Abstract
Background and study aims Recently, a growing body of evidence has been amassed on evaluation of artificial intelligence (AI) known as deep learning in computer-aided diagnosis of gastrointestinal lesions by means of convolutional neural networks (CNN). We conducted this meta-analysis to study pooled rates of performance for CNN-based AI in diagnosis of gastrointestinal neoplasia from endoscopic images.
Methods Multiple databases were searched (from inception to November 2019) and studies that reported on the performance of AI by means of CNN in the diagnosis of gastrointestinal tumors were selected. A random effects model was used and pooled accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated. Pooled rates were categorized based on the gastrointestinal location of lesion (esophagus, stomach and colorectum).
Results Nineteen studies were included in our final analysis. The pooled accuracy of CNN in esophageal neoplasia was 87.2 % (76–93.6) and NPV was 92.1 % (85.9–95.7); the accuracy in lesions of stomach was 85.8 % (79.8–90.3) and NPV was 92.1 % (85.9–95.7); and in colorectal neoplasia the accuracy was 89.9 % (82–94.7) and NPV was 94.3 % (86.4–97.7).
Conclusions Based on our meta-analysis, CNN-based AI achieved high accuracy in diagnosis of lesions in esophagus, stomach, and colorectum.
Introduction
Early detection of gastrointestinal neoplasia by endoscopy is a widely adopted strategy to prevent cancer-related morbidity and/ or mortality. The disease prognosis greatly depends on the stage of cancer at diagnosis. Gastrointestinal neoplastic conditions are frequently detected by direct endoscopic visualization by a trained endoscopist and endoscopists use their knowledge, gathered from experience of endoscopic appearance, to detect these lesions.
To maximize detection and/or differentiation of a lesion, a clean mucosal surface and a meticulous mechanical exploration are paramount. Apart from detecting a lesion, predicting its potential to be carcinogenic is difficult. In addition, both lesion detection and its assessment are subject to substantial operator dependence. To improve detection of lesion by human eye, various optical enhancements of the endoscope have been made. High-definition white light endoscopy with or without chromo-endoscopy, narrow-band imaging (NBI) with or without magnification, confocal laser endomicroscopy, and endocytoscopic imaging system are some of the examples.
Recently, a growing body of evidence has been amassed on use of artificial intelligence (AI) known as deep learning in computer-aided diagnosis (CAD) of health-related conditions based on medical imaging 1 . A convolutional neural network (CNN) is a type of deep learning method that enables machines to analyze various training images and extract specific clinical features using a back-propagation algorithm. CNN data-driven systems are trained on datasets containing large numbers of images with their corresponding labels. CNN can be seen as a system that first extracts relevant features from the input images and it subsequently uses those learned features to classify a given image. The network uses convolutions of the input image to extract the most relevant information that helps to classify the image into different entities. Based on the accumulated data features, machine algorithms can diagnose newly acquired clinical images prospectively 2 3 4 .
CNN-based CAD has been reported as being highly beneficial in the field of endoscopy, including EGD, colonoscopy and capsule endoscopy. 2 5 6 CNN has transformed the field of computer vision and has been shown to work in real-time with raw, unprocessed frames from the video sequence. 2 In this systematic review and meta-analysis, we aim to quantitatively appraise the current reported data on the diagnostic performance of CNN based computer aided diagnosis of gastrointestinal neoplasia.
Methods
Search strategy
The literature was searched by a medical librarian for the concepts of AI with endoscopy for gastrointestinal lesions. The search strategies were created using a combination of keywords and standardized index terms. Searches were run in November 2019 in ClinicalTrials.gov, Ovid EBM Reviews, Ovid Embase (1974 +), Ovid Medline (1946 + including epub ahead of print, in-process & other non-indexed citations), Scopus (1970 +) and Web of Science (1975 +). Results were limited to English language. All results were exported to Endnote X9 (Clarivate Analytics) where obvious duplicates were removed leaving 4245 citations. Search strategy is listed in Appendix 1 . The MOOSE checklist was followed and is listed in Appendix 2 . Reference lists of evaluated studies were examined to identify other potential studies of interest.
Study selection
In this meta-analysis, we included studies that developed or validated a deep CNN learning model for diagnosis of neoplasia of the gastrointestinal tract (esophagus, stomach, and colorectum) using either one or a combination of white-light endoscopy (WLE), narrow-band imaging (NBI) endoscopy (magnifying and/ or non-magnifying), and chromoendoscopy. Study selection was restricted to only those that used CNN-based deep machine learning models. Studies were included irrespective of inpatient/outpatient setting, study sample-size, follow-up time, abstract/ manuscript status, and geography as long as they provided the appropriate data needed for the analysis.
Our exclusion criteria were as follows: (1) studies that used non-CNN-based machine learning algorithms (like support vector machine etc); (2) studies that used endoscopic optics other than standard WLE and/or NBI-based images as their training and testing platform; and (3) studies not published in English language. In cases of multiple publications from a single research group reporting on the same patient cohort and/or overlapping cohorts, each reported contingency table was treated as being mutually exclusive. When needed, authors were contacted via email for clarification of data and/or study-cohort overlap.
Data abstraction and analysis
Data on study-related outcomes from the individual studies were abstracted independently onto a predefined standardized form by at least two authors (BPM, SRK). Disagreements were resolved by consultation with a senior author (GK). Diagnostic performance data were extracted and contingency tables were created at the reported thresholds. Contingency tables consisted of reported accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). The results from testing of the algorithm were collected for the pooled analysis.
Definitions are as follows: (1) Accuracy: number of lesions detected by CNN/total number of lesions; (2) Sensitivity: detected number of correct neoplastic lesions by CNN (true positives)/histologically confirmed number of neoplastic lesions (total positives); (3) Specificity: detected number of correct non-neoplastic lesions by CNN (true negatives)/number of histologically proven non-neoplastic lesions (total negatives); (4) PPV: detected number of correct neoplastic lesions by CNN (true positives)/number of neoplastic lesions diagnosed by CNN (true positives + false positives); and (5) NPV: number of lesions correctly diagnosed as non-neoplastic lesions by CNN (true negatives)/number of lesions diagnosed as non-neoplastic by CNN (true negatives + false negatives).
If a study provided multiple contingency tables for the same or for different algorithms, we assumed these to be independent from each other. This assumption was accepted, as the goal of the study was to provide an overview of the pooled rates of various studies rather than providing precise point estimates. This methodology has been used and reported in literature 1 . A formal assessment of study quality was not done, due to the non-clinical nature of the studies.
We used meta-analysis techniques to calculate the pooled estimates in each case following the random-effects model 8 . We assessed heterogeneity between study-specific dom-effects model 8 . We assessed heterogeneity between study-specific estimates by using Cochran Q statistical test for heterogeneity, 95 % prediction interval (PI), which deals with dispersion of the effects, and the I 2 statistics 9 10 . In this, values < 30 %, 30 %–60 %, 61 %–75 %, and > 75 % were suggestive of low, moderate, substantial, and considerable heterogeneity, respectively. A formal publication bias assessment was not done due to the nature of the pooled results being derived from the studies.
All analyses were performed using Comprehensive Meta-Analysis (CMA) software, version 3 (BioStat, Englewood, New Jersey, United States).
Results
Search results and study characteristics
The literature search resulted in 4245 study hits (study search and selection flowchart: Supplementary Fig. 1 ). All 4245 studies were screened and 106 full-length articles and/or abstracts were assessed. Nineteen studies 2 5 6 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 reported on the detection and/ or classification of gastrointestinal neoplastic lesions by CNN. Among the 19 studies, five 6 12 15 17 26 reported on efficacy of CNN in diagnosing esophageal neoplasia, eight 5 14 16 18 19 22 23 25 reported on use of CNN in neoplasia of the stomach and six 2 11 13 20 21 24 evaluated use of CNN in diagnosing colorectal neoplasia. Seven studies 5 11 12 14 19 20 25 used standard WLE, eight used NBI (magnifying and/ or non-magnifying) 2 6 13 15 18 22 23 26 and four 16 17 21 24 used a combination of standard WLE and/or NBI and/or chromo-endoscopy images ( Table 1 ).
Table 1. Study characteristics.
| Study | Aim | Endoscopy technique | Machine learning model | Training strategy | Testing strategy | Accuracy | Sensitivity | Specificity | PPV | NPV | Remarks |
| Ahmad, 2019 11 | Colorectal adenoma detection | Standard colonoscopy | CNN | Multicenter colonoscopy images and videos of 4664 polyp test frames | 17 video datasets of complete colonoscopy withdrawal with 83 polyps consisting of 83716 frames (14634 polyp & 69082 non-polyp) | 92.5 | 84.5 | 92.5 | NR | NR | Conference abstract |
| Byrne, 2019 2 | Colorectal polyp detection in real-time endoscopic video images | NBI endoscopy | CNN | Unaltered video frames | 125 videos of consecutively encountered diminutive polyps | 94 | 98 | 83 | 90 | 97 | – |
| Cai, 2019 12 | Detect early ESCC under conventional endoscopic white light imaging | White light endoscopy | CNN | 1332 abnormal and 1096 normal images | 187 images from 57 patients | 91.4 | 97.8 | 85.4 | 86.4 | 97.6 | – |
| Chen, 2018 13 | Colorectal polyp detection | NBI endoscopy | CNN (TensorFlow algorithm) | 1476 images of neoplastic and 681 of hyperplastic polyps | 96 hyperplastic and 188 neoplastic polyps smaller than 5 mm | 90.1 | 96.3 | 78.1 | 89.6 | 91.5 | – |
| Cho, 2019 14 | Classify gastric neoplasms based on endoscopic white-light images | White light endoscopy | CNN (Inception-v4, Resnet-152, Inception-Resnet-v2) | 5017 images from 1269 individuals | 200 images from 200 patients | 93 | 60.7 | 98.3 | 85 | 93.9 | Advanced gastric cancer |
| 74.5 | 28.3 | 88.3 | 41.9 | 80.5 | Early gastric cancer | ||||||
| 86.4 | 0 | 99.4 | 0 | 86.9 | High-grade dysplasia | ||||||
| 78.5 | 6.7 | 91.2 | 11.8 | 84.7 | Low-grade dysplasia | ||||||
| 66.5 | 95.7 | 50.8 | 51.1 | 95.7 | Non-neoplasm | ||||||
| Guo, 2019 15 | Real time automated diagnosis of precancerous and early ESCC in both non-magnifying and magnifying settings | NBI endoscopy | CNN (SegNet architecture) | 2770 images of precancerous lesions and early ESCC in 191 cases and 3703 images of non-cancerous lesions in 358 cases | 1480 malignant images in 59 cases, 5191 non-cancerous images in 2004 cases, 27 precancerous and early ESCC videos, and 33 normal videos | nr | 98.04 | 95.03 | NR | NR | – |
| NBI endoscopy videos | nr | 60.8 | 99.9 | NR | NR | – | |||||
| Hirasawa, 2018 16 | Detect early and advanced gastric cancer | Standard white-light, chromoendoscopy, NBI | CNN (Single Shot MultiBox Detector) | 13584 EGD images for 2639 histologically proven gastric cancer | 2296 images from 77 gastric cancer lesions of 69 patients | nr | 92.2 | nr | 30.6 | NR | – |
| Horie, 2018 17 | Detect esophageal cancer | White-light, NBI | CNN (Single Shot MultiBox Detector) | 8428 histologically proven EGD images of cancer in 384 patients | 162 images of cancer and 376 images without cancer from 47 patients with 49 cancer lesions. 573 images of non-cancerous areas from 50 patients with no cancer | nr | 77 | 79 | 39 | 95 | – |
| Horiuchi, 2019 18 | Differentiate gastric cancer from gastritis | Magnifying NBI endoscopy | CNN (GoogLeNet) | 1492 cancer and 1078 gastritis images | 151 cancer and 107 gastritis images | 85.3 | 95.4 | 71 | 82.3 | 91.7 | – |
| Ikenoyama, 2019 19 | Detect gastric cancer | Standard EGD | CNN | 13584 images from 2639 lesions | 2940 images from 140 cases (209 early cancer images, 2731 non-neoplastic images) | nr | 65.6 | NR | 14.6 | NR | Conference abstract |
| Ito, 2018 20 | Assist in cT1b colorectal cancer diagnosis | White-light colonoscopy | CNN (AlexNet & Caffe) | Group 1: 2520 cTis + cT1a, 2418 cT1b images; Group 2: 2604 cTis + cT1a, 2400 cT1b images; Group 3: 2604 cTis + cT1a, 2418 cT1b images | 190 conventional white-light images | 81.2 | 67.5 | 89 | NR | NR | – |
| Komeda, 2019 21 | Colorectal polyp classification | White-light, NBI, chromo-endoscopy | CNN | 29572 adenoma images, 62999 non-adenoma images | 60 cases of colon polyps | nr | 97.5 | 97.9 | NR | NR | Conference abstract, white light |
| nr | 94.8 | 96.5 | NR | NR | Conference abstract, NBI | ||||||
| nr | 90.1 | 99.5 | NR | NR | Conference abstract, chromo-endoscopy | ||||||
| Li, 2019 22 | Early gastric cancer detection | Magnifying NBI endoscopy | CNN (InceptionV3-Keras framework) | 386 images of non-cancer lesions, 1702 images of early cancer | 341 images (170 early cancer & 171 non-cancer lesions) | 90.91 | 91.18 | 90.64 | 90.64 | 91.18 | – |
| Liu, 2018 23 | Early gastric cancer detection | Magnifying NBI endoscopy | CNN (VGG16, InceptionV3, InceptionResNetV2) | Magnifying NBI of normal gastric images and early gastric cancer images | Images (number not mentioned) | 98.5 | 98.1 | 98.9 | NR | NR | Conference abstract |
| Ozawa, 2018 24 | Automatic endoscopic detection and classification of colorectal polyps | White-light & NBI colonoscopy | CNN (Single Shot MultiBox Detector) | 16418 images of 4752 histologically proven colorectal polyps and 4013 images of normal colorectum | 3533 images | nr | 92 | nr | 93 | NR | Conference abstract |
| Sakai, 2018 25 | Detect early gastric cancer | White light endoscopy | CNN (Single Shot MultiBox Detector) | 1000 images of 0-I, 0-IIa, 0-IIc lesions | 228 cancer images | 82.8 | 73.6 | 98.8 | NR | NR | – |
| Wu, 2019 5 | Detect early gastric cancer | EGD | CNN (VGG-16, ResNet-50, Google's TensorFlow) | 2204 early cancer, 326 advanced cancer, 4791 control | 170 images | 92.5 | 94 | 91 | 91.26 | 93.81 | – |
| Zhang, 2017 26 | Early esophageal neoplasia | NBI and magnifying endoscopy | CNN | 218 endoscopic images generated 218000 patches, 90 % used for training and rest for testing | 79.38 | 73.41 | 83.54 | 72.09 | 84.44 | Conference abstract | |
| Zhao, 2019 6 | Early ESCC using magnifying NBI | Magnifying NBI endoscopy | VGG16 | three test groups with 463, 438 & 449 images | 1350 images in 219 patients | 89.2 | 87 | 84.1 | 81.9 | 90.4 | – |
CNN, convolutional neural networks; PPV, positive predictive value; NPV, negative predictive value; ESCC, esophageal squamous cell carcinoma; NBI, narrow band imaging; EGD, esophagogastroduodenoscopy; PET, positron emission tomography; ESD, endoscopic submucosal dissection; VGG, visual geometry group; NR, not reported
From all the included studies, we were able to extract a total of 26 contingency table datasets for CNN performance in diagnosing gastrointestinal lesions ( Table 1 ).
Meta-analysis outcomes
CNN performance by gastrointestinal location:
Esophageal neoplasia:
The pooled accuracy of CNN in the computer-aided diagnosis of esophageal neoplasia was 87.2 % (95 % CI 76–93.6). The sensitivity was 87.1 % (95 % CI 69.4–95.3), specificity was 87.3 % (95 % CI 74.3–94.2), PPV was 72.3 % (95 % CI 41.7–90.5) and NPV was 92.1 % (95 % CI 85.9–95.7).
Neoplastic lesions in stomach:
The pooled accuracy of CNN in the computer-aided diagnosis of neoplastic lesions of the stomach was 85.8 % (95 % CI 79.8–90.3). The sensitivity was 75.1 % (95 % CI 57.9–86.9), specificity was 91.4 % (95 % CI 84.3–95.4), PPV was 51 % (95 % CI 30.9–70.8) and NPV was 92.1 % (95 % CI 85.9–95.7).
Colorectal neoplasia:
The pooled accuracy of CNN in the computer-aided diagnosis of colorectal neoplasia was 89.9 % (95 % CI 82–94.6). The sensitivity was 92.6 % (95 % CI 82.8–97), specificity was 92.4 % (95 % CI 84.5–96.4), PPV was 91 % (95 % CI 68.8–97.9) and NPV was 94.3 % (95 % CI 86.4–97.7).
Results are summarized in Table 1 . Forest plots are shown in Fig. 1, Fig. 2, , Fig. 3, Fig. 4 , and Fig. 5 .
Fig. 1.
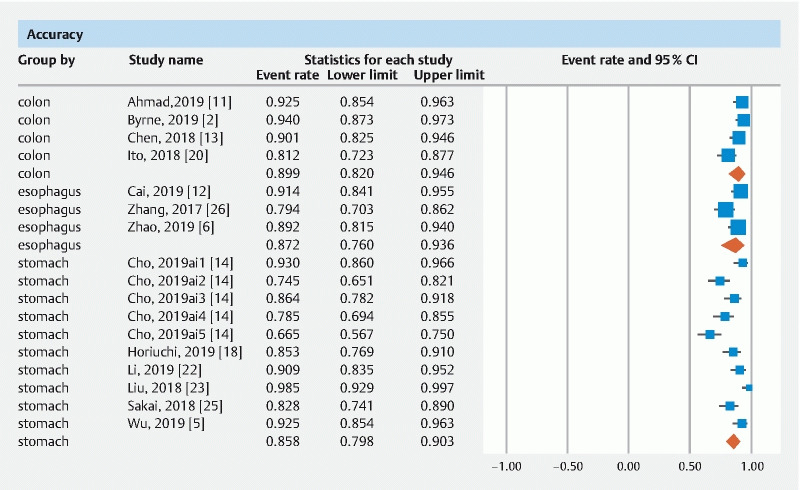
Forest plot, accuracy.
Fig. 2 .
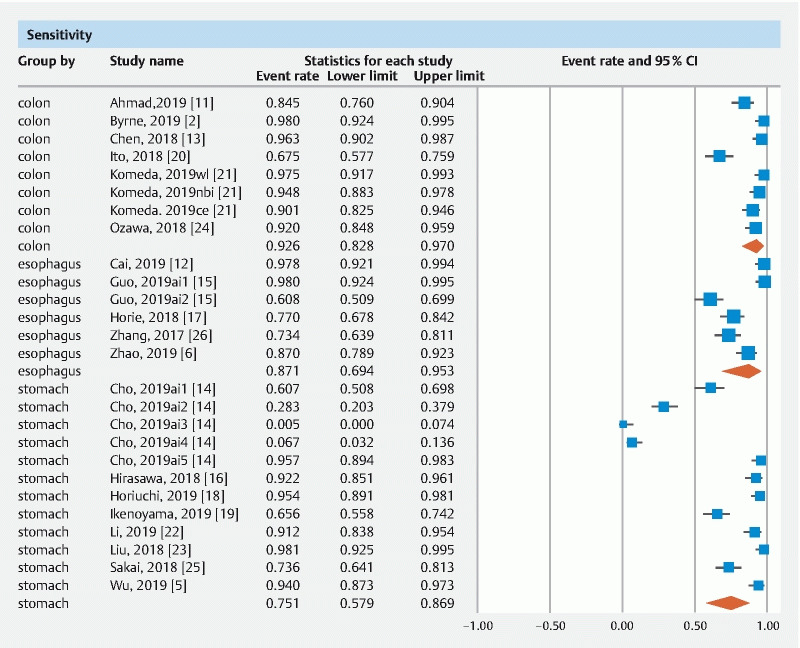
Forest plot, sensitivity.
Fig. 3.
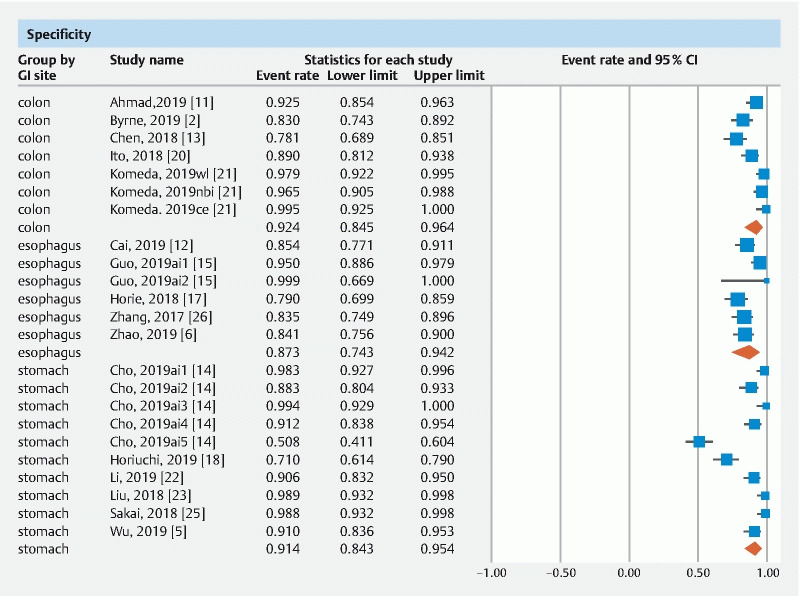
Forest plot, specificity.
Fig. 4.
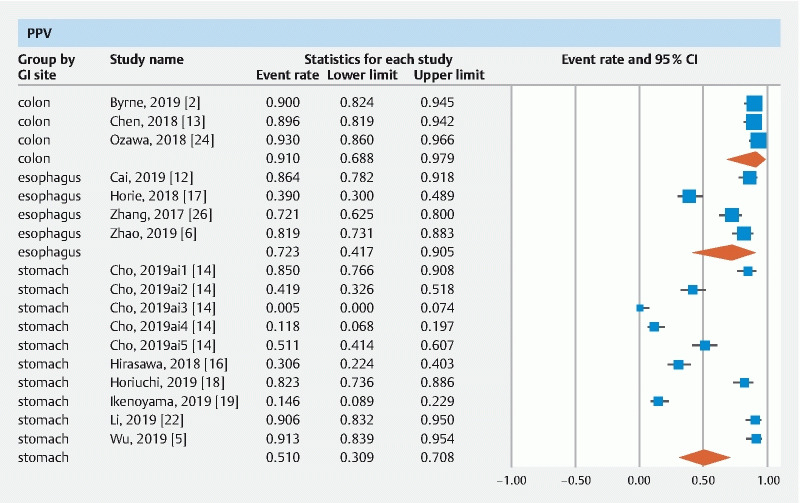
Forest plot, PPV.
Fig. 5.
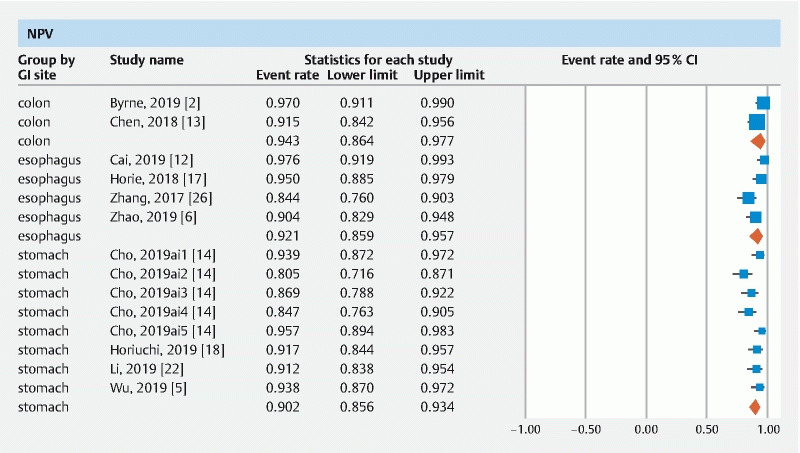
Forest plot, NPV.
Validation of meta-analysis results
Sensitivity analysis
To assess whether any one study had a dominant effect on the meta-analysis, we excluded one study at a time and analyzed its effect on the main summary estimate. On this analysis, no single study significantly affected the outcome or the heterogeneity.
Heterogeneity
A large degree of between-study heterogeneity was expected due to the broad nature of machine learning algorithms and endoscopic optics included in this study. This is reflected in our I 2 % values ( Table 2 ). Our subgroup analysis based on tumor location did not affect the observed I 2 % values and therefore it can be said that tumor location was not a contributory factor. Prediction interval statistics was not calculated due to the expected large degree of heterogeneity and the fact that the goal was not to provide precise point estimates.
Table 2. Summary of results.
| Pooled rates | Accuracy | Sensitivity | Specificity | PPV | NPV |
| Esophagus | 87.2 (76–93.6) I 2 = 70 3 datasets | 87.1 (69.4–95.3) I 2 = 90 6 datasets | 87.3 (74.3–94.2) I 2 = 60 6 datasets | 72.3 (41.7–90.5) I 2 = 95 4 datasets | 92.1 (85.9–95.7) I 2 = 74 4 datasets |
| Stomach | 85.8 (79.8–90.3) I 2 = 83 10 datasets | 75.1 (57.9–86.9) I 2 = 96 12 datasets | 91.4 (84.3–95.4) I 2 = 92 10 datasets | 51 (30.9–70.8) I 2 = 97 10 datasets | 90.2 (85.6–93.4) I 2 = 64 8 datasets |
| Colorectal | 89.9 (82–94.6) I 2 = 69 4 datasets | 92.6 (82.8–97) I 2 = 88 8 datasets | 92.4 (84.5–96.4) I 2 = 81 7 datasets | 91 (68.8–97.9) I 2 = 0 3 datasets | 94.3 (86.4–97.7) I 2 = 61 2 datasets |
CNN, convolutional neural network, PPV, positive predictive value; NPV,negative predictive value
Publication bias
Publication bias assessment largely depends on the sample size and the effect size. A publication bias assessment was deferred in this study due to the fact that the reported effects were independent of the sample size. We, however, do not rule out the possibility of potential publication bias in terms of negative studies being less frequently published.
Quality of evidence
The quality of evidence was rated for results from the meta-analysis according to the GRADE working group approach 27 . Observational studies begin with a low-quality rating, and based on the risk of bias and heterogeneity, the quality of this meta-analysis would be considered as low-quality evidence.
Discussion
To the best of our knowledge, this is the first systematic review and meta-analysis assessing the accuracy parameters of convolutional neural network (CNN) based computer aided diagnosis of gastrointestinal lesions that includes esophageal, gastric and colorectal data. Based on our analysis, CNN-based deep machine learning demonstrates high accuracy in image-based diagnosis of lesions in esophagus, stomach and colorectum.
A key finding of our study is that CNN achieved > 90 % NPV in diagnosis of esophageal, gastric and colorectal lesions. The majority of the included studies evaluated performance of CNN in experimental conditions and not in a real-life clinical scenario. Prospective studies and real-time video analysis of endoscopic images are lacking. Only high-quality images were used to train the CNN. In a real clinical setting, less insufflation of air, post-biopsy bleeding, halation, blur, defocus or mucus can all affect an accurate CAD. There was variability in the choice of threshold used to report sensitivity and specificity. There was lack in uniformity of validating the training process of the algorithm before using it for testing.
A recent meta-analysis published by Liu et al 28 reported similar diagnostic accuracy results for use of AI in prediction and detection of colorectal polyps. They reported better performance for AI under NBI and performance superior to that of non-expert endoscopists. This study primarily differs in the reported AI parameters for esophageal, gastric, and colorectal lesions. In addition, we did not include studies that primarily assessed the nuances of mathematical formulae behind the CNN algorithm and we did not include studies that used support vector machine-based algorithm.
The strengths of this review lie in careful selection of studies reporting on machine-based learning that is solely based on CNN-based algorithms and avoiding other redundant studies. The American Society of Gastrointestinal Endoscopy (ASGE) in its second Preservation Incorporation of Valuable Endoscopic Innovations (PIVI-2) declaration proposed a NPV threshold of 90 % or greater for real-time optical diagnosis of diminutive colorectal polyps using advanced endoscopic imaging 29 . We have demonstrated that CNN achieves this threshold in CAD of gastrointestinal lesions regardless of their location.
There are limitations to this study. The included studies were not representative of the general population and community practice, with studies being performed in an experimental environment. Our analysis had studies that were retrospective in nature contributing to selection bias. To capture maximum available data, we included six conference abstracts that have not been published as full manuscripts yet. We were unable to formally conduct a quality assessment, as there is no guidance on how to appropriately score and report quality on items pertaining to machine-based learning. Moreover, we considered individual accuracy tables as independent of each other, which does not reflect real-life case scenario.
Our analysis has the limitation of heterogeneity. We were unable to statistically ascertain a cause for the observed heterogeneity. We hypothesize, however, that the observed heterogeneity is primarily due to the following variables: threshold cut-off used, different training algorithm as well as the training methodology employed, and the variability in endoscopic optics (standard white-light, NBI, chromo-endoscopy). In addition, endoscopic optics differ in their diagnostic accuracy based on the underlying gastrointestinal lesion being assessed. In terms of algorithm training and testing, not all studies employed a validation step to fine-tune the algorithm. Therefore, the possibility of over-fitting in the reported accuracy data is possible.
We only included studies that evaluated the performance of CNN-based algorithms and not others, such as support vector machine algorithms (SVM). This is due to the inherent mathematical differences in the algorithms that make CNNs highly unique and superior performers when compared to SVMs, and due to the fact that SVMs are less likely to be used for image classification in the near future. Although the technology is rapidly advancing in AI, we do not anticipate that CNN-based deep learning will become obsolete before further real-life prospective studies are reported. We do, however, anticipate rapid technical improvements in CNN algorithms in terms of faster processing times despite an increase in number of deep hidden learning layers, and the implementation of positive reinforcement in CNN learning that allows the algorithm to learn from its errors and encourages it to execute a correct neuron while inhibiting a wrong one.
Conclusions
In conclusion, based on our meta-analysis, deep machine learning by means of CNN -based algorithms demonstrates high accuracy in diagnosis of gastrointestinal lesions. Deep learning in gastroenterology is in its infancy and is witnessing a rapid, steep growth in terms of learning as well as technological development. Future studies are needed to streamline the machine-learning process and define its role in the CAD of gastrointestinal neoplastic conditions in real-life clinical scenarios.
Acknowledgement
The authors thank Dana Gerberi, MLIS, Librarian, Mayo Clinic Libraries, for help with the systematic literature search, and Unnikrishnan Pattath, BTECH, MBA, Artificial intelligence solutions, Bangalore, India, for help with technical details on convolutional neural network algorithms.
Footnotes
Competing interests Dr. Dulai received an American Gastroenterology Association Research Scholar Award. He is a consultant for Takeda, Janssen, Pfizer, and Abbvie. He has also received grant support from Takeda, Janssen, Pfizer, and Abbvie.
Supplementary material :
References
- 1.Liu X, Faes L, Kale A U et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Health. 2019;1:e271–e97. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 2.Byrne M F, Chapados N, Soudan F et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100. doi: 10.1136/gutjnl-2017-314547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krizhevsky A, Sutskever I, Hinton G E. Neural Information Processing Systems Foundation; 2012. Advances in neural information processing systems. p. 1269.
- 4.Szegedy C, Vanhoucke V, Ioffe S Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. [DOI]
- 5.Wu L, Zhou W, Wan X et al. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. 2019;51:522–531. doi: 10.1055/a-0855-3532. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y Y, Xue D X, Wang Y L et al. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333–341. doi: 10.1055/a-0756-8754. [DOI] [PubMed] [Google Scholar]
- 7.Stroup D F, Berlin J A, Morton S C et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 8.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 9.Higgins J P, Thompson S G, Deeks J J et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan B P, Adler D G. Heterogeneity in systematic review and meta-analysis: how to read between the numbers. Gastrointest Endosc. 2019;89:902–903. doi: 10.1016/j.gie.2018.10.036. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad O F, Brandao P, Sami S S et al. Artificial intelligence for real-time polyp localisation in colonoscopy withdrawal videos. Gut. 2019;68:A2–A3. [Google Scholar]
- 12.Cai S L, Li B, Tan W M et al. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video) Gastrointest Endosc. 2019;90:745–5300. doi: 10.1016/j.gie.2019.06.044. [DOI] [PubMed] [Google Scholar]
- 13.Chen P J, Lin M C, Lai M J et al. Accurate classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Cho B J, Bang C S, Park S W et al. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy. 2019:23. doi: 10.1055/a-0981-6133. [DOI] [PubMed] [Google Scholar]
- 15.Guo X, Zhang N, Guo J et al. Automated polyp segmentation for colonoscopy images: A method based on convolutional neural networks and ensemble learning. Med Phys. 2019;46:5666–5676. doi: 10.1002/mp.13865. [DOI] [PubMed] [Google Scholar]
- 16.Hirasawa T, Aoyama K, Tanimoto T et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653–660. doi: 10.1007/s10120-018-0793-2. [DOI] [PubMed] [Google Scholar]
- 17.Horie Y, Yoshio T, Aoyama K et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25–32. doi: 10.1016/j.gie.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 18.Horiuchi Y, Aoyama K, Tokai Y et al. Convolutional neural network for differentiating gastric cancer from gastritis using magnified endoscopy with narrow band imaging. Digest Dis Sci. 2020;65:1355–1363. doi: 10.1007/s10620-019-05862-6. [DOI] [PubMed] [Google Scholar]
- 19.Ikenoyama Y, Hirasawa T, Ishioka M et al. 379 comparing artificial intelligence using deep learning through convolutional neural networks and endoscopist's diagnostic ability for detecting early gastric cancer. Gastrointest Endosc. 2019;89:AB75. [Google Scholar]
- 20.Ito N, Kawahira H, Nakashima H et al. Endoscopic diagnostic support system for cT1b colorectal cancer using deep learning. Oncology (Switzerland) 2018;96:44–50. doi: 10.1159/000491636. [DOI] [PubMed] [Google Scholar]
- 21.Komeda Y, Handa H, Matsui R et al. Computer-aided diagnosis based on convolutional neural network system using artificial intelligence for colorectal polyp classification. Gastrointest Endosc. 2019;89:AB631. [Google Scholar]
- 22.Li L, Chen Y, Shen Z et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging images. Gastric Cancer. 2020;23:126–132. doi: 10.1007/s10120-019-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Wang C, Hu Y 25th IEEE International Conference on Image Processing; 2018. Transfer learning with convolutional neural network for early gastric cancer classification on magnifying narrow-band imaging images. [DOI]
- 24.Ozawa T, Ishihara S, Fujishiro M et al. Automated endoscopic detection and classification of colorectal polyps using convolutional neural networks. Gastrointest Endosc. 2018;87:AB271. doi: 10.1177/1756284820910659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai Y, Takemoto S, Hori K Annual International Conference of the IEEE Engineering in Medicine & Biology Society; 2018. Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. pp. 4138–4141. [DOI] [PubMed]
- 26.Zhang C, Ma L, Uedo N et al. The use of convolutional neural artificial intelligence network to aid the diagnosis and classification of early esophageal neoplasia a feasibility study. Gastrointest Endosc. 2017;85:AB581–AB2. [Google Scholar]
- 27.Puhan M A, Schunemann H J, Murad M H et al. A GRADE Working Group approach for rating the quality of treatment effect estimates from network meta-analysis. BMJ. 2014;349:g5630. doi: 10.1136/bmj.g5630. [DOI] [PubMed] [Google Scholar]
- 28.Lui T K, Guo C G, Leung W K. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: A systematic review and meta-analysis. Gastrointest Endosc. 2020;92:1.1022E10. doi: 10.1016/j.gie.2020.02.033. [DOI] [PubMed] [Google Scholar]
- 29.Abu Dayyeh B K, Thosani N, Konda V et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015;81:5020–e16. doi: 10.1016/j.gie.2014.12.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


