Highlights
-
•
Development of indirect IgG ELISA using β-propiolactone-inactivated SARS-CoV-2.
-
•
Detection of anti-SARS-CoV-2 IgG during early disease phase.
-
•
Higher IgG seropositivity and OD values in COVID-19 patients with severe disease.
Keywords: COVID-19, SARS-CoV-2, IgG, Antibody, ELISA, Inactivated virus
Abstract
Background
Coronavirus disease 2019 (COVID-19) pandemic caused by infection with severe acute respiratory syndrome – coronavirus-2 (SARS-CoV-2) continues to affect many countries and large populations. Serologic assays for antibody detection aid patient diagnosis and seroepidemiologic investigations.
Methods
An indirect IgG ELISA was developed indigenously using β-propiolactone (BPL) inactivated SARS-CoV-2. This assay was used for screening 200 healthy donor sera collected prior to COVID-19 emergence (2017–2019), 185 serum/plasma samples of confirmed COVID-19 patients (n = 137) and 57 samples of viral RNA positive asymptomatic contacts (n = 51). The IgG response was studied in relation to duration and severity of illness.
Results
The ELISA demonstrated 97 % specificity and IgG detection in >50 %, 80 %, 93.8 % and 100 % of the patients respectively during the first, second, third and fourth week of illness. IgG detection rate was higher in patients with severe disease (SD, 90.9 %) than those with mild disease (MD, 68.8 %) during the second week of illness (P = 0.027). IgG seropositivity among asymptomatic contacts was 64.7 %. IgG ELISA absorbance values were higher in SD than MD patients during the first 2 weeks of illness (P < 0.05). No significant difference was observed between the absorbance values of asymptomatic subjects and MD patients (P = 0.94).
Conclusion
The BPL inactivated virus-based ELISA could detect IgG antibodies early and in a significant proportion of COVID-19 patients suggesting its potential utility as a supplement to the currently used viral RNA detection tests in patient diagnosis and contact screening algorithms.
1. Introduction
Coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome – coronavirus-2 (SARS-CoV-2) has affected 216 countries with 16,523,815 confirmed cases and 655,112 deaths reported till 29th July 2020 (World Health Organization, 2020a). In the absence of vaccines and antivirals, the control strategy for COVID-19 involves patient isolation, contact tracing and quarantine of suspects, social distancing for limiting virus spread. Thus, timely and accurate diagnosis remains the mainstay of COVID-19 management. Moreover, estimation of disease prevalence, extent of exposure and immunity to the virus in a particular population assumes importance for planning and implementation of control measures.
Currently, viral RNA detection in respiratory tract samples by RT-PCR is the method of choice for COVID-19 patient diagnosis and contact screening (World Health Organization, 2020b). Although highly sensitive, the RT-PCR has some limitations due to its dependence on sampling technique, sample type and quality, virus genetic variability (Tahamtan and Ardebili, 2020). Also, viral RNA is detectable for a limited period post disease onset, making the timing of sample collection a crucial factor affecting the performance of RT-PCR (Zhao et al., 2020a; 2020b; Guo et al., 2020; To et al., 2020). Need for sensitive and specific antibody detection tests to supplement molecular diagnosis is thus obvious. Studies conducted so far have indicated limited utility of IgM in detection of recent infections (To et al., 2020; Long et al., 2020a; Zhang et al., 2020a, b; Wu et al., 2020; Van Elslande et al., 2020). This underscores the importance of IgG detection. Moreover, IgG remains an important marker of past exposure to the virus.
Efforts have been made to develop ELISAs for detection of IgM and IgG antibodies using recombinant viral proteins. The viral nucleoprotein (NP) and spike (S) protein have been exploited for the purpose (Zhao et al., 2020a; 2020b; Zhang et al., 2020b; Liu et al., 2020a; Sun et al., 2020; Kohmer et al., 2020a, b). Within S protein, the S1 component and receptor binding domain (RBD), responsible for induction of neutralizing antibodies and T cell response, have also been explored (Van Elslande et al., 2020; Liu et al., 2020a; Kohmer et al., 2020a, b; Zhao et al., 2020a; 2020b; Perera et al., 2020; Serrano et al., 2020). However, use of whole virus offers the advantage of simultaneous detection of antibodies generated against majority of the surface epitopes.
In the present study, we report development of ELISA for detection of anti-SARS-CoV-2 IgG antibodies using β-propiolactone (BPL) inactivated SARS-CoV-2. Further, we present data on IgG response among Indian COVID-19 patients with respect to duration of illness and severity of disease.
2. Materials and methods
2.1. Clinical samples
This study was approved by the institutional ethics committee of Bharati Vidyapeeth Medical College. A total of 242 blood samples were collected from RT-PCR confirmed COVID-19 patients (n = 137, 1–8 samples per patient) admitted at Bharati Hospital, Pune, and their SARS-CoV-2 RNA-positive asymptomatic contacts (n = 51) following informed consent. The asymptomatic contacts were identified by contact tracing and admitted at Bharati Hospital as per the government guidelines at that time. Serum/plasma samples were stored at −80 °C till testing. In addition, 200 serum/plasma samples collected before the emergence of SARS-CoV-2 (during 2017–2019) from healthy blood donors, and stored at −80 °C were included as negative controls.
2.2. Isolation, propagation and inactivation of SARS-CoV-2
SARS-CoV-2 was isolated in Vero CCL81 cells from nasopharyngeal swab of an acute-phase SARS-CoV-2 RNA positive patient from Pune, India (8004/IND/2020, Genbank Accession No: MT416726). The isolated virus was propagated and then inactivated using BPL. Briefly, Vero cells maintained using minimum essential medium (MEM, Gibco, USA), 10 % fetal bovine serum (FBS, Gibco, USA), 100 IU/mL penicillin and streptomycin were infected at 100 % confluency with 0.01 MOI SARS-CoV-2. After 48 or 72 h, cell suspension was harvested and centrifuged at 4816x g for 30 min at 4 °C. Cell supernatant was collected and inactivated using 0.2 %, 0.1 %, 0.05 %, 0.025 %, 0.0125 %, 0.00625 % BPL diluted in phosphate buffer saline (PBS), pH 7.2 for 24 h at 4 °C. Following BPL treatment, the virus was aliquoted and stored at -80 °C, until inactivation was confirmed. For inactivation check, 1 mL BPL treated virus was centrifuged in 30,000 molecular weight cutoff tubes (Vivaspin6, Sartorius, Germany) to remove BPL. The concentrate was resuspended in 1 mL MEM with 2% FBS and added onto 80–90 % confluent Vero cells in a 6-well plate. After 1 h adsorption, the virus suspension was removed and cells were maintained in MEM with 2% FBS for 5 days and observed for any cytopathic effect (CPE). After 5 days, the cell supernatant was collected and subjected to two successive blind passages with observation of any CPE.
2.3. Inactivated SARS-CoV-2 based IgG detection ELISA
The BPL-inactivated SARS-CoV-2 was diluted in carbonate-bicarbonate buffer, pH 9.2 and used to coat ELISA microwell strips (Nunc Maxisorp, Thermofisher Scientific, USA). Coated wells were incubated overnight at 4 °C and washed six times using PBS containing 0.05 % tween 20 (Sigma-Aldrich, USA). Washing protocol was kept constant for further washing steps. Plates were blocked for 1 h at 37 °C using PBS containing 10 % FBS, followed by washing. 100 μL of serum/plasma samples diluted 1:100 in ELISA diluent (PBS containing 10 % FBS, 1% uninfected Vero cell supernatant and 0.1 % tween 20) was added to plates and incubated for 1 h at 37 °C. Wells were washed and 100 μL of anti-human-IgG-HRP (Sigma-Aldrich, USA) diluted 1:20,000 in ELISA diluent was added. After incubation for 30 min at 37 °C, 100 μL tetramethylbenzidine (TMB) substrate containing hydrogen peroxide (Clinical Science Products Inc., USA) was added and reaction was stopped after 10 min using 100 μL 2 N H2SO4. Plates were read at 450 nm.
The performance of this in-house developed ELISA was compared with the commercially available Anti-SARS-CoV-2 IgG ELISA (Euroimmun IgG ELISA, Euroimmun Medizinische Labordiagnostika AG, Germany), purchased from the market. The Euroimmun IgG ELISA employs the recombinant spike protein (S1 domain) of SARS-CoV-2 as the coating antigen.
2.4. Statistical analysis
Percent IgG positivity and ELISA absorbance values between different groups were compared respectively using chi square test for proportions and Mann-Whitney U test. P values <0.05 were considered significant. All analyses were conducted using GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Standardization of inactivated SARS-CoV-2 based indirect IgG ELISA
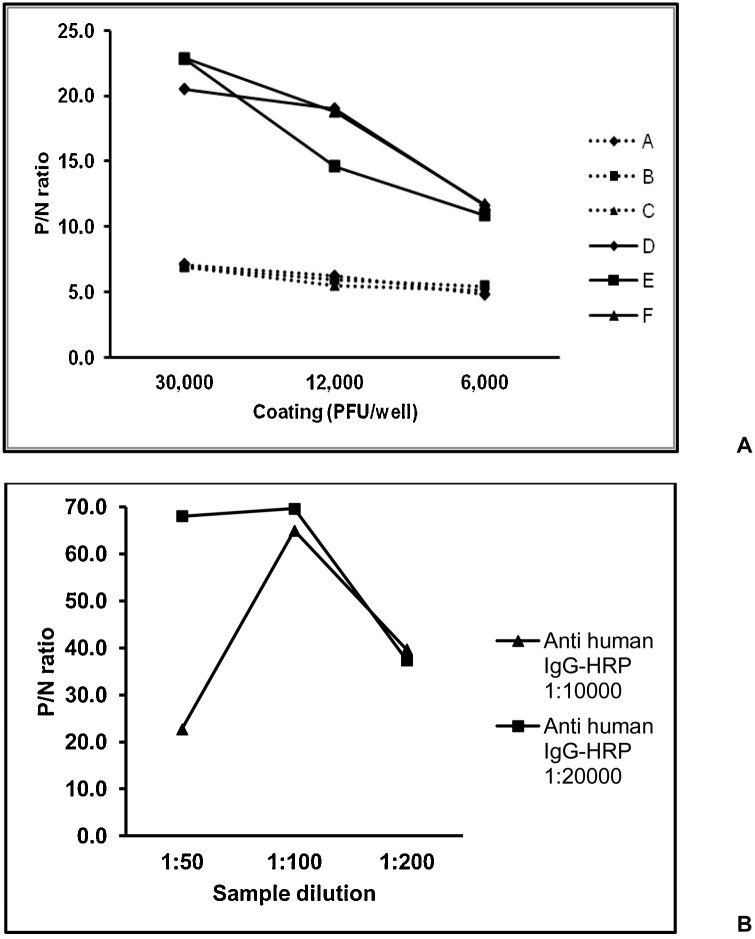
Complete inactivation of SARS-CoV-2 was achieved after treatment with 0.0125 %-0.2 % BPL. SARS-CoV-2 infected Vero cell supernatants harvested at 48 and 72 h post infection and inactivated with 0.025 %, 0.05 % and 0.1 % BPL were assessed for their suitability as coating antigen at 30,000 PFU/well, 12,000 PFU/well and 6000 PFU/well in indirect IgG ELISA (Fig. 1 A). Convalescent serum sample from a confirmed COVID-19 patient was used as positive control and healthy donor serum collected during 2017 was used as negative control. The ratio of absorbance of positive control to negative control i.e. P/N ratio was higher for viruses harvested at 72 h, but comparable for viruses inactivated with different concentrations of BPL. The P/N ratio was highest at 30,000 PFU/well. Therefore, SARS-CoV-2 harvested at 72 h post infection and inactivated with 0.1 % BPL was chosen as the coating antigen at 30,000 PFU/well. Serum dilution of 1:100 and anti-human IgG-HRP conjugate diluted at 1:20000 were found to be optimum for the ELISA (Fig. 1B).
Fig. 1.
Standardization of inactivated SARS-CoV-2 based indirect IgG ELISA.Fig. 1A shows comparison of different coating antigens (Harvesting at 48 h and inactivation with 0.025 % (A), 0.05 % (B), 0.1 % (C) BPL, harvesting at 72 h and inactivation with 0.025 % (D), 0.05 % (E), 0.1 % (F) BPL) for IgG ELISA, in terms of the ratio of absorbance of positive control to negative control i.e. P/N ratio. Fig. 1B shows comparison of different serum and conjugate dilutions with coating of antigen F at 30,000 PFU/well.
To determine the cut-off value, we screened 100 blood donor samples. The cut-off value (0.504) of mean absorbance of the negative controls (Mean NC) + 3 standard deviations (SD) corresponded to ∼2 times the mean NC (0.254). Therefore, the ELISA cut-off for further testing was decided as 2.5 times the Mean NC. We then screened additional 100 blood donors. With 6/200 donor samples testing positive, the specificity of the ELISA was 97 %. All the 6 samples scored negative in plaque reduction neutralization test (data not shown).
3.2. Comparison of IgG detection using in-house ELISA and commercial Euroimmun ELISA
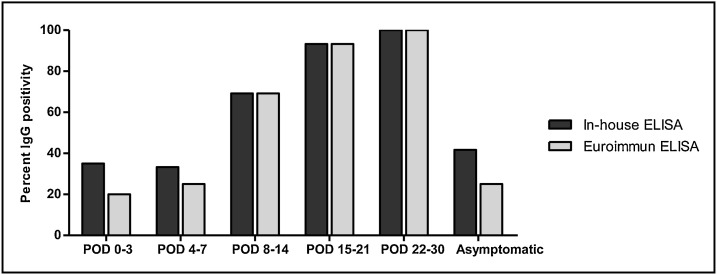
Next, we compared the efficiency of the in-house ELISA (inactivated whole virus-based) with the widely used Euroimmun IgG ELISA kit (recombinant S1 protein-based). For this purpose, 125 serum/plasma samples from confirmed COVID-19 patients were screened simultaneously by both the ELISAs; 68 (54.4 %) tested positive by in-house ELISA, while 60 (48 %) were positive by Euroimmun ELISA. During the first 3 days post onset of illness (POD 0−3), IgG detection by in-house ELISA was 35 % (7/20) while that of the Euroimmun ELISA was 20 % (4/20) (Fig. 2 ). At POD 4−7, in-house ELISA showed 33.3 % (12/36) positivity, while Euroimmun test showed 25 % (9/36) positivity. However, this difference between both the tests was not significant (P > 0.05). After the first week of illness, percent IgG detection by both the tests was exactly equal – POD 8−14: 69.2 % (27/39), POD 15−21: 93.3 % (14/15), POD 22−30: 100 % (3/3). Among the 12 samples from SARS-CoV-2 positive, asymptomatic subjects, 5 (41.7 %) tested positive for IgG by in-house ELISA, while 3 (25 %) were positive by Euroimmun test. In view of the comparable performance of both ELISAs for IgG detection (in-house ELISA performing better during early days post disease onset), the in-house ELISA was used further for investigation of the IgG response among COVID-19 patients.
Fig. 2.
Comparison of SARS-CoV-2 IgG detection by in-house ELISA (inactivated whole virus-based) and commercial Euroimmun ELISA (recombinant S1 protein-based). Percent IgG positivity by both the ELISAs among 125 samples is shown – 113 samples collected at different post onset day (POD) of symptoms from COVID-19 patients, 12 samples from asymptomatic subjects.
3.3. IgG antibody response among COVID-19 patients
We screened 242 samples collected at different intervals from confirmed COVID-19 patients using our in-house IgG ELISA. IgG was detected in >50 % of the RT-PCR confirmed COVID-19 patients within 7 days post onset of symptoms (POD 0−7), with the earliest detection at POD 2 (n = 3) (Table 1 ). During the second week (POD 8−14), IgG positivity rose to 80 %, increasing further to 93.8 % during the third week (POD 15−21). At 22−30 days post disease onset, IgG positivity was 100 %, although the number of samples tested was small (n = 4). Only 1 sample collected beyond POD30 was IgG-positive.
Table 1.
IgG seroprevalence at different post onset day (POD) of symptoms among COVID-19 patients.
| POD | Total no. of samples tested | No. of samples positive for IgG | Percent IgG positivity |
|---|---|---|---|
| 0−3 | 30 | 16 | 53.3 |
| 4−7 | 53 | 27 | 50.9 |
| 8−14 | 65 | 52 | 80.0 |
| 15−21 | 32 | 30 | 93.8 |
| 22−30 | 4 | 4 | 100.0 |
| >30 | 1 | 1 | 100.0 |
Eight patients in this study were admitted as asymptomatic contacts of confirmed COVID-19 patients, and developed symptoms after hospitalization. Of these, 3 patients were already IgG-positive on the day of onset of symptoms (POD 0), while 2 were positive even prior to disease onset.
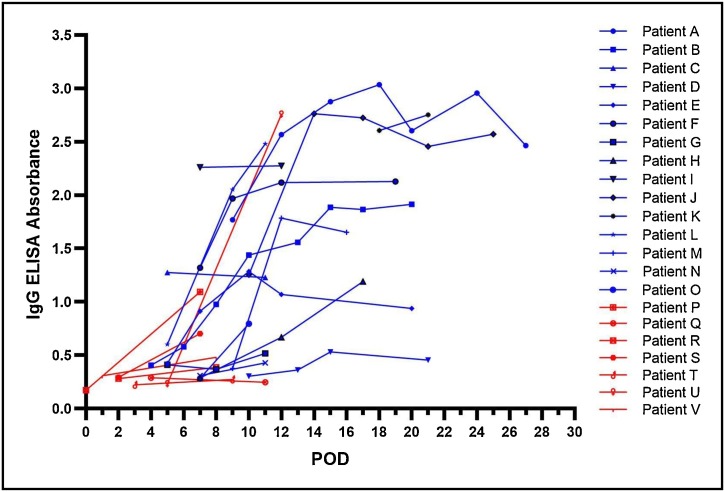
Further, longitudinal samples available from 22 patients were analyzed to understand the kinetics of IgG response (Fig. 3 ). Eight patients tested positive for IgG at the time of first sample collection (POD 5–18). Seroconversion was observed for 11 patients at POD 6−13. The remaining 3 patients remained negative for IgG and did not seroconvert till the time of last sample collection (POD 8–11). Among these 22 patients, the IgG ELISA absorbance values showed steady increase with increasing POD. Peak absorbance values were observed around POD 15 for majority of the patients.
Fig. 3.
Comparison of IgG ELISA absorbance values for longitudinal samples collected at different post onset day (POD) of symptoms for 22 COVID-19 patients. Blue colour denotes severe disease patients, red colour denotes mild disease patients (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.4. Comparison of IgG seropositivity in relation to outcomes of SARS-CoV-2 infection
Of the 51 asymptomatic subjects, 33 were reactive for IgG (64.7 %). The IgG detection rate was low (40.7 %, 11/27) during the early days (0–4) of hospitalization, but increased gradually later – 63.2 % (12/19) at 6–10 days, 83.3 % (5/6) at 13–15 days and 100 % (5/5) at 30 days post hospital admission. For 6 subjects, a follow-up sample was available; IgG seroconversion was observed on day 7 and 14 respectively for 3 and 1 patients. The remaining 2 patients remained negative for IgG till 6/7 days post hospitalization.
Table 2 presents IgG seropositivity in patients with mild (MD) or severe disease (SD). During the first 7 days of illness (POD ≤ 7), no significant difference was observed in the IgG positivity between MD (50 %) and SD patients (57.9 %). However, at 8−14 days post disease onset, IgG positivity among SD patients (90.9 %) was significantly higher than MD patients (68.8 %, P = 0.027). At 15−21 days post disease onset and beyond, the IgG detection rate was comparable between the two patient groups (P > 0.05).
Table 2.
Comparison of IgG seropositivity among COVID-19 patients with mild (MD) and severe disease (SD) at different post onset day (POD) of symptoms.
| POD | MD | SD | P-value |
|---|---|---|---|
| No. positive/ No. tested (%) | No. positive/ No. tested (%) | ||
| 0−7 | 32/64 (50.0 %) | 11/19 (57.9 %) | 0.55 |
| 8−14 | 22/32 (68.8 %) | 30/33 (90.9 %) | 0.027 |
| 15−21 | 13/15 (86.7 %) | 17/17 (100 %) | 0.13 |
| ≥22 | 1/1 (100 %) | 4/4 (100 %) | – |
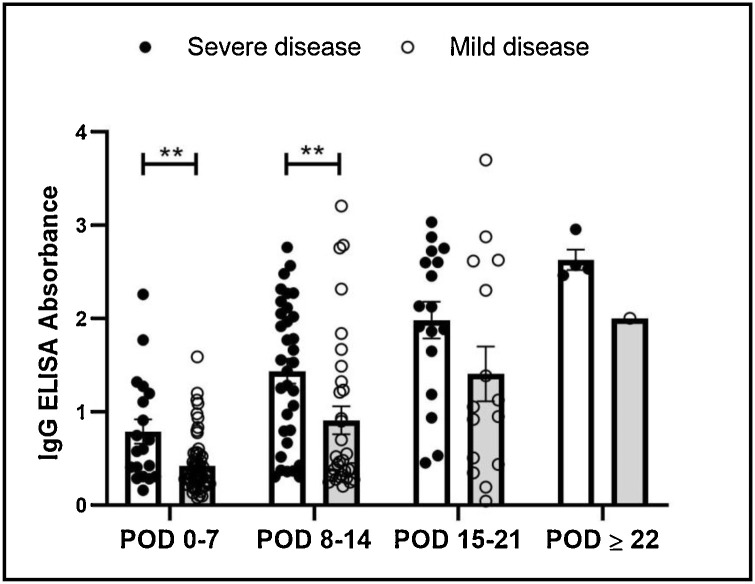
Further, ELISA absorbance values were compared among the asymptomatic subjects, MD and SD patients. The ELISA absorbance values among the asymptomatic subjects were comparable to that of the MD patients (P = 0.94), however, were lower than the SD patients (P < 0.0001). Within the symptomatic group, absorbance values were significantly higher in SD patients during the first 2 weeks of illness (Fig. 4 ). On analysis of longitudinal samples of 22 patients, ELISA absorbance values appeared lower among the MD as compared to SD patients around POD 7–10 (Fig. 3).
Fig. 4.
Comparison of IgG ELISA absorbance values for samples collected from patients with mild and severe disease at different post onset day (POD) of symptoms.
4. Discussion
Sensitive and specific serologic antibody detection assays are of immense value in understanding any disease. Their importance magnifies exponentially when one deals with an unprecedented pandemic caused by a novel agent such as SARS-CoV-2. To avoid handling of the highly infectious SARS-CoV-2 and for ease of working, researchers have preferred recombinant viral proteins for the development of antibody detection immunoassays. However, for this novel virus, it would be important to understand the dynamics of antibody response in infected individuals using the whole virus, in comparison with the antibodies detected against specific viral proteins. Though in vitro tests for detection of neutralizing antibodies have been developed and used, ELISA, with its intrinsic advantages of rapidity, ability to test large number of samples and detection of non-neutralizing antibodies as well, remains a test of choice for seroepidemiologic studies.
In this study, we evaluated inactivated virus as an antigen source for IgG ELISA. For inactivation, BPL, an agent used for preparation of inactivated viral vaccines was selected. The inactivated virus was expected to have higher probability of retaining antigenic properties, and thus detection of conformation dependent antibodies would be possible. We did try different concentrations of BPL, and chose 0.1 % as optimum for inactivation. Based on screening of 200 healthy donor sera/plasma collected before SARS-CoV-2 emergence, specificity of our ELISA was 97 %, and thus comparable with the several recombinant protein based ELISAs (95.2–100 %) (Tahamtan and Ardebili, 2020; Van Elslande et al., 2020; Kohmer et al., 2020a, b; Zhao et al., 2020a; 2020b; Perera et al., 2020; Serrano et al., 2020) and a gamma irradiated whole virus based test (97.9 %) (Sapkal et al., 2020) reported so far. Importantly, the performance of our ELISA was at par with a US-FDA approved, widely-used commercial test (Euroimmun ELISA) (Fig. 2). In fact, our in-house ELISA detected IgG among higher proportion of COVID-19 patients during the first week of illness, although the difference was not statistically significant (P > 0.05) probably because of the small sample numbers. The observed difference may reflect availability of more epitopes with whole virus than the S1 recombinant protein coating used in Euroimmun ELISA.
We then used our in-house IgG ELISA to analyse IgG seropositivity in relation to the disease duration among COVID-19 patients (Table 1). Of note, >50 % of the patients sampled during the first week of illness were circulating IgG antibodies. Importantly, as many as 16 (53.3 %) of the 30 patients sampled during the first 3 days of illness showed presence of IgG. Clearly, BPL-inactivated virus-based ELISA was able to detect IgG early during the disease phase. The only other study employing whole virus (gamma irradiated) based IgG ELISA used samples collected after the second week of disease onset, hence data regarding the performance of this ELISA in early disease phase is unavailable (Sapkal et al., 2020). We would like to point out here that use of different recombinant proteins such as NP, RBD, S1 and whole S led to variable IgG detection rates during the early disease phase. For NP-based ELISAs, early IgG seropositivity of 19.1 % (POD ≤ 7), 31.8 % (POD ≤ 5), 41.7 % (POD ≤ 7) has been reported, with one study demonstrating 100 % detection (POD ≤ 5) among the limited number of patients studied (16/16) (Zhao et al., 2020a; 2020b; Zhang et al., 2020b; Liu et al., 2020a; Sun et al., 2020). Using RBD-based ELISA, IgG has been detected in 30–40.9 % patients during the first 5–10 days of illness (Liu et al., 2020a; Sun et al., 2020; Kohmer et al., 2020a, b; Zhao et al., 2020a; 2020b; Perera et al., 2020). Evaluation of a recombinant S1-based ELISA employing a limited number of samples (n = 69) revealed 100 % IgG seropositivity (21/21) in the first week of illness (Zhao et al., 2020a; 2020b). Our results with the S1-based Euroimmun ELISA (23.2 % positivity at POD 0−7, Fig. 2) are in agreement with several other studies demonstrating lower seropositivity of this test (21.6–53.5 %) during the first week of illness (Van Elslande et al., 2020; Serrano et al., 2020). The use of S protein resulted in an IgG detection rate of 58.3 % within one week of disease onset (Sun et al., 2020). Taken together, early IgG seropositivity varied with the antigens used. Unless the different ELISAs, including ours, are subjected to testing of a large panel of samples, true performance comparison is not possible. Nonetheless, high IgG seropositivity using BPL-inactivated virus-based ELISA during the early disease phase is promising.
Further, we compared the IgG response among different categories of SARS-CoV-2 infected individuals – asymptomatic, mild and severe. While the IgG detection rate was comparable among the MD and SD patients during the early stage of illness (POD 0−7), a subsequent increase was seen in the SD patients (POD 8−14) (Table 2). Likewise, the IgG ELISA absorbance values were higher among the SD patients during the first 2 weeks of illness (Fig. 4). This data confirms previous observations of earlier appearance of IgG and a more vigorous antibody response among severe COVID-19 patients (Zhao et al., 2020a; 2020b; Wu et al., 2020; Perera et al., 2020; Serrano et al., 2020; Liu et al., 2020b). However, contradictory to a previous report suggesting a weaker immune response among asymptomatic individuals (Long et al., 2020b), IgG ELISA absorbance values of the asymptomatic contacts in our study were comparable to those of the MD patients (P = 0.94).
So far, IgM seropositivity has not been reported among Indian COVID-19 patients. In our study, the BPL-inactivated virus was not useful in IgM detection in the indirect ELISA format (data not shown). In this context, it should be noted that anti-SARS-CoV-2 IgM and IgG antibodies arise nearly simultaneously in serum, with IgG appearance even preceding IgM in some cases (To et al., 2020; Long et al., 2020a; Zhang et al., 2020a, b). This indicates limited utility of IgM testing, with reports suggesting no additional value of IgM over IgG detection in patient diagnosis (Wu et al., 2020; Van Elslande et al., 2020). Till a seromarker of recent infection becomes available, IgG can be used as a surrogate marker during the current situation. However, with rapid spread of the disease and asymptomatic infections in a population, IgG may not be able to discriminate between present and past SARS-CoV-2 infections at different times of the pandemic. Nonetheless, utility in contact screening, seroepidemiologic studies and probably, assessment of antibody response to vaccines remain promising.
In summary, our study demonstrates that BPL-inactivated SARS-CoV-2 based ELISA can efficiently detect IgG antibodies in a significant proportion of patients. Early detection of IgG suggests potential utility of this ELISA as a supplement to RT-PCR in patient diagnosis and contact screening algorithms, particularly, when large number of subjects are to be screened. The need for such serologic assays will become more pronounced when seasonal viruses such as influenza, dengue, chikungunya and other agents causing similar symptoms will start affecting large populations in different countries.
Funding
This work was supported by a grant from Department of Biotechnology-Biotechnology Industry Research Assistance Council (DBT-BIRAC), India, under the “Innovate in India (i3)” program of National Biopharma Mission (Grant No. BIRAC/BT/NBM0095/02/18).
CRediT authorship contribution statement
Ruta Kulkarni: Conceptualisation, Formal analysis, Methodology, Writing - original draft. Harshad P. Patil: Formal analysis, Methodology. Sonali Palkar: Resources. Sanjay Lalwani: Resources. Akhilesh Chandra Mishra: Conceptualization, Funding acquisition. Vidya Arankalle: Conceptualization, Formal analysis, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The technical assistance provided by Mr. Tushar Bhosale and Mr. Pravin Kore is gratefully acknowledged.
References
- Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q., Xu S., Zhu H., Xu Y., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;(March):21. doi: 10.1093/cid/ciaa310. ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020;(March):28. doi: 10.1093/cid/ciaa344. ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. 2020;(June):129. doi: 10.1016/j.jcv.2020.104480. 104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J. Med. Virol. 2020;(June):8. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of Nucleocapsid and Spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58(May (6)) doi: 10.1128/JCM.00461-20. e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.H. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg. Microbes Infect. 2020;9(December(1)):1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu LH Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.L., Xiang J.L., Du HX Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li Z.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(June(6)):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;(June):18. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Perera R.A., Mok C.K., Tsang O.T., Lv H., Ko R.L., Wu N.C., Yuan M., Leung W.S., Chan J.M., Chik T.S., Choi C.Y., Leung K., Chan K.H., Chan K.C., Li K.C., Wu J.T., Wilson I.A., Monto A.S., Poon L.L., Peiris M. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 2020;25(April(16)):2000421. doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Li M., Song H., Chen J., Ren W., Feng Y., Gao G.F., Song J., Peng Y., Su B., Guo X., Wang Y., Chen J., Li J., Sun H., Bai Z., Cao W., Zhu J., Zhang Q., Sun Y., Sun S., Mao X., Su J., Chen X., He A., Gao W., Jin R., Jiang Y., Sun L. Early detection of SARS-CoV-2 antibodies in COVID-19 patients as a serologic marker of infection. Clin. Infect. Dis. 2020;(May) doi: 10.1093/cid/ciaa523. 1:ciaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkal G., Shete-Aich A., Jain R., Yadav P.D., Sarkale P., Lakra R., Baradkar S., Deshpande G.R., Mali D., Tilekar B.N., Majumdar T., Kaushal H., Gurav Y., Gupta N., Mohandas S., Deshpande K., Kaduskar O., Salve M., Patil S., Gaikwad S., Sugunan A.P., Ashok M., Giri S., Shastri J., Abraham P., Gangakhedkar R.R. Development of indigenous IgG ELISA for the detection of anti-SARS-CoV-2 IgG. Indian J. Med. Res. 2020;151(May(5)):444–449. doi: 10.4103/ijmr.IJMR_2232_20. doi: 10.4103/ijmr. IJMR_2232_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M.M., Rodríguez D.N., Palop N.T., Arenas R.O., Córdoba M.M., Mochón M.D.O., Cardona C.G. Comparison of commercial lateral flow immunoassays and ELISA for SARS-CoV-2 antibody detection. J. Clin. Virol. 2020;129(June):104529. doi: 10.1016/j.jcv.2020.104529. Epub ahead of print. PMID: 32659710; PMCID: PMC7323682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P., Peng P., Liu X., Chen Z., Huang H., Zhang F., Luo W., Niu X., Hu P., Wang L., Peng H., Huang Z., Feng L., Li F., Zhang F., Li F., Zhong N., Chen L. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020;9(December(1)):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(May(5)):453–454. doi: 10.1080/14737159.2020.1757437. Epub 2020 Apr 22. PMID: 32297805; PMCID: PMC7189409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(May(5)):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E., Van Ranst M., Lagrou K., Vermeersch P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020;26(August(8)):1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/, 2020 (accessed 22 July 2020).
- World Health Organization . 2020. Laboratory Testing for Coronavirus Disease (COVID-19) in Suspected Human Cases: Interim Guidance. 19 March 2020. [Google Scholar]
- Wu J.L., Tseng W.P., Lin C.H., Lee T.F., Chung M.Y., Huang C.H., Chen S.Y., Hsueh P.R., Chen S.C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020;(June):15. doi: 10.1016/j.jinf.2020.06.023. S0163-4453(20)30404-30407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Nie S., Zhang Z., Zhang Z. Longitudinal Change of Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies in Patients with Coronavirus Disease 2019. J. Infect. Dis. 2020;222(June (2)):183–188. doi: 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du RH Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9(February (1)):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]