Abstract
The p15Ink4b gene is frequently hypermethylated in myeloid neoplasia and has been demonstrated to be a tumor suppressor. Since it is a member of the INK4b family of cyclin-dependent kinase inhibitors, it was initially presumed that its loss in leukemic blasts caused a dysregulation of the cell cycle. However, animal model experiments over the last several years have produced a very different picture of how p15Ink4b functions in hematopoietic cells and how its loss contributes to myelodysplastic syndrome and myeloid leukemia. It is clear now, that in early hematopoietic progenitors, p15Ink4b functions outside of its canonical role as a cell cycle inhibitor. Its functions are involved in signal transduction and influence the development of erythroid, monocytic and dendritic cells.
Keywords: leukemia, tumor suppressor, hematopoiesis, methylation, immune surveillance, erythropoiesis
p15INK4b deletion in human cancers
P15INK4b is a member of the INK4 family of cyclin-dependent kinase inhibitors. The gene, p15INK4b, also known as CDKN2b and MTS2, is located on chromosome 9q21 and is often deleted in conjunction with sequences encoding two other gene products p16INK4a and p14ARF [1; 2; 3]. This has been shown to occur in lymphomas as well as carcinomas and sarcomas [4; 5; 6; 7]. P15INK4b and p16INK4a are both capable of inducing cell cycle arrest in G1 by inhibiting cyclin-dependent kinases 4 and 6 (cdk4/6), whereas p14ARF is an activator of TRP53 [2; 8]. p15INK4b probably has an important backup function for p16INK4a in senescence which may explain why the genes encoding both are often deleted in human tumors. Experiments in mice show that deletions prohibiting expression of all three INK-ARF proteins promotes a wider spectrum of tumors than deletions affecting only p16Ink4a and p14ARF [9].
p15INK4b methylation in human MDS and AML
Loss of p16INK4a and p15INK4b expression in hematopoietic neoplasms also occurs through DNA methylation of the genes encoding these proteins [4; 10; 11]. Importantly, however, p15INK4b is the only INK4 family gene member that is methylated in MDS and AML suggesting a specific function in the myeloid lineage. This gene is hypermethylated in 50–60 % of patients with myelodysplastic syndrome and 70–80 % of patients with AML [4]. Aberrant p15INK4b methylation has been generally associated with poor prognosis in AML [12; 13] and in MDS patients its methylation is associated with increased risk of transformation to AML [14; 15]. p15INK4b hypermethylation is found in almost all FAB subtypes, although higher frequencies are seen in M1, M2, M3, and M4 [12; 16; 17; 18]. In patients with therapy-induced AML (t-AML) the frequency of methylation is over 90% [13]. The aberrant hypermethylation is found in the CpG islands extending throughout the promoter region, exon1 and part of intron 1 and is associated with signatures of polycomb repression and reduced H3K4 trimethylation [11; 19]. A study that looked at whether methylation occurred in all the common cytogenetic subtypes found that a strikingly low level of methylation occurred in AML with inv(16) [20]. This led to the finding that the inv(16)-encoded CBFβ-SMMHC silenced p15INK4b by displacing the transcription factor RUNX1 from the promoter.
Demonstration of tumor suppressor function in murine models
A mouse model was developed to define a role for p15INK4b as a tumor suppressor. p15Ink4b(−/−) mice, in which the second coding exon of the gene was eliminated by homologous recombination [21]. These mice showed no increased susceptibility to AML over wild-type mice although the mice had extramedullary hematopoiesis and a low incidence of angiosarcomas. Furthermore, the p15Ink4b-deficient background in transgenic mice expressing Rgr oncogene under a CD4 promoter significantly decreased incidence of thymic lymphomas when compared with CD4-Rgr progeny with wild-type or heterozygous expression of p15Ink4b [22].
Although these experiments failed to show that loss of p15Ink4b directly results in AML, there was reason to believe that p15Ink4b was a tumor suppressor for AML in mice. First, retrovirus-induced leukemias of the myelomonocytic phentype were found to have undergone hypermethylation of the 5′ CpG island of the p15Ink4b gene. In addition, when the knockout was redeveloped on a 129/sv background using the same target vector as Latres et al. [21], it was possible to show that loss of p15INK4b results in increased numbers of myeloid progenitors in the bone marrow. Furthermore, bone marrow from these mice had a competitive advantage in myeloid cell formation in a repopulating assay [23; 24].
The mouse model that has been most informative as to p15Ink4b’s role as a tumor suppressor for myeloid malignancy is a conditional one in which p15Ink4b is knocked out specifically in myeloid cells [25]. For this model, a floxed exon2 was introduced and the mice were crossed with transgenic mice expressing Cre under the control of a LysM promoter that ensured a myeloid cell-specific expression pattern [26]. These mice, called p15Ink4bfl/flLysMcre, develop monocytosis with increased circulating monocytes in the blood starting at 5 to 7 months of age, but have normal levels of neutrophils, lymphocytes, platelets and red blood cells. In addition, an expansion of myelomonocytic cells in the bone marrow was observed in these mice as evidence by increases in mature myeloid (Gr-1+/Mac-1+) and monocytic cells (Gr-1lo/Mac-1+) as well as immature myeloid cells (Mac-1+/cKit+). Immunohistochemical staining of spleen tissue revealed a small increase in myelomonocytic cells in the red pulp which was confirmed by FACS analysis. A small number of mice spontaneously developed a myeloproliferative neoplasm characterized as the advanced form of chronic myelomonocytic leukemia with a significantly increased number of cKit+ immature cells in bone marrow and peripheral blood [25]. Since the p15Ink4bfl/flLysMcre mice did not develop acute disease, they were subjected to retroviral insertional mutagenesis to determine whether they had increased susceptibility to acute leukemia when provided with additional oncogenic hits. Mice were inoculated shortly after birth with MOL4070LTR retrovirus [27] and monitored for 15 months for disease development. Control mice developed leukemia with low penetrance, whereas the incidence of retrovirus-induced leukemia was statistically highly significant for the p15Ink4bfl/flLysMcre mice. Furthermore, there was a much higher incidence of myeloid tumors, mostly monocytic and myelomonocytic in p15Ink4bfl/flLysMcre (92%) and p15Ink4bfl/wtLysMcre (79%) mice than in control, p15Ink4bwt/wtLysMcre mice, where there was an equal distribution of myeloid and lymphoid leukemia. This model has, therefore, provided evidence that loss of p15Ink4b can itself promote preleukemic conditions and demonstrated experimentally that this gene is a tumor suppressor for AML [25].
p15Ink4b and erythroid/myeloid cell fate
Since p15Ink4b is the only member of the INK4 family that is inactivated by DNA methylation in AML, it is of interest to determine if there is any other function specific to the myeloid lineage that can be assigned to this gene. Initial investigations of hematopoiesis in Ink4b−/− mice showed that these mice have greater numbers of bi-potent granulocyte-macrophage progenitors (GMP) and this characteristic was found, in competitive repopulating studies in vivo, to be intrinsic to the cells. Interestingly, Ink4b-deficient GMPs did not cycle more frequently than wild-type progenitors and showed no differences in apoptosis or self-renewal potential. However, Ink4b was shown to affect differentiation of common myeloid progenitor (CMP) cells toward GMPs, resulting in an imbalance of down-stream progenitors. In vitro analysis of progenitor cells from knockout mice demonstrated that loss of p15Ink4b causes increased differentiation toward GMPs and decrease in differentiation toward megakaryocyte-erythroid progenitors (MEP) [23; 28].
Based upon the data obtained from the knockout mice, it was hypothesized that p15Ink4b is required for efficient erythropoiesis. Subsequently, p15Ink4b was shown to promote erythroid differentiation and suppresses myeloid differentiation of hematopoietic progenitors under both normal and stress conditions. First, it was demonstrated that p15Ink4b is expressed more highly in committed MEPs than GMPs. More importantly, mice lacking p15Ink4b have lower numbers of primitive red cell progenitors and a severely impaired response to induced hematopoietic stress caused by 5-fluorouracil or phenylhydrazine. When p15Ink4b was re-introduced into the bone marrow progenitors from p15Ink4b−/− mice, which had a low erythroid to myeloid balance, it corrected the observed skewing in hematopoietic cell differentiation. In support of a new function for this gene outside its canonical function as a CDKI, evidence was provide that pRB was not required for alterations in the balance of myeloid/erythroid cells in p15Ink4b−/− mice. When p15Ink4b was introduced into a multi-potential progenitor cell line (EML,) it produced changes at the molecular level, including activation of MEK/ERK signaling, increased GATA-1, EpoR, and decreased PU.1 and GATA-2 expression. These changes rendered cells more permissive to erythroid commitment and less permissive to myeloid commitment, as demonstrated by an increase in early burst forming unit erythroid (BFU-E) formation with a concomitant decrease in myeloid progenitors [29]. Therefore, p15Ink4b functions in erythropoiesis, by maintaining proper lineage commitment of progenitors and assisting in rapid red blood cell replenishment following stress.
Role of p15Ink4b in dendritic cell development
Recently, another cell fate function of p15Ink4b was found in driving dendritic cells (DCs) maturation. It was shown that mice lacking a functional p15Ink4b in the myeloid lineage had a significantly reduced numbers of common DC progenitor cells (CDPs) [30] that can give rise to all DCs (Figure 1). Concomitantly, numbers of conventional (classical) dendritic cells (cDCs), but not plasmacytoid dendritic cells (pDCs), were significantly decreased in the spleen [31]. In vitro experiments with bone marrow-derived DCs (BM-DCs) from hematopoietic progenitors lacking p15Ink4b cultured in granulocytes-macrophages colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4) confirmed a decreased capacity of myeloid progenitors lacking p15Ink4b to differentiate into DCs. Furthermore, the maturity of LPS-activated BM-DCs from knockout mice was also significantly compromised as compared to wt-derived BM-DCs. In accordance with these data, p15Ink4b-deficient BM-DCs expressed significantly lower levels of cell surface molecules MHCI, MHCII and co-stimulatory molecules CD80, CD86 known to be necessary for an efficient stimulation of naïve T-cells [31]. The immaturity of cDCs from knockout mice was functionally confirmed by their limited ability to stimulate allogeneic T cells. Importantly, enforced expression of p15Ink4b restored differentiation and maturation defects in DCs derived from mice with silenced endogenous p15Ink4b, confirming a positive role for p15Ink4b in development and maturation of cDCs.
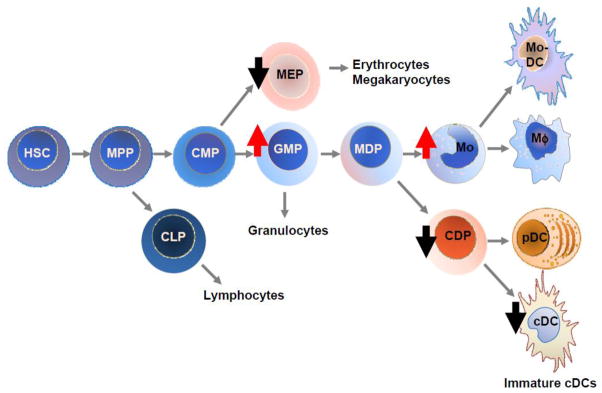
Figure 1. Impaired homeostasis of myeloid cells in p15Ink4b-deficient mice.
A simplified scheme of hematopoietic stem cells (HSC) differentiating toward myeloid lineages is presented. Increased production (red arrow pointing up) of granulocyte-macrophage progenitors (GMP) and monocytes (Mo), as well as decreased production (black arrow pointing down) of megakaryocyte-erythrocyte progenitors (MEP), common DC progenitors (CDP), and conventional DCs (cDC) in p15Ink4-deficient mice are indicated. Multipotent progenitors (MPP), common myeloid progenitors (CMP), common lymphoid progenitors (CLP), macrophage/dendritic cell progenitors (MDP), monocyte-derived DCs (Mo-DC), macrophages (Mφ), plasmacytoid DCs (pDC).
These defects in development and maturation of cDCs in the absence p15Ink4b also have important implications regarding its role as a tumor suppressor for AML, because pre-leukemic myeloid cells differentiating into cDCs may have an impaired ability to provide competent immune surveillance. DCs are the critical cells that initiate immune surveillance and at the same time maintain appropriate self-tolerance. The scheme of dendritopoiesis suggests that myeloid precursors of CDPs include pre-leukemic/leukemic cells with acquired genetic/epigenetic changes. These abnormalities that include silencing of p15Ink4b expression are required for transformation of hematopoietic cells into leukemic cells and also they affect immune system through modulation of development and/or maturation of cDCs [31].
Insight into how p15Ink4b might control maturation at the molecular level has been obtained. p15Ink4b potentiates transcriptional activity of the transcription factor PU.1, an important downstream target that directly regulates differentiation and maturation of cDCs, through increased phosphorylation of ERK kinases [31]. Interestingly, PU.1 is absolutely critical for development not only cDCs, but also plasmacytoid DCs (pDCs). However, higher activity of this transcription factor is required for development of cDCs than pDCs [32; 33], which is in a good agreement that p15Ink4b regulates mainly development of cDCs, but not pDCs. Increased expressions of many Pu.1 targets genes, such as CD80, CD86 [34], CD40 [35] and MHCII [36; 37] in BM-DCs with restored p15Ink4b confirmed further that this tumor suppressor regulates maturation of cDCs through the transcription factor PU.1. Since expression of p15Ink4b is also activated by PU.1 in myeloid cell [38], and in turn p15Ink4b positively regulates PU.1 [31], it is hypothesized that this mutual cooperation creates a positive feedback loop that amplifies differentiation and maturation of cDCs.
Concluding remarks
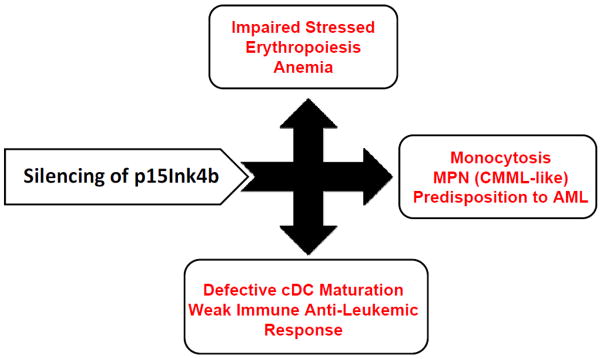
Experiments, in animal model systems with embryonal and conditional knockout of p15Ink4b in mice, reveal that, in addition to its canonical function as regulator of cell cycle, p15Ink4b has an important cell cycle-independent role in regulation of myeloid cell differentiation. It was shown that its expression in early myeloid progenitors contributes to proper homeostatic development of monocytic, erythroid, and dendritic cells. In early stages of myeloid cells differentiation, p15Ink4b modulates activities of specific instructive transcription factors such as PU.1 [33; 39], and GATA-1, and GATA-2 [40] that drive myeloid progenitors into specific cell lineages. Thus silencing of p15Ink4b by DNA methylation in early myeloid lineage, as observed frequently in pre-leukemic and leukemic stages of AML, has multiple consequences that contribute to leukemia development (summarized in Figure 2). This suggests that loss of p15Ink4b expression not only favors production of immature myeloid cells that are targets of additional mutations during leukemogenesis, but also impairs the differentiation and maturation of cDCs from these precursors, which may have negative effect on the immune system to recognize and clear cancerous cells. In support of this hypothesis, immature, tolerogenic DCs are frequently detected in patients with MDS and AML [41; 42; 43]. Since DCs modulate the nature and intensity of the adaptive immune response, they provide an attractive target for cancer immunotherapy. Therefore, successful targeting of p15Ink4b re-expression in clinical treatment regimens may not only restore control over the of cancer cell production, but it may also improve the anti-leukemic function of the immune system through generation of more mature, immunostimulatory DCs that prime naive T-cells to recognize and remove leukemic cells. These recent data may have also a translational implication in improved patient-specific anti-leukemic DCs immunotherapy to fight minimal residual disease that still remains a critical obstacle in the successful treatment of AML.
Figure 2. Silencing of p15Ink4b in myeloid cells affects development of several hematopoietic cell lineages that may cumulatively contribute to AML development.
MPN (myeloproliferative neoplasm), CMML (chronic myelomonocytic leukemia), AML (acute myeloid leukemia), cDC (conventional dendritic cells)
Acknowledgments
This work was supported by the intramural research program of the National Cancer Institute, Center for Cancer Research, NIH
Footnotes
Author’s Contributions.
L.W. and J.B. wrote the article.
Disclosure.
The authors declare no conflict of interest. All authors have approved the final article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 2.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 3.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–77. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 4.Drexler HG. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia. 1998;12:845–59. doi: 10.1038/sj.leu.2401043. [DOI] [PubMed] [Google Scholar]
- 5.Krug U, Ganser A, Koeffler HP. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002;21:3475–95. doi: 10.1038/sj.onc.1205322. [DOI] [PubMed] [Google Scholar]
- 6.Murao K, Kubo Y, Ohtani N, Hara E, Arase S. Epigenetic abnormalities in cutaneous squamous cell carcinomas: frequent inactivation of the RB1/p16 and p53 pathways. Br J Dermatol. 2006;155:999–1005. doi: 10.1111/j.1365-2133.2006.07487.x. [DOI] [PubMed] [Google Scholar]
- 7.Orlow I, Drobnjak M, Zhang ZF, et al. Alterations of INK4A and INK4B genes in adult soft tissue sarcomas: effect on survival. J Natl Cancer Inst. 1999;91:73–9. doi: 10.1093/jnci/91.1.73. [DOI] [PubMed] [Google Scholar]
- 8.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 9.Krimpenfort P, Ijpenberg A, Song JY, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–6. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Civin CI, Issa JP, et al. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–41. [PubMed] [Google Scholar]
- 11.Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–7. [PubMed] [Google Scholar]
- 12.Shimamoto T, Ohyashiki JH, Ohyashiki K. Methylation of p15(INK4b) and E-cadherin genes is independently correlated with poor prognosis in acute myeloid leukemia. Leuk Res. 2005;29:653–9. doi: 10.1016/j.leukres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2003;17:1813–9. doi: 10.1038/sj.leu.2403054. [DOI] [PubMed] [Google Scholar]
- 14.Tien HF, Tang JH, Tsay W, et al. Methylation of the p15(INK4B) gene in myelodysplastic syndrome: it can be detected early at diagnosis or during disease progression and is highly associated with leukaemic transformation. Br J Haematol. 2001;112:148–54. doi: 10.1046/j.1365-2141.2001.02496.x. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113:1315–25. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggerholm A, Guldberg P, Hokland M, Hokland P. Extensive intra- and interindividual heterogeneity of p15INK4B methylation in acute myeloid leukemia. Cancer Res. 1999;59:436–41. [PubMed] [Google Scholar]
- 17.Tsellou E, Troungos C, Moschovi M, et al. Hypermethylation of CpG islands in the promoter region of the p15INK4B gene in childhood acute leukaemia. Eur J Cancer. 2005;41:584–9. doi: 10.1016/j.ejca.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Wong IH, Ng MH, Huang DP, Lee JC. Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood. 2000;95:1942–9. [PubMed] [Google Scholar]
- 19.Paul TA, Bies J, Small D, Wolff L. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood. 2010;115:3098–108. doi: 10.1182/blood-2009-07-233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markus J, Garin MT, Bies J, et al. Methylation-independent silencing of the tumor suppressor INK4b (p15) by CBFbeta-SMMHC in acute myelogenous leukemia with inv(16) Cancer Res. 2007;67:992–1000. doi: 10.1158/0008-5472.CAN-06-2964. [DOI] [PubMed] [Google Scholar]
- 21.Latres E, Malumbres M, Sotillo R, et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19:3496–506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osei-Sarfo K, de Castro IP, Pellicer A. p15(INK4b) plays a crucial role in murine lymphoid development and tumorigenesis. Carcinogenesis. 2012;33:708–13. doi: 10.1093/carcin/bgs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosu-Myles M, Taylor BJ, Wolff L. Loss of the tumor suppressor p15Ink4b enhances myeloid progenitor formation from common myeloid progenitors. Exp Hematol. 2007;35:394–406. doi: 10.1016/j.exphem.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Wolff L, Garin MT, Koller R, et al. Hypermethylation of the Ink4b locus in murine myeloid leukemia and increased susceptibility to leukemia in p15(Ink4b)-deficient mice. Oncogene. 2003;22:9265–74. doi: 10.1038/sj.onc.1207092. [DOI] [PubMed] [Google Scholar]
- 25.Bies J, Sramko M, Fares J, et al. Myeloid-specific inactivation of p15Ink4b results in monocytosis and predisposition to myeloid leukemia. Blood. 2010;116:979–87. doi: 10.1182/blood-2009-08-238360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 27.Wolff L, Koller R, Hu X, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J Virol. 2003;77:4965–71. doi: 10.1128/JVI.77.8.4965-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosu-Myles M, Wolff L. p15Ink4b: dual function in myelopoiesis and inactivation in myeloid disease. Blood Cells Mol Dis. 2008;40:406–9. doi: 10.1016/j.bcmd.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 29.RR-M, Humeniuk M, Fares J, Koller R, Bies J, Wolff L. The role of tumor suppressor p15Ink4b in the regulation of hematopoietic progenitor cell fate. Blood Cancer Journal. 2013 doi: 10.1038/bcj.2012.44. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onai N, Obata-Onai A, Schmid MA, et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8:1207–16. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 31.Fares J, Koller R, Humeniuk R, Wolff L, Bies J. The tumor suppressor p15Ink4b regulates the differentiation and maturation of conventional dendritic cells. Blood. 2012;119:5005–15. doi: 10.1182/blood-2011-10-387613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerriero A, Langmuir PB, Spain LM, Scott EW. PU.1 is required for myeloid-derived but not lymphoid-derived dendritic cells. Blood. 2000;95:879–85. [PubMed] [Google Scholar]
- 33.Carotta S, Dakic A, D’Amico A, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–41. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Kanada S, Nishiyama C, Nakano N, et al. Critical role of transcription factor PU.1 in the expression of CD80 and CD86 on dendritic cells. Blood. 2011;117:2211–22. doi: 10.1182/blood-2010-06-291898. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen VT, Benveniste EN. Involvement of STAT-1 and ets family members in interferon-gamma induction of CD40 transcription in microglia/macrophages. J Biol Chem. 2000;275:23674–84. doi: 10.1074/jbc.M002482200. [DOI] [PubMed] [Google Scholar]
- 36.Ito T, Nishiyama C, Nakano N, et al. Roles of PU.1 in monocyte- and mast cell-specific gene regulation: PU.1 transactivates CIITA pIV in cooperation with IFN-gamma. Int Immunol. 2009;21:803–16. doi: 10.1093/intimm/dxp048. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura N, Yokoyama H, Yashiro T, et al. Role of PU.1 in MHC class II expression through transcriptional regulation of class II transactivator pI in dendritic cells. J Allergy Clin Immunol. 2012;129:814–824. e6. doi: 10.1016/j.jaci.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt M, Bies J, Tamura T, Ozato K, Wolff L. The interferon regulatory factor ICSBP/IRF-8 in combination with PU.1 up-regulates expression of tumor suppressor p15(Ink4b) in murine myeloid cells. Blood. 2004;103:4142–9. doi: 10.1182/blood-2003-01-0285. [DOI] [PubMed] [Google Scholar]
- 39.Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24:1249–57. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- 40.Bresnick EH, Katsumura KR, Lee HY, Johnson KD, Perkins AS. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 2012;40:5819–31. doi: 10.1093/nar/gks281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma L, Delforge M, van Duppen V, et al. Circulating myeloid and lymphoid precursor dendritic cells are clonally involved in myelodysplastic syndromes. Leukemia. 2004;18:1451–6. doi: 10.1038/sj.leu.2403430. [DOI] [PubMed] [Google Scholar]
- 42.Mohty M, Jarrossay D, Lafage-Pochitaloff M, et al. Circulating blood dendritic cells from myeloid leukemia patients display quantitative and cytogenetic abnormalities as well as functional impairment. Blood. 2001;98:3750–6. doi: 10.1182/blood.v98.13.3750. [DOI] [PubMed] [Google Scholar]
- 43.Rickmann M, Krauter J, Stamer K, et al. Elevated frequencies of leukemic myeloid and plasmacytoid dendritic cells in acute myeloid leukemia with the FLT3 internal tandem duplication. Ann Hematol. 2011;90:1047–58. doi: 10.1007/s00277-011-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]