Abstract
A family of polypeptide GalNAc-transferases (GalNAc-Ts) initiates mucin-type O-glycosylation, transferring GalNAc onto hydroxyl groups of Ser and Thr residues of target substrates. The 20 GalNAc-T isoenzymes in humans are classified into nine subfamilies according to sequence similarity. GalNAc-Ts select their sites of glycosylation based on weak and overlapping peptide sequence motifs, as well prior substrate O-GalNAc glycosylation at sites both remote (long-range) and neighboring (short-range) the acceptor. Together, these preferences vary among GalNAc-Ts imparting each isoenzyme with its own unique specificity. Studies on the first identified GalNAc-Ts showed Thr acceptors were preferred over Ser acceptors; however studies comparing Thr vs. Ser glycosylation across the GalNAc-T family are lacking. Using a series of identical random peptide substrates, with single Thr or Ser acceptor sites, we determined the rate differences (Thr/Ser rate ratio) between Thr and Ser substrate glycosylation for 12 isoenzymes (representing 7 GalNAc-T subfamilies). These Thr/Ser rate ratios varied across subfamilies, ranging from ~2 to ~18 (for GalNAc-T4/GalNAc-T12 and GalNAc-T3/GalNAc-T6, respectively), while nearly identical Thr/Ser rate ratios were observed for isoenzymes within subfamilies. Furthermore, the Thr/Ser rate ratios did not appreciably vary over a series of fixed sequence substrates of different relative activities, suggesting the ratio is a constant for each isoenzyme against single acceptor substrates. Finally, based on GalNAc-T structures, the different Thr/Ser rate ratios likely reflect differences in the strengths of the Thr acceptor methyl group binding to the active site pocket. With this work, another activity that further differentiates substrate specificity among the GalNAc-Ts has been identified.
Keywords: GALNT, glycosyltransferase, mucin-type O-glycosylation, O-glycan biosynthesis, peptide acceptor
Introduction
Polypeptide GalNAc-transferases (GalNAc-Ts) comprise a large family of glycosyltransferases that initiate mucin-type O-glycosylation in eukaryotes by adding GalNAc onto Ser and Thr residues of membrane and secreted proteins. In man, this family consists of 20 different isoenzymes (Bennett et al. 2012) with similar numbers found in most mammals. Interestingly, the number of isoenzymes decreases in the lower metazoans with 14 in Drosophila, 9 in Caenorhabditis elegans and smaller numbers in multicellular and single-cell eukaryotes and protists (Schwientek et al. 2002; Bennett et al. 2012; Raman et al. 2012; Tomita et al. 2017; DeCicco RePass et al. 2018; Zhang and Ten Hagen 2019). This suggests that over evolution the GalNAc-Ts may have taken on more diverse and/or specialized biological roles which presently remain poorly understood.
Although the biological roles of these transferases and their targets are not fully elucidated, it is clear that individual isoenzymes play critical roles in development in both the fly (Tian and Ten Hagen 2006, 2007; Tran et al. 2012) and the mouse (Tian et al. 2015). The expression of the GalNAc-Ts is highly regulated, and their dysregulation is associated with complex disorders and disease states (Joshi et al. 2018; Reily et al. 2019) including coronary heart disease (Willer et al. 2008; Holleboom et al. 2011; Tian et al. 2015; Khetarpal et al. 2016; Wang et al. 2018) and many human cancers (Kohsaki et al. 2000; Guo et al. 2004; Berois et al. 2006; Freire et al. 2006; Wood et al. 2007; Wu et al. 2010; Gill et al. 2013; Horynová et al. 2013; Remmers et al. 2013; Niang et al. 2016; Kurita et al. 2017; Lin et al. 2017; Balcik-Ercin et al. 2018; Sheta et al. 2019). The number of O-glycosylation sites in proteins spans from single sites of glycosylation to hundreds of closely spaced sites, the latter comprising the so-called mucin domains. Such heavily glycosylated mucin domains are highly resistant to proteases and serve to protect cell surfaces and modulate cell–cell interaction. Similarly single sites of glycosylation modulate proprotein processing, thereby regulating important biological processes (Topaz et al. 2004; Kato et al. 2006; Christensen et al. 2008; Semenov et al. 2009; Schjoldager et al. 2010; van der Wel et al. 2011; Schjoldager and Clausen 2012; Nishikimi et al. 2015; Goth et al. 2018; King et al. 2018; Hansen et al. 2019; Nakamura and Kurosaka 2019). Clearly, to fully elucidate both the biological and molecular roles of O-glycosylation in health and disease, we must first understand how individual GalNAc-T isoenzymes choose their sites of glycosylation.
Our laboratory’s focus has been on understanding the underlying principles of substrate selection and site preferences for the individual GalNAc-T isoenzyme. Toward this goal, we have developed a series of random peptide and glycopeptide substrate approaches to quantify isoenzyme specificity. From such work, we have obtained “soft” peptide sequence motifs for about half of the GalNAc-T isoenzymes revealing both their unique and overlapping specificities and in particular the (T/S)PXP motif which is common to most but not all isoenzymes (Gerken et al. 2006, 2008, 2011). We have further shown that nearly all GalNAc-T isoenzymes possess strong rate enhancing preferences for prior glycosylated substrates having GalNAc-O-Ser/Thr 6–17 residues N- or C-terminal from the acceptor Thr or Ser (Revoredo et al. 2016). We call this a long-range glycosylation or remote, glycopeptide activity which further modulates a GalNAc-T’s specificity. Furthermore, we discovered a subgroup of isoenzymes (i.e., GalNAc-T4, GalNAc-T7, GalNAc-T10 and GalNAc-T12) that recognized GalNAc-O-Ser/Thr 1 or 3 residues N- or C-terminal of the acceptor site (Perrine et al. 2009; Revoredo et al. 2016) an activity we term as neighboring, or short-range, glycosylation activity. Thus, these latter isoenzymes prefer to glycosylate sites close to previously glycosylated sites and serve the so-called “filling in” activity that is important for glycosylating heavily glycosylated mucin and mucin-like domains. Several of these “filling in” isoenzymes also possess remote glycopeptide activity and therefore recognize both long- and short-range prior glycosylation—showing di-glycopeptide preferences (Revoredo et al. 2016; de las Rivas, Coelho, et al. 2018; de las Rivas, Paul Daniel, et al. 2018; Fernandez et al. 2019). The presence of the long- and short-range glycopeptide preferences of these transferases suggests that O-glycosylation may actually be a highly orchestrated and perhaps controlled process in vivo.
Structurally all GalNAc-Ts (except h-GalNAc-T20) consist of an N-terminal catalytic domain linked via a short flexible linker to a C-terminal ricin-like lectin domain, which is tethered to the Golgi lumen via an N-terminal transmembrane domain and variable length “stem” domain (Fritz et al. 2004). (Note that we will be using the transferase numbering nomenclature of Bennett and coworkers 2012 for consistency.) We have shown that the catalytic domain is responsible for recognizing peptide sequence motifs and prior glycosylated sites, ~<±5 residues of the acceptor site (Gerken et al. 2006, 2008, 2011; Perrine et al. 2009; Revoredo et al. 2016). We have further shown that the lectin domain binds remote prior O-GalNAc sites (~≥±5 residues of the acceptor) and is responsible for the long-range enhancing glycopeptide activity (Raman et al. 2008; Gerken et al. 2013). X-ray crystallographic studies have confirmed these notions and have provided structural insights of how these domains work together to select peptide and glycopeptide substrates (Lira-Navarrete et al. 2015; de las Rivas et al. 2017; de las Rivas, Paul Daniel, et al. 2018; Fernandez et al. 2019) including the recent finding that charge residues composing loops protruding from the lectin domain can alter substrate specificity (de las Rivas, Paul Daniel, et al. 2018; Ji et al. 2018). Thus each isoenzyme possesses a different combination of substrate recognition modes, achieved by its “naked” peptide specificity (recognizing sequence motifs) and glycopeptide specificity (recognizing prior glycosites) (de las Rivas et al. 2019). It is likely, therefore, that the GalNAc-Ts have evolved their unique peptide and glycopeptide specificities to achieve their specific and unique biological function(s).
One aspect of GalNAc-T specificity that has largely gone unstudied is whether and by how much do the rates of glycosylation of Thr vs. Ser acceptors vary among isoenzymes. Early kinetic studies of GalNAc-T1, GalNAc-T2 and GalNAc-T3 showed that they preferred Thr over Ser acceptors by factors of ~10 to ~100 (Elhammer et al. 1999); however we know of no other such studies for other GalNAc-T isoenzymes. Consistent with this are the observations of multiple in vivo glycoproteomic studies that report about twice as many Thr residues being glycosylated over Ser residues (Steentoft et al. 2013; Kong et al. 2015; Lavrsen et al. 2018). This behavior has also been found in other distant glycosyltransferases such as PoFUT2 (Valero-González et al. 2016) and the ppGlcNAcTs of the protists which are thought to represent evolutionary predecessors of the animal GalNAc-Ts (Ercan and West 2005; Heise et al. 2009; Mendonca-Previato et al. 2013). We now report a systematic characterization of the Thr vs. Ser glycosylation rates (Thr/Ser rate ratios) for 12 of the 20 GalNAc-T isoenzymes spanning 7 of the 9 GalNAc-T subfamilies. Interestingly we show that the Thr/Ser ratios are largely similar within subfamilies but can vary between subfamilies, ranging from ~2- to ~18-fold, further adding to GalNAc-T isoenzyme substrate diversity. We further show that these Thr/Ser rate ratios are relatively independent of acceptor peptide sequence within an isoenzyme. Finally, the inclusion of these rate ratios will help refine the predictive approaches for O-glycosylation, such as the isoform specific O-glycosylation prediction (ISOGlyP) website (http://isoglyp.utep.edu), M-Y Leung, J. Mohl, and G. Cardenas, Boarder Biomedical Research Center, Univ. Texas at El Paso and T. A. Gerken, Dept. Biochemistry, Case Western Reserve Univ., Cleveland OH).
Results
The goal of this work was to compare the rates of glycosylation of Thr and Ser acceptors for a series of representative GalNAc-T isoenzymes from across subfamilies. Rather than utilize acceptors with fixed sequences, we utilized the random peptides that we have previously exploited for determining the GalNAc-T peptide substrate preferences (Gerken et al. 2006, 2011). These random peptides, given in Table I, consist of the general sequence GAGAXn (T/S) XnAGAG where the X positions consist of randomized residues and where n = 3 or 5. We reasoned that the random peptide substrates would serve as universal substrates and would be active against most GalNAc-T isoenzymes even if they preferred different peptide motifs. By using the three different sets of Thr/Ser random peptides, each with a different length and/or amino acid composition, we could further confirm that local peptide sequence did not affect the obtained Thr/Ser rate ratios. Note that all three of these peptide sets contained Pro, Gly and Ala as these residues are common around sites of O-glycosylation (Elhammer et al. 1993; Gerken et al. 2006). As described in the methods, simultaneous reactions under identical reaction conditions and incubation times were performed for each Ser/Thr peptide pair against each GalNAc-T isoenzyme. As the GalNAc-Ts commonly possess a UDP-GalNAc hydrolysis activity in addition to their transferase activity, the reaction products (free 3H-GalNAc and 3H-GalNAc-peptide, respectively) were separated on Sephadex G10 chromatography (see Figure 1 and Supplemental Figure 1). Final transferase activity (i.e., productive transfer of GalNAc to peptide) was determined based on GalNAc incorporation into the peptide substrate as described in the Methods. Note that we take the extent of UDP-GalNAc hydrolysis, based on the ratio of the free GalNAc and peptide-GalNAc peaks, as an indicator of how well a peptide substrate binds to the active site of the catalytic domain, where weakly bound peptide substrates typically give elevated rates of UDP-GalNAc hydrolysis, i.e., the glycopeptide preferring transferases GalNAc-T4, GalNAc-T7, GalNAc-T10 and GalNAc-T12 in the figures. Importantly, for each isoenzyme (except for GalNAc-T12), the extent of UDP-GalNAc hydrolysis was found to be the same for both the Ser and Thr acceptors within a peptide pair (Figure 1, Supplemental Figure 1), suggesting both reactions were performed identically and that their transferase binding affinities were similar. Thr/Ser rate ratios for each substrate pair of transferase reactions were obtained as described below.
Table I.
Random peptide Thr and Ser acceptor pairs
| Peptide set 1 | Rxn1 | GAGAXXXXXTXXXXXAGAGK |
| Rxn2 | GAGAXXXXXSXXXXXAGAGK | |
| X = G,A,P,R,E,V,Y,N | ||
| Peptide set 2 | Rxn1 | GAGAXXXTXXXAGAGK |
| Rxn2 | GAGAXXXSXXXAGAGK | |
| X = G,A,P,R,E,V,Y,Q,H,L | ||
| Peptide set 3 | Rxn1 | GAGAXXXTXXXAGAGK |
| Rxn2 | GAGAXXXSXXXAGAGK | |
| X = G,A,P,D,H,Q,I,M,K,F,W |
Fig. 1.
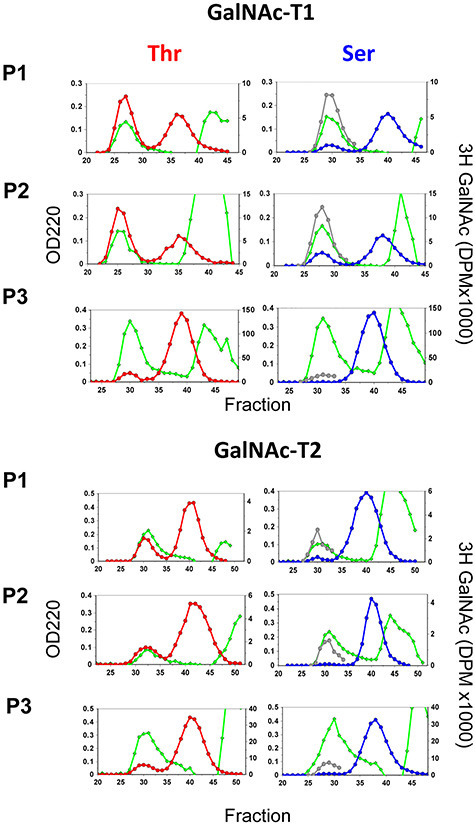
Product analysis of Thr and Ser random peptides glycosylated by GalNAc-T1 and -T2. Displayed are representative Sephadex G10 size exclusion chromatograms showing the separation of the 3H-GalNAc glycopeptide and UDP-3H-GalNAc hydrolysis products for transferase reactions against the three random peptide sets in Table I. Plotted are OD220 nm (green diamonds) representing peptide and buffers and 3H-DPM (red and blue circles, for the Thr and Ser random peptides, respectively) representing 3H-GalNAc glycosylated peptide (first peaks) and free 3H-GalNAc (middle peaks). Note that for the Ser peptides (right panels), the 3H-GalNAc DPM values for the glycopeptide peaks were also multiplied by the obtained Thr/Ser ratios (gray circle) for comparison to the Thr 3H-GalNAc plots (left panels). The identical OD220 nm plots for the random peptide peaks, and the identical free 3H-GalNAc DPM plots, for each of the Thr and Ser random peptide pairs suggests that each reaction pair was performed under identical conditions. Representative chromatograms of the remaining GalNAc-T reactions are given in Supplemental Figure 1.
GalNAc-T Thr/Ser substrate preference determination
We initially observed variability in our determined Thr/Ser rate ratios which upon examination inversely correlated with the extent of substrate peptide glycosylation. When these ratios from all three substrate pairs were plotted against the extent of the Thr peptide glycosylation, linear plots were obtained as shown in Figure 2, which were fit using least square analysis. This initially unexpected inverse correlation with substrate glycosylation is understood if one considers the random nature of the peptide substrates. As the glycosylation of the random peptide progresses, its rate of glycosylation will steadily slow as the transferase’s optimal peptide sequences are glycosylated first (i.e., most rapidly), leaving behind the less optimal and slower to glycosylate sequences. In addition the more rapidly reacting Thr acceptor substrates will deplete their optimal sequences faster than the Ser acceptor substrates; therefore together the extent of glycosylation (and apparent rate) of the Thr acceptor substrate will decrease more quickly than the Ser acceptor, leading to the lowering of the Thr/Ser ratio as glycosylation progresses. On this basis we have taken the value of this ratio extrapolated to zero percent conversion as the intrinsic Thr/Ser rate value, whose values are given in Table II. The magnitude of the slopes of the plots in Figure 2 (also given in Table II) is generally consistent with this explanation, where transferases with high Thr/Ser ratios (where the Thr acceptor would be glycosylated more rapidly than the Ser acceptor) commonly have steeper slopes (Table II) than those transferases that have low Thr/Ser values (where the rates are less different). In addition, these slopes may also reflect on a transferase’s peptide sequence specificity, where steeper slopes would suggest a stricter or narrower motif, while a less steep slope would reflect a transferase with a more relaxed or broader motif. Alternatively, the Thr/Ser ratios may depend on peptide sequence, where one might expect the optimal most active substrates to likely show the largest Thr/Ser rate differences compared to the poorer least active substrates. This possibility is addressed below.
Fig. 2.
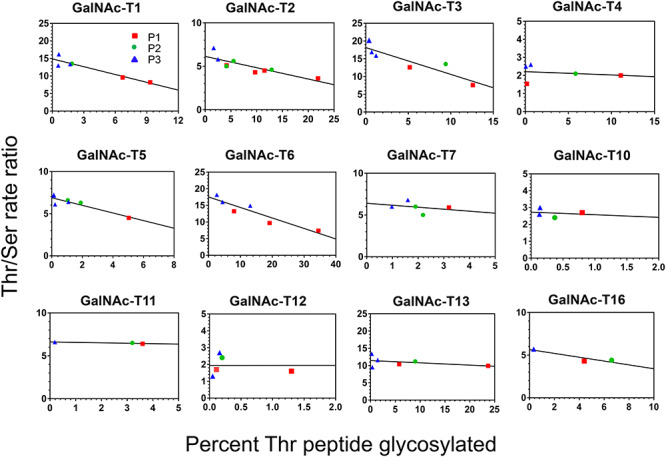
Individual Thr/Ser rate ratios inversely vary with extent of random peptide glycosylation. Shown are the individual Thr/Ser rate ratios plotted against the percent glycosylation of the Thr random peptide for 12 GalNAc-T isoenzymes. Note that the data for the three different random peptide sets (Set 1, P1 red squares; Set 2, P2 green circles; Set 3, P3 blue triangles) fit on the same linear plot which when extrapolated to zero percent glycosylation gives the intrinsic Thr/Ser rate ratio for each isoenzyme. The results of the linear regression analysis for the plots are given in Table II.
Table II.
Random peptide derived GalNAc-T isoenzyme Thr/Ser acceptor rate ratiosa
| Isoenzyme | Phylogenetic family | Intrinsic Thr/Ser ratiob | Slope | r 2 |
|---|---|---|---|---|
| GalNAc-T1 | Family Ia | 14.9 ± 0.7 | −0.7 ± 0.1 | 0.87 |
| GalNAc-T2 | Family Ib | 6.1 ± 0.3 | −0.1 ± 0.03 | 0.70 |
| GalNAc-T3 | Family Ic | 18.1 ± 1.2 | −0.7 ± 0.1 | 0.81 |
| GalNAc-T4 | Family IIa | 2.2 ± 0.3 | −0.01 ± 0.04 | n.ac |
| GalNAc-T5 | Family Id | 6.9 ± 0.2 | −0.4 ± 0.1 | 0.82 |
| GalNAc-T6 | Family Ic | 17.5 ± 1.1 | −0.3 ± 0.06 | 0.86 |
| GalNAc-T7 | Family IIb | 6.4 ± 0.9 | −0.2 ± 0.4 | n.ac |
| GalNAc-T10 | Family IIb | 2.7 ± 0.2 | −0.1 ± 0.5 | n.ac |
| GalNAc-T11 | Family If | 6.6 ± 0.1 | −0.04 ± 0.02 | 0.84 |
| GalNAc-T12 | Family IIa | 1.94 ± 0.3 | −0.009 ± 0.005 | n.ac |
| GalNAc-T13 | Family Ia | 11.90 ± 0.7 | −0.08 ± 0.05 | 0.4 |
| GalNAc-T16 | Family Ib | 5.65 ± 0.4 | −0.2 ± 0.09 | 0.83 |
Role of acceptor sequence/activity on Thr/Ser rate ratios
The extrapolated intrinsic Thr/Ser rate ratios obtained from our randomized peptide substrates assume that the Thr/Ser ratios do not change with acceptor peptide sequence or whether the acceptor is a good (i.e., optimal) or a poor (i.e., suboptimal) substrate. To confirm this, we performed assays using GalNAc-T3 and GalNAc-T12 against three nonrandom peptide substrates, displaying a range of activities against these transferases (Table III). These peptides were designed based on ISOGlyP predictions to be optimal GalNAc-T2, GalNAc-T3 and GalNAc-T12 substrates (see de las Rivas et al. 2020). The ISOGlyP predictions (i.e., enhancement value products, EVPs in Table III) for these three peptides against GalNAc-T3 and GalNAc-T12 suggest that each peptide would show a significantly different specific activity, which was borne out experimentally (Table III). Furthermore, the Thr/Ser rate ratios derived from the different peptides for both GalNAc-T3 and GalNAc-T12 do not greatly change with peptide specific activity and are consistent with the intrinsic Thr/Ser ratios obtained from the random peptide data (Table II), particularly when taking into account the experimental scatter within each data set (compare Figure 3 and Figure 2). For GalNAc-T3 the average Thr/Ser rate ratio from the peptides is ~21, while the random peptide derived intrinsic Thr/Ser ratio is ~18, and for GalNAc-T12 the peptide average Thr/Ser ratio is ~2.5, while its intrinsic Thr/Ser ratio is ~1.9 (Table III). These findings suggest that the Thr/Ser rate ratios remain remarkably constant over a range of substrate specific activity irrespective of peptide sequence for a given GalNAc-T isoenzyme. Furthermore, since we find that the Thr/Ser rate values are relatively constant within isoenzymes displaying the maximum and minimum observed Thr/Ser rate ratios, we will assume that the remaining isozymes will behave similarly. Therefore, we will take the extrapolated intrinsic Thr/Ser rate ratios to represent the Thr/Ser rate ratio for each of the GalNAc-T isoenzymes characterized (Figure 2 and Table II).
Table III.
GalNAc-T3 and GalNAc-T12 Thr/Ser rate ratios against fixed substrate peptides
| Optimal T3a | Rxn1 | GAGAYAVTPGPGAGA | |||||
| Rxn2 | GAGAYAVSPGPGAGA | ||||||
| Optimal T12 | Rxn1 | GAGAYYITPRPGAGA | |||||
| Rxn2 | GAGAYYISPRPGAGA | ||||||
| Optimal T2 | Rxn1 | GAGAPGPTPGPGAGA | |||||
| Rxn2 | GAGAPGPSPGPGAGA | ||||||
| Isoenzyme | Substrate | ISOGlyP prediction (EVP) b | Exp. specific activity c (min−1) | Peptide T/S rate ratio d | Avg Peptide T/S ratio e | Intrinsic T/S ratio (random peptide) | |
| GalNAc-T3 | Optimal substrate | Opt-T3 (T) | 106 | 335 ± 30 | 17.9 ± 1.7 | 21.5 ± 4.4 | 18.1 ± 1.2 |
| Opt-T3 (S) | 19 ± 3.1 | ||||||
| Sub optimal substrate | Opt-T12 (T) | 45 | 80 ± 8.8 | 26.8 ± 3.6 | |||
| Opt-T12 (S) | 3.0 ± 0.6 | ||||||
| Poor substrate | Opt-T2 (T) | 18 | 6.8 ± 0.8 | 20.1 ± 2.0 | |||
| Opt-T2 (S) | 0.34 ± 0.05 | ||||||
| GalNAc-T12 | Optimal substrate | Opt-T12 (T) | 74 | 6.4 ± 1.0 | 2.9 ± 0.3 | 2.6 ± 1.0 | 1.9 ± 0.3 |
| Opt-T12 (S) | 2.2 ± 0.6 | ||||||
| Sub optimal substrate | Opt-T3 (T) | 21 | 0.50 ± 0.05 | 3.6 ± 0.4 | |||
| Opt-T3 (S) | 0.14 ± 0.01 | ||||||
| Poor substrate | Opt-T2 (T) | 4.0 | 0.35 ± 0.12 | 1.3 ± 0.3 | |||
| Opt-T2 (S) | 0.33 ± 0.15 | ||||||
aGalNAc-T isoenzyme specific optimal peptide sequences predicted by the ISOGlyP predictor: http//isoglyp.utep.edu.
bISOGlyP Enhancement Value Products (EVP) that represent a substrates’ ability to be glycosylated.
cExperimental rates obtained using 0.7 mM peptides as described in the Methods. See Figure 3 for dot plots of the original data. Note units on rate are (mmole product/mmole enzyme)/min.
dAverage Thr/Ser rate ratios for the individual peptides shown in the dot plots of Figure 3.
eAverage of all of the Thr/Ser rate ratios plotted in the dot plots of Figure 3.
Fig. 3.
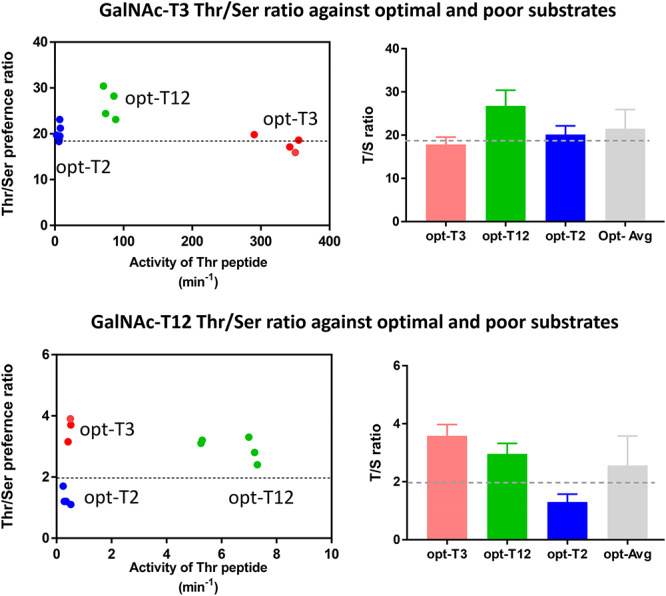
Thr/Ser rate ratios do not appreciably change with peptide sequence or intrinsic substrate activity for GalNAc-T3 or GalNAc-T12. Left Panels: Dot plots showing individually obtained Thr/Ser rate ratios plotted against relative substrate activity (in (moles product/mole transferase)/min) for the three unique peptides of different relative activity (see Table III). The dotted line through the plots represents the intrinsic Thr/Ser ratio obtained from the random peptide substrates (see Figure 2 and Table II). Note that for GalNAc-T12 the large difference in the Thr/Ser rate ratios for the opt-T2 and opt-T3 peptides is likely due to their very low activities and high rates of UDP-GalNAc hydrolysis. Right panels: Bar graphs showing the average Thr/Ser ratio obtained for each peptide substrate, and the average of all three, again a line representing the random peptide derived intrinsic Thr/Ser rate ratio for each transferase is also plotted. Averaged Thr/Ser rate ratios obtained from Table III. Data for the peptide substrates (see Table III) are color coded as red for opt-T3, green for opt-T12 and blue for opt-T2. Note the “opt” peptides refer to the transferase specific optimal peptides given in Table III as predicted by the ISOGlyP predictor http//isoglyp.utep.edu.
Thr/Ser ratios vary between GalNAc-T isoenzyme subfamily
We next compared the intrinsic Thr/Ser rate ratios with the GalNAc-T phylogenetic tree showing its subfamilies in Figure 4 (Bennett et al. 2012). It is noteworthy that the Thr/Ser rate ratios are similar within subfamilies but vary between distant subfamilies with values ranging from ~2 to ~18, a nearly 9-fold range in preferences for Thr over Ser. Interestingly, GalNAc-T1 and GalNAc-T13 in the Ia subfamily give ratios of ~12–15, while GalNAc-T3 and GalNAc-T6 in the closely related Ic subfamily give ratios of ~18, the highest that we have observed (Figure 4 and Table II). In contrast, GalNAc-T4 and GalNAc-T12 in the IIa subfamily, which are glycopeptide preferring transferases, give nearly identical ratios of ~2 which are the lowest observed ratios. GalNAc-T2, GalNAc-T16 (in subfamily Ib), GalNAc-T5 (in Id) and GalNAc-T11 (in 1f), all have nearly identical ratios of ~6. The only variation in Thr/Ser ratios within a subfamily is observed within the glycopeptide preferring subfamily IIb, where GalNAc-T7 has a value 6.4 while GalNAc-T10 is 2.7 (Figure 4 and Table I classes of GalNAc-T isoenzymes having different ranges of Thr/Ser rate ratios, ~13–18 (GalNAc-T1, GalNAc-T3, GalNAc-T6 and GalNAc-T13), ~6 (GalNAc-T2, GalNAc-T6, GalNAc-T7, GalNAc-T11 and GalNAc-T16) and ~2.0 (GalNAc-T4, GalNAc-T10 and GalNAc-T12).
Fig. 4.
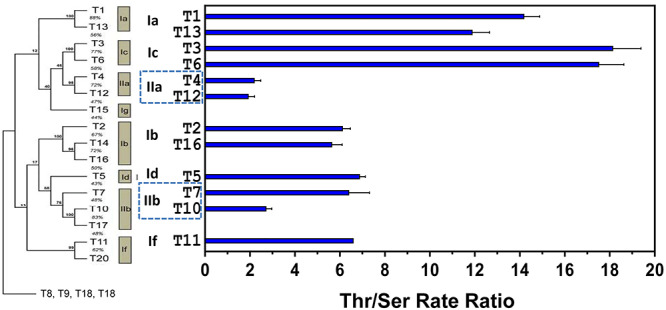
Intrinsic Thr/Ser rate ratios are similar within GalNAc-T subfamilies but can vary between GalNAc-T subfamilies. Shown is the human GalNAc-T phylogenetic tree (left) along with a bar graph plot (right) representing the obtained intrinsic Thr/Ser ratios for each of the studied GalNAc-T isoenzymes. The plot shows that the intrinsic Thr/Ser rate ratios are similar for isoenzymes belonging to same subfamily but vary between families. Note that the GalNAc-T phylogenetic tree excluded the subfamily 1e (at bottom), which contain the so-called “Y series” of GalNAc-Ts, (GalNAc-T8, GalNAc-T9, GalNAc-T18 and GalNAc-T19) that do not readily glycosylate Thr or Ser substrates.
Discussion
Our study confirms that as a class the GalNAc-T isoenzymes possess an inherent preference for Thr acceptor residues over Ser. Such Thr over Ser residue preferences have been observed for the original GalNAc-T preparations isolated from bovine colostrum and salivary glands where it was originally thought that separate transferases glycosylated Ser and Thr residues (O’Connell et al. 1992; Wang et al. 1992). This was corrected by subsequent studies that demonstrated the purified recombinant transferases could indeed glycosylate both Ser and Thr residues although at different rates (O’Connell and Tabak 1993; Wang et al. 1993). Elhammer and coworkers (Elhammer et al. 1999) using recombinant GalNAc-T1, GalNAc-T2 and GalNAc-T3 showed that these isoenzymes glycosylate the Ser EPO substrate (PPDAA (T/S)AAPL) at much lower rates than the Thr EPO substrate giving Vmax (and Vmax/Km) Thr/Ser ratios of ~11 (~99), ~6 (~30) and ~2 (~56) for GalNAc-T1, GalNAc-T2 and GalNAc-T3 respectively. Interestingly their Thr/Ser Vmax ratios are very similar to our intrinsic Thr/Ser rate ratios for GalNAc-T1 and GalNAc-T2 but significantly different for GalNAc-T3. Their findings on the multiple acceptor peptide pair PPAS (T/S) SAPG, however, gave much larger differences between Thr and Ser acceptors, which did not correlate with their EPO Thr/Ser peptide ratios. These differences likely arise from the presence of the multiple acceptor sites in the peptide competing nonproductively with binding at the catalytic domain, as the central Thr/Ser were shown predominantly glycosylated in both substrates. Thus, there is only partial agreement with our random peptide derived intrinsic Thr/Ser rate ratios and the kinetic parameters (i.e., Vmax) reported by Elhammer et al. (Elhammer et al. 1999). We do not fully understand the cause for this discrepancy, however there is the possibility that the GalNAc-T3 used by Elhammer et al. 1999 may have actually been GalNAc-T4, as we show GalNAc-T4 to have an intrinsic Thr/Ser ratio of 2.2. Since there are no methods reported in the Elhammer paper describing the origins or sequences of the transferases used in their work, we are unable to confirm this possibility. Nevertheless, our finding that the individual Thr/Ser ratios, obtained for all three of our random peptide substrates fit on the same line (which extrapolates to an “intrinsic” Thr/Ser rate ratio (Figure 2)), and from our studies with nonrandom peptides showing the Thr/Ser rate ratios are similar to the intrinsic values (and do not significantly change with the relative substrate activity (Figure 3)), strongly suggests that our intrinsic Thr/Ser values may be taken as a constant for each GalNAc-T isoenzyme against simple single acceptor peptide substrates.
Molecular basis of Thr/Ser preferences
Elhammer et al. (1999) have shown that allo-Thr acceptors are inactive against GalNAc-T1, from which they suggested the elevated catalytic efficiency for Thr residues was due to its methyl group binding to a specific pocket on the transferase catalytic domain. This binding would specifically orient the Thr acceptor hydroxyl for optimal transfer of GalNAc from the UDP-GalNAc donor, while the allo-Thr acceptor hydroxyl would be inappropriately oriented for efficient transfer (Elhammer et al. 1999). With this model a Ser acceptor would lack the conformational steering of the methyl group and possess an intermediate rate between Thr and allo-Thr acceptors. These workers further concluded that the Thr/Ser rate differences were not due to any significant increased binding affinity of the Thr substrate compared to the Ser substrates based on inhibition studies with nonacceptor peptides. Recent studies of Hurtado-Guerrero and coworkers (Lira-Navarrete et al. 2015) have suggested that the GalNAc-T methyl-binding pocket may further reduce the entropy of the acceptor, thereby enabling tighter substrate binding. Finally, Turupcu and coworkers (Turupcu et al. 2019) using comparative molecular dynamics studies of the PTP and PSP peptides in solution have shown that the Thr peptide acceptor possesses a constrained conformation that may be better oriented for GalNAc transfer when bound to a GalNAc-T than the Ser peptide acceptor. Thus both hydrophobic and steric interactions of the Thr methyl group likely enable Thr acceptors to adopt a more stable and optimally bound conformation for GalNAc transfer as compared to Ser acceptors.
In an attempt to understand how the GalNAc-T isoenzymes possess different Thr/Ser rates, we examined the available X-ray crystal structures of GalNAc-T2 (PDB: 2FFU) (Fritz et al. 2006), GalNAc-T3 (PDB: 6S24) (de las Rivas et al. 2020), GalNAc-T4 (PDB: 5QNA) (de las Rivas, Paul Daniel, et al. 2018) and GalNAc-T12 (PDB: 6PSU) (Fernandez et al. 2019) with tightly bound Thr acceptor substrates in catalytically active, flexible loop “closed”, conformations (Lira-Navarrete et al. 2015; Liu et al. 2017; de Las Rivas, Coelho, et al. 2018). An examination of these structures (Figure 5) reveals differences in polar contacts (hydrogen bonds) between the acceptor Thr and bound UDP and, more importantly, differences in the distances between the Thr methyl group and the hydrophobic residues of the transferase active site binding pocket thought to be responsible for the common (T/S)PXP motif (Revoredo et al. 2016) (see Table IV). As shown in Table IV, the Thr acceptor methyl groups are more tightly bound to the hydrophobic pocket of GalNAc-T3 and GalNAc-T2 compared to GalNAc-T4 and GalNAc-T12, based on the distances between the Thr methyl group and the side chain Phe and Trp residues making the hydrophobic pocket (Table IV and Figure 5). Thus the average hydrophobic interaction distances for GalNAc-T3 and GalNAc-T2 are 4.5 and 4.4 Å and for GalNAc-T4 and GalNAc-T12, 4.8 and 4.9 Å, respectively. Interestingly, the distance between the Thr methyl and the Phe located in the catalytic flexible loop (Phe335 in GalNAc-T3) most closely correlates with the observed Thr/Ser ratio for all four transferases, ranging from 3.8 Å for GalNAc-T3 to 5.1 Å for GalNAc-T12 (Table IV). On this basis we can reasonably suggest that the strength of the Thr methyl group binding to the active site pocket of the transferase largely accounts for the different Thr/Ser rate ratios observed for the GalNAc-Ts.
Fig. 5.
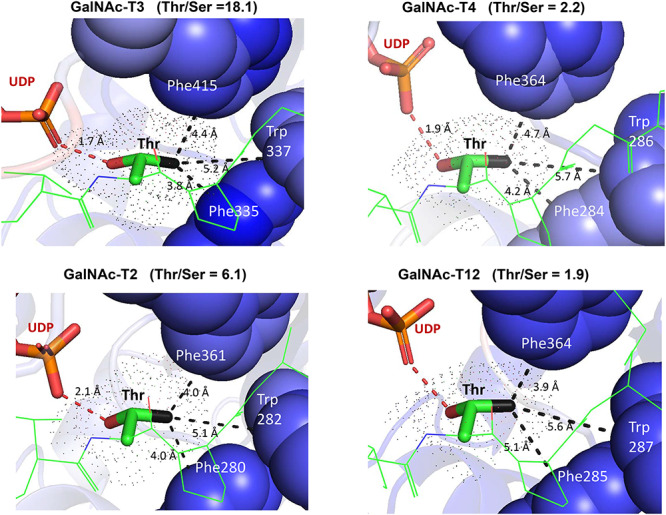
Thr acceptor-GalNAc-T catalytic domain contacts. Displayed are the zoomed in structures of GalNAc-T2 (PDB: 2FFU), GalNAc-T3 (PDB: 6S24), GalNAc-T4 (PDB: 5QNA) and GalNAc-T12 (PDB: 6PSU) with their bound Thr acceptor peptide substrate (green). The bound Thr residue is displayed in tube format (hydroxyl oxygen in red, methyl carbon in black) and its space filled surface is shown as dots. Shown (dashed lines) are hydrogen bond interactions of the Thr (OH group) with UDP; and hydrophobic contacts of the Thr methyl group with the Phe and Trp residues (space filled) of the transferase hydrophobic groove. See Table IV for bound substrate sequences and acceptor Thr contact distances to both UDP and transferase.
Table IV.
Thr acceptor interactions with UDP and the GalNAc-T catalytic domain peptide binding site correlate with intrinsic Thr/Ser rate ratios
| GalNAc-T substrate complex with Thr acceptor sequence (PBD accession) | Intrinsic Thr/Ser rate ratio | Acceptor Thr hydrogen bonds with UDPa | Hydrophobic interactions with acceptor Thr methyl groupb | |||
|---|---|---|---|---|---|---|
| GalNAc-T3 + ATGAGAGAGTTPGP (PDB: 6S24) | 18.1 | O-H:O=P 1.7 Å | Phe335 (ring C-2) 3.8 Å | Trp337 (ring C-3) 5.2 Å | Phe415 (ring C-5) 4.4 Å | Avg: 4.5 Å |
| GalNAc-T2 + STTPAPTTK (PDB: 2FFU) | 6.1 | N-H:O-P 2.1 Å | Phe228 (ring C-2) 4.0 Å | Trp282 (ring C-3) 5.1 Å | Phe361 (ring C-3) 4.0 Å | Avg: 4.4 Å |
| GalNAc-T4 + TGAGAGAGTTPGPG (PDB: 5QNA) | 2.2 | O-H:O-P 1.9 Å | Phe284 (ring C-2) 4.2 Å | Trp286 (ring C-3) 5.7 Å | Phe364 (ring C-5) 4.7 Å | Avg: 4.8 Å |
| GalNAc-T12 + GYYITPRTPGAGA (PDB: 6PSU) | 1.9 | O-H:O=P 2.9 Å | Phe285 (ring C-6) 5.1 Å | Trp287 (ring C-3) 5.6 Å | Phe364 (ring C-3) 3.9 Å | Avg: 4.9 Å |
aClosest polar interaction of the UDP terminal phosphate, P=O, to either the acceptor Thr OH or NH backbone. Hydrogen bonding (<2.1 Å) with UDP is likely for all isoenzymes except GalNAc-T12.
bClosest hydrophobic interactions (in Å) between the acceptor Thr methyl and GalNAc-T aromatic peptide binding residues. Note that these residues are responsible for the common GalNAc-T (T/S)PXP peptide binding motif (Revoredo et al. 2016).
Based on our prior studies, we have shown that each GalNAc-T isoenzyme contains a unique combination specificities based on peptide sequence and prior GalNAc glycosylation which imparts each isoenzyme with unique as well as overlapping properties. We have now shown, in the present work, that the isoenzymes also prefer Thr and Ser acceptor residues differently (i.e., Thr/Ser rate ratio) irrespective of the surrounding peptide sequence. This preference adds to the ever-increasing levels of specificity observed for the GalNAc-T family of isoenzymes and will be included into the next versions of the ISOGlyP O-glycosylation predictor. With this study we have further contributed to the understanding of why there is such a large family of GalNAc-Ts, which we believe is to have a diverse repertoire of isoenzymes capable of performing multiple unique and specific biological functions. Collectively our studies further suggest that mucin-type O-glycosylation may be a highly ordered and orchestrated process, whereby acceptor glycosylation by one isoenzyme produces a glycopeptide substrate specific for another isoenzyme. In this manner the altered expression or activity of a single transferase could cause downstream alterations in O-glycosylation which could potentially lead to disease.
Materials and methods
Reagents and random peptide substrates
Random peptide substrate pairs (Table I) containing Thr or Ser acceptors were custom synthesized by Quality Controlled Biochemicals (QCB) (Hopkinton, MA) and Sussex Research (Ottawa, ON, Canada). Each random peptide substrate pair was synthesized at the same time to ensure identical compositions which were confirmed by Edman amino acid sequencing. Fixed sequence peptide pairs were synthesized by RS Synthesis (Louisville, KY). Stock solutions of 50 mg/mL (~20 mM) of each (random) peptide substrate were prepared after lyophilizing from water several times and adjusting to pH 7 with dilute NaOH/HCl. Fully N-acetylated UDP-3H-GalNAc was purchased from American Radiolabeled Chemicals, Inc. (St Louis, MO), while nonlabeled UDP-GalNAc was obtained from Sigma Aldrich (St Louis, MO). Stock solutions of radiolabeled 20 mM UDP-GalNAc were prepared by adding stock UDP-3H-GalNAc to cold 20 mM UDP-GalNAc giving ~6 × 108 DPM/μmole. Liquid scintillation counting was performed using a Beckman Model LS650. Dowex 1 × 8 anion exchange resin (100–200 mesh) was purchased from Acros Organics (FisherScientific), and Sephadex G10 was obtained from Sigma Aldrich (St Louis, MO).
Transferases
As in our previous work, human GalNAc-Ts were obtained from multiple sources as soluble N-terminal truncated purified enzymes (Revoredo et al. 2016). GalNAc-T1, GalNAc-T2, GalNAc-T3, GalNAc-T5, GalNAc-T7, GalNAc-T11, GalNAc-T12, GalNAc-T13 and GalNAc-T16 were expressed from High Five insect cells (Wandall et al. 1997; Bennett et al. 1996) a gift of Henrik Clausen, University of Copenhagen. Purified GalNAc-T4 and GalNAc-T10 were expressed in mammalian HEK293f (Gerken et al. 2013; Revoredo et al. 2016) gift of Kelley Moremen, Complex Carbohydrate Research Center, University of Georgia. GalNAc-T2 expressed in Pichia pastoris (Gerken et al. 2011) was a gift from Larry Tabak, NIDCR National Institutes of Health, Bethesda, MD. We also obtained purified GalNAc-T2, GalNAc-T3, GalNAc-T4 and GalNAc-T6 expressed in P. pastoris (de las Rivas et al. 2017, 2020; de las Rivas, Paul Daniel, et al. 2018) as a gift of Ramon Hurtado-Guerrero, Zaragoza, Spain. Note that experiments completed using transferases from different sources gave identical results.
Glycosylation reactions
For the random substrate assays, each Thr and Ser peptide of a given peptide pair (Table I) was glycosylated at the same time under identical reaction conditions. Reactions contained the following: 50 mM sodium cacodylate buffer, pH 6.8, 1 mM β-mercaptoethanol, 0.1% Triton X-100, 10 mM MnCl2, 2 mM UDP-[3H]GalNAc (~6 × 108 DPM/μmole), 2.5 mM peptide (~75 nMol) and 5–20 uL GalNAc-T to a final volume of 50 μL to 70 μL in capped Eppendorf tubes. Reaction mixtures were incubated at 37 °C in a shaking microincubator. The amount of transferase to use was determined by trial and error based on the activity and concentration of each GalNAc-T isoform stock. Reaction times ranged from 2 h to overnight and were typically optimized for less than 10% glycosylation of the Thr peptide. Following incubation, reactions were quenched by the addition of 1 volume of 250 mM EDTA. Typically, reactions were performed with all three sets of peptide substrates at a given time with the same transferase concentrations and UDP-[3H]-GalNAc stock. Reactions were typically repeated two or more times with each peptide set; however, due to limited amounts of GalNAc-T11 and GalNAc-T16 and random peptide Set 2, fewer than two determinations per peptide set were performed for these.
Product purification by anion exchange and gel filtration chromatography
UDP and nonhydrolyzed UDP-GalNAc were removed from the quenched reactions (after dilution to 3.0–5.0 mL with water) by passage over a column of ~3 mL of Dowex 1 × 8 anion exchange resin in the Cl− form. Total transferase activity (i.e., 3H-GalNAc transfer to substrate and water) was determined from the difference in 3H-GalNAc content before and after passing over the Dowex column. As many GalNAc-T reactions show nonproductive transfer to water (i.e., UDP-GalNAc hydrolysis) (Gerken et al. 2013; Revoredo et al. 2016), productive 3H-GalNAc transferred to peptide was determined after isolating the glycopeptide on Sephadex G10 gel filtration chromatography (1.5 × 105 cm column, in 50 mM acetic acid pH 4.5 with NH4OH) as shown in Figure 1 and Supplemental Figure 1. The G10 column fractions were monitored for 3H-DPM and OD220 and OD280 nm and the 3H-glycopeptide (productive transfer) peaks pooled and lyophilized. The percent 3H-peptide formation was determined from the summed 3H-content of the peptide product peak and the summed 3H-content of the free GalNAc peak. The ratio of the Thr/Ser 3H-peptide product was then taken as the Thr/Ser rate ratio. The Thr/Ser rate ratio was also calculated from the lyophilized glycopeptide products. In this case lyophilized peaks were dissolving in 1.00 mL water and their OD220 measured and a 0.5 mL aliquot taken for measuring 3H-GalNAc content by scintillation counting. These GalNAc-3H-DPM/OD220 ratios were then used to obtain another measure of the Thr/Ser rate ratio for each reaction pair. The Thr/Ser rate ratios obtained via these two methods were then averaged to obtain the values plotted in Figure 2. The difference between the two calculation methods did not differ by more than 0.7 for any given peptide reaction pair. Finally, for each GalNAc-T, the obtained Thr/Ser rate ratios were plotted vs. the percent Thr peptide glycosylation (Figure 2) from which the intrinsic Thr/Ser rate ratios were obtained from the y intercepts (representing 0% glycosylation). The data were linear least square fit and plotted by GraphPad Prism 7.05. The obtained intrinsic Thr/Ser rate ratios and additional fitting parameters are given in Table II.
For the nonrandom peptide substrates, each Thr and Ser peptide of a given peptide pair were glycosylated under identical conditions (50 mM sodium cacodylate buffer, pH 6.8, 1 mM β-mercaptoethanol, 0.1% Triton X-100, 10 mM MnCl2, 2 mM UDP-[3H]GalNAc (~6 × 108 DPM/μmole), 0.7 mM peptide in 20 uL reactions) using the same enzyme mix but reacted for different time points to compensate for their different rates of reaction. Reaction rates were calculated by dividing the moles of glycopeptide product (based on 3H DPM from Sephadex G10 chromatography) by the incubation time. Peptide substrate specific activity was obtained by further normalizing the rate to enzyme concentration. Triplicate or greater determinations were performed for GalNAc-T3 and GalNAc-T12 against each peptide set.
Molecular modeling
GalNAc-T crystal structures with Thr acceptor substrate bound of GalNAc-T2 (PDB: 2FFU), GalNAc-T3 (PDB: 6S24), GalNAc-T4 (PDB: 5QNA) and GalNAc -T12 (PDB: 6PSU) were compared and analyzed (i.e., atom to atom distances) using Pymol 2.3 version (64 bit).
Abbreviations
- GalNAc
N-Acetylgalactosamine
- GalNAc-T
UDP-GalNAc:polypeptide N-α-acetylgalactosaminyl-transferases
- UDP
uridine diphosphate
Supplementary Material
Acknowledgements
We would like to thank the following individuals for providing the transferases utilized in this work: Malene Bech Vester-Christensen, Yun Kong and Shengjun Wang (H. Clausen Laboratory) and Roy W. Johnson, Shuo Wang and Heather A. Moniz (K. Moremen Laboratory).
Funding
This work was supported by grants from the National Institutes of Health: R01-GM113534 (to T.A.G.), P41-GM103390 (to K. Moremen, Complex Carbohydrate Research Center, University of Georgia), P01-GM107012 (to G.J. Boons, Complex Carbohydrate Research Center, University of Georgia) and NIDCR intramural program grant 1-ZIA-DE000739-05 (to L. Tabak, National Institutes of Health, Bethesda MD). Funding by the Danish National Research Foundation (DNRF107) is also acknowledged (to H. Clausen, Univ. Copenhagen). We thank ARAID, MEC (BFU2016-75633-P to R. Hurtado-Guerrero) and Gobierno de Aragón (E34_R17 and LMP58_18 to R. Hurtado-Guerrero) with FEDER (2014–2020) funds for “Building Europe from Aragón” for financial support.
Conflict of interest statement
None declared.
References
- Balcik-Ercin P, Cetin M, Yalim-Camci I, Odabas G, Tokay N, Sayan AE, Yagci T. 2018. Genome-wide analysis of endogenously expressed ZEB2 binding sites reveals inverse correlations between ZEB2 and GalNAc-transferase GALNT3 in human tumors. Cell Oncol (Dordr). 41:379–393. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Clausen H. 1996. cDNA cloning and expression of a novel human UDP-N-acetyl-alpha-d-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-t3. J Biol Chem. 271:17006–17012. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. 2012. Control of Mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 22:736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berois N, Mazal D, Ubillos L, Trajtenberg F, Nicolas A, Sastre-Garau X, Magdelenat H, Osinaga E. 2006. UDP-N-acetyl-d-Galactosamine:polypeptide N-acetylgalactosaminyltransferase-6 as a new Immunohistochemical breast cancer marker. J Histochem Cytochem. 54:317–328. [DOI] [PubMed] [Google Scholar]
- Christensen H, Christiansen MS, Petersen J, Jensen HH. 2008. Direct formation of beta-glycosides of N-acetyl glycosamines mediated by rare earth metal triflates. Org Biomol Chem. 6:3276–3283. [DOI] [PubMed] [Google Scholar]
- de las Rivas M, Coelho H, Diniz A, Lira-Navarrete E, Compa I, Barbero J, Schjoldager KT, Bennett EP, Vakhrushev SY, Clausen H et al. 2018. Structural analysis of a GalNAc-T2 mutant reveals an induced-fit catalytic mechanism for GalNAc-Ts. Chem Eur J. 24:8382–8392. [DOI] [PubMed] [Google Scholar]
- de las Rivas M, Lira-Navarrete E, Paul Daniel EJ, Companon I, Coelho H, Diniz A, Jimenez-Barbero J, Peregrina JM, Clausen H, Corzana F et al. 2017. The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences. Nat Commun. 8: Article number 1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Rivas M, Lira-Navarrete E, Gerken TA, Hurtado-Guerrero R. 2019. Polypeptide GalNAc-Ts: From redundancy to specificity. Curr Opin Struct Biol. 56:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Rivas M, Paul Daniel EJ, Coelho H, Lira-Navarrete E, Raich L, Companon I, Diniz A, Lagartera L, Jimenez-Barbero J, Clausen H et al. 2018. Structural and mechanistic insights into the catalytic-domain-mediated short-range glycosylation preferences of GalNAc-T4. ACS Cent Sci. 4:1274–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Rivas M, Paul Daniel EJ, Narimatsu Y, Compañón I, Kato K, Hermosilla P, Thureau A, Ceballos-Laita L, Coelho H, Bernadó P et al. 2020. Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3. Nat Chem Biol. 16:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCicco RePass MA, Bhat N, Heimburg-Molinaro J, Bunnell S, Cummings RD, Ward HD. 2018. Molecular cloning, expression, and characterization of UDP N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 from Cryptosporidium parvum. Mol Biochem Parasitol. 221:56–65. [DOI] [PubMed] [Google Scholar]
- Elhammer AP, Poorman RA, Brown E, Maggiora LL, Hoogerheide JG, Kezdy FJ. 1993. The specificity of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase as inferred from a database of in vivo substrates and from the in vitro glycosylation of proteins and peptides. J Biol Chem. 268:10029–10038. [PubMed] [Google Scholar]
- Elhammer ÅP, Kézdy FJ, Kurosaka A. 1999. The acceptor specificity of UDP-GaINAc:polypeptide N-acetylgalactosaminyltransferases In: Berger EG, Clausen H, Cummings RD, editors. Glycotechnology. Boston, MA: Springer US. [DOI] [PubMed] [Google Scholar]
- Ercan A, West CM. 2005. Kinetic analysis of a Golgi UDP-GlcNAc:polypeptide-Thr/Ser N-acetyl-{alpha}-glucosaminyltransferase from Dictyostelium. Glycobiology. 15:489–500. [DOI] [PubMed] [Google Scholar]
- Fernandez AJ, Paul Daniel EJ, Mahajan SP, Gray JJ, Gerken TA, Tabak LA, Samara NL. 2019. The structure of the colorectal cancer-associated enzyme GalNAc-T12 reveals how nonconserved residues dictate its function. Proc Natl Acad Sci. 116:20404–20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire T, Nora B, Sonora C, Varangot M, Barrios E, Osinaga E. 2006. UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a potential new marker for detection of bone marrow-disseminated breast cancer cells. Int J Cancer. 6:1383–1388. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Hurley JH, Trinh LB, Shiloach J, Tabak LA. 2004. The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-T1. Proc Natl Acad Sci USA. 101:15307–15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA. 2006. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. J Biol Chem. 281:8613–8619. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA. 2011. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J Biol Chem. 286:14493–14507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Raman J, Fritz TA, Jamison O. 2006. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 281:32403–32416. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Revoredo L, Thome JJ, Tabak LA, Vester-Christensen MB, Clausen H, Gahlay GK, Jarvis DL, Johnson RW, Moniz HA et al. 2013. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J Biol Chem. 288:19900–19914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen, Jamison O. 2008. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology. 18:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DJ, Tham KM, Chia J, Wang SC, Steentoft C, Clausen H, Bard-Chapeau EA, Bard FA. 2013. Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proc Natl Acad Sci. 110:E3152–E3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goth CK, Vakhrushev SY, Joshi HJ, Clausen H, Schjoldager KT. 2018. Fine-tuning limited proteolysis: A major role for regulated site-specific O-glycosylation. Trends Biochem Sci. 43:269–284. [DOI] [PubMed] [Google Scholar]
- Guo JM, Chen HL, Wang GM, Zhang YK, Narimatsu H. 2004. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-12 in gastric and colonic cancer cell lines and in human colorectal cancer. Oncology. 67:271–276. [DOI] [PubMed] [Google Scholar]
- Hansen LH, Madsen TD, Goth CK, Clausen H, Yang C, Dzhoyashvili N, Iyer SR, Jeson Sangaralingham S, Burnett JC, Rehfeld JF et al. 2019. Discovery of O-glycans on atrial natriuretic peptide (ANP) that affect both its proteolytic degradation and potency at its cognate receptor. J Biol Chem. 294:12567–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N, Singh D, Wel H, Sassi SO, Johnson JM, Feasley CL, Koeller CM, Previato JO, Mendonca-Previato L, West CM. 2009. Molecular analysis of a UDP-GlcNAc:polypeptide-N-acetylglucosaminyltransferase implicated in the initiation of mucin-type O-glycosylation in Trypanosoma cruzi. Glycobiology. 19:918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleboom A, Helen Karlsson G, Lin RS, Thomas MB, Jeroen AS, Daniel SH, Erik SS, Johannes MA, John JK, Mohammad MM et al. 2011. Heterozygosity for a loss-of-function mutation in GALNT2 improves plasma triglyceride clearance in man. Cell Metab. 14:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horynová MS, Raška M, Clausen H, Novak J. 2013. Aberrant O-glycosylation and anti-glycan antibodies in an autoimmune disease IgA nephropathy and breast adenocarcinoma. Cell Mol Life Sci. 70:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Samara NL, Revoredo L, Zhang L, Tran DT, Muirhead K, Tabak LA, Ten Hagen. 2018. A molecular switch orchestrates enzyme specificity and secretory granule morphology. Nat Commun. 9:3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi HJ, Hansen L, Narimatsu Y, Freeze HH, Henrissat B, Bennett E, Wandall HH, Clausen H, Schjoldager KT. 2018. Glycosyltransferase genes that cause monogenic congenital disorders of glycosylation are distinct from glycosyltransferase genes associated with complex diseases. Glycobiology. 28:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. 2006. Polypeptide GaINAc-transferase T3 and familial tumoral calcinosis: Secretion of FGF23 requires O-glycosylation. J Biol Chem. 281:18370–18377. [DOI] [PubMed] [Google Scholar]
- Khetarpal S, Katrine T Schjoldager A, Christoffersen C, Raghavan A, Andrew CE, Reutter HM, Ahmed B, Ouazzani R, Peloso GM, Vitali C et al. 2016. Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SL, Goth CK, Eckhard U, Joshi HJ, Haue AD, Vakhrushev S, Schjoldager K, Overall CM, Wandall HH. 2018. TAILS N-Terminomics and proteomics reveal complex regulation of proteolytic, cleavage by O-glycosylation. J Biol Chem. 293:7629–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaki T, Nishimori I, Nakayama H, Miyazaki E, Enzan H, Nomoto M, Hollingsworth MA, Onishi S. 2000. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isozymes T1 and T2 in human colorectal cancer. J Gastroenterol. 35:840–848. [DOI] [PubMed] [Google Scholar]
- Kong Y, Joshi HJ, Schjoldager KT-BG, Madsen TD, Gerken TA, Vester-Christensen MB, Wandall HH, Bennett EP, Levery SB, Vakhrushev SY et al. 2015. Probing polypeptide GalNAc-Transferase isoform substrate specificities by in vitro analysis. Glycobiology. 25:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita T, Thi TN, Koi CHIH, Murakami MIDO, Kagami SEIJ, Izumi HIRO, Hachisuga TORU. 2017. Expression of N-acetylgalactosaminyltransferase-6 is related to expression of cell adhesion molecules in endometrial cancer. Anticancer Res. 37:3905–3910. [DOI] [PubMed] [Google Scholar]
- Lavrsen K, Dabelsteen S, Vakhrushev SY, Levann AMR, Haue AD, Dylander A, Mandel U, Hansen L, Frodin M, Bennett EP et al. 2018. De novo expression of human polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) in colon adenocarcinoma inhibits the differentiation of colonic epithelium. J Biol Chem. 293:1298–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-C, Chen S-T, Huang M-C, Huang J, Hsu C-L, Juan H-F, Lin H-H, Chen C-H. 2017. GALNT6 expression enhances aggressive phenotypes of ovarian cancer cells by regulating EGFR activity. Oncotarget. 8:42588–42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Navarrete E, Matilde las Rivas IC, Pallares MC, Kong Y, Iglesias-Fernandez J, Bernardes GJL, Peregrina JM, Rovira C, Bernado P, Bruscolini P et al. 2015. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat Commun. 6: Article number: 6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xu K, Xu Z, Rivas M d l, Li X, Lu J, Delso I, Merino P, Hurtado-Guerrero R, Zhang Y. 2017. The small molecule luteolin inhibits N-acetyl alpha-galactosaminyltransferases and reduces mucin-type O-glycosylation of amyloid precursor protein. J Biol Chem. 292:21304–21319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca-Previato L, Penha L, Garcez TC, Jones C, Previato JO. 2013. Addition of O-GlcNAc to threonine residues define the post-translational modification of mucin-like molecules in Trypanosoma cruzi. Glycoconj J. 30:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Kurosaka A. 2019. Mucin-type glycosylation as a regulatory factor of amyloid precursor protein processing. J Biochem. 165:205–208. [DOI] [PubMed] [Google Scholar]
- Niang B, Jin L, Chen X, Guo X, Zhang H, Wu Q, Padhiar AA, Xiao M, Fang D, Zhang J. 2016. GalNAc-T4 putatively modulates the estrogen regulatory network through FOXA1 glycosylation in human breast cancer cells. Mol Cell Biochem. 411:393–402. [DOI] [PubMed] [Google Scholar]
- Nishikimi T, Nakagawa Y, Minamino N, Ikeda M, Tabei K, Fujishima A, Takayama K, Akimoto K, Yamada C, Nakao K et al. 2015. Pro-B-type natriuretic peptide is cleaved intracellularly: Impact of distance between O-glycosylation and cleavage sites. Am J Physiol Regul Integr Comp Physiol. 309:R639–R649. [DOI] [PubMed] [Google Scholar]
- O'Connell BC, Hagen FK, Tabak LA. 1992. The influence of flanking sequence on the O-glycosylation of threonine in vitro. J Biol Chem. 267:25010–25018. [PubMed] [Google Scholar]
- O'Connell BC, Tabak LA. 1993. A comparison of serine and threonine O-glycosylation by UDP-GaINAc:polypeptide N-acetylgalactosaminyltransferase. J Dent Res. 72:1554–1558. [DOI] [PubMed] [Google Scholar]
- Perrine C, Tongzhong J, Cummings RD, Gerken TA. 2009. Systematic determination of the peptide acceptor preferences for the human UDP-Gal:glycoprotein-alpha-GalNAc beta 3 galactosyltranferase (T-synthase). Glycobiology. 19:321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Lu M, Tabak LA. 2008. The catalytic and lectin domains of UDP- GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J Biol Chem. 283:22942–22951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Yu G, Perrine CL, Gerken TA, Tabak LA. 2012. UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases: Completion of the family tree. Glycobiology. 22:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily C, Stewart TJ, Renfrow MB, Novak J. 2019. Glycosylation in health and disease. Nat Rev Nephrol. 15:346–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers N, Anderson JM, Linde E, DiMaio DJ, Lazenby A, Hans W, Mandell U, Clausen H, Yu F, Hollingsworth M. 2013. Aberrant expression of mucin core proteins and O-linked glycans associated with progression of pancreatic cancer. Clin Cancer Res. 18:1981–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revoredo L, Wang S, Bennett EP, Clausen H, Moremen KW, Jarvis DL, Ten Hagen, Tabak LA, Gerken TA. 2016. Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology. 26:360–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Vester-Christensen MB, Bennett EP, Levery SB, Schwientek T, Yin W, Blixt O, Clausen H. 2010. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J Biol Chem. 285:36293–36303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT-B, Clausen H. 2012. Site-specific protein O-glycosylation modulates proprotein processing -deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta Gen Subj. 1820:2079–2094. [DOI] [PubMed] [Google Scholar]
- Schwientek TJ, Bennett EP, Flores C, Thacker J, Hollman M, Reis CA, Behrens J, Mandel U, Keck B, Schafer MA et al. 2002. Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, C. elegans and mammals: One subfamily comprised of l(2)35Aa is essential in Drosophila. J Biol Chem. 277:22623–22638. [DOI] [PubMed] [Google Scholar]
- Semenov AG, Postnikov AB, Tamm NN, Seferian KR, Karpova NS, Bloshchitsyna MN, Koshkina EV, Krasnoselsky MI, Serebryanaya DV, Katrukha AG. 2009. Processing of pro-brain natriuretic peptide is suppressed by O-glycosylation in the region close to the cleavage site. Clin Chem. 55:489–498. [DOI] [PubMed] [Google Scholar]
- Sheta R, Bachvarova M, Macdonald E, Gobeil S, Vanderhyden B, Bachvarov D. 2019. The polypeptide GALNT6 displays redundant functions upon suppression of its closest homolog GALNT3 in mediating aberrant O-glycosylation, associated with ovarian cancer progression. Int J Mol Sci. 20:2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-B, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L et al. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32:1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E, Stevens SR, Guan Y, Springer DA, Anderson SA, Starost MF, Patel V, Ten Hagen, Tabak LA. 2015. Galnt1 is required for normal heart valve development and cardiac function. PLoS One. 10:e0115861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E, Ten Hagen. 2006. Expression of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology. 16:83–95. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen. 2007. O-linked glycan expression during Drosophila development. Glycobiology. 17:820–827. [DOI] [PubMed] [Google Scholar]
- Tomita T, Sugi T, Yakubu R, Vincent T, Ma Y, Weiss LM. 2017. 'Making home sweet and sturdy: Toxoplasma gondii ppGalNAc-Ts glycosylate in hierarchical order and confer Cyst Wall rigidity. mBio. 8:e02048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V et al. 2004. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet. 36:579–581. [DOI] [PubMed] [Google Scholar]
- Tran DT, Zhang L, Ying Z, Tian E, Earl LA, Ten Hagen. 2012. Multiple members of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J Biol Chem. 287:5243–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turupcu A, Diem M, Smith L, Oostenbrink C. 2019. Structural aspects of the O-glycosylation linkage in glycopeptides via MD simulations and comparison with NMR experiments. ChemPhysChem. 20:1527–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero-González J, Leonhard-Melief C, Lira-Navarrete E, Jiménez-Osés G, Hernandez-Ruiz C, Pallares MC, Yruela I, Vasudevan D, Lostao A, Corzana F et al. 2016. A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat Chem Biol. 12:240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wel H, Johnson JM, Xu Y, Karunaratne CV, Wilson KD, Vohra Y, Boons GJ, Taylor CM, Bendiak B, West CM. 2011. Requirements for Skp1 processing by cytosolic Prolyl 4(trans)-hydroxylase and α-N-acetylglucosaminyltransferase enzymes involved in O2 signaling in Dictyostelium. Biochemistry. 50:1700–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall HH, Hassan H, Mirgorodskaya E, Kristensen AK, Roepstorff P, Bennett EP, Nielsen PA, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J et al. 1997. Substrate specificities of three members of the human UDP-N-acetyl- alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J Biol Chem. 272:23503–23514. [DOI] [PubMed] [Google Scholar]
- Wang S, Yang M, Narimatsu Y, Ye Z, Tian W, Goth CK, Lira-Navarrete E, Pedersen NB, Benito-Vicente A, Martin C et al. 2018. Site-specific O-glycosylation of members of the low-density lipoprotein receptor superfamily enhances ligand interactions. J Biol Chem. 293:7408–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Abernethy JL, Eckhardt AE, Hill RL. 1992. Purification and characterization of a UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase specific for glycosylation of threonine residues. J Biol Chem. 267:12709–12716. [PubMed] [Google Scholar]
- Wang Y, Agrwal N, Eckhardt AE, Stevens RD, Hill RL. 1993. The acceptor substrate specificity of porcine submaxillary UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is dependent on the amino acid sequences adjacent to serine and threonine residues. J Biol Chem. 268:22979–22983. [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 40:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Williams Parsons D, Jones S, Lin J, Sjoblom T, Leary RJ, Dong S, Boca SM, Barber T, Ptak J et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science. 318:1108–1113. [DOI] [PubMed] [Google Scholar]
- Wu C, Guo X, Wang W, Wang Y, Shan YJ, Zhang B, Song W, Ma S, Ge J, Deng H et al. 2010. N-Acetylgalactosaminyltransferase-14 as a potential biomarker for breast cancer by immunohistochemistry. BMC Cancer. 10:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ten Hagen. 2019. O-Linked glycosylation in Drosophila melanogaster. Curr Opin Struct Biol. 56:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


