Abstract
Several risk stratification tools were developed to predict disease progression in coronavirus disease 2019, with no external validation to date. We attempted to validate three previously published risk-stratification tools in a multicenter study. Primary outcome was a composite outcome of development of severe coronavirus disease 2019 disease leading to ICU admission or death censored at hospital discharge or 30 days. We collected data from 169 patients. Patients were 73 years old (59–82 yr old), 66 of 169 (39.1%) were female, 57 (33.7%) had one comorbidity, and 80 (47.3%) had two or more comorbidities. Area under the receiver operating characteristic curve (95% CI) for the COVID-GRAM score was 0.636 (0.550–0.722), for the CALL score 0.500 (0.411–0.589), and for the nomogram 0.628 (0.543–0.714).
Keywords: coronavirus disease 2019, calibration, risk stratification, validation
To the Editor:
We found that three risk tools (COVID-GRAM, CALL-TOOL, and a nomogram) developed in small, homogenous patient populations showed poor discrimination in a multicenter study and are unlikely to be clinically useful in different settings. Re-evaluation of these tools is urgently needed using international datasets.
Since the outbreak of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic in January 2020, there has been a rush to develop clinical risk prediction scores, particularly at the first epicenter of the pandemic in China (1–3). Predicting risk of deterioration or severe coronavirus disease 2019 (COVID-19)–related illness is of significant interest, both as part of guidance for clinical treatment and resource allocation as well as to highlight the likely groups benefitting from novel disease modifying therapies. The RECOVERY trial demonstrated that dexamethasone treatment offers mortality advantage in patients needing oxygen therapy or mechanical ventilation, whereas it might be harmful in patients who do not need supplemental oxygen (4). These results emphasize the need for a reliable risk prediction scores, which might help providers to decide about therapeutic approaches. Notwithstanding the urgency, we must not forget past lessons learnt during developing risk scores for diseases and conditions encompassing a wide range of clinical risk. We have shown in sepsis that conflicting definitions with ill-calibrated tools lead to the overprovision of medical therapy, with potential for associated harm (5).
Although many of the risk prediction tools were developed in multicenter studies, their external clinical utility and face validity have not been established in independent cohorts (6). There are significant differences between the respective populations affected by SARS-CoV-2 in China compared with the United Kingdom, and we attempted to validate three previously published risk-stratification tools in a multicenter study.
Anonymized patient data were collected as part of a service evaluation project by the Secondary Care Group Members of the Welsh Government COVID-19 response from patients admitted to the University Hospital of Wales, a tertiary academic center, and to the two district general hospitals in Aneurin Bevan UHB during the first 6 weeks of the pandemic in Wales, between March 9, 2020, and April 19, 2020. Due to the anonymized nature of the data collection, formal written consent was waived by the institutional review board.
Data were collected to enable to calculate the CALL score, the COVID-GRAM risk score, and a nomogram developed by Gong et al (1–3). Primary outcome was a composite outcome of development of severe COVID-19 disease leading to ICU admission or death, in line with the definitions and outcomes used in the original publications. The outcome was censored at hospital discharge or 30 days.
For statistical analysis, receiver operating characteristics (ROC) curves were used to establish predictive ability. We planned to use calibration plot to assess calibration of the prediction tools if area under the area under the ROC (AUROC) curve for any of the tools was found to be above 0.8 (good discrimination ability). Data are presented as n (%), median (interquartile range), or ROC (95% CI) as appropriate.
We collected data from 169 patients. Patients were 73 years old (59–82 yr old), 66 of 169 (39.1%) were female, 32 (18.9%) had no significant comorbidities, 57 (33.7%) had one comorbidity, and 80 (47.3%) had two or more comorbidities. Most prevalent comorbid conditions were diabetes in 42 patients (24.9%), chronic obstructive pulmonary disease in 32 patients (19.0%), and ischemic heart disease in 26 patients (15.4%). Patients presented after 9 days (2–12 d) of symptom onset to the hospital. Eighty-one patients (47.9%) had reached the composite outcome of ICU admission or death, the hospital mortality was 33.7%. Neither of the three risk-prediction tools were able to accurately predict outcome. AUROC (95% CI) for the COVID-GRAM score was 0.636 (0.550–0.722) p value equals to 0.003, for the CALL score 0.500 (0.411–0.589) p value equals to 0.997, and for the nomogram 0.628 (0.543–0.714) p value equals to 0.005 (Fig. 1). As none of the tools exhibited good discrimination characteristics, we have not performed formal assessment of calibration. The COVID-GRAM tool underperformed in the medium risk category: 40% of our patients experienced the predicted composite outcome versus 7.3% in the original cohort. The CALL score underpredicted the outcome in the 7–9 points (medium risk) category (10–40% predicted vs 52% actual occurrence) and overpredicted in the 10–13 (high risk) category (over 50% predicted vs 46% actual occurrence). The nomogram overpredicted the outcome: out of the 108 patients where the nomogram predicted over 90% chance for the composite outcome to manifest, only 61 (56.5%) experienced it.
Figure 1.
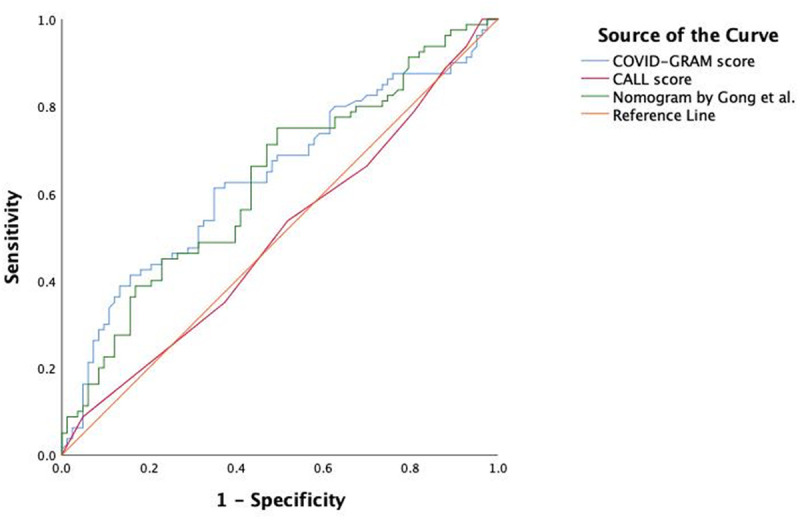
Receiver operating characteristic curves to describe predictive capabilities of the three risk stratification tools.
All three clinical risk prediction scores, the CALL score, COVID-GRAM risk score, and nomogram, had poor discriminative value for the composite outcome of ICU admission or death within our cohort. The COVID-GRAM risk score (derived from n = 2300 patients) performed better than the CALL score (n = 208) and narrowly better than the nomogram developed by Gong et al (3) (n = 372), as evidenced by the AUROC.
Our findings highlight the difficulties of predictive tool development for a new disease with uncertain and potentially changing outcomes (7). The discriminatory performance of the three different models was well below the performance compared with their derivation or validation cohort, in line with recent findings of a large U.K. dataset (8). This questions if the proposed models could transfer over to a different setting and could offer reasonable performance. The difference observed between the precision of these tools in the original publications and in our independent cohort in a different location could be explained by several factors. Some of this might be population based, as were significant differences between the patient characteristics of these three studies and ours. Importantly, the mean or median age was below 50 years in the development and validations cohorts of the original publications, whereas it was above 70 in our study in line with observed characteristics in the United Kingdom (9). Han Chinese patients in the original studies had low comorbidity burden with 70–75% of patients without any significant comorbidities, whereas four of five in our largely Caucasian cohort had one or more comorbidities, again in line with the data from the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) study (9). As age and comorbidities are established risk factors for disease progression and adverse outcome in COVID-19, it is unsurprising that the predictive scores developed in a young and relatively healthy population do not perform well in an older cohort with significant comorbidities (7, 8). It is also possible that there were differences in standard of care; however, our cohort was admitted to the hospital at the very beginning of the U.K. phase of the pandemic, when there were no established treatment options (10).
All three tools overestimated the morbidity and mortality, especially in those with a higher comorbidity burden. This issue highlights significant questions about the development of such tools that use small sample sizes. The original studies used regression analysis to derive the significant variables incorporated in their scores. Although the least absolute shrinkage and selection operator regression used by two groups is regarded as more appropriate than the Cox-regression used in the third study, with their low event rate, these models suffer a significant reduction in predictive capabilities when used in a relatively small sample size. The small number of events in the three studies compared with the number of variables used makes overfitting a real possibility (7). This is illustrated by our finding that we observed underprediction in the low-risk and overprediction in the high-risk groups, both recognized features of an overfitting model (11).
Furthermore, predictive accuracy of all three tools was only assessed by ROC analysis without any other alternative method such as a discrimination slope (12).
Generally, when the discrimination of a clinical risk prediction tool is satisfactory, it is necessary to investigate the quality of the calibration to ensure there is acceptable agreement between the observed occurrence of ICU admission and death and the risk predicted by the score. Liang et al 12) did not provide any data on the calibration of their COVID-GRAM model. The use of a calibration plot would have the added benefit of assessing the overfitting of the model, allowing for the fine-tuning of regression coefficients if indicated for better clinical utility (12). On the other hand, the nomogram developed by Gong et al (3) and the CALL score both had adequate discrimination with respect to their training and validation cohorts as well as calibration data, in the form of a calibration curve, for the probability of developing severe COVID-19 disease. Their calibration curves showed well-fitted agreement between the nomogram and CALL score prediction and actual benefit according to their datasets. However, part of the reason for the poor discriminative value in both cases concerning our cohort is likely due to the small and homogenous training and validation cohorts used in its development.
Very recently, using the currently largest clinical dataset of almost 60,000 patients from the ISARIC 4C (Coronavirus Clinical Characteristics Consortium) group published the development and validation of the 4C score (8). Their model showed good discrimination, excellent calibration, and resilient to imputation of missing values (8). They also noted that the more elaborate scores, such as we have investigated, could be applied to a smaller subset of patients, as some physiologic variables or laboratory values are not routinely recorded. Although it was not an issue in our population, collection of diverse clinical information might not be feasible in the pandemic, even in a developed healthcare system, increasing the fragility of the prediction model.
There is a temptation to use more sophisticated tools for outcome prediction, such as artificial intelligence–based methods or using electronic healthcare records when individual patient data are not available; however, there is a significant question about the clinical usefulness of such models (13, 14). It is unclear if the effort to improve discriminatory capability by adding new variables and more complicated methods or the use of just historical data to sort patients into broad high and low risk categories actually changes the answer to the clinical question and of benefit.
In the fast changing landscape of the pandemic, with emerging and rapidly adopted new treatment options and more sophisticated phenotyping of the immune response, plus possible changes in the characteristics of the virus, it is unlikely that one prediction model will be able to answer all the questions (4, 15, 16). However, any new prediction models should be developed and reported in accordance with the guidance of best practice (7).
There are significant limitations to our study, most importantly the small sample size. To limit the number of patients recruited was a pragmatic decision, so we could rapidly assess the face validity of these tools. However, we have recruited all consecutive patients admitted to the three hospitals, reducing selection bias. Our patient cohort showed strong similarities to the U.K.-wide ISARIC dataset, and arguably, our patients who were recruited from a tertiary center, from a medium and small district general hospital, give appropriate cross-sectional view to evaluate the usefulness of these scoring tools (9). The relatively late presentation to hospital following the onset of symptoms could also be seen as a limitation, as it is possible, that by adhering to the national guidance of “Stay at home”, patients presented with significantly more advanced disease, than in the development cohorts in China. Further analysis of this potential effect would be possible in the large international datasets.
In summary, our results show that the early tools developed in a relatively small, homogenous patient population are unlikely to be clinically useful in different settings. There is a continued need to develop reliable, easy-to-use risk stratifications tools, from commonly available clinical variables, according to the guiding principles for predictive model development. We must scrutinize new tools using international datasets to achieve better and more universal discrimination and calibration of risk prediction tools for patients with COVID-19.
ACKNOWLEDGMENTS
We would like to thank the Gwent Coronavirus Disease 2019 Group members for their support in data collection: A. Allen-Ridge, E. Baker, C. Bailey, T. Baumer, S. Beckett, S. Champanerkar, Y. Cheema, S. Cherian, E. Cocks, S. Cutler, E. Dawe, A. Dhadda, S. Dumont, N. Duric, S. Elgarf, T. Evans, E. Godfrey, L. Harding, D. Hepburn, A. E. Heron, L. Jesani, P. Jones, C. Killick, C. King, A. Kiss, J. Lavers, J. Lloyd-Evans, N. Mason, L. McClelland, B. Muthuswamy, J. Parry-Jones, E. Phillips, S. Pooley, B. Radford, O. Richards, A. Rimmer, G. Roberts, A. Roynon-Reed, A. Sieunarine, K. Sullivan, T. Szakmany, C. Thomas, E. Thomas, J. Tozer, T. West, and M. Winstanley.
Haamed Al Hassan, MB BCh, Cardiff and Vale University Health Board, Cardiff, United Kingdom, and Department of Anaesthesia, Intensive Care and Pain Medicine, Division of Population Medicine, Cardiff University, Cardiff, United Kingdom; Eve Cocks, DN, Lara Jesani, MB BCh, Critical Care Directorate, Aneurin Bevan University Health Board, Newport, United Kingdom; Sally Lewis, MRCGP, Value Based Healthcare Team, Aneurin Bevan University Health Board, Newport, United Kingdom; Tamas Szakmany, MD, PhD, Department of Anaesthesia, Intensive Care and Pain Medicine, Division of Population Medicine, Cardiff University, Cardiff, United Kingdom and Critical Care Directorate, Aneurin Bevan University Health Board, Newport, United Kingdom; on behalf of the Gwent COVID-19 Group
Footnotes
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Drs. Al Hassan, Cocks, Jesani, and Lewis. The first draft of the article was written by Drs. Al Hassan and Szakmany, and all authors commented on previous versions of the article. All authors read and approved the final article.
The authors have not disclosed any potential conflicts of interest.
Contributor Information
Collaborators: A. Allen-Ridge, E. Baker, C. Bailey, T. Baumer, S. Beckett, S. Champanerkar, Y. Cheema, S. Cherian, E. Cocks, S. Cutler, E. Dawe, A. Dhadda, S. Dumont, N. Duric, S. Elgarf, T. Evans, E. Godfrey, L. Harding, D. Hepburn, A. E. Heron, L. Jesani, P. Jones, C. Killick, C. King, A. Kiss, J. Lavers, J. Lloyd-Evans, N. Mason, L. McClelland, B. Muthuswamy, J. Parry-Jones, E. Phillips, S. Pooley, B. Radford, O. Richards, A. Rimmer, G. Roberts, A. Roynon-Reed, A. Sieunarine, K. Sullivan, T. Szakmany, C. Thomas, E. Thomas, J. Tozer, T. West, and M. Winstanley
REFERENCES
- 1.Ji D, Zhang D, Xu J, et al. Prediction for progression risk in patients with COVID-19 pneumonia: The CALL score. Clin Infect Dis. 2020; 71:1393–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang W, Liang H, Ou L, et al. ; China Medical Treatment Expert Group for COVID-19. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020; 180:1081–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong J, Ou J, Qiu X, et al. A tool for early prediction of severe coronavirus disease 2019 (COVID-19): A multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis. 2020; 71:833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with Covid-19 — preliminary report. N Engl J Med. 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopczynska M, Sharif B, Unwin H, et al. Real world patterns of antimicrobial use and microbiology investigations in patients with sepsis outside the critical care unit: Secondary analysis of three nation-wide point prevalence studies. J Clin Med. 2019; 8:1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. BMJ. 2020; 369:m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leisman DE, Harhay MO, Lederer DJ, et al. Development and reporting of prediction models: Guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med. 2020; 48:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight SR, Ho A, Pius R, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: development and validation of the 4C mortality score. BMJ. 2020; 370:m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Docherty AB, Harrison EM, Green CA, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: Prospective observational cohort study. BMJ. 2020; 369:m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumer T, Phillips E, Dhadda A, et al. Epidemiology of the first wave of COVID-19 ICU admissions in South Wales – the interplay between ethnicity and deprivation. Front Med. 2020; 7:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavlou M, Ambler G, Seaman SR, et al. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015; 351:h3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010; 21:128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong DL, Kempsell KE, Szakmany T, et al. Development of a bioinformatics framework for identification and validation of genomic biomarkers and key immunopathology processes and controllers in infectious and non-infectious severe inflammatory response syndrome. Front Immunol. 2020; 11:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barda N, Riesel D, Akriv A, et al. Developing a COVID-19 mortality risk prediction model when individual-level data are not available. Nat Commun. 2020; 11:4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha P, Calfee CS, Cherian S, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: A prospective observational study. Lancet Respir Med. 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber B, Fischer WM, Gnanakaran S, et al. ; Sheffield COVID-19 Genomics Group. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020; 182:812–827.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]


