Highlights
-
•
CircRNAs are formed by pre-mRNA through “back-splicing”.
-
•
CircRNAs regulate host immune response and virus replication.
-
•
CircRNAs have potential as diagnostic markers or treatment targets for viral infection.
Keywords: Virus, Circular RNAs, Diagnostic marker, Treatment targets
Abstract
Circular RNAs (circRNAs) are a class of non-coding RNAs with a special covalently closed circular structure, which is formed by precursor mRNA (pre-mRNA) through “back-splicing”. CircRNAs are more stable than linear RNAs because they are resistant to exoribonucleases. Viral infections often cause abnormal expression of circRNAs, which could serve as novel biomarkers for the diagnosis of viral infections by detecting specific circRNAs in cells, body fluids, or tissues. CircRNAs also play a critical role in regulating host immune response and virus replication. Here, we reviewed the production and function of circRNAs, mainly focusing on their regulation on virus infection, to provide novel insights into the potential role of circRNAs as diagnostic marker or treatment targets for viral infection.
1. Introduction
CircRNAs were originally discovered in plant viroids by Sanger et al. in 1976 (Sanger et al., 1976). With the development of high-throughput sequencing technology, circRNAs have been widely identified in humans (Memczak et al., 2013; Wang et al., 2018), animals (Capel et al., 1993; Gupta et al., 2018; He et al., 2017; Memczak et al., 2013; Qiu et al., 2018) and plants (Han et al., 2020; Sablok et al., 2016), not only in cells, but also in various body fluids (Liu et al., 2019a) and exosomes (Shi et al., 2020; Wang et al., 2019). CircRNA, without the 5′-cap structure and 3′ poly(A) tail (Eger et al., 2018), is generated by pre-mRNA through “back-splicing” (Starke et al., 2015). Therefore, they are more stable and less susceptible to degradation by exoribonucleases than linear RNAs (Jeck et al., 2013). CircRNAs are mainly located in the cytoplasm in eukaryotic cells (Memczak et al., 2013; WR and biotechnology, 2014) and play critical roles in gene regulation. Some circRNAs have been found to express at a specific stage of some human tissues (Hansen et al., 2013; Salzman et al., 2012; Xu et al., 2017) and they are enriched in cells that proliferate slowly, such as in nerve cells (Rybak-Wolf et al., 2015). In 2017, Piwecka et al. reported a circRNA, CDR1as, could regulate brain sensorimotor gating and synaptic transmission through interacting with miR-7 and miR-671, which is the first study with conclusive evidence for the biological function of a particular circRNA (Piwecka et al., 2017).
Virus infection and the related disease endanger the health of humans, animals, and plants. Studies have shown that the virus-generated circRNAs or differentially expressed host circRNAs can be used as candidate biomarkers for viral infection (Wang et al., 2018; Yu et al., 2020; Zhao et al., 2019a). Meanwhile, circRNAs are involved in regulating antiviral immune response (Chen et al., 2017; Li et al., 2017), viral replication (Lu et al., 2020; Sekiba et al., 2018; Tagawa et al., 2018; Yu et al., 2019; Zhang and Wang, 2020), and pathogenesis of infectious diseases (Zhang et al., 2019; Zhao et al., 2019a, b). In this review, we summarized the current understanding of circRNAs, mainly focused on the circRNAs in the regulation of viral infections.
2. Formation and function of circRNAs
2.1. Formation mechanism of circRNAs
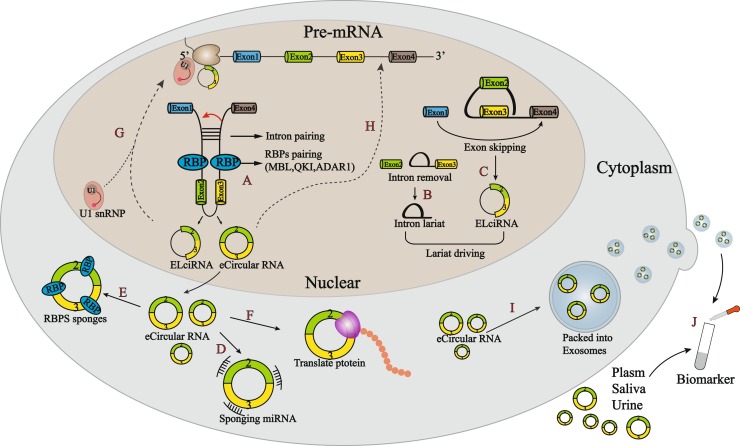
CircRNA originates from pre-mRNA “back-splicing”, which links the 3`-end of an exon to the 5`-end of an upstream exon by a covalent bond. This process requires the donor splice site of an exon not connected with the receptor splice site of the downstream exon as observed in a linear RNA splicing, but it connects with the upstream receptor site (Aufiero et al., 2019). There are three types of circRNAs: exon-intron circular RNAs (EIciRNAs) (Li et al., 2015b; Salzman et al., 2013), exonic circular RNAs (eCircular RNAs) (Salzman et al., 2012) and circular intronic RNAs (ciRNAs) (Zhang et al., 2013b), which are formed through intron pairing-driven circularization, RNA binding proteins (RBPs)-driven circularization, and lariat-driven circularization, respectively (Fig. 1 ).
Fig. 1.
Formation and function of circRNAs. (A) The cis-acting elements and trans-acting factors participate in circularization. The cis-acting element allows the introns flanking exons directly base pair into circularization through SINEs. Trans-acting factor driving circularization by binding to specific sites of introns flanking exons. Among them, MBL and QKI promote cyclization, while ADAR1 inhibits circRNAs formation. (B, C) Lariat-driven circularization mode includes exon skipping and intron removal toform EIciRNAs and ciRNAs respectively. (D) CircRNAs act as miRNA sponges to inhibit the interaction between miRNAs and their target mRNAs. (E) Circ RNAs directly bind RBPs to affect gene expression. (F) CircRNAs directly bind RBPs to affect gene expression. (G) CircRNAs interact with U1 snRNP to form circRNAs-U1 snRNP complexes and regulate gene transcription in the nucleus. (H) RNA-DNA hybrid strand regulates its parental DNA transcription by a negative feedback loop. (I) CircRNAs are packed into exosomes and involved in cell-to-cell signal transmission. (J) Some circRNAs exists freely in various body fluids such as blood, saliva, and urine, together with exosomes circuRNAs can be used as biomarkers for disease diagnosis.
In intron pairing-driven circularization, cis-acting elements flanking back-spliced exons enable the flanking introns base-paired directly through complementary sequences (Fig. 1A) (Aufiero et al., 2019). The elements involved in pairing include short interspersed nuclear elements (SINEs; such as complementary Alu repeat sequences) and non-repetitive complementary sequences (Jeck et al., 2013; Liang and Wilusz, 2014; Zhang et al., 2014). The competitive complementary pairing between introns can result in the production of multiple circRNAs on a parental gene locus (Zhang et al., 2014). It has been reported that double-stranded RNA(dsRNA)-specific adenosine deaminase (ADAR) inhibit circRNAs formation through binding to dsRNA and interfering RNA base pairing on introns (Fig. 1A) (Ivanov et al., 2015). Immune factors NF90/NF110, which contain a dsRNA binding region, promote circRNAs production in nucleus by binding with intronic complementary RNA pairs flanking the back-spliced exons (Li et al., 2017).
In RBPs-driven circularization, trans-acting factors, the proteins binding to cis-acting elements, initiate circRNAs formation by docking on specific sites of the introns flanking back-spliced exons (Ashwal-Fluss et al., 2014a; Conn et al., 2015). Several proteins, such as muscleblind-like protein 1 (MBNL1) (Ashwal-Fluss et al., 2014b) and protein quaking (QKI) (Conn et al., 2015), have been reported to facilitate circRNAs formation through interacting with flanking introns (Fig. 1A).
Lariat driving circularization including lariat removal (Zhang et al., 2013a) that forms ciRNAs during linear pre-mRNA splicing (Fig. 1B), and exon skipping (Zaphiropoulos, 1996) that forms exon-containing lariat (also EIciRNAs) (Fig. 1C).
In summary, circRNA formation is a complicated process. Circularization process is regulated by many elements including cis-acting elements, trans-acting factors, and spliceosomes (Chen, 2016; Yu and Kuo, 2019).
2.2. Biological function of circular RNAs
Although some circRNAs exist in nucleus (Li et al., 2015a), various secretions (Bahn et al., 2015; Liu et al., 2019a; Song et al., 2020; Yu et al., 2020) and exosomes (Shi et al., 2020; Wang et al., 2019), most circRNAs are mainly located in the cytoplasm (Memczak et al., 2013; WR and biotechnology, 2014). The biological function of circRNAs varies according to their physical location, though most of them are still poorly recognized.
2.2.1. CircRNAs localized in the cytoplasm
CircRNAs located in the cytoplasm function in three patterns: 1) function as microRNA (miRNA) sponge (Hansen et al., 2013; Memczak et al., 2013); 2) affect gene expression by binding RBPs (Abdelmohsen et al., 2017; Ashwal-Fluss et al., 2014a; Du et al., 2017, 2016), and 3) translate into proteins directly (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017, Yang et al., 2018). Although circRNAs have many biological functions, sponge miRNA is the first biological function discovered, which prevents the interaction between certain miRNAs and their target mRNAs in the cytoplasm (Fig. 1D) (Hansen et al., 2013; Memczak et al., 2013). CircRNAs have also been found to form specific circRNPs with some cytoplasmic proteins. CircRNPs can affect gene expression and function of these protein, therefore serving as RBPs sponges (Fig. 1E) (Abdelmohsen et al., 2017; Ashwal-Fluss et al., 2014a; Du et al., 2017, 2016). Some cytoplasmic circRNAs have internal ribosome entry site (IRES) elements, which can be translated into proteins or peptides after binding to ribosomes (Fig. 1F) (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2018). Other circRNAs without IRESs but contain m6A modification sites can also initiate the protein translation process (Yang et al., 2017). Recently, a study reported that circSfl was translated into a 25-kDa protein by sharing the N-terminus with the 110-kDa SFL protein encoded by host sulfateless (sfl) gene. Both proteins encoded by circSfl and linear SFL transcripts can regulate the life-span of flies, while the circSfl protein lacks the enzymatically active domain at the C terminus (Weigelt et al., 2020).
2.2.2. CircRNAs localized in the nucleus
CiRNAs and EIciRNAs are mainly located in the nucleus, and their functions have been reported to regulate gene transcription and alternative splicing through interacting with U1 snRNP to form circular RNAs-U1 snRNP complexes (Fig. 1G) (Li et al., 2015a). These circRNAs can also affect the transcription efficiency of many genes by interaction with the RNA polymerase II (Pol II) transcription complex of pre-mRNA promoter region (Zhang et al., 2013a). Most recently, it is reported that circRNAs can be used as a template for de novo transcription under Pol Ⅱ catalysis (Dissanayaka Mudiyanselage and Wang, 2020). The stranded circRNAs in the nucleus are able to form an RNA-DNA hybrid strand with its parental dsDNA, which negatively regulates the parental DNA transcription (Fig. 1H) (Conn et al., 2017).
2.2.3. CircRNAs secreted into the extracellular
Some circRNAs can be secreted into various body fluids such as blood, saliva, and urine. Studies have reported that circRNAs can be packed into exosomes to function in cell-to-cell signal transmission and regulation (Fig. 1I) (Shi et al., 2020; Wang et al., 2019). These secreted circRNAs are promising biomarkers for disease diagnosis and progression (Fig. 1J) (Bahn et al., 2015; Liu et al., 2019a; Song et al., 2020; Yu et al., 2020).
3. CircRNAs associated with virus infection
Virus-derived circRNAs and/or differentially-expressed host circular RNAs have been observed following various virus infections. These results indicated that circRNAs may play important roles in virus infection and host defense. In this section, we summarize the potential use of the viroid circRNAs, viral-derived circRNAs, and differentially expressed host circRNAs as biological markers for virus infection and related diseases progression.
3.1. Viroid circRNAs
Viroid replicates autonomously in plants to induce disease. The genome is a single-stranded RNA molecule, which can form a “naked” rod-like conformation (Lopez-Carrasco and Flores, 2017). The special circular genomic structure of viroid makes it replication by a rolling-circle way in the nucleus and chloroplast of host cells (Flores et al., 2009, 2011; Gago-Zachert, 2016).
In 1976, Sanger et al. found a single-stranded covalently closed circular RNA molecule in plant viroid, which can base complementary pairing to form a rod-like structure (Sanger et al., 1976). In 1988, three kinds of viroid circRNAs were found in nucleic acids of grapevine analyzed by two-dimensional gel electrophoresis (Rezaian et al., 1988). In 1997, a cherry small circRNA (csc RNA1) was discovered as a new viroid-like satellite circRNA (Di Serio et al., 1997). The small circRNA (sc-RNA) associated with rice yellow mottle sobemovirus (RYMV) was discovered in 1988, which contains only 220 nucleotides and is the smallest viroid circRNA identified so far (Collins et al., 1998). This sc-RNA is the only viroid circRNA that can encode a 16-kDa protein, which uses novel modalities for coding, translation, and gene expression (AbouHaidar et al., 2014).
3.2. Virus-derived circRNAs
Hepatitis delta virus (HDV) is the first mammal virus that has been found to have a circRNA genome (Kos et al., 1986). Compared with viroid, HDV uses host cell RNA polymerase to replicate its circular genomic RNA by rolling-circle model (Macnaughton et al., 2002). With the development and wide application of high-throughput sequencing technologies, a variety of circRNAs derived from viruses, including human papillomavirus (HPV) (Zhao et al., 2019a), hepatitis B virus (HBV) (Sekiba et al., 2018), blackcurrant leaf chlorosis associated virus (BCLCaV) (James et al., 2018), Epstein-Barr virus (EBV) (Huang et al., 2019; Toptan et al., 2018; Ungerleider et al., 2018, 2019), murine gamma herpesvirus 6868 (MHV68) (Ungerleider et al., 2019) and Kaposi sarcoma herpesvirus (KSHV) (Tagawa et al., 2018; Toptan et al., 2018; Ungerleider et al., 2019) have been identified and confirmed (Table 1 ). Interestingly, some studies have reported that the terminal regions of flavivirus genomes contain inverted complementary sequences, which can mediate long-range RNA interactions and RNA cyclization. This circular conformation of genomes is essential for flavivirus RNA replication (Alvarez et al., 2005; Khromykh et al., 2001; Lo et al., 2003; Villordo and Gamarnik, 2009).
Table 1.
Virus-derived circRNAs.
| Virus | Sample type | Virus-derived circRNAs | Reference |
|---|---|---|---|
| HPV | TCGA RNA-Seq data from HPV-positive cancers | HPV16 circE7 | (Zhao et al., 2019a) |
| HBV | HBV-infected HepG2 cells, HepAD38 cells and human primary hepatocytes | HBV circRNA | (Sekiba et al., 2018) |
| BCLCaV | BCLCaV-infected Ribes nigrum and Nicotiana benthamiana tissue | circular forms of BCLCaV RNA-2 and RNA-3 | (James et al., 2018) |
| rLCV SIV | SIV/LCV-infected rhesus lymphoma model | rLCV circRPMS1_E5_E3a EBV circRPMS1_E4_E3a circRPMS1_E2b_E1b circEBNA_U | (Ungerleider et al., 2019) |
| MHV68 | MHV68-infected NIH 3T12 cells | MHV68 circM11_ORF69 | |
| KSHV | KSHV positive BCBL-1 cells | KSHV circvIRF4 | |
| KSHV | KSHV-infected HUVECs or infected B cell line, MC116 | KSHV circvIRF4 | (Tagawa et al., 2018) |
| EBV | EBV-infected cell lines, including SNU-719, AGS-EBV, C666-1 and Akata | EBV circ_RPMS1 | (Huang et al., 2019) |
| EBV | Represent type I, II, and III latency transcription programs and during reactivation period lymphocyte gastric cancer cell models infected with EBV | circEBNA_U、circBHLF1 circRPMS1_E4_E3a circRPMS1_E4_E2 circEBNA_W1_C1 circEBNA_W2_C1 circLMP2_E8_E2 | (Ungerleider et al., 2018) |
| EBV | EBV-positive PTLDs | circBARTs | (Toptan et al., 2018) |
| KSHV | DMSO-treated or NaB/TPA-induced KSHV-infected PEL cell lines BCBL1 and BC-1 | circvIRF4, circPANs/K7.3 |
TCGA, The Cancer Genome Atlas; rLCV, rhesus macaque lymphocryptovirus; SIV, simian immunodeficiency virus; HUVECs, HUVEC-human umbilical vein endothelial cells; PTLD, posttransplant lymphoproliferative disease.
High-risk HPV infection stimulates the development of cervical, oropharyngeal, anal, vulvovaginal, and penile cancers. An HPV16-derived circRNA, circE7 has been identified in HPV-positive cancers. Knocking down of HPV16-derived circE7 in CaSki cells reduces the translation of E7 protein and inhibits the growth of HPV-related cancer cells. These findings indicated that HPV16-derived circE7 may be used as a screening marker for high-risk HPV infection. It also illustrated that virus-encoded circRNA can be translated into proteins (Zhao et al., 2019a).
3.3. Differentially-expressed host circRNAs following virus infection
Virus infection significantly changes the expression patterns of host circRNAs (Table 2 ). 226 differentially-expressed circRNAs have been identified in hepatocellular carcinoma (HCC) patients by circRNA microarray. Of these 226 circRNAs, 189 are up-regulated and 37 down-regulated. Further functonal studies demonstrated that some of these circRNAs are involved in the pathogenesis of HCC. One such example is the upregulated circRNA_100338 regulated the cancer cells invasion through sponging miR-1413p (Huang et al., 2017). Similarly, 99 differently-expressed circRNAs were identified in liver biopsies from chronic hepatitis B (CHB) patients using RNA sequencing. Computational analysis of the circRNA-miRNA-mRNA pathways revealed that four pathways may be involved in HBV infection and progression of HBV-associated liver disease. Of these four pathways, hsa_circ_0000650 has been found to regulate transforming growth factor-β (TGFβ) pathway by sponging miR-6873−3p using a regression analysis of gene expression (Zhou et al., 2018).
Table 2.
Differentially expressed host circRNAs in human following virus infection.
| Virus | Sample type | Name of circRNAs with differentially expressed | Expression level | Reference |
|---|---|---|---|---|
| HTNV | HTNV-infected HUVECs | hsa_circ_0000479 | Up-regulated | (Lu et al., 2020) |
| HBV | Blood and liver tissues from CHB patients, HBV-infected hepatoma cells | hsa_circ_0004812 | Up-regulated | (Zhang and Wang, 2020) |
| HBV | Liver biopsies from untreated CHB patients | hsa_circ_0005389 and hsa_circ_0000038 |
Upregulated | (Zhou et al., 2018) |
| hsa_circ_0000650 | Down-regulated | |||
| HIV | PBMC of HARRT-naive EHI patients | hsa_circ_0000711 hsa_circ_0006968 | EHI group compared with HC group: up-regulated | (Zhang et al., 2018) |
| PBMCs of HARRT-naive CHI patients | hsa_circ_0003863 hsa_circ_0049083 | CHI group compared with EHI group: up-regulated | ||
| IAV H1N1 | H1N1-infected A549 cells | hsa_circ-GATAD2A | Up-regulated | (Yu et al., 2019) |
| HCMV | HCMV latent-infected THP-1 cells | hsa_circ_0001445 hsa_circ_0001206 | Down-regulated | (Lou et al., 2019) |
| KSHV | KSHV-infected HUVECs, B cell line, MC116 | hsa_circ_0001400 hsa_circ_0001741 hsa_circ_0008311 hsa_circ_0005145 | Up-regulated | (Tagawa et al., 2018) |
| HSV-1 | HSV-1 infected KMB17 cells | hsa_circRNA3046 hsa_circRNA3683 hsa_circRNA6783 hsa_circRNA7752 hsa_circRNA7231 | Up-regulated | (Shi et al., 2018) |
HTNV, hantaan virus; HIV-1, human immunodeficiency virus type 1; IAV, influenza A virus; HSV-1, herpes simplex virus; HCs, healthy controls; PBMC, Peripheral blood mononuclear cell; HARRT, Highly Active Antiretroviral Therapy; EHI, early HIV infection; CHI, chronic HIV infection.
CircRNAs differentially expressed in plasma are expected to be potential biomarkers for HBV-related HCC. In a recent research, three circRNAs, hsa_circ_0000976, hsa_circ_0007750, hsa_circ_0139897 were found to be significantly up-regulated in the plasma of HBV-related HCC patients compared with those of healthy controls, CHB, and HBV-related liver cirrhosis patients (Yu et al., 2020). Another study screened the differentially expressed plasma circRNAs from 10 patients with HBV-related HCC and 5 patients with HBV-related liver cirrhosis using microarray. They verified that the expression of hsa_circ_0027089 was significantly altered in HCC patients and can be used to distinguish HCC patients from cirrhotic patients and healthy participants (Zhu et al., 2020). In addition, circRNA profiles were also investigated in HBV-related HCC and their paired adjacent non-tumorous (NT) tissues. They found 24 significantly up-regulated and 23 down-regulated circRNAs tissues in HBV−HCC tissues. Among them, up-regulation of hsa_circ_100381, hsa_circ_103489 and down-regulation of hsa_circ_101764 were verified in another 10 validation HCC tissues. Further study also indicated that circRNA_101764 may play an important role in the development of HCC (Wang et al., 2018). However, although these studies are all focused on HBV-related HCC, there is none overlap of the differently expressed circRNAs, which may arise from different sample types and control groups used. More investigation on the association between specific circRNAs and HBV-related HCC are warranted in future research.
Similarly, the change of circRNA profiles can also be detected in animals infected with viruses. Tumors from subgroup J avian leucosis virus infected (ALV-J-infected) chickens lead to high mortality. Several studies have investigated the circRNAs related to ALV-J infection in chickens. It is found that circ-Vav3 was significantly up-regulated in liver tumor tissues of ALV-J-infected chickens. Overexpression of circ-Vav3 in DF-1 cells can sponge gga-miR-375 and increase the Yes-associated protein 1 (YAP1) expression to promote epithelial-mesenchymal transition (EMT) and tumorigenesis. These findings suggest that the circ-Vav3/gga-miR-375/YAP1 axis is a regulator of ALV-J infection-induced tumorigenesis (Zhang et al., 2019). In addition, 106 up-regulated circRNAs and 46 down-regulated circRNAs were identified in the spleen tissues of ALV-J-infected chicken by using RNA sequencing (Qiu et al., 2018). 32 differentially expressed circRNAs were identified in liver tissues from ALV-J-resistant and ALV-J-susceptible chickens (Zhang et al., 2017). Furthermore, a novel circHRH4 is highly expressed and stably existed in various tissues of chicken (Qiu et al., 2018).These results provide new insight into circRNAs regulation on tumor induction and development in chicken.
Piglets infected by transmissible gastroenteritis coronavirus (TGEV) suffer from severe vomiting and diarrhea with symptoms of transmissible gastroenteritis (TGE). Research revealed that circEZH2 expression level was significantly down-regulated in piglets infected with TGEV. Overexpression of circEZH2 in IPEC-J2 cells inhibited TGEV-induced mitochondrial permeability transition pore (mPTP) opening via attaching miR-22 to regulate hexokinase 2 (HK2) and interleukin 6 (IL-6), while silencing of IL-6 supressed activation of NF-κB pathway. This study provides potential drug targets for the prevention and treatment of TGE (Zhao et al., 2019b). Studies have also examined circRNA expression profiles by using high-throughput sequencing in MBDK cells infected with bovine viral diarrhea virus (BVDV) (Li et al., 2019) and in the silkworm following bombyx mori nuclear polyhedrosis virus (BmNPV) infection (Hu et al., 2018). All these findings implied that circRNAs play critical roles in regulating virus infection both in animals and plants.
4. CircRNAs regulate viral replication and induce antiviral immune response
Research on the role of circRNAs in various diseases (such as tumors (Vo et al., 2019), nervous system diseases (Piwecka et al., 2017), and cardiovascular diseases (Aufiero et al., 2019), etc) is moving rapidly in recent years. Studies also demonstrated that circRNAs play important roles in virus replication and host antiviral immune response, though the detailed mechanisms are still unclear. Here we summarize some of the reports on circRNAs regulation of viral replication and related diseases (Table 3 ).
Table 3.
Mechanisms of circRNAs regulation in viral replication and related disease.
| CircRNA | Effect on virus | Regulated pathway | Refer-ence |
|---|---|---|---|
| circ_0000479 | Overexpression promotes the expression of RIG-I and hampering HTNV replication | circ_0000479/ miR-149-5p/ RIG-I pathway | (Lu et al., 2020) |
| hsa_circ_0004812 | Knockdown expression promotes the IFN-α/β, and inhibits HBV replication. | circ_0004812/ miR-1287-5p/ FSTL1 axis to regulate IFN-induced immune response | (Zhang and Wang, 2020) |
| circRNA_100338 | Overexpression enhanced migratory and invasive ability of cancer cells | circRNA_100338 / miR-1413p axis to regulate invasive potential in liver cancer cells | (Huang et al., 2017) |
| circ-GATAD2A | Knockdown expression enhances autophagy and inhibits IAV H1N1 replication, reverse after overexpression | circ-GATAD2A/ VPS34-dependent autophagy pathway | (Yu et al., 2019) |
| circ-Vav3 | Overexpression induces EMT, promote ALV-J related tumorigenesis | circ-Vav3/gga-miR-375/YAP1 axis induces EMT pathway | (Zhang et al., 2019) |
| circEZH2 | Overexpression inhibits TGEV-induced mPTP opening and regulate NF-κB activation | circEZH2/ miR-22/ HK2 axis, circEZH2 /miR-22/IL-6/ NF-Kb pathway to regulate mPTP opening | (Zhao et al., 2019b) |
4.1. CircRNAs regulate virus replication through mediating host-virus interaction
Studies have showen that viral-derived circRNAs were produced during some virus infection such as HBV and EBV. It was found that HBV-derived circRNA was regulated by the host DHX9 (DEAH-box helicase 9) protein. Specific disruption of DHX9 protein in HepG2 cells, HepAD38 cells, and human primary hepatocytes increased HBV-derived circRNA levels, while decreased the HBV protein levels. The author therefore concluded that DHX9 functions as an RNA binding factor to regulate expression levels of viral-derived circRNAs or viral proteins(Sekiba et al., 2018). Using a R-resistance RNA-seq, EBV-encoded circRNAs have been investigated. They identified the miRNAs sponged by EBV-encoded circRNAs and their interactions with target mRNAs, which regulate the EBV replication and its related tumorigenesis (Qiao et al., 2019). It has been shown that KSHV infection increased hsa_circ_0001400 expression level in HUVECs cells. While ectopic expression of this circRNA in SLK cells suppressed the expression of key viral latent gene LANA and lytic gene RTA in KSHV de novo infections, siRNA knockdown of hsa_circ_0001400 stimulated the expression of KSHV genes (LANA and RTA), suggesting that hsa_circ_0001400 has an antiviral effect and may serve as a new therapeutic target for antiviral infection (Tagawa et al., 2018).
4.2. Circular RNAs induces antiviral immune response
CircRNAs are involved in the host anti-viral immune response. dsRNA-binding proteins are important components of host antiviral innate immune system. Studies have reported that some host and virus-derived circRNAs regulate virus infection through interacting with these dsRNA binding antiviral proteins (Awan et al., 2020; Liu et al., 2019b). Retinoic acid-inducible gene I (RIG-I) is a cytosolic RNA-sensing protein that recognizes viral dsRNA and elicit innate immune response. Recently, it has been reported that RIG-I can also sense exogenous circRNAs and stimulates innate immunity to inhibit viral infection (Chen et al., 2017). In addition to the 5 verified circRNAs upregulated by HSV-1 infection (shown in Table 2), Shi et al. analyzed the circRNA-miRNA-genes regulatory axis using the top-9 dysregulated circRNAs induced by HSV-1 infection (hsa_circRNA1450, hsa_circRNA14514, hsa_circRNA14515, hsa_circRNA14189, hsa_circRNA15655, hsa_circRNA15950, hsa_circRNA14556, hsa_circRNA15053 and hsa_circRNA15906), they found that the a large number of immunity-related genes, mainly enriched in NOD-like receptor/JAK-STAT signaling pathway, were potentially regulated by HSV-1-induced circRNAs (Shi et al., 2018).
Immune factors NF90/NF110 can promote circRNAs production in the nucleus and interact with mature circRNAs in the cytoplasm. Upon viral infection, partly due to the nuclear export of NF90/NF110 to the cytoplasm, the expression level of circRNA is reduced. In addition, NF90/NF110 released by the circRNP complexes binds to the viral mRNA and inhibits viral translation to play an important role in antiviral immune response (Li et al., 2017). HTNV infection in humans causes high mortality of hemorrhagic fever with renal syndrome (HFRS). Circ_0000479 expression in HUVEC has been found to increase after HTNV infection, which in turn to inhibit viral replication by sponging miR-149−5p and promoting RIG-I expression (Lu et al., 2020). The expression of hsa_circ_0004812 in liver cancer tissues of CHB patients is significantly up-regulated. Knockdown of hsa_circ_0004812 stimulated the expression of IFN-α/β to inhibit viral replication, while overexpression of hsa_circ_0004812 promoted HBV-induced immunosuppression through sponging miR-1287−5p. Therfore, hsa_circ_0004812 was speculated as a potential therapeutic target for the treatment of HBV infection (Zhang and Wang, 2020). IAV H1N1 infection is easy to cause seasonal epidemic acute respiratory disease. The expression level of circ-GATAD2A is significantly up-regulated in A549 cells infected with H1N1 virus. Overexpression of circ-GATAD2A promoted H1N1 replication by blocking VPS34-dependent autophagy, while the virus replication was inhibited after knockdown of circ-GATAD2A (Yu et al., 2019).
In addition, some studies revealed that the cytoplasmic endonuclease RNase L is activated after viral infection, which would restrict circRNAs expression and thereby releasing NF90, NF110, and PKR to promote antiviral immune responses activities (Chen, 2020; Li et al., 2017; Liu et al., 2019b).
5. Conclusions and prospects
5.1. Prospects
Early detection is the key to treat virus infections. CircRNAs can exist in cells, body fluids (Bahn et al., 2015; Liu et al., 2019a; Song et al., 2020; Yu et al., 2020), and exosomes (Shi et al., 2020; Wang et al., 2019), which makes them more stable than linear RNAs. Based on these characteristics, circRNAs are potential candidate biomarkers for the diagnosis of viral infections in a noninvasive manner. However, little is known in using specific circRNAs as biomarkers for virus diagnosis, due to the lack of standard methods for circRNA detection.
In addition, circRNAs also have a therapeutic potential for virus-related diseases. Recently, a circRNA constructed artificially in vitro can successfully inhibit HCV replication by sponging miR-122, which is required for HCV replication in HuH-7.5 or HuH-7 cells (Jost et al., 2018).
However, some challenges and problems still need to be solved. Firstly, circRNAs contain almost the same sequence as its linear parental gene except for the sequence at the back-splice junction. Therefore, it is difficult to specifically detect circRNAs from its linear parental gene. Secondly, the study on circRNAs is still in the early stage. For example, other mechanisms except for sponging miRNA are not clear, besides most studies on circRNAs still stay at the cellular level. Further investigations in animal models and patients are needed. A unified and standardized circRNAs naming system should also be established as soon as possible. Last but not least, since circRNAs can resist the degradation of RNase and remain stable in cells, how can it be degraded safely is an important question when it was considered as a drug.
5.2. Conclusions
In summary, circRNA as an emerging regulatory RNA molecule, its existence and function expand the complexity and diversity of eukaryotic transcriptomes. The special circular structure endows the circRNAs with good stability and different functions from those of their parental linear RNAs (Li et al., 2018). Here, we discussed the formation and function of circRNAs, as well as the molecular mechanism by which circRNAs regulate virus replication and disease occurrence. Future study is required to shed the potential role of circRNAs in the diagnosis and treatment of infectious diseases.
Funding
This work is supported by the National Key Research and Development Program of China (2018YFE0107500 to L.C.); and Sichuan Provincial Science and Technology Department Funding, China (2019YJ0281 to X.D.).
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- Abdelmohsen K. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbouHaidar M.G. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. U.S.A. 2014;111(40):14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D.E. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339(2):200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Aufiero S. Circular RNAs open a new chapter in cardiovascular biology. Nature reviews. Cardiology. 2019 doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- Awan F.M. The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: possible implication as theranostic agents. RNA Biol. 2020:1–15. doi: 10.1080/15476286.2020.1790198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J.H. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61(1):221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Chen L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020 doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- Chen Y.G. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67(2) doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R.F. Self-cleaving circular RNA associated with rice yellow mottle virus is the smallest viroid-like RNA. Virology. 1998;241(2):269–275. doi: 10.1006/viro.1997.8962. [DOI] [PubMed] [Google Scholar]
- Conn S.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Conn V.M. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- Di Serio F. A 451-nucleotide circular RNA from cherry with hammerhead ribozymes in its strands of both polarities. J. Virol. 1997;71(9):6603–6610. doi: 10.1128/jvi.71.9.6603-6610.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissanayaka Mudiyanselage S.D., Wang Y. Evidence Supporting That RNA Polymerase II Catalyzes De Novo Transcription Using Potato Spindle Tuber Viroid Circular RNA Templates. Viruses. 2020;12(4) doi: 10.3390/v12040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.W. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.W. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- Eger N. Circular RNA splicing. Adv. Exp. Med. Biol. 2018;1087:41–52. doi: 10.1007/978-981-13-1426-1_4. [DOI] [PubMed] [Google Scholar]
- Flores R. Viroid replication: rolling-circles, enzymes and ribozymes. Viruses. 2009;1(2):317–334. doi: 10.3390/v1020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 2011;8(2):200–206. doi: 10.4161/rna.8.2.14238. [DOI] [PubMed] [Google Scholar]
- Gago-Zachert S. Viroids, infectious long non-coding RNAs with autonomous replication. Virus Res. 2016;212:12–24. doi: 10.1016/j.virusres.2015.08.018. [DOI] [PubMed] [Google Scholar]
- Gupta S.K. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122(2):246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. Identification, characterization, and functional prediction of circular RNAs in maize. Mol. Genet. Genomics. 2020;295(2):491–503. doi: 10.1007/s00438-019-01638-9. [DOI] [PubMed] [Google Scholar]
- Hansen T.B. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- He L. Deep circular RNA sequencing provides insights into the mechanism underlying grass carp reovirus infection. Int. J. Mol. Sci. 2017;18(9) doi: 10.3390/ijms18091977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X. Circular RNA alterations in the Bombyx mori midgut following B. Mori nucleopolyhedrovirus infection. Mol. Immunol. 2018;101:461–470. doi: 10.1016/j.molimm.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Huang X.Y. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 2017;7(1):5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T. Identification of virus-encoded circular RNA. Virology. 2019;529:144–151. doi: 10.1016/j.virol.2019.01.014. [DOI] [PubMed] [Google Scholar]
- Ivanov A. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- James D. Blackcurrant leaf chlorosis associated virus: evidence of the presence of circular RNA during infections. Viruses. 2018;10(5) doi: 10.3390/v10050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost I. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018;15(8):1032–1039. doi: 10.1080/15476286.2018.1435248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75(14):6719–6728. doi: 10.1128/jvi.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Legnini I. Circ-ZNF609 is a circular RNA that can Be translated and functions in Myogenesis. Mol. Cell. 2017;66(1) doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Li Z. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Li X. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 2017;67(2):214–227. doi: 10.1016/j.molcel.2017.05.023. e217. [DOI] [PubMed] [Google Scholar]
- Li X. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Li C. Comprehensive analysis of circRNAs expression profiles in different periods of MDBK cells infected with bovine viral diarrhea virus. Res. Vet. Sci. 2019;125:52–60. doi: 10.1016/j.rvsc.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. Characterization of tissue-specific biomarkers with the expression of circRNAs in forensically relevant body fluids. Int. J. Legal Med. 2019;133(5):1321–1331. doi: 10.1007/s00414-019-02027-y. [DOI] [PubMed] [Google Scholar]
- Liu C.X. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–880. doi: 10.1016/j.cell.2019.03.046. e821. [DOI] [PubMed] [Google Scholar]
- Lo M.K. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3’ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77(18):10004–10014. doi: 10.1128/jvi.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Carrasco A., Flores R. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: a "naked" rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 2017;14(8):1046–1054. doi: 10.1080/15476286.2016.1223005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y.-Y. Differential circRNA expression profiles in latent human cytomegalovirus infection and validation using clinical samples. Physiol. Genomics. 2019;51(2):51–58. doi: 10.1152/physiolgenomics.00096.2018. [DOI] [PubMed] [Google Scholar]
- Lu S. RNA-seq revealed a circular RNA-microRNA-mRNA regulatory network in hantaan virus infection. Front. Cell. Infect. Microbiol. 2020;10:97. doi: 10.3389/fcimb.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnaughton T.B. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 2002;76(8):3920–3927. doi: 10.1128/JVI.76.8.3920-3927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Pamudurti N.R. Translation of CircRNAs. Mol. Cell. 2017;66(1) doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357) doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- Qiao Y. Epstein-Barr virus circRNAome as host miRNA sponge regulates virus infection, cell cycle, and oncogenesis. Bioengineered. 2019;10(1):593–603. doi: 10.1080/21655979.2019.1679698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L. Circular RNA and mRNA profiling reveal competing endogenous RNA networks during avian leukosis virus, subgroup J-induced tumorigenesis in chickens. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0204931. e0204931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaian M.A. Isolation of three viroids and a circular RNA from grapevines. J. Gen. Virol. 1988;69(Pt 2):413–422. doi: 10.1099/0022-1317-69-2-413. [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Sablok G. Plant circular RNAs (circRNAs): transcriptional regulation beyond miRNAs in plants. Mol. Plant. 2016;9(2):192–194. doi: 10.1016/j.molp.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Salzman J. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger H.L. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiba K. DHX9 regulates production of hepatitis B virus-derived circular RNA and viral protein levels. Oncotarget. 2018;9(30):20953–20964. doi: 10.18632/oncotarget.25104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. Deep RNA sequencing reveals a repertoire of human fibroblast circular RNAs associated with cellular responses to herpes simplex virus 1 infection. Cell. Physiol. Biochem. 2018;47(5):2031–2045. doi: 10.1159/000491471. [DOI] [PubMed] [Google Scholar]
- Shi X. circRNAs and exosomes: a mysterious frontier for human Cancer. Molecular therapy. Nucleic acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. Identification of urinary hsa_circ _0137439 as potential biomarker and tumor regulator of bladder cancer. Neoplasma. 2020;67(1):137–146. doi: 10.4149/neo_2018_181214N970. [DOI] [PubMed] [Google Scholar]
- Starke S. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Tagawa T. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc. Natl. Acad. Sci. U.S.A. 2018;115(50):12805–12810. doi: 10.1073/pnas.1816183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toptan T. Circular DNA tumor viruses make circular RNAs. Proc. Natl. Acad. Sci. U.S.A. 2018;115(37):E8737–e8745. doi: 10.1073/pnas.1811728115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerleider N. The Epstein Barr virus circRNAome. PLoS Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007206. e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerleider N.A. Comparative analysis of gammaherpesvirus circular RNA repertoires: conserved and unique viral circular RNAs. J. Virol. 2019;93(6) doi: 10.1128/jvi.01952-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villordo S.M., Gamarnik A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139(2):230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo J.N. The landscape of circular RNA in Cancer. Cell. 2019;176(4) doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Screening and bioinformatics analysis of circular RNA expression profiles in hepatitis B-related hepatocellular carcinoma. Cancer Biomark. 2018;22(4):631–640. doi: 10.3233/cbm-170910. [DOI] [PubMed] [Google Scholar]
- Wang Y. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer. 2019;18(1):116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt C.M. An insulin-sensitive circular RNA that regulates lifespan in Drosophila. Mol. Cell. 2020;79(2):268–279. doi: 10.1016/j.molcel.2020.06.011. e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WR, J., biotechnology, S.N.J.N., 2014. Detecting and characterizing circular RNAs. 32(5), 453-461. https://doi.org/10.1038/nbt.2890. [DOI] [PMC free article] [PubMed]
- Xu T. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Extensive translation of circular RNAs driven by N-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110(3) doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-Y., Kuo H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019;26(1):29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 2019;231:238–245. doi: 10.1016/j.vetmic.2019.03.012. [DOI] [PubMed] [Google Scholar]
- Yu J. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: a large-scale, multicenter study. Int. J. Cancer. 2020;146(6):1754–1763. doi: 10.1002/ijc.32647. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. U.S.A. 1996;93(13):6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang Z. Circular RNA hsa_circ_0004812 impairs IFN-induced immune response by sponging miR-1287-5p to regulate FSTL1 in chronic hepatitis B. Virol. J. 2020;17(1):40. doi: 10.1186/s12985-020-01314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Zhang X.-O. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang X. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. 2017;8(21):34961–34970. doi: 10.18632/oncotarget.16442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J. Transl. Med. 2018;16(1):332. doi: 10.1186/s12967-018-1706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Circular RNA Vav3 sponges gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol. 2019;16(1):118–132. doi: 10.1080/15476286.2018.1564462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019;10(1):2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Circular RNA CircEZH2 suppresses transmissible gastroenteritis coronavirus-induced opening of mitochondrial permeability transition pore via targeting MiR-22 in IPEC-J2. Int. J. Biol. Sci. 2019;15(10):2051–2064. doi: 10.7150/ijbs.36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T.C. Differential expression profile of hepatic circular RNAs in chronic hepatitis B. J. Viral Hepat. 2018;25(11):1341–1351. doi: 10.1111/jvh.12944. [DOI] [PubMed] [Google Scholar]
- Zhu K. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2020;41(3):296–302. doi: 10.1093/carcin/bgz154. [DOI] [PMC free article] [PubMed] [Google Scholar]