Abstract
The stria vascularis (SV) in the cochlea generates and maintains the endocochlear potential, thereby playing a pivotal role in normal hearing. Knowing transcriptional profiles and gene regulatory networks of SV cell types establishes a basis for studying the mechanism underlying SV-related hearing loss. While we have previously characterized the expression profiles of major SV cell types in the adult mouse, transcriptional profiles of rare SV cell types remained elusive due to the limitation of cell capture in single-cell RNA-Seq. The role of these rare cell types in the homeostatic function of the adult SV remain largely undefined. In this study, we performed single-nucleus RNA-Seq on the adult mouse SV in conjunction with sample preservation treatments during the isolation steps. We distinguish rare SV cell types, including spindle cells and root cells, from other cell types, and characterize their transcriptional profiles. Furthermore, we also identify and validate novel specific markers for these rare SV cell types. Finally, we identify homeostatic gene regulatory networks within spindle and root cells, establishing a basis for understanding the functional roles of these cells in hearing. These novel findings will provide new insights for future work in SV-related hearing loss and hearing fluctuation.
Subject terms: Computational biology and bioinformatics, Molecular biology, Auditory system, Computational neuroscience, Ion channels in the nervous system
Introduction
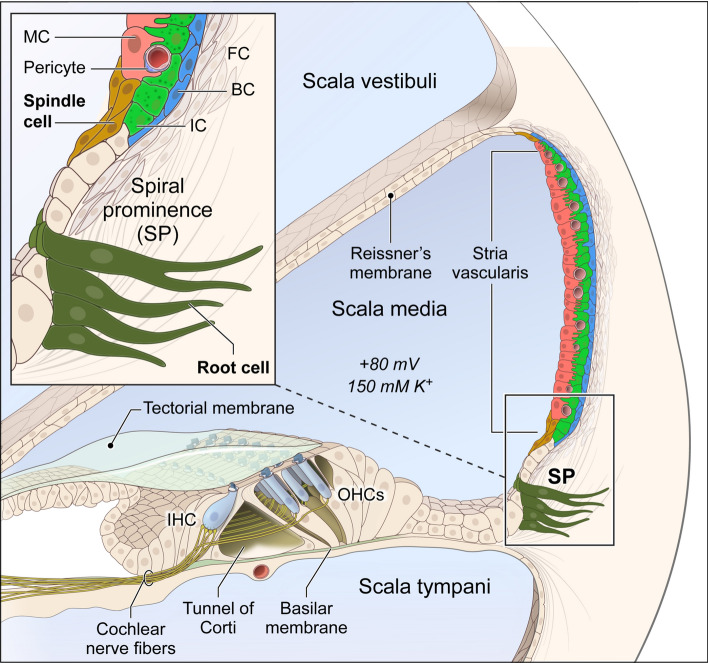
The stria vascularis (SV) is a heterogenous tissue located in the lateral wall of the cochlear duct. It is a stratified epithelium consisting of three major cell types (marginal, intermediate and basal cells), as well as other rare cell types, including spindle cells, macrophages, endothelial cells, and pericytes (Fig. 1). Marginal cells face the endolymph, while the basal cells abut the connective tissue of the spiral ligament. The marginal and basal cell layers sandwich the layer of intermediate cells, which interdigitate with these two cell types1,2. These cells work together to produce and maintain the endocochlear potential (EP), which is necessary for normal hearing3–6. Mutations in genes expressed in major SV cell types, such as Kcnq1, Kcne1, Kcnj10 and Cldn11, are known to cause deafness and/or SV dysfunction including loss of EP6–10. Spindle cells, a rare SV cell type, are found at the superior and inferior borders of the SV and have been implicated in hearing loss and hearing fluctuation11–13. Other rare SV cell types including macrophages, pericytes, and endothelial cells11,14,15, as well as rare cell types adjacent to the SV including root cells16,17, have roles that remain incompletely defined as they relate to EP generation and ion homeostasis in the cochlea. Therefore, understanding the transcriptional profiles and underlying gene regulatory networks in rare SV cell types from the unperturbed adult SV will be critical for mechanistic studies of the role that rare cell types play in EP generation and ion homeostasis in the cochlea.
Figure 1.
Illustration of adult mouse cochlea. Major stria vascularis cell types (marginal (MC), intermediate (IC) and basal (BC) cells) are colored as red, green and blue, respectively. Rare root cells just outside the stria vascularis and rare spindle cells in the stria vascularis are in magnified view, and colored as dark olive green and dark gold, respectively.
Our group has previously characterized transcriptional profiles of the major cell types in the adult mouse SV by single-cell RNA-Seq (scRNA-Seq) and single-nucleus RNA-Seq (snRNA-Seq)2. However, due to the low number of rare cell types captured, transcriptional profiles for rare SV cell types, specifically spindle and root cells, remained undefined. Previously, we and others have shown that that snRNA-Seq decreases cell size and shape heterogeneity, enabling more comprehensive transcriptional profiling of the tissues with heterogeneity in cell size and shape2,18–22. Despite the more comprehensive picture provided by snRNA-Seq, transcriptional profiles of rare SV cell types, specifically spindle and root cells, remained poorly distinguished in our previous study2. Recently in other organ systems, sample preservation methods have been utilized to improve capture of rare cell types and we hypothesized that a similar approach might facilitate transcriptional profiling of rare SV cell types23–25. In this study, we utilize two sample preservation methods, methanol fixation and RNAlater treatment, in snRNA-Seq to characterize rare cell transcriptional profiles in the adult SV. We define spindle cell transcriptional profiles and distinguish them clearly from those of root cells in the lateral wall of the cochlea. In the process, we compare these methods to the previously published adult SV snRNA-Seq. Finally, we define gene regulatory networks involved in homeostatic functions, referring to functions of the unperturbed adult SV, of spindle and root cells which was not possible with our previously published dataset2.
Results
Sample preservation process improves snRNA-Seq data quality
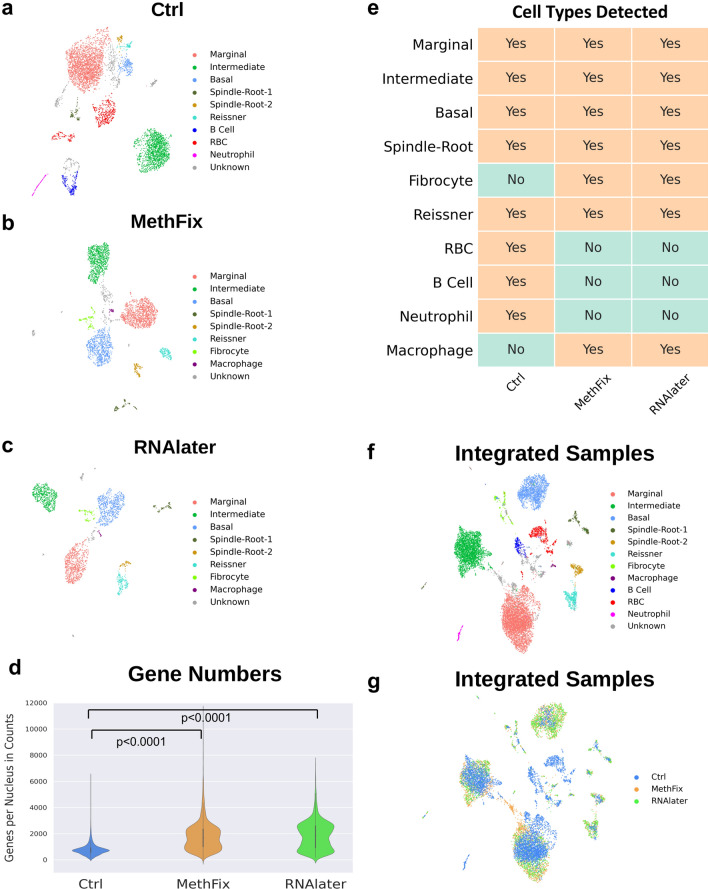
As a first step towards analyzing rare cell types in the SV, we utilized two forms of sample preservation, methanol fixation of nuclei (MethFix) and RNAlater treatment (RNAlater) of tissue prior to isolation of nuclei, to generate single nucleus transcriptional profiles from the P30 mouse SV. A total of 5681 nuclei were isolated and analyzed from the P30 mouse SV utilizing these sample preservation methods (3371 nuclei in the MethFix dataset, 2310 nuclei in the RNAlater dataset). We compared our previously published adult SV single-nucleus transcriptional profiles (Ctrl) consisting of 5176 nuclei2 to these two datasets (MethFix, RNAlater) (Fig. 2a–c, respectively). The schematic for the different sample preparation steps for all three data sets is shown in Supplementary Figure S1a. Comparison of the three datasets demonstrated an increased number of genes detected per nucleus in the sample preservation datasets with 727, 1682, and 1857 median number of genes per nuclei in the Ctrl, MethFix, and RNAlater datasets (one-way ANOVA p < 0.0001). Post hoc t-tests demonstrated a significant difference between Ctrl and either MethFix or RNAlater datasets (both p < 0.0001), but not between MethFix and RNAlater datasets (p = 0.12) (Fig. 2d). The percentage of nuclei remaining after bioinformatic preprocessing, which included accounting for mitochondrial gene expression and removal of doublet nuclei, was increased in the sample preservation datasets (MethFix, RNAlater) compared to the previously published dataset (Ctrl) (Suppl. Fig. S1b).
Figure 2.
Comparison of Ctrl, MethFix and RNAlater datasets. snRNASeq datasets. (a) Ctrl, (b) MethFix and (c) RNAlater, were clustered by modularity-based clustering method with Leiden optimization algorithm, and visualized by 2D UMAP embedding. (d) Increased number of genes detected per nucleus in sample preservation datasets (1682 median genes per nuclei in MethFix, 1857 median genes per nuclei in RNAlater) compared to Ctrl dataset (727 median genes per nuclei) (one-way ANOVA p < 0.0001), while no significant difference between MethFix and RNAlater dataset (post hoc t-test, p = 0.88). (e) Major SV cell types (marginal, intermediate, basal), spindle-root and Reissners’ membrane cells are detected in all three datasets, but other small populations of cells (fibrocyte,macrophage, RBC, B cell and neutrophil ) are detected in either Ctrl or MethFix/RNAlater datasets. Ctrl, MethFix and RNAlater datasets are integrated by Harmony and visualized by 2D UMAP embedding. Integrated cells are colored based on (f) cell identities or (g) original dataset (Ctrl, MethFix, RNAlater).
Stria vascularis cell type clusters remain consistent in control and sample preservation snRNA-Seq datasets of the adult SV
Despite these differences, UMAP (Uniform Manifold Approximation and Projection) plots depicting clustering for the previously published adult SV dataset (Fig. 2a)2 and the two datasets where sample preservation methods was employed (MethFix, RNAlater) (Fig. 2b,c, respectively) demonstrated similar clusters of SV cell types. SV cell type clusters including marginal, intermediate, basal, spindle/root cells, as well as, cells of Reissner’s membrane, were identified in all 3 datasets (Ctrl, MethFix, RNAlater). Furthermore, other small populations of cells including fibrocytes and macrophages were identified as unique clusters in both sample preservation datasets (MethFix, RNAlater) (Fig. 2b,c). In contrast, red blood cells (RBCs), B cells, and neutrophils were only detected in the Ctrl dataset (Fig. 2a). A summary of cell types identified in the three datasets is shown (Fig. 2e).
To understand how the proportions of cell types detected in each of the datasets (Ctrl, MethFix, RNAlater) reflected in vivo proportions, we compared the relative percentages of major SV cell types (marginal, intermediate, basal, and spindle/root cells) in all three datasets to in vivo percentages calculated from cell counts from mid-modiolar cochlear cross-sections (Suppl. Fig. S1c-f). Relative in vivo percentages of SV marginal, intermediate, basal and spindle/root cells (N = 6 adult mice) are shown with a representative mid-modiolar cochlear cross-section of the SV (Suppl. Fig. S1c and S1d, respectively). By comparison to the Ctrl dataset (Suppl. Fig. S1e), sample preservation datasets (Suppl. Fig. S1f. and S1g) demonstrated percentages of major SV cell types that were more similar to in vivo major SV cell type percentages (Suppl. Fig. S1c). Furthermore, the percentage of rare spindle/root cells in the MethFix and RNAlater datasets were 6.2% and 6.3%, respectively, nearly doubling the 3.4% of these cells in the control dataset.
In order to visualize SV cells from all three datasets, the datasets were integrated with batch correction utilizing Harmony as previously described26,27. All SV cell types remained clustered together with cell identities based on clustering of individual datasets (Fig. 2f). The distribution of the three datasets (Ctrl, MethFix, RNAlater) within each of the clusters is shown in Fig. 2g. Major SV cell types remained in cell type-specific clusters after dataset integration. The expression pattern of representative marker genes used for cluster labeling are shown in Supplementary Figures S2-1 and S2-2. While, fibrocyte gene (Coch) expression was detected in the Ctrl dataset (Suppl. Fig. S2-2), these cells partially overlapped with other SV cell type clusters and as a result were not annotated as a fibrocyte cluster in the Ctrl dataset. Interestingly, in all three P30 SV data sets, two sub-clusters of spindle-root cells are revealed, and they are labeled as Spindle-Root-1 and Spindle-Root-2, respectively.
Differential expression (DE) analysis reveals transcriptional differences between rare populations of spindle and root cells
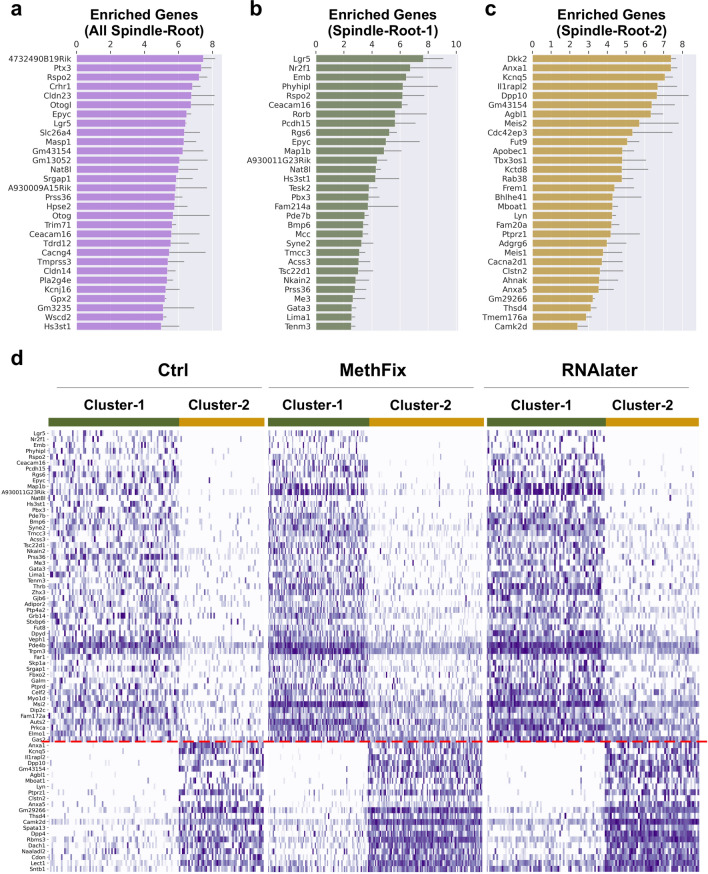
Cell type-specific gene expression across all three snRNA-Seq datasets for SV cell types (Ctrl, MethFix, RNAlater) is shown in Fig. 3. The top 30 up-regulated genes (p value < 0.05, and sorted by fold change) in Spindle-Root cells obtained by comparing to other cell types are listed in Fig. 3a. Mean fold-change values were calculated based on DE analysis determined within each dataset (Ctrl, MethFix, RNAlater). Among the top 30 up-regulated genes in Spindle-Root cells were Slc26a4 and Kcnj16, which are expressed in both sub-clusters of Spindle-Root cells and is consistent with our previously published spindle-root cell clustering2. Based on the observation of two sub-clusters of Spindle-Root cells, we took advantage of this opportunity to perform a more detailed analysis of the transcriptional differences between the Spindle-Root sub-clusters. The top 30 significant (p < 0.05) up-regulated genes in each Spindle-Root sub-cluster are shown in Fig. 3b,c, respectively.
Figure 3.
Differential expression analysis on Ctrl, MethFix and RNAlater datasets. Top 30 enriched genes in (a) spindle-root cells compared to other cell types, (b) Spindle-Root-1 compared to Spindle-Root-2 and (c) Spindle-Root-2 compared to Spindle-Root-1 are shown in barplot. The logarithm (base 2) of the fold-change (logFC) are displayed along the horizontal axis, and gene names are displayed along the vertical axis. Standard deviation of logFC among three datasets is shown with the error bar. (d) Normalized counts of unique enriched genes in Spindle-Root-1 (above red dashed line) and Spindle-Root-2 (below red dashed line) are shown as heatmaps for each dataset (Ctrl, MethFix, RNAlater). Nuclei are displayed along the horizontal axis and genes are displayed along the vertical axis.
To identify uniquely expressed genes in the each of the Spindle-Root sub-clusters, the DE gene lists for Spindle-Root-1 (Fig. 3b) and Spindle-Root-2 (Fig. 3c) were cross-referenced against genes expressed by other SV cell types to identify differentially expressed genes expressed by Spindle-Root-1 and Spindle-Root-2 (Fig. 3d). The resulting list of DE genes were compared across all 3 datasets and the list of commonly expressed genes between the 3 datasets were displayed in heatmaps for the Ctrl (Fig. 3d, left), MethFix (Fig. 3d, middle), and RNAlater (Fig. 3d, right) datasets. The resulting lists of differentially expressed genes demonstrate that the Spindle-Root-1 and 2 exhibit distinct gene expression patterns.
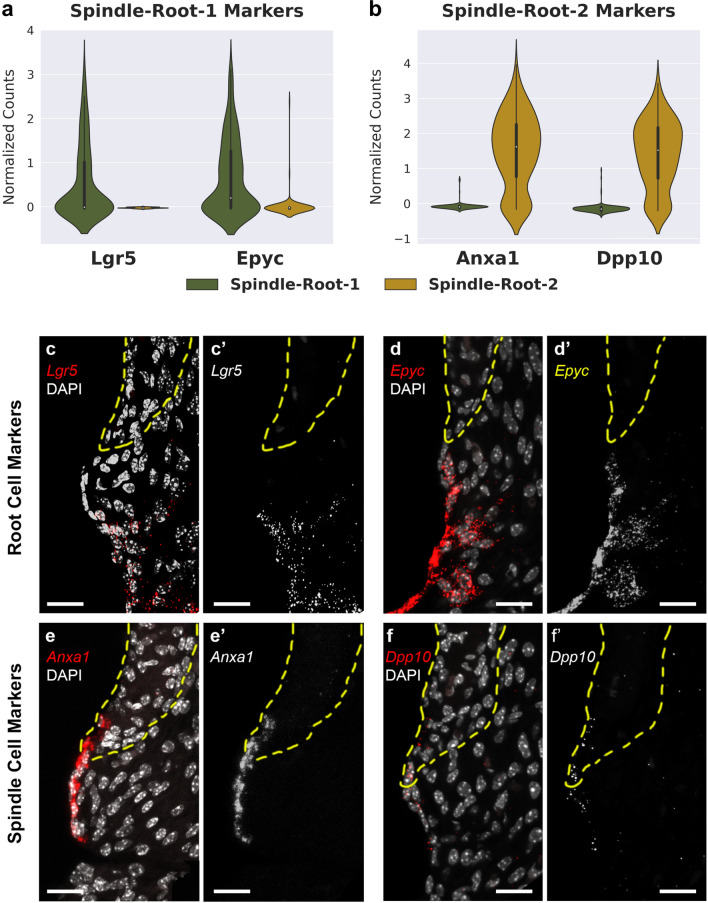
To investigate the expression pattern of identified Spindle-Root sub-cluster markers in Fig. 3, select candidate genes were visualized on violin plots and single-molecule fluorescent in situ hybridization (smFISH) was performed for selected marker genes (Fig. 4). Violin plots, demonstrating expression level in normalized counts, for Spindle-Root-1 candidate genes, Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 (Lgr5), and Epiphycan (Epyc) are shown in Fig. 4a. Violin plots for Spingle-Root-2 candidate genes, Annexin A1 (Anxa1) and Dipeptidyl Peptidase Like 10 (Dpp10), are shown in Fig. 4b. Interestingly, we found that Spindle-Root-1 enriched genes Lgr5 (Fig. 4c, c’) and Epyc (Fig. 4d, d’) are expressed in root cells, while Spindle-Root-2 enriched genes Anxa1 (Fig. 4e, e’) and Dpp10 (Fig. 4f, f’) are expressed in SV spindle cells. Since KCNJ10 protein has been previously shown to be expressed in root cells16,17 in addition to SV intermediate cells2,9,28, we co-localized Lgr5 RNA with Kcnj10 RNA in root cells (Suppl. Fig. S3a, a’). While Anxa1 was expressed by the spiral prominence surface epithelial cells in continuity with the spindle cells, we did not identify a distinct transcriptional cluster of these surface epithelial cells in the spiral prominence. Neither Anxa1 nor Dpp10 RNA was expressed in other regions of the cochlea (data not shown). Lgr5 RNA expression was also observed in Deiters cells and cells of the outer sulcus (Suppl. Fig. S3b). In addition to root cells, Epyc RNA expression was localized to the outer sulcus and the greater epithelial ridge (Suppl. Fig. S3c). smFISH validation of Lgr5 and Epyc expression (Fig. 4c,d, respectively) appears to be consistent with previous publications that demonstrated expression of these genes in the region of the outer sulcus or spiral prominence29,30. Therefore, our data demonstrate that Spindle-Root cells can be further distinguished into spindle and root cell clusters by previously uncharacterized marker gene expression. Based on marker gene expression confirmed by smFISH, Spindle-Root-1 cells and Spindle-Root-2 cells will be referred to as root and spindle cells, respectively.
Figure 4.
Validation of identified markers in Spindle-Root sub-clusters by single-molecule fluorescent in situ hybridization. Expression level (normalized counts) of (a) Spindle-Root-1 candidate markers (Lgr5 and Epyc) and (b) Spindle-Root-2 candidate markers (Anxa1 and Dpp10) are shown in violin plots. (c–d’) smFISH demonstrates expression of Lgr5 (c, c’) and Epyc (d, d’) in root cells, while expression of Anxa1 (e, e’) and Dpp10 (f, f’) are detected in spindle cells. Grayscale images of smFISH probe are shown in single channel images (c’–f’). Scalebars are all 20 μm. Yellow dotted lines indicate location of stria vascularis. DAPI labels cell nuclei.
To determine the difference between the three datasets, we combined the datasets on their mutual genes without applying any data merging algorithm. A detectable batch effect, as demonstrated by the cells from each dataset clustering into distinct clusters, was observed between the Ctrl data set and the MethFixed (Suppl. Fig. S4a) and RNAlater (Suppl. Fig. S4b) data sets, respectively. However, a batch effect was not detected between MethFix and RNAlater datasets as suggested by the overlapping distributions amongst cell types between the two sample preservation datasets (Suppl. Fig. S4c) as well as when all three datasets are combined together (Suppl. Fig. S4d). Comparison of the combined MethFix and RNAlater datasets without batch correction (Suppl. Fig. S4e) to the combined MethFix and RNAlater datasets integrated with batch correction using Harmony as previously described26,27 demonstrated minimal impact on the clustering of SV cell types (Suppl. Fig. S4f.). Therefore, MethFix and RNAlater datasets were combined (MethFix-RNAlater) (Suppl. Fig. S4e) for downstream gene regulatory network analysis without using additional data merging algorithms.
Gene regulatory network landscape in SV root and spindle cells
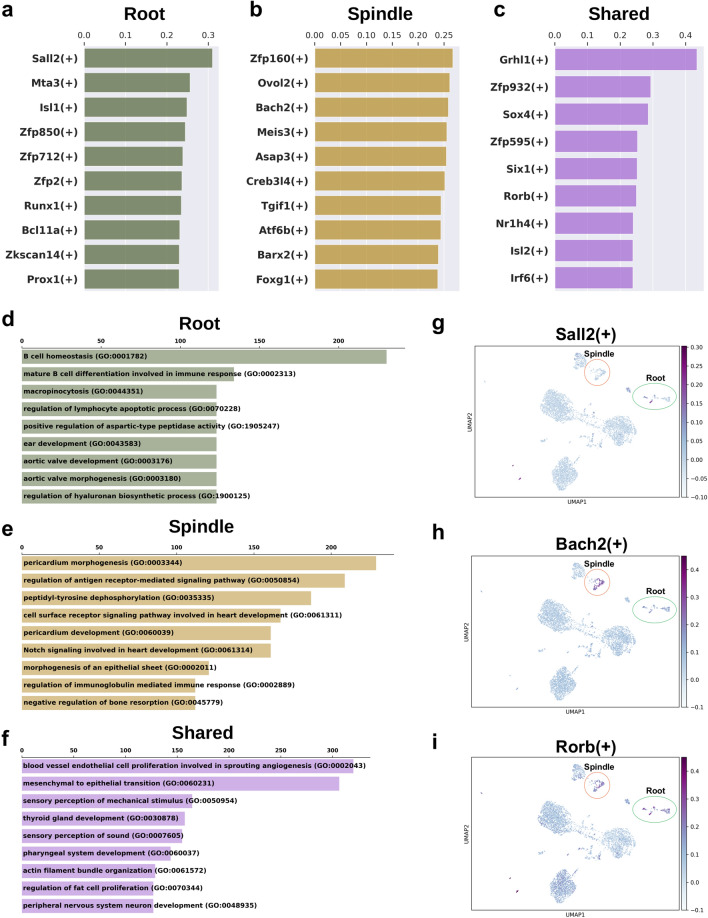
To further explore potential homeostatic functional differences and similarities between spindle and root cells, we applied SCENIC as previously described by our labs and others2,31 to the combined sample preservation (MethFix-RNAlater) P30 mouse SV datasets (Suppl. Fig. S4c), which demonstrated overlapping distributions amongst cell types clusters without batch correction (Suppl. Fig. S4e). The top 10 regulons for root and spindle cells as determined by the regulon specificity score (RSS) along the horizontal axis are shown in Fig. 5a,b, respectively. The top shared regulons between root and spindle cells are shown in Fig. 5c. The higher the RSS, the more specific a given regulon is to the spindle and root cells, respectively32. The top enriched GO biological process terms for root, spindle, and shared root and spindle regulons were B cell homeostasis (GO:0001782), pericardium morphogenesis (GO:0003344), and blood vessel endothelial cell proliferation involved in sprouting angiogenesis (GO:0002043), respectively (Fig. 5d–f). Sall2 and Bach2 regulon activity are shown for root and spindle cells, respectively (Fig. 5g,h, respectively). Both regulons represent novel gene regulatory networks that have not been related previously to homeostatic function in the inner ear. Putative Sall2 target genes include Sall2 itself and Nr2f1 with UMAP plots in Supplementary Figure S5 (Suppl. Fig. S5a and S5b, respectively) demonstrating expression in root cell predominantly. Nr2f1 also expressed by cells of Reissner’s membrane and an unknown population of cells in close proximity to root cells on the UMAP plot. While Bach2 has been previously shown to be expressed in chick otic epithelium33 and is known for its role in the Bcl6-Bcl2-p53 axis which controls hair cell apoptosis (reviewed by Morill and colleagues34), its role in the inner ear, either in inner ear development or in hearing remain incompletely characterized. Expression of putative Bach2 target genes including Anxa1 and Dpp10 are shown with UMAP plots (Suppl. Fig. S5c and S5d, respectively) and are validated by smFISH (Fig. 4e–e’ and f–f’) and colocalized with Bach2 (Suppl. Figure 6a–a’).
Figure 5.
Regulon analysis by pyScenic on combined MethFix-RNAlater dataset. Top non-shared regulons by regulon specificity score (RSS) for (a) root cells and (b) spindle cells and (c) Top shared regulons are shown in bar plots. X-axis is the RSS. The top enriched GO biological process terms for genes in (d) the top 10 non-shared root regulons, (e) the top 10 non-shared spindle regulons and (f) the top shared regulons are ranked by combined score from Enrichr. Regulon activity (AUC score) of (g) Sall2, (h) Bach2 and (i) Rorb are shown in the 2D UMAP of MethFix-RNAlater combined dataset. Root and spindle clusters are highlighted by green and red circle, respectively.
The Rorb regulon, which is shared between root and spindle cells, and its regulon activity plot are shown in Fig. 5i. Putative Rorb target genes include Otog, Cldn14, and Pde4b. While mutations in Otog35,36 and Cldn1437–39 have been linked to hearing loss, mutations in Pde4b has not been previously linked with hearing loss. UMAP plots of gene expression for Rorb, Otog, Cldn14, and Pde4b are provided in the supplement (Suppl. Fig. S5e-h, respectively). While Rorb is expressed in the region of the future root cells as well as the organ of Corti in the apical cochlea at E15.5 with expression decreasing towards the basal turn of the cochlea, its expression is noted to be more widespread in the adult mouse cochlea40. Our data identifies Rorb expression in both root and intermediate cells of the adult SV (Suppl. Fig. S5e). Otog expression (Suppl. Fig. S5f) is consistent with previously published expression in root cells in the perinatal mouse cochlea35. Cldn14 expression (Suppl. Fig. S5g) is seen in both root and spindle cells of the adult SV and is consistent with previously reported expression by Ben-Yosef and colleagues37. While not previously described in the inner ear, Pde4b is expressed by both root and spindle cells as well as intermediate cells of the adult SV (Suppl. Fig. S5h).
Discussion
In this study, we utilize two sample preservation methods with snRNA-Seq to characterize rare cell transcriptional profiles in the adult SV. We compare these transcriptional profiles to previously published adult snRNA-Seq, demonstrating not only comparability, but also demonstrate the advantages of sample preservation in increasing the yield of rare cell types. Compared to our previous study which focused on transcriptional profiles of major SV cell types including marginal, intermediate and basal cells2, this current study utilizes novel methodologies to identify transcriptional profiles of rare SV cell types, notably spindle and root cells. While the small numbers of these rare cell types in our previously published control single nucleus dataset2 limited resolution of their transcriptional identities, we hypothesized that sample preservation methods might enable isolation of greater numbers of these particular rare cell types to resolve their transcriptional profiles. One caveat is that different sample isolation methods may result in preferential capture of certain cell types, necessitating that methods be tailored to the objectives of a given study24. Notably, Denisenko and colleagues noted that immune cells were detected at lower rates across all single nucleus RNA-sequencing experiments24. While the capture of circulating immune cell types including B cells and neutrophils was less robust with these sample preservation methods, our goal of isolating rare intrinsic cell types of the SV and adjacent tissues including spindle and root cells was achieved. In addition, sample preservation methods also allowed the capture of other rare cell types including a larger number of fibrocytes and macrophages, which were not detected in the control snRNA-Seq dataset.
We demonstrate that use of RNAlater as a sample preservation method for snRNA-Seq is viable and comparable to methanol fixation. Sample preservation techniques led to a higher percentage of rare cell types by the near doubling of root and spindle cells captured while the median genes per nuclei was increased in the sample preservation datasets compared to control dataset. Alterations in our bioinformatic processing pipeline (see Supplementary Note) from our published analysis2 may have contributed to the ability to resolve rare cell populations. While use of methanol fixation is employed after nuclei isolation and requires fairly rapid isolation of nuclei from tissue, the use of RNAlater allows for the placement of tissue at room temperature allowing for some flexibility in tissue and nuclei isolation. These results suggest that this method might be utilized with precious difficult-to-obtain tissue (i.e. human pathological tissue) for the purposes of snRNA-Seq, potentially facilitating collaborations across institutions, leading to further application of these technologies to human disease. Furthermore, regulatory network identification for spindle and root cells was not possible with the control dataset and was entirely based on data derived from the sample preservation datasets possibly due to the higher percentage of rare cell types captured. While these newly identified potential regulatory networks will need to be validated, examined and perturbed in future studies, our study establishes a basis for perturbing these regulatory networks.
Despite work elucidating the cyto-architecture and structural components involved in membrane physiology of root cells reviewed by Jagger and Forge17, the functional roles of root cells remain largely undefined. Proposed functions for root cells include cation absorption from the endolymph, potassium transport between junctional compartments, and involvement in the inflammatory response of the cochlea to pathological stress16,17. The involvement of root cells in cochlear ionic homeostasis is supported by previously published root cell expression of the α1 and β1 subunits of the Na, K-ATPase antiporter in the rat41, expression of the AE2 Cl−/HCO3− exchanger in root cell processes invading the spiral ligament in the guinea pig42, expression of SLC26A4, an anion exchanger for chloride and bicarbonate, in the root cell processes in the adult mouse43 in addition to its expression in spindle cells12, and the expression of Kir4.1, encoded by Kcnj10, in root cells16 in addition to its expression in intermediate cells in the SV2,3,44.
Neonatal and adult Epyc knockout mice, while possessing normal-appearing cochleae including hair cells and supporting cells, have elevated hearing thresholds above 16 kHz on ABR, suggesting a role for Epyc in hearing30. Despite work that has established Lgr5, a cell membrane receptor of the Wnt signaling pathway, as a marker of potential inner ear stem cells45–49, its role in hearing is uncertain. Both Lgr5 and its ligand, R-Spondin 2 (Rspo2), are differentially expressed by root cells when compared to spindle cells (Fig. 3). Rspo2 has been shown to promote clustering of acetylcholine receptors through its interaction with Lgr5 at the neuromuscular junction50 and the expression of acetylcholine receptors has been previously demonstrated in the region of the root cells51. While the role of these acetylcholine receptors is generally thought to be inhibitory51, their role is largely undefined. Wangemann and colleagues have previously shown that potassium secretion in SV marginal cells is negatively regulated by stimulation of muscarinic acetylcholine receptors52, suggesting the possibility that Lgr5 and its ligand, Rspo2, may play some role in regulating the undefined function of these receptors in root cells. Jagger and Forge have suggested that root cells “act as a continuous K+ sink” likely from the endolymph and we suggest that the ability to inhibit potassium entry may be protective by possibly preventing potassium loss from the endolymph in situations where potassium homeostasis is disrupted in the endolymph17. However, this is highly speculative with the functional roles in root cells of Lgr5 and Rspo2 remaining largely undefined. Our study establishes a possible rationale for future experiments which will define the functional roles of these genes in root cells.
Similar to root cells, the role of spindle cells in hearing have remained poorly defined. In this study, DE analysis identifies spindle cell-specific transcriptional profiles for the first time and validation of two candidate genes, Anxa1 and Dpp10, consistently distinguishes these cells from adjacent root cells (Fig. 4e,f). Dpp10 is a previously uncharacterized transmembrane channel protein in the inner ear, with an undefined role in SV function and hearing, that may play a role in modulating the activity of voltage-gated potassium channels53. While Anxa1 RNA expression in other regions of the organ of Corti notably Hensen cells (data not shown) as described by Kalinec and colleagues54 was not seen, the existence of secreted forms and observations of ANXA1 in punctate form around lipid droplets leave open the possibility that other cochlear cell types may store ANXA1 after production. Alternatively, glucocorticoid stimulation could result in production of Anxa1 in other cochlear cell types not included in our unstimulated snRNA-Seq datasets. Anxa1 expression by smFISH (Fig. 4e,e’) in spindle cells and the surface epithelial cells of the spiral prominence suggests that these cell types, if distinct, may share transcriptional similarities. Alternatively, it is possible that a limited collection of these spiral prominence surface epithelial cells in our dataset limits the ability to transcriptionally distinguish them. Nonetheless, Anxa1 expression distinguishes spindle cells from root cells.
Closer examination of spindle cell-specific gene regulatory networks supports the potential role of spindle cells in responses to inflammation. Specifically, the Bach2 regulon (Fig. 5h), a spindle cell-specific regulon, includes Anxa1 and Dpp10. Bach2 is a transcription factor belonging to the BTB and Cap’n’collar (CNC) gene family that functions within multiple innate and adaptive lineages to control immune response. Bach2 functions as a transcriptional repressor and is a known susceptibility gene for a number of autoimmune diseases55–57. Bach2 appears to promote a shift from myeloid to lymphoid programs by suppressing myeloid genes in B cells58 as well as directing T helper (Th) cell differentiation, homeostasis, and effector functions while preventing full effector differentiation within Th cells in vitro57. Aberrant expansion of T follicular helper (Tfh) cells results in pathogenic autoantibodies and is frequently associated with autoimmune diseases57,59,60. Recently, Zhang and colleagues demonstrated that the loss of Bach2 expression led to increased Tfh cell accumulation with a shift towards an IL-4-producing subset57. These data suggest that Bach2 prevents humoral autoimmunity, at least in part by inhibiting the generation of pathogenic Tfh cells. Bach2 also appears to regulate differentiation and effector functions of other T cell subsets including Treg and Th17 cells, which play prominent roles in autoimmunity when deregulated61,62. Finally, Bach2 appears to be necessary for an appropriate macrophage responses to T cell-induced inflammation63. Thus, Bach2 regulon activity in spindle cells suggests that these cells may play a role in regulating autoimmune responses in the inner ear. Furthermore, gene regulatory networks in root cells corroborate the suggested role of root cells in endolymph ion homeostasis as reviewed by Jagger and Forge16,17. The Sall2 regulon (Fig. 5g), a root cell-specific regulon, includes target genes Nr2f1 and Sall2. While Sall2 has not previously been characterized in the inner ear, mutations in Nr2f1 result in hearing loss64–66. Nr2f1 expression is present in the cells which are fated to become the root cells in the cochlea66,67. Tarchini and colleagues demonstrate that long-range downregulation of Nr2f1 through a mutation in Mctp1 in the deaf wanderer (Mctp1dwnd) mice results in hearing loss with minimal alterations to cochlear structure67. Intriguingly, Bergeron and colleagues demonstrate that overexpression of Nr2f1 in the SpotTg/Tg mutant mouse results in expansion of the endolymph-containing scala media with melanocytes failing to migrate to their proper locations in the vestibule but not the cochlea68. Despite, the apparent absence of hair cell degeneration in the face of Nr2f1 downregulation, hearing loss associated with changes in the endolymph ion homeostasis may occur in a delayed fashion69–71. The difficulty with SpotTg/Tg mutant mouse model is that the vast majority of offspring die at birth with few that survive to young adulthood, thus making auditory testing inconclusive. Nonetheless, in combination with previously published expression of ion channel proteins expressed by root cells, including Na, K-ATPase antiporter, AE2 Cl−/HCO3− exchanger, SLC26A4, and Kir4.117,41–43, our observations of root cell regulons corroborate the idea that root cells may be involved in regulation of endolymph ion homeostasis.
Finally, shared root and spindle cell regulons implicate these cells in Meniere’s disease, an inner ear disease with a poorly understood pathophysiology and with no implicated cell types72–74. The retinoid-related orphan receptor β (Rorb) regulon is a shared between root and spindle cells. Rorb expression has been previously demonstrated to be involved in the differentiation of neuronal cell types and in regulating circadian activity75–77. In addition to previously validated expression of Slc26a42, spindle and root cells express potential Rorb target genes, Otog and Cldn14 (Suppl. Figure 5f and g, respectively). Recently, missense mutations in Slc26a4, Otog, and Cldn14 have been characterized in patients with Meniere’s disease38,78,79. These authors suggest that acquisition of additional mutations over time in the form of missense mutations may underlie the onset of Meniere’s disease. Single cell transcriptional data provides an opportunity to identify potentially involved cell types in human disease80. This is particularly relevant to diseases where the organ of interest is difficult to access and has rare opportunities for tissue sampling in humans, as is the case for the inner ear. Thus, these data suggest that root and spindle cells may be involved in the pathophysiologic mechanisms underlying Meniere’s disease.
In conclusion, we define distinct transcriptional profiles for rare SV spindle cells and root cells in the spiral prominence. We characterize putative gene regulatory networks for these rare cell types and in doing so, identify potential roles that these cell types may play in the cochlea, including ion homeostasis and regulation of immune responses in the cochlea. Furthermore, we implicate these rare cell types in both genetic and acquired hearing loss. Finally, we provide some initial evidence that dysfunction in spindle and root cells may be related to Meniere’s disease.
Methods
Animal model and experimental design
CBA/J mice were purchased from JAX (Stock No. 000656). Postnatal day 30 (P30) mice were used for snRNA-Seq experiments and single molecule RNA fluorescent in situ hybridization (smFISH).
Adult mouse stria vascularis preparation
The method of adult mouse SV preparation has been previously described2. Briefly, the lateral wall of the cochlea was microdissected from the bony wall of adult mouse cochlea and the pigmented strip in the cochlea lateral wall denoting the SV was microdissected from the spiral ligament using fine forceps. SV from all turns of the cochlea were collected. Samples were collected at the same time of day across individual mice and batches. For each collection, less than 1 h was spent prior to single nucleus capture on the 10 × Genomics Chromium platform. Sexes of mice were generally mixed for each experiment. 5 mice (2 female, 3 male P30 mice) were used for the methanol-fixed single nucleus capture and 6 mice (3 female, 3 male P30 mice) were used for the RNAlater-treated single nucleus capture. For the methanol-fixed sample, isolated cell nuclei obtained as previously described2,23,81. Briefly, nuclei were suspended in 200 μL Dulbecco’s phosphate buffered saline (DPBS), then 800 μL of ice-cold methanol was slowly added drop-by-drop to the single nuclei suspension while gently stirring the nuclei suspension. Nuclei were moved to the freezer and incubated 30 min at − 20 °C. Subsequently, cells were rehydrated in wash and resuspension buffer (1 × PBS with 1% BSA and 0.2 U/ul RNase Inhibitor). Nuclei suspension underwent centrifugation (100 rcf, 5 min, 4 °C) and supernatant was removed and cells were resuspended in 50 μL of wash and resuspension buffer to obtain 700–1200 cells/μL prior to nuclei isolation and sequencing. For the RNAlater-treated sample, freshly dissected adult SV tissues were submerged in and stored in 0.7 mL of RNAlater solution (Catalog No. AM7020, ThermoFisher, Waltham, MA) at room temperature in a 1.5 mL Eppendorf tube and then stored at 4 °C overnight. After incubation, DPBS was added in equal volumes (0.7 mL) to the tube and gently mixed, then centrifuged at 500 g for 5 min at room temperature. Supernatant was removed and replaced with lysis buffer before previously described nuclei isolation and sequencing.
Single nucleus suspension
Isolation of nuclei from the adult mouse SV has been previously described2. Briefly, SV from ~ 5 to 6 P30 animals were isolated and collected in 3 ml DMEM F-12 media. Following collection, the media was replaced with 3 ml chilled lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.005% Nonidet P40 in Nuclease free water) and the tissue were lysed at 4 °C for 25 min. The lysis buffer was then replaced with 1.5 ml DMEM F-12 media. The tissues were triturated and filtered through a 20um filter (pluriSelect Life Science, El Cajon, CA). The filtrate was centrifuged at 500rcf for 5 min at 4 °C. The supernatant was removed, and the cell pellet was resuspended in 1 ml nuclei wash and resuspension buffer (1 × PBS with 1% BSA and 0.2 U/μl RNase Inhibitor). The cells were filtered through a 10um filter (pluriSelect Life Science, El Cajon, CA) and centrifuged at 500rcf for 5 min at 4 °C. The supernatant was removed, and pellet resuspended in 50 μl of nuclei wash and resuspension buffer. Nuclei were counted in a Luna cell counter (Logos Biosystems, Annandale, VA) and a nuclear density of 1 × 106 cells/ml was used to load onto the 10× genomics chip.
10× Chromium genomics platform
Single nuclei captures were performed following manufacturer's recommendations on a 10× Genomics Controller device (Pleasanton, CA). The targeted number of captured nuclei ranged from 6000 to 7000 per run. Library preparation was performed according the instructions in the 10× Genomics Chromium Single Cell 3′ Chip Kit V2. Libraries were sequenced on a Nextseq 500 instrument (Illumina, San Diego, CA) and reads were subsequently processed using 10× Genomics CellRanger analytical pipeline using default settings and 10× Genomics downloadable mm10 genome as previously described2.
Single-nucleus RNA-seq data preprocessing
Quality Control—snRNA-Seq data preprocessing was conducted by Scanpy (v1.4.5)82. Genes were filtered based on number of cells. Only genes detected in at least 3 cells are kept. Low-quality cells were filtered out when: (1) less than 200 genes were detected; (2) more than 8000 counts in total; and (3) more than 10% of mitochondria genes were detected.
Doublet detection—Transcriptional profiles of doublet nuclei in the snRNA-Seq dataset were computationally predicted by Scrublet (v0.2.1)83, and excluded from downstream analysis. Default parameters were utilized including an expected doublet rate of 0.1, number of principle components set to 30, and minimal gene variability set to 85.
Clustering and data visualization
To cluster the cells by their expression, we used modularity-based clustering with Leiden algorithm implemented in Scanpy (v1.4.5) for each dataset separately. In brief: (1) raw counts were normalized by total with parameter exclude_highly_expressed set as True, and scaled by the function pp.log1p; (2) principal component analysis (PCA) was performed on top 4000 high variable genes, which were generated with default threshold of the mean expression and dispersion by the function pp.highly_variable_genes; (3) KNN graph was constructed based on the euclidean distance in top 30 PCA dimensions by the function pp.neighbors with parameter n_neighbors set as 10; (4) Cells were clustered by the function tl.leiden with the resolution Ctrl = 1.0, MethFix = 1.0 and RNAlater = 1.5. Clustered cells were visualized by Uniform Manifold Approximation and Projection (UMAP) embedding with 2 components. Heatmaps or violin plots were constructed as previously described2.
Dataset integration
The integration of all three datasets (Ctrl, MethFix, RNAlater) and the sample preservation datasets (MethFix, RNAlater) was performed by utilizing the Harmony algorithm, which projects cells into a shared embedding26. The package used for data integration was harmony-pytorch (v0.1.3). Default parameters were utilized.
Cell cycle and dissociation effect calibration
Biological effect calibration is conducted by Scanpy (v1.4.5).
Cell cycle effect calibration—Cell cycle heterogeneity in snRNA-Seq data sets was calibrated by calculating cell cycle phase scores based on identified cell cycle markers as previously described84. Briefly, cell cycle phase score is calculated by the difference of mean expression of the list of cell cycle genes85 and the mean expression of reference genes. The reference genes are randomly selected, which match the distribution of the expression of the given list. Detailed steps can be found in Scanpy tutorial documents (https://nbviewer.jupyter.org/github/theislab/scanpy_usage/blob/master/180209_cell_cycle/cell_cycle.ipynb).
Dissociation effect calibration—Dissociation effect calibration in snRNA-Seq data sets was performed similarly to cell cycle effect calibration based on identified dissociation related genes86. We have previously detailed the application of this technique to the single nucleus RNA-Seq data from the adult SV2. Cell cycle and sample dissociation effects minimally impact cluster composition in major SV cell types (Supplementary Note, Suppl. Fig. S7).
Differential expression (DE) analysis
Methanol-fixed and RNAlater-treated data sets are combined on their mutual genes without any further data merging algorithms. DESingle (v1.6.0) was utilized to perform DE analysis between known SV cell types with default settings87. DESingle is specifically designed for single cell DE analysis and employs a zero-inflated negative binomial model to estimate the proportion of dropout and real zeros, allowing for a more accurate representation of differential gene expression at the single cell or single nucleus level87.
Regulatory network inference
Gene regulatory network inference using single cell regulatory network inference and clustering (SCENIC) has been previously described2,31. It is a computational method for inferring GRN based on the expression level of transcriptional factors and their conserved motif-enriched cis-regulatory sequences. Briefly, SCENIC identifies potential gene regulatory networks by identifying regulons, defined as transcription factors and their downstream motif-enriched target genes, and by determining the activity of each of these regulons within each cell. From these analyses, a regulon activity matrix is constructed that can be utilized to cluster cells on the basis of shared regulatory networks and may identify cell types and cell states on the basis of shared activity of a regulatory subnetwork. SCENIC was implemented utilizing pySCENIC (v0.10.0). Downstream visualization and plots are created by Matplotlib (v3.2.0) and Seaborn (v0.10.0).
Gene ontology and gene-set enrichment analysis
Gene ontology analyses and gene enrichment analyses were performed using Enrichr (https://amp.pharm.mssm.edu/Enrichr/) as previously described2,88–91. The combined score approach where enrichment score is calculated from the combination of the p-value computed using the Fisher exact test and the z-score was utilized. Top gene ontology (GO) terms were chosen by utilizing the combined score approach as described.
Single-molecule fluorescent in situ hybridization (smFISH)
Fluorescent in situ hybridization was performed as previously described2,92 using the following RNAscope probes: Lgr5 (Catalog No. 312171), Epyc (Catalog No. 572901), Anxa1 (Catalog No. 509291), Dpp10 (Catalog No. 553331), Bach2 (Catalog No. 887121-C3), and Kcnj10 (Catalog No. 458831-C3). RNAscope probes were obtained from Advanced Cell Diagnostics (Newark, CA, United States) and used with sections of cochleae from CBA/J mice at P30. Adult cochleae were dissected from the head and fixed over night at 44 °C in 4% paraformaldehyde (PFA) in 1 × PBS. Fixed adult mouse inner ears were decalcified in 150 mM EDTA for 5–7 days, transferred to 30% sucrose, and then embedded and frozen in SCEM tissue embedding medium (Section-Lab Co, Ltd., Hiroshima, Japan). Adhesive film (Section-Lab Co, Ltd., Hiroshima, Japan) was fastened to the cut surface of the sample to support the section and cut slowly with a blade to obtain thin mid-modiolar sections. The adhesive film with section attached was submerged in 100% EtOH for 60 s, then transferred to distilled water. Frozen tissues were sectioned (10 μm thickness) with a CM3050S cryostat microtome (Leica, Vienna, Austria). Sections were mounted with SCMM mounting media (Section-Lab Co, Ltd., Hiroshima, Japan) and imaged using a 1.4 N.A. objective.
Ethical approval
All animal experiments and procedures were performed according to protocols approved by the Animal Care and Use Committee of the National Institute of Neurological Diseases and Stroke and the National Institute on Deafness and Other Communication Disorders, National Institutes of Health. All experimental protocols were approved by the Animal Care and Use Committee of the National Institute of Neurological Diseases and Stroke and the National Institute on Deafness and Other Communication Disorders, National Institutes of Health. All methods were carried out in accordance with relevant guidelines and regulations of the Animal Care and Use Committee of the National Institute of Neurological Diseases and Stroke and the National Institute on Deafness and Other Communication Disorders, National Institutes of Health.
Supplementary information
Acknowledgements
This research was supported (in part) by the Intramural Research Program of the NIH, NIDCD to M.H. (DC000088) and R.J.M. (DC000086). The authors would like to acknowledge Matthew W. Kelley and Doris Wu who provided helpful feedback and review of this paper. The authors acknowledge Alan Hoofring for his illustrations. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (https://biowulf.nih.gov).
Author contributions
S.G. performed snRNA-Seq data analysis, interpretation, drafted the manuscript, and contributed to fluorescent in situ hybridization. R.O. performed stria vascularis dissection and single nucleus isolation, fluorescent in situ hybridization and imaging. I.T. contributed to stria vascularis dissection and single nucleus isolation, imaging, and snRNA-Seq data analysis. Z.W. contributed to single nucleus capture. D.M. and R.M. were responsible for sequencing and alignment of snRNA-Seq datasets. M.H. conceptualized, designed, and supervised the study, performed data interpretation, and drafted and revised the manuscript. All authors reviewed the manuscript.
Data availability
All scripts for this study can be found in (https://github.com/Hoa-Lab/2020_Spindle-Root). All data generated in these studies have been deposited in the Gene Expression Omnibus (GEO) database (GEO Accession ID: GSE152551) and can be found on GEO [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152551]). The data has also been uploaded into the gene Expression Analysis Resource (gEAR), a website for visualization and comparative analysis of multi-omic data, with an emphasis on hearing research (https://umgear.org/p?l=58911b5d)93.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75238-8.
References
- 1.Kikuchi K, Hilding DA. The development of the stria vascularis in the mouse. Acta Otolaryngol. 1966 doi: 10.3109/00016486609119573. [DOI] [PubMed] [Google Scholar]
- 2.Korrapati S, et al. Single cell and single nucleus RNA-Seq reveal cellular heterogeneity and homeostatic regulatory networks in adult mouse stria vascularis. Front. Mol. Neurosci. 2019 doi: 10.3389/fnmol.2019.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wangemann P. K+ cycling and the endocochlear potential. Hear. Res. 2002 doi: 10.1016/S0378-5955(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 4.Wangemann P. Comparison of ion transport mechanisms between vestibular dark cells and strial marginal cells. Hear. Res. 1995 doi: 10.1016/0378-5955(95)00157-2. [DOI] [PubMed] [Google Scholar]
- 5.Marcus DC, Wu T, Wangemann P, Kofuji P. KCNJ10 (Kir4.1) potassium channel knockout abolishes endocochlear potential. Am. J. Physiol. Physiol. 2013;1:2. doi: 10.1152/ajpcell.00312.2001. [DOI] [PubMed] [Google Scholar]
- 6.Gow A. Deafness in claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J. Neurosci. 2004 doi: 10.1523/jneurosci.1640-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Q, et al. Virally mediated Kcnq1 gene replacement therapy in the immature scala media restores hearing in a mouse model of human Jervell and Lange-Nielsen deafness syndrome. EMBO Mol. Med. 2015 doi: 10.15252/emmm.201404929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faridi R, et al. Mutational and phenotypic spectra of KCNE1 deficiency in Jervell and lange-Nielsen syndrome and Romano-Ward syndrome. Hum. Mutat. 2019 doi: 10.1002/humu.23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wangemann P, et al. Loss of KCNJ10 protein expression abolishes endocochlear potential and causes deafness in Pendred syndrome mouse model. BMC Med. 2004 doi: 10.1186/1741-7015-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitajiri SI, et al. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear. Res. 2004 doi: 10.1016/S0378-5955(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 11.Ito T, et al. Slc26a4-insufficiency causes fluctuating hearing loss and stria vascularis dysfunction. Neurobiol. Dis. 2014 doi: 10.1016/j.nbd.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishio A, et al. Slc26a4 expression prevents fluctuation of hearing in a mouse model of large vestibular aqueduct syndrome. Neuroscience. 2016 doi: 10.1016/j.neuroscience.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Nathans J. Estrogen-related receptor β/NR3B2 controls epithelial cell fate and endolymph production by the stria vascularis. Dev. Cell. 2007;13:325–337. doi: 10.1016/j.devcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Neng L, Zhang F, Kachelmeier A, Shi X. Endothelial cell, pericyte, and perivascular resident macrophage-type melanocyte interactions regulate cochlear intrastrial fluid-blood barrier permeability. JARO J. Assoc. Res. Otolaryngol. 2013 doi: 10.1007/s10162-012-0365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X. Pathophysiology of the cochlear intrastrial fluid-blood barrier (review) Hear. Res. 2016 doi: 10.1016/j.heares.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagger DJ, Nevill G, Forge A. The membrane properties of cochlear root cells are consistent with roles in potassium recirculation and spatial buffering. JARO J. Assoc. Res. Otolaryngol. 2010 doi: 10.1007/s10162-010-0218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagger DJ, Forge A. The enigmatic root cell—emerging roles contributing to fluid homeostasis within the cochlear outer sulcus. Hear. Res. 2013 doi: 10.1016/j.heares.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 2019 doi: 10.1681/asn.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaublomme JT, et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun. 2019 doi: 10.1038/s41467-019-10756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng W, et al. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucl. Acids Res. 2016 doi: 10.1093/nar/gkw739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu P, et al. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018 doi: 10.1101/gad.316802.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alles J, et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 2017;15:44. doi: 10.1186/s12915-017-0383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denisenko E, et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. bioRxiv. 2019 doi: 10.1101/832444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, et al. PBMC fixation and processing for chromium single-cell RNA sequencing. J. Transl. Med. 2018 doi: 10.1186/s12967-018-1578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019 doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran HTN, et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020 doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hibino H, Nin F, Tsuzuki C, Kurachi Y. How is the highly positive endocochlear potential formed? The specific architecture of the stria vascularis and the roles of the ion-transport apparatus. Pflugers Arch. 2010 doi: 10.1007/s00424-009-0754-z. [DOI] [PubMed] [Google Scholar]
- 29.Shi F, Kempfle JS, Edge ASB. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J. Neurosci. 2012 doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanada Y, et al. Epiphycan is specifically expressed in cochlear supporting cells and is necessary for normal hearing. Biochem. Biophys. Res. Commun. 2017 doi: 10.1016/j.bbrc.2017.08.092. [DOI] [PubMed] [Google Scholar]
- 31.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017 doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suo S, et al. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, et al. A systems-level approach reveals new gene regulatory modules in the developing ear. Development. 2017 doi: 10.1242/dev.148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrill S, He DZZ. Apoptosis in inner ear sensory hair cells. J. Otol. 2017 doi: 10.1016/j.joto.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen-Salmon M, Mattei MG, Petit C. Mapping of the otogelin gene (OTGN) to mouse Chromosome 7 and human chromosome 11p14.3: a candidate for human autosomal recessive nonsyndromic deafness DFNB18. Mamm. Genome. 1999 doi: 10.1007/s003359901033. [DOI] [PubMed] [Google Scholar]
- 36.Simmler MC, et al. Targeted disruption of Otog results in deafness and severe imbalance. Nat. Genet. 2000 doi: 10.1038/72793. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Yosef T, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum. Mol. Genet. 2003 doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 38.Gallego-Martinez A, Lopez-Escamez JA. Genetic architecture of Meniere’s disease. Hear. Res. 2019 doi: 10.1016/j.heares.2019.107872. [DOI] [PubMed] [Google Scholar]
- 39.Wilcox ER, et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001 doi: 10.1016/S0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- 40.Li C, et al. Fate-mapping analysis using Rorb-IRES-Cre reveals apical-to-basal gradient of Rorb expression in mouse cochlea. Dev. Dyn. 2020 doi: 10.1002/dvdy.111. [DOI] [PubMed] [Google Scholar]
- 41.Peters TA, Kuijpers W, Curfs JHAJ. Occurrence of NaK-ATPase isoforms during rat inner ear development and functional implications. Eur. Arch. Oto-Rhino-Laryngol. 2001 doi: 10.1007/s004050000304. [DOI] [PubMed] [Google Scholar]
- 42.Stanković KM, Brown D, Alper SL, Adams JC. Localization of pH regulating proteins H+ATPase and Cl-/HCO3/- exchanger in the guinea pig inner ear. Hear. Res. 1997 doi: 10.1016/S0378-5955(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 43.Royaux IE, et al. Localization and functional studies of pendrin in the mouse inner ear provide insight about the etiology of deafness in pendred syndrome. JARO J. Assoc. Res. Otolaryngol. 2003 doi: 10.1007/s10162-002-3052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hibino H, et al. An ATP-dependent inwardly rectifying potassium channel, K AB-2 (Kir4.1), in cochlear stria vascularis of inner ear: its specific subcellular localization and correlation with the formation of endocochlear potential. J. Neurosci. 2018 doi: 10.1523/jneurosci.17-12-04711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, et al. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1415901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi F, Hu L, Edge ASB. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl. Acad. Sci. 2013;110:13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groves AK. The challenge of hair cell regeneration. Exp. Biol. Med. 2010 doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge ASB. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2014 doi: 10.1016/j.stemcr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lenz DR, et al. Applications of Lgr5-positive cochlear progenitors (LCPs) to the study of hair cell differentiation. Front. Cell Dev. Biol. 2019 doi: 10.3389/fcell.2019.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakashima H, et al. R-spondin 2 promotes acetylcholine receptor clustering at the neuromuscular junction via Lgr5. Sci. Rep. 2016 doi: 10.1038/srep28512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan KM, Drescher MJ, Hatfield JS, Khan AM, Drescher DG. Muscarinic receptor subtypes are differentially distributed in the rat cochlea. Neuroscience. 2002 doi: 10.1016/S0306-4522(02)00020-9. [DOI] [PubMed] [Google Scholar]
- 52.Wangemann P, et al. Muscarinic receptors control K+ secretion in inner ear strial marginal cells. J. Membr. Biol. 2001 doi: 10.1007/s00232-001-0042-0. [DOI] [PubMed] [Google Scholar]
- 53.McNicholas K, Chen T, Abbott CA. Dipeptidyl peptidase (DP) 6 and DP10: Novel brain proteins implicated in human health and disease. Clin. Chem. Lab. Med. 2009 doi: 10.1515/CCLM.2009.061. [DOI] [PubMed] [Google Scholar]
- 54.Kalinec F, et al. Glucocorticoid-stimulated, transcription-independent release of annexin A1 by cochlear Hensen cells. Br. J. Pharmacol. 2009 doi: 10.1111/j.1476-5381.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richer MJ, Lang ML, Butler NS. T cell fates zipped up: how the Bach2 basic leucine zipper transcriptional repressor directs T cell differentiation and function. J. Immunol. 2016 doi: 10.4049/jimmunol.1600847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Wu H, Zhao M, Chang C, Lu Q. The bach family of transcription factors: a comprehensive review. Clin. Rev. Allergy Immunol. 2016 doi: 10.1007/s12016-016-8538-7. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, et al. Bach2 deficiency leads to spontaneous expansion of IL-4-producing T follicular helper cells and autoimmunity. Front. Immunol. 2019 doi: 10.3389/fimmu.2019.02050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itoh-Nakadai A, et al. The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nat. Immunol. 2014 doi: 10.1038/ni.3024. [DOI] [PubMed] [Google Scholar]
- 59.Crotty S. T Follicular helper cell differentiation, function, and roles in disease. Immunity. 2014 doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueno H. T follicular helper cells in human autoimmunity. Curr. Opin. Immunol. 2016 doi: 10.1016/j.coi.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Roychoudhuri R, et al. BACH2 represses effector programs to stabilize T reg-mediated immune homeostasis. Nature. 2013 doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Chen S, Zhao Q, Sun Y, Nie H. The critical role of Bach2 in shaping the balance between CD4+ T cell subsets in immune-mediated diseases. Mediat. Inflamm. 2019 doi: 10.1155/2019/2609737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ebina-Shibuya R, et al. Inflammatory responses induce an identity crisis of alveolar macrophages, leading to pulmonary alveolar proteinosis. J. Biol. Chem. 2017 doi: 10.1074/jbc.M117.808535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown KK, et al. NR2F1 deletion in a patient with a de novo paracentric inversion, inv(5)(q15q33.2), and syndromic deafness. Am. J. Med. Genet. Part A. 2009 doi: 10.1002/ajmg.a.32764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen CA, et al. The expanding clinical phenotype of Bosch-Boonstra-Schaaf optic atrophy syndrome: 20 new cases and possible genotype-phenotype correlations. Genet. Med. 2016 doi: 10.1038/gim.2016.18. [DOI] [PubMed] [Google Scholar]
- 66.Tang LS, Alger HM, Lin F, Pereira FA. Dynamic expression of COUP-TFI and COUP-TFII during development and functional maturation of the mouse inner ear. Gene Exp. Patterns. 2005 doi: 10.1016/j.modgep.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 67.Tarchini B, et al. A spontaneous mouse deletion in Mctp1 uncovers a long-range cis-regulatory region crucial for NR2F1 function during inner ear development. Dev. Biol. 2018 doi: 10.1016/j.ydbio.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bergeron KF, et al. Upregulation of the Nr2f1-A830082K12Rik gene pair in murine neural crest cells results in a complex phenotype reminiscent of Waardenburg syndrome type 4. DMM Dis. Model. Mech. 2016 doi: 10.1242/dmm.026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huebner AK, et al. Early hearing loss upon disruption of Slc4a10 in C57BL/6 Mice. JARO J. Assoc. Res. Otolaryngol. 2019 doi: 10.1007/s10162-019-00719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morell RJ, et al. Noncoding microdeletion in mouse Hgf disrupts neural crest migration into the stria vascularis, reduces the endocochlear potential, and suggests the neuropathology for human nonsyndromic deafness DFNB39. J. Neurosci. 2020 doi: 10.1523/JNEUROSCI.2278-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu H, et al. Organ of corti and stria vascularis: Is there an interdependence for survival? PLoS ONE. 2016 doi: 10.1371/journal.pone.0168953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishiyama G, López IA, Ishiyama A. Aquaporins and Meniere’s disease. Curr. Opin. Otolaryngol. Head Neck Surg. 2006 doi: 10.1097/01.moo.0000244191.51560.22. [DOI] [PubMed] [Google Scholar]
- 73.Crossley J, Hussaini AS, Kim HJ, Hoa M. Ménière’s disease clinical subtypes in a population from the USA. J. Laryngol. Otol. 2020 doi: 10.1017/S002221511900255X. [DOI] [PubMed] [Google Scholar]
- 74.Hoa M, Friedman RA, Fisher LM, Derebery MJ. Prognostic implications of and audiometric evidence for hearing fluctuation in Meniere’s disease. Laryngoscope. 2015 doi: 10.1002/lary.25579. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Aramaki M, Fu Y, Forrest D. Retinoid-related orphan receptor β and transcriptional control of neuronal differentiation. Curr. Top. Dev. Biol. 2017 doi: 10.1016/bs.ctdb.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Lin E, et al. Effects of circadian clock genes and healthrelated behavior on metabolic syndrome in a Taiwanese population: evidence from. PLoS ONE. 2017 doi: 10.1371/journal.pone.0173861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamphuis W, Cailotto C, Dijk F, Bergen A, Buijs RM. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem. Biophys. Res. Commun. 2005 doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- 78.Gallego-Martinez A, Requena T, Roman-Naranjo P, Lopez-Escamez JA. Excess of rare missence variants in hearing loss genes in sporadic meniere disease. Front. Genet. 2019 doi: 10.3389/fgene.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roman-Naranjo P, et al. Burden of rare variants in the OTOG gene in familial meniere’s disease. Ear Hear. 2020 doi: 10.1097/aud.0000000000000878. [DOI] [PubMed] [Google Scholar]
- 80.Skene NG, Grant SGN. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 2016 doi: 10.3389/fnins.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenberg AB, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018;360:176–182. doi: 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018 doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019 doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;80:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Den Brink SC, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017 doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 87.Miao Z, Deng K, Wang X, Zhang X. DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty332. [DOI] [PubMed] [Google Scholar]
- 88.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucl. Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pazhouhandeh M, et al. Comparative network analysis of patients with non-small cell lung cancer and smokers for representing potential therapeutic targets. Sci. Rep. 2017;7:13812. doi: 10.1038/s41598-017-14195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jagannathan R, et al. Systems biology genetic approach identifies serotonin pathway as a possible target for obstructive sleep apnea: results from a literature search review. Sleep Disord. 2017;2017:1–8. doi: 10.1155/2017/6768323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoa M, et al. Characterizing adult cochlear supporting cell transcriptional diversity using single-cell RNA-seq: validation in the adult mouse and translational implications for the adult human cochlea. Front. Mol. Neurosci. 2020 doi: 10.3389/fnmol.2020.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orvis, J. et al. gEAR: gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts for this study can be found in (https://github.com/Hoa-Lab/2020_Spindle-Root). All data generated in these studies have been deposited in the Gene Expression Omnibus (GEO) database (GEO Accession ID: GSE152551) and can be found on GEO [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152551]). The data has also been uploaded into the gene Expression Analysis Resource (gEAR), a website for visualization and comparative analysis of multi-omic data, with an emphasis on hearing research (https://umgear.org/p?l=58911b5d)93.