extended-β and disordered structure which was confirmed by circular dichroism (CD) spectral analysis (Figure 2D). The high-performance liquid chromatography (HPLC) chromatograms of the synthesized proteins are shown in the Supporting Information (Figure S1). The observed mass was found to be equivalent to the calculated mass (Figure S2). UV–vis absorption spectra were recorded to confirm the presence of AzAla in the designed proteins, and they showed the absorption peak of the proteins BP01, BP02, and BP03 at 280 and 342 nm and the absorption peak of VG01 and Trp-cage at 280 nm (Figure S3). N-Acetyl-l-tryptophanamide (NATA), a Trp analogue, has been used as a reference for Trp-containing proteins (Figure 3). At an excitation wavelength of 280 nm, the emission of NATA was at 356 nm, which shows the solvent-exposed condition of the Trp residue.36 The Trp side chain in NATA is more exposed to the solvent compared to other synthesized proteins, Trp-cage and VG01. The relative low Trp fluorescence intensity indicates the inability of the VG01 mini protein sequence to sustain a stable hydrophobic core under ambient conditions. AzAla-containing peptides show emission at 363 and 382 nm, which is the characteristic emission wavelength of AzAla. At the excitation wavelength of 342 nm, only proteins BP01, BP02, and BP03 show emission at 364 nm and 382 nm. Emission spectra of proteins BP01, BP02, and BP03 were compared with that of N-acetyl azulenylalanine (Nazu). Nazu has the AzAla fluorophore which is almost fully exposed to the solvent. The fluorescence intensity of AzAla-containing proteins is expected to have a much more prominent fluorescence peak compared to Nazu. This will confirm the formation of a well-defined hydrophobic core, which is an important criterion for a stable functional foldamer. At the excitation of 280 nm, the fluorescence intensity estimate of BP02 is significantly high, compared to that of Nazu. This observation indicates the burial of AzAla in the hydrophobic core of the mini protein, as envisaged in the design. However, BP01 and BP03 do not show a significant difference in fluorescence intensity compared to Nazu. The low intensity of proteins BP01 and BP03 suggests quenching of AzAla due to the solvent. The excitation spectrum of AzAla-containing peptides shows two major absorption maxima at 280 and 342 nm, respectively, which can be used for the excitation of the nonalternating aromatic ring system to yield emission at 382 nm.28 AzAla, an unnatural amino acid containing azulene in its side chain, shows a quite strong 1La absorption band centered at 320–380 nm (ε ∼ 4200 cm–1 M–1)37 and a weak 1Lb absorbance at 600 nm (ε ∼ 400 cm–1 M–1). Excitation at this wavelength does not result in a measurable fluorescence signal, while excitation in the 1La band yields an emission band at ∼380 nm with a quantum yield comparable to that of Trp.27 AzAla can be excited selectively at 342 nm to get the emission at 382 nm. Although the relative fluorescence intensity of AzAla when excited at 342 nm is lower than that of Trp, this amino acid could represent an interesting fluorescence probe in peptides and proteins, as a potential blue-colored Trp analogue.28 The UV illumination of BP02 shows a visible bright blue color (Figure 3C), compared to that of BP03, while the intensity of BP01 was significantly less. An empty microcentrifuge tube was taken as the control. BP02 thus qualifies to be a de novo designed heterochiral protein foldamer, pregnant with a fluorophore in a well-defined hydrophobic core.
Figure 1.
Schematic representation of (A) generation of folds by AR-SAMD; (B) flowchart illustrating the sequence optimization steps involved in the IDeAS program suite.
Figure 2.
Representation of (A) surface model, (B) C-alpha traces, and (C) dihedral angles of the protein foldamer BP02, where d-amino acids are shown in red circles and l-amino acids are shown in black circles. The representation in all four quadrants of the Ramachandran map confirms the heterochiral nature of the designed protein. (D) CD spectra of the designed proteins indicating the disordered conformation of the designed proteins.
Figure 3.
Fluorescence emission spectra of synthesized protein foldamers: (A) at an excitation wavelength of 342 nm, BP01 (red), BP02 (green), and BP03 (blue) showed emission at 363 and 382 nm; (B) at an excitation wavelength of 280 nm, NATA (cyan), Trp-cage (black), and VG01 (orange) showed emission at 356, 355, and 348 nm, respectively. (C) UV illumination of synthesized proteins. BP02 shows a more visible bright blue color compared to BP03.
The formation of a well-defined hydrophobic core facilitating spontaneous folding to the designed conformation may be indirectly reflected in the thermal stability of the designed proteins. This was characterized by differential scanning calorimetry (DSC). The heat capacity curves obtained from DSC measurements (Figure 4) show several characteristic features. Starting from room temperature, the apparent protein heat capacity gradually increases until the onset of thermal unfolding, accompanied by a sharp peak in Cp corresponding to an endothermic unfolding transition. The peak at the transition gives the midpoint temperature (Tm), where the folded and unfolded forms of the protein are at equilibrium (i.e., ΔG = 0),38 and the area under the transition gives the enthalpy of unfolding (ΔHunf) at this temperature.39 The higher Tm value of proteins indicates higher thermal stability.40 Under the experimental conditions, the thermally induced transition of proteins BP02 and BP03 occurs at 66 and 42 °C, respectively. The DSC thermogram of the protein BP02 shows more thermal stability compared to BP03. BP01, being completely nonfluorescent, was omitted from thermodynamic stability measurements.
Figure 4.
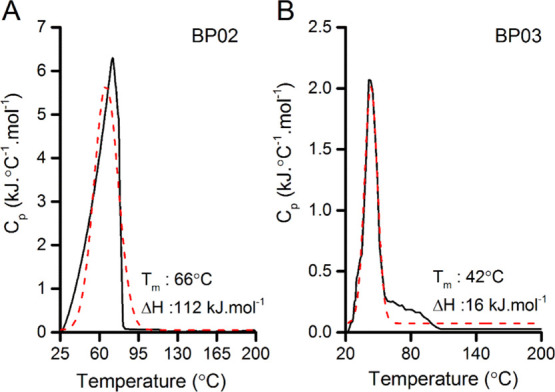
DSC thermogram of proteins (A) BP02 and (B) BP03. The Tm value of the protein BP02 (66 °C) is greater than that of BP03 (42 °C). The solid black line represents experimental data, while the dashed red line represents the curve fit (the nonlinear curve fit).
Secondary structures of the designed protein were analyzed by CD and Fourier transform infrared (FT-IR) spectroscopy. The spectra of synthesized proteins in deionized water were further investigated using CD spectroscopy (Figure 2D). CD spectra of Trp-cage show negative peaks at 208 and 223 nm, with a positive peak at 195 nm (the spectrum is not shown) majorly forming an α-helix as previously reported.21,41 Similarly, BP03 (a mutant of Trp-cage) shows a negative peak at 208 nm and a positive peak at 195 nm, which constitutes majorly an α-helix. VG01 shows a positive peak at ∼219 nm which might be due to the contribution of Trp to the far-UV CD spectra. Previous work by Chakrabartty et al. showed that it arises when the aromatic group is fixed in a particular structure.42 This spectral feature has also been observed in folded peptides containing a well-ordered Trp side chain.43,44
In the spectrum of BP02, the positive peak shows a blue shift of around 209 nm. The far-UV CD spectrum of VG01, BP01, and BP02 is not suggestive of any specific conformation because analysis of the CD spectra of heterochiral proteins cannot be compared using the observations usually deduced from poly l-homochiral proteins, and therefore interpreted with caution. However, we further analyzed and deconvoluted the spectrum using the CONTIN-LL algorithms on the Dichroweb online server using the SP175 reference set.45
It shows that proteins BP01, BP02, and VG01 majorly constitute the β-extended and disordered structures, while BP03 constitutes mainly an α-helix structure (74.7%) and a disordered structure (16.9%) (Table 2). It clearly indicates that the extended-β content increases in the mutant protein BP02 (W21Z) from 36.1 to 59.9% and in the protein BP01 from 36.1 to 49.1% when compared with that in the control protein VG01, which may contribute to the blue shift. The substitution of W21Z in BP02 makes it more structured compared to VG01. The secondary structure of proteins was also verified by FT-IR (Figure S4). Information on the secondary structure of proteins can be derived from the analysis of a strong amide I band. The amide I band arises predominantly from C=O stretching vibrations of the amide group46 and shows absorption in the region 1600–1700 cm–1. Amide I bands in the spectral range of 1620–1640 cm–1 are attributed to the β-sheet structure.47 The C=O stretching frequencies of proteins VG01, BP01, and BP02 are similar to those of β-structures, although they cannot be confirmed by standard comparisons, because of the heterochiral stereochemical sequence of the polypeptide foldamer.48
Table 2. Secondary Structure Content (in Percentage) of Synthesized Proteins.
| algorithm | conformation | BP01 | BP02 | BP03 | VG01 | Trp-cage |
|---|---|---|---|---|---|---|
| CONTIN-LL | α-helix | 0.8 | 1.9 | 74.7 | 5.3 | 68.7 |
| β-extended | 49.1 | 59.9 | 0 | 36.6 | 0 | |
| turns | 6 | 7.8 | 8.3 | 17.9 | 9.0 | |
| disordered | 44.2 | 30.4 | 16.9 | 40.1 | 22.2 |
Conclusions and Future Possibilities
In this study, we have shown the possibility of the designed heterotactic protein sequence, pregnant with an unnatural amino acid, AzAla, as a minimal fluorescent protein. Using UV–vis and fluorescence spectroscopy, we have shown that the designed protein has blue fluorescence. On comparing the fluorescence intensity of Nazu with those of other proteins, at the excitation wavelength of 342 nm, we found that the protein BP02 has the maximum intensity, indicating that AzAla is buried inside a stable core, excluding the solvent. The secondary structure reveals the presence of the extended-β and disordered structures. DSC complements well with fluorescence experiments, indicating higher thermal stability of the protein BP02. Our observations complement the primary objectives of our work in designing a heterochiral mini protein. The design and experimental protocols employed in the BP02 prototype construct may be helpful in the generation of future peptide-based biomaterials beyond the limits of natural alphabets.
Experimental Section
Protein Design
The design and sequence optimization of the de novo protein were performed employing the protocol described elsewhere.11,34,35 First, the amino acids were folded in the space by using the AR-SAMD protocol. In brief, energy-minimized polypeptides were subjected to AR-SAMD cycles with the customized GROMOS-96 parameter. A fairly large family of structures was generated sequentially, simulating a long high-temperature MD trajectory and performing simulated annealing periodically. The starting structure was first energy-minimized, followed by molecular dynamics simulation with the parameter set customized for randomization of either the stereochemical structure or the conformation of a polypeptide, using the GROMOS-96 force field. The final conformer resulting from each SAMD cycle was stored for later processing. The possibility of redesigning stable folds to a possible functional protein molecule was further evaluated by an automated sequence design tool, IDeAS. The input files for IDeAS were Cartesian coordinates of the most stable fold specifically in the PDB format. The coordinates over l-side-chain rotamers were applied inversion-symmetry transform, generating the database of d-enantiomers.
Materials
All chemicals and solvents used for experiments are of reagent grade. Amino acids, solvents, trifluoroacetic acid (TFA), thioanisole, and 1,2-ethanedithiol were purchased from Sigma-Aldrich. Diethyl ether and m-cresol were purchased from Merck. AzAla was purchased from Iris Biotech GMBH, Germany. Ultrapure water was used throughout the experiments.
Peptide Synthesis and Characterization
Peptides were synthesized manually by the SPSS method using Fmoc chemistry. Peptides were cleaved from the resin using a cleavage cocktail comprising TFA, ethanedithiol, thioanisole, and m-cresol (20:1:2:2). Purification was done using a Shimadzu Prominence Modular HPLC instrument with a reversed-phase C18 column using a linear gradient of water and acetonitrile (10–100%) containing 0.1% TFA. Peptides were further characterized by verifying the mass using a Bruker Autoflex Speed MALDI TOF/TOF spectrometer.
UV–vis Absorption Spectroscopy
A Cary 60 UV–vis spectrophotometer (Agilent Technologies) was used to record the UV–vis absorption of synthesized peptides. Absorption spectra were scanned from 200 to 700 nm, and the data were recorded at the medium mode using a quartz cuvette of a 10 mm path length.
Fluorescence Spectroscopy
The fluorescence of the synthesized protein samples was measured using a Horiba FluoroMax-4 spectrofluorimeter at room temperature in a quartz cuvette at a 10 mm path length. An equimolar concentration (10 μM) was prepared by using the molar absorption coefficient of Trp (5690 M–1 cm–1), tyrosine (1490 M–1 cm–1), and AzAla (4212 M–1 cm–1).
CD Spectroscopy
CD spectra were recorded in a Jasco 700 CD spectropolarimeter (Jasco Labor-und Datentechnik GmbH, Groß-Umstadt, Germany). Scans were recorded at 25 °C between 190 and 260 nm as an average of five scans and smoothed using the Savitzky–Golay algorithm to obtain the final data. Spectra were collected at 1.0 nm intervals and a bandwidth of 1 nm in a buffer containing a 10 mM phosphate buffer. Spectra of all peptides were recorded at a concentration of 100 μM and recorded in a 1 mm quartz cuvette. CD spectra were presented as a plot of mean residue ellipticities.
FT-IR Spectroscopy
Peptide samples were cast on potassium bromide pellets, and FT-IR spectra were recorded on a micro FTIR-200 (Jasco Co., Japan) equipped with an MCT detector at a 4 cm–1 resolution between 4000 and 400 cm–1. The 16 scan data were collected and processed using Spectra Manager software (Jasco Co., Tokyo, Japan).
DSC
DSC scans were carried out using a Netzsch (model: STA449F3A00 DSC/TGA) instrument. Peptides were dissolved in deionized water with a concentration of 1 mg/mL. The same solution but without the protein was used in the reference cell. Both the sample and reference were scanned from room temperature to 200 °C at a scan rate of 1 K/min.
Acknowledgments
We acknowledge Professor Susheel Durani of IIT Bombay for the core idea of this manuscript; K. Dharmalingam and Vinay Kumar Belwal of IIT Guwahati for experimental support. The Council of Scientific and Industrial Research (grant no. 37(1649)/15/EMR-II), the Department of Biotechnology (grant no. BT/565/NE/U-Excel/2016), and the Board of Research in Nuclear Sciences (grant no. 35/14/07/2017-BRNS/35090), Government of India, for funding. We thank the Central Instruments Facility, IIT Guwahati, for instrumentation support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02574.
Purification and characterization of proteins, UV–vis absorption spectroscopy, and secondary structure determination (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Novotny M.; Kleywegt G. J. A Survey of Left-handed Helices in Protein Structures. J. Mol. Biol. 2005, 347, 231–241. 10.1016/j.jmb.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Ramakrishnan V. Creating novel protein scripts beyond natural alphabets. Syst. Synth. Biol. 2010, 4, 247–256. 10.1007/s11693-011-9068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedge J. F.Evolution: Possible Or Impossible?: Molecular Biology and the Laws of Chance in Nontechnical Language; a New Approach to the Subject, Based on Exciting Recent Discoveries Involving Proteins and DNA, the “Golden Molecule” of Heredity; Zondervan, 1973. [Google Scholar]
- Durani S. Protein Design with l- and d-α-Amino Acid Structures as the Alphabet. Acc. Chem. Res. 2008, 41, 1301–1308. 10.1021/ar700265t. [DOI] [PubMed] [Google Scholar]
- Hazam P. K.; Jerath G.; Kumar A.; Chaudhary N.; Ramakrishnan V. Effect of tacticity-derived topological constraints in bactericidal peptides. Biochim. Biophys. Acta, Biomembr. 2017, 1859, 1388–1395. 10.1016/j.bbamem.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Bada J. L. Origins of homochirality. Nature 1995, 374, 594–595. 10.1038/374594a0. [DOI] [PubMed] [Google Scholar]
- Flory P. J.; Volkenstein M. Statistical mechanics of chain molecules. Biopolymers 1969, 8, 699–700. 10.1002/bip.1969.360080514. [DOI] [Google Scholar]
- Brant D. A.; Flory P. J. The Configuration of Random Polypeptide Chains. II. Theory. J. Am. Chem. Soc. 1965, 87, 2791–2800. 10.1021/ja01091a003. [DOI] [Google Scholar]
- Gordon M. Statistical mechanics of chain molecules. P. J. Flory, pp. xix + 432, 1969. New York: Interscience. 164s. Br. Polym. J. 1970, 2, 302–303. 10.1002/pi.4980020511. [DOI] [Google Scholar]
- Ramakrishnan V.; Ranbhor R.; Kumar A.; Durani S. The Link between Sequence and Conformation in Protein Structures Appears To Be Stereochemically Established. J. Phys. Chem. B 2006, 110, 9314–9323. 10.1021/jp056417e. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V.; Ranbhor R.; Durani S. Simulated folding in polypeptides of diversified molecular tacticity: Implications for protein folding and de novo design. Biopolymers 2005, 78, 96–105. 10.1002/bip.20241. [DOI] [PubMed] [Google Scholar]
- Davis L. K. Intelligent Design of 14-3-3 Docking Proteins Utilizing Synthetic Evolution Artificial Intelligence (SYN-AI). ACS Omega 2019, 4, 18948–18960. 10.1021/acsomega.8b03100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S.; Boyken S. E.; Baker D. The coming of age of de novo protein design. Nature 2016, 537, 320–327. 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Sood D.; Tomar R.; Chandra R. Antimicrobial Peptide Designing and Optimization Employing Large-Scale Flexibility Analysis of Protein-Peptide Fragments. ACS Omega 2019, 4, 21370–21380. 10.1021/acsomega.9b03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A. W.; Anderson D. M.; Tessmer M. H.; Frank D. W.; Feix J. B.; Meiler J. Structure and Dynamics of Type III Secretion Effector Protein ExoU As determined by SDSL-EPR Spectroscopy in Conjunction with De Novo Protein Folding. ACS Omega 2017, 2, 2977–2984. 10.1021/acsomega.7b00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras S. C.; Bertolani S. J.; Siegel J. B. A Benchmark for Homomeric Enzyme Active Site Structure Prediction Highlights the Importance of Accurate Modeling of Protein Symmetry. ACS Omega 2019, 4, 22356–22362. 10.1021/acsomega.9b02636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj G.; Mulligan V. K.; Bahl C. D.; Gilmore J. M.; Harvey P. J.; Cheneval O.; Buchko G. W.; Pulavarti S. V. S. R. K.; Kaas Q.; Eletsky A.; Huang P.-S.; Johnsen W. A.; Greisen P. J. Jr.; Rocklin G. J.; Song Y.; Linsky T. W.; Watkins A.; Rettie S. A.; Xu X.; Carter L. P.; Bonneau R.; Olson J. M.; Coutsias E.; Correnti C. E.; Szyperski T.; Craik D. J.; Baker D. Accurate de novo design of hyperstable constrained peptides. Nature 2016, 538, 329–335. 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinzadeh P.; Bhardwaj G.; Mulligan V. K.; Shortridge M. D.; Craven T. W.; Pardo-Avila F.; Rettie S. A.; Kim D. E.; Silva D.-A.; Ibrahim Y. M.; Webb I. K.; Cort J. R.; Adkins J. N.; Varani G.; Baker D. Comprehensive computational design of ordered peptide macrocycles. Science 2017, 358, 1461–1466. 10.1126/science.aap7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J.; Vorobieva A. A.; Sheffler W.; Doyle L. A.; Park H.; Bick M. J.; Mao B.; Foight G. W.; Lee M. Y.; Gagnon L. A.; Carter L.; Sankaran B.; Ovchinnikov S.; Marcos E.; Huang P.-S.; Vaughan J. C.; Stoddard B. L.; Baker D. De novo design of a fluorescence-activating β-barrel. Nature 2018, 561, 485–491. 10.1038/s41586-018-0509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers S. A.; Karanicolas J.; Chang H. W.; Zhao A.; Jiang L.; Zirafi O.; Stevens J. T.; Münch J.; Baker D.; Eisenberg D. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 2011, 475, 96–100. 10.1038/nature10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidigh J. W.; Fesinmeyer R. M.; Andersen N. H. Designing a 20-residue protein. Nat. Struct. Biol. 2002, 9, 425. 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- Juraszek J.; Bolhuis P. G. Sampling the multiple folding mechanisms of Trp-cage in explicit solvent. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 15859–15864. 10.1073/pnas.0606692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W.; Gallicchio E.; Deng N.; Andrec M.; Levy R. M. Kinetic Network Study of the Diversity and Temperature Dependence of Trp-Cage Folding Pathways: Combining Transition Path Theory with Stochastic Simulations. J. Phys. Chem. B 2011, 115, 1512–1523. 10.1021/jp1089596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy; Springer Science & Business Media, 2013. [Google Scholar]
- Royer C. A. Probing Protein Folding and Conformational Transitions with Fluorescence. Chem. Rev. 2006, 106, 1769–1784. 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- Callis P. R. Binding phenomena and fluorescence quenching. II: Photophysics of aromatic residues and dependence of fluorescence spectra on protein conformation. J. Mol. Struct. 2014, 1077, 22–29. 10.1016/j.molstruc.2014.04.051. [DOI] [Google Scholar]
- Shao J.; Korendovych I. V.; Broos J. Biosynthetic incorporation of the azulene moiety in proteins with high efficiency. Amino Acids 2015, 47, 213–216. 10.1007/s00726-014-1870-4. [DOI] [PubMed] [Google Scholar]
- Loidl G.; Musiol H.-J.; Budisa N.; Huber R.; Poirot S.; Fourmy D.; Moroder L. Synthesis of β-(1-azulenyl)-L-alanine as a potential blue-colored fluorescent tryptophan analog and its use in peptide synthesis. J. Pept. Sci. 2000, 6, 139–144. . [DOI] [PubMed] [Google Scholar]
- Ridgway Z.; Picciano A. L.; Gosavi P. M.; Moroz Y. S.; Angevine C. E.; Chavis A. E.; Reiner J. E.; Korendovych I. V.; Caputo G. A. Functional characterization of a melittin analog containing a non-natural tryptophan analog. Biopolymers 2015, 104, 384–394. 10.1002/bip.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa N.; Paramita Pal P. Designing novel spectral classes of proteins with a tryptophan-expanded genetic code. Biol. Chem. 2004, 385, 893. 10.1515/bc.2004.117. [DOI] [PubMed] [Google Scholar]
- Creasey R. C. G.; Mostert A. B.; Solemanifar A.; Nguyen T. A. H.; Virdis B.; Freguia S.; Laycock B. Biomimetic Peptide Nanowires Designed for Conductivity. ACS Omega 2019, 4, 1748–1756. 10.1021/acsomega.8b02231. [DOI] [Google Scholar]
- Tang J. D.; Lampe K. J. From de novo peptides to native proteins: advancements in biomaterial scaffolds for acute ischemic stroke repair. Biomed. Mater. 2018, 13, 034103. 10.1088/1748-605x/aaa4c3. [DOI] [PubMed] [Google Scholar]
- Zanna N.; Focaroli S.; Merlettini A.; Gentilucci L.; Teti G.; Falconi M.; Tomasini C. Thixotropic Peptide-Based Physical Hydrogels Applied to Three-Dimensional Cell Culture. ACS Omega 2017, 2, 2374–2381. 10.1021/acsomega.7b00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranbhor R.; Kumar A.; Patel K.; Ramakrishnan V.; Durani S. Automated design evolution of stereochemically randomized protein foldamers. Phys. Biol. 2018, 15, 036001. 10.1088/1478-3975/aaac9a. [DOI] [PubMed] [Google Scholar]
- Ranbhor R.; Kumar A.; Tendulkar A.; Patel K.; Ramakrishnan V.; Durani S. IDeAS: automated design tool for hetero-chiral protein folds. Phys. Biol. 2018, 15, 066005. 10.1088/1478-3975/aacdc3. [DOI] [PubMed] [Google Scholar]
- Alston R. W.; Lasagna M.; Grimsley G. R.; Scholtz J. M.; Reinhart G. D.; Pace C. N. Peptide sequence and conformation strongly influence tryptophan fluorescence. Biophys. J. 2008, 94, 2280–2287. 10.1529/biophysj.107.116921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz Y. S.; Binder W.; Nygren P.; Caputo G. A.; Korendovych I. V. Painting proteins blue: [small beta]-(1-azulenyl)-l-alanine as a probe for studying protein-protein interactions. Chem. Commun. 2013, 49, 490–492. 10.1039/c2cc37550h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. A.Protein Stability and Folding: Theory and Practice; Springer, 1995; Vol. 21. [Google Scholar]
- Cooper A. Protein Heat Capacity: An Anomaly that Maybe Never Was. J. Phys. Chem. Lett. 2010, 1, 3298–3304. 10.1021/jz1012142. [DOI] [Google Scholar]
- Durowoju I. B.; Bhandal K. S.; Hu J.; Carpick B.; Kirkitadze M. Differential Scanning Calorimetry—A Method for Assessing the Thermal Stability and Conformation of Protein Antigen. J. Visualized Exp. 2017, (121), 55262. 10.3791/55262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culik R. M.; Serrano A. L.; Bunagan M. R.; Gai F. Achieving secondary structural resolution in kinetic measurements of protein folding: a case study of the folding mechanism of Trp-cage. Angew. Chem., Int. Ed. 2011, 50, 10884–10887. 10.1002/anie.201104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabartty A.; Kortemme T.; Padmanabhan S.; Baldwin R. L. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry 1993, 32, 5560–5565. 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- Diez-García F.; Pantoja-Uceda D.; Jiménez M. Á.; Chakrabartty A.; Laurents D. V. Structure of a simplified β-hairpin and its ATP complex. Arch. Biochem. Biophys. 2013, 537, 62–71. 10.1016/j.abb.2013.06.009. [DOI] [PubMed] [Google Scholar]
- Cochran A. G.; Skelton N. J.; Starovasnik M. A. Tryptophan zippers: Stable, monomeric β-hairpins. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 5578–5583. 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L.; Wallace B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser K.; Engelhard M.; Friedman N.; Sheves M.; Siebert F. Interpretation of Amide I Difference Bands Observed during Protein Reactions Using Site-Directed Isotopically Labeled Bacteriorhodopsin as a Model System. J. Phys. Chem. A 2002, 106, 3553–3559. 10.1021/jp012926e. [DOI] [Google Scholar]
- Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta, Bioenerg. 2007, 1767, 1073–1101. 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Haris P. I.; Chapman D. Does Fourier-transform infrared spectroscopy provide useful information on protein structures?. Trends Biochem. Sci. 1992, 17, 328–333. 10.1016/0968-0004(92)90305-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.