Abstract
The aim of the present study was to investigate the neuroprotective roles and mechanisms of the circular RNA circSHOC2 in ischemic-preconditioned astrocyte-derived exosomes (IPAS-EXOs) against ischemic stroke. We established an ischemia model based on oxygen glucose deprivation (OGD) in vitro and isolated resultant exosomes from astrocytes. Neuronal viability and apoptosis were determined by Cell Counting Kit-8 (CCK-8) assays and TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) staining, respectively. Autophagy-related proteins were analyzed by western blotting. We found that exosomes derived from IPAS-preconditioned medium (IPAS-CM) exerted neuroprotection. Furthermore, circSHOC2 expression was significantly upregulated in exosomes released from IPAS-CM. Overexpression of circSHOC2 in neurons yielded the same protective effects as those from IPAS-EXOs in vitro, and similar results were also observed in the middle cerebral artery occlusion (MCAO) mouse model. Mechanistically, circSHOC2 reduced neuronal apoptosis via regulating autophagy. Furthermore, circSHOC2 was found to sponge miR-7670-3p, which regulated SIRT1 expression. Transfection with an miR-7670-3p small interfering RNA (siRNA) (siRNA-7670-3p) and incubation with circSHOC2 extracellular vesicles attenuated ischemia-induced neuronal apoptosis in vivo and in vitro, while silencing of SIRT1 reversed the protective effects of exosomal circSHOC2 on hypoxic cerebral neurons. Taken together, our findings indicate that circSHOC2 in IPAS-EXOs suppressed neuronal apoptosis and ameliorated neuronal damage by regulating autophagy and acting on the miR-7670-3p/SIRT1 axis, which might contribute to a therapeutic strategy for ischemic stroke treatment.
Keywords: circRNA, ischemic-preconditioned astrocyte-derived exosomes, OGD, MCAO, miR-7670-3p, autophagy, neuroprotection, SIRT1
Graphical Abstract
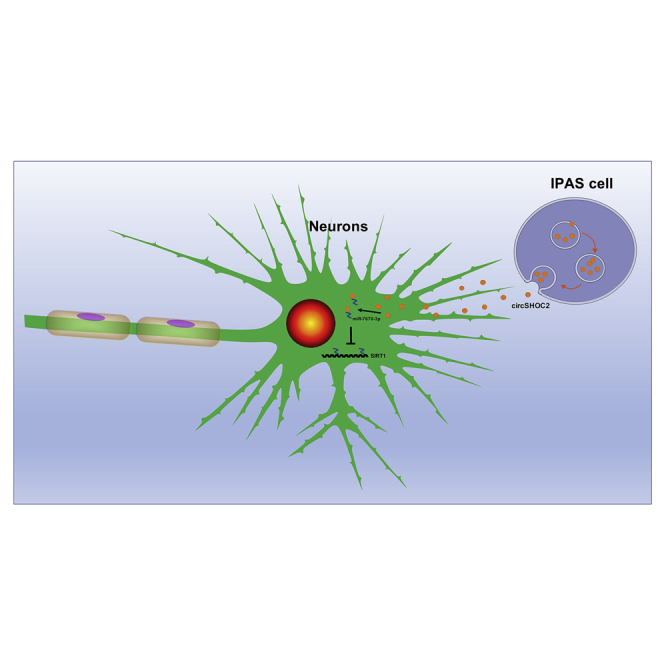
Chen and colleagues found that exosomes from ischemic-preconditioned astrocytes could alleviate the severity of ischemic stroke. Exosome-delivered circSHOC2 exerts neuroprotective effects via modulating the miR-7670-3p/SIRT1 pathway in ischemic stroke. These findings may serve as a novel treatment therapeutic strategy in patients with ischemic stroke.
Introduction
Ischemic stroke is a central nervous system disease and one of the main causes of permanent morbidity and disability worldwide.1,2 Therefore, there is a continued need to elucidate novel molecular mechanisms and identify therapeutic targets to control the severity of ischemic stroke. To develop ideal therapeutic methods and expand therapeutic targets, therapeutic approaches that are beneficial to a variety of cell types must be considered. Among the variety of cell types that could be targeted, targeting astrocytes in ischemic stroke is particularly promising.3 It is unclear whether astrocytes that are activated after ischemic stroke exert harmful or beneficial effects on neuronal survival. However, recent studies have shown that treatments that reduce infarct size are usually accompanied by a weakened astrocytic response.4,5 Therefore, discovering ways to promote the beneficial aspects of astrocytes and/or reduce their deleterious aspects may represent an effective strategy for treating ischemic stroke.
In the past decade, exosomes have been used as a treatment strategy for ischemic stroke.6,7 Exosomes are extracellular vesicles with a diameter of 30–100 nm that are secreted by all cell types and are able to cross the blood-brain barrier (BBB).8,9 Exosomes contain various circular RNAs (circRNAs), proteins, mRNAs, and DNA that can function locally or be stably transferred to recipient cells.10, 11, 12 For example, astrocytes exposed to amyloid β release exosomes with heat shock protein B1 have a protective effect on neurodegeneration.13 The neural-derived exosome miR-132 has been reported to act as an intercellular signal that mediates neurovascular communication.14,15 Collectively, these findings indicate the important role of exosomes in neurological diseases; however, the roles and mechanisms of exosomes in ischemic stroke remain poorly understood.
Autophagy is a highly conserved catabolic pathway in cells that can degrade and recover long-lived proteins and damaged organelles through lysosomal digestion.16,17 Autophagy is active at basal levels under normal conditions but rapidly increases after exposure to stress factors (e.g., nutritional/hormonal deficiencies, hypoxic stress, misfolded protein accumulation, and bacterial invasion).18 In recent years, increasing evidence has demonstrated that autophagy is activated after ischemic stroke and may be involved in the occurrence and development of ischemic stroke.19 However, the mechanisms regulating autophagy remain unclear. Many studies have indicated that circRNAs play an important role in the regulation of autophagy.20,21 circRNAs are endogenous non-coding RNAs that have covalently closed circular structures. circRNAs are highly homologous and are usually more stable than their linear counterparts because they lack accessible ends and are consequently resistant to exonucleases.22 Although the biological importance of circRNAs has been revealed, little is known about their expression patterns and biological functions in the pathogenesis of ischemic stroke.
In this study, we investigated the protective role of ischemic-preconditioned astrocyte-derived exosomes (IPAS-EXOs) in ischemic stroke and determined whether they regulate autophagy. Using circRNA microarray analysis, we found the top five upregulated circRNAs in IPAS-EXOs. Among these candidates, we found that after ischemic treatment, a circRNA transcribed from the SHOC2 gene, named as circSHOC2 (circ_0092670), had significantly increased expression in IPAS-EXOs. Additionally, silencing circSHOC2 promoted neuronal apoptosis. We further demonstrated that circSHOC2 acted as a sponge for miR-7670-3p, thereby upregulating sirtuin 1 (SIRT1) levels and subsequently inhibiting apoptosis of gliomas. Our findings reveal a novel role of circSHOC2 in regulating neuronal apoptosis and suggest that the circSHOC2/miR-7670-3p/SIRT1 axis may represent a therapeutic target in patients with ischemic brain injury.
Results
IPAS-Preconditioned Medium Provides Neuroprotection in Primary Neurons In Vitro
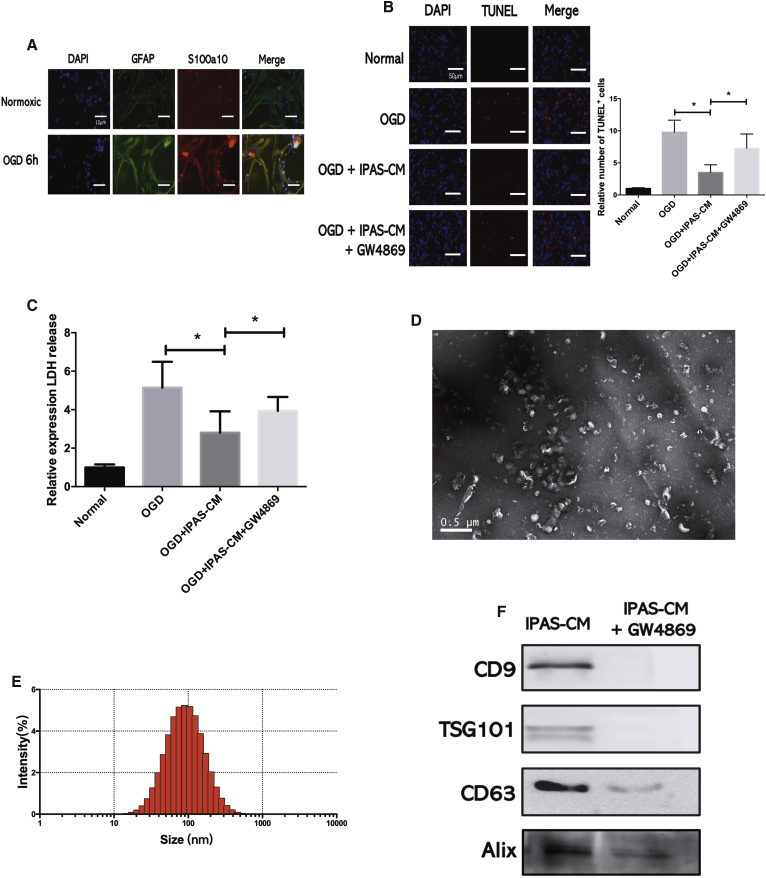
To investigate whether IPAS-preconditioned medium (IPAS-CM) confers neuroprotection, astrocytes were subjected to oxygen glucose deprivation (OGD) for 6 h. The expression levels of the astrocytic markers, glial fibrillary acidic protein (GFAP) and S100A10, were significantly increased in the IPAS group (Figure 1A). IPAS-CM was collected at 24 h after reoxygenation. Then, TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) staining was performed to determine the amount of apoptosis that occurred after OGD in neurons treated with IPAS-CM. IPAS-CM significantly mitigated OGD-induced loss of neuronal viability, whereas culture medium from control astrocytes had no effect on neuronal survival (Figure 1B). To quantify the protective effect of IPAS-CM on neurons that underwent OGD, lactate dehydrogenase (LDH) assays were performed for rapid quantification of cellular injury. OGD treatment increased cellular injury 6.5-fold, whereas the addition of IPAS-CM decreased OGF-induced cellular injury to 3.0-fold that of the control group. Moreover, GW4869, a small-molecule inhibitor that reduces extracellular vesicle secretion, partly reversed the neuroprotective effects of IPAS-CM (Figure 1C). These results suggested that IPAS-CM-derived extracellular vesicles protected neurons from lethal ischemic injury. Furthermore, exosomes were isolated from IPAS-CM via ultracentrifugation and were subsequently identified via transmission electron microscopy (TEM) (Figure 1D). As shown in Figure 1D, typical exosomal morphology—observed and analyzed via TEM and the Zetasizer Nano-ZS—revealed that the diameter of particles ranged from 50 to 150 nm (Figure 1E). Additionally, typical exosomal markers (i.e., CD9, ALIX, tumor susceptibility gene 101 [TSG101], and CD63) were detected by western blotting in the collected vesicular fractions (Figure 1F). Hence, these isolated vesicles were considered to be IPAS-EXOs.
Figure 1.
IPAS-Preconditioned Medium Provides Neuroprotection in Primary Neurons In Vitro
(A) Immunofluorescence of astrocytes under normoxic conditions or after 6 h of OGD. (B) TUNEL staining was performed to evaluate neuronal death and apoptosis as follows: (1) neurons under normal culture conditions (normal); (2) neurons following OGD treatment (OGD); (3) neurons pretreated with IPAS-preconditioned medium (IPAS-CM) and exposed to OGD (OGD+CM); and (4) neurons pretreated with IPAS-CM plus GW4869 and exposed to OGD injury (OGD+CM+GW4869). ∗p < 0.05. (C) LDH release of neurons treated with IPAS-CM plus GW4869 after OGD injury. ∗p < 0.05. (D) Exosomes were observed under a transmission electron microscope. (E) The size of exosomes was assessed by Nano-ZS analysis. (F) Expression levels of exosomal markers (CD9, TSG101, CD63, and Alix) were measured by western blotting.
IPAS-EXOs Provide Neuroprotection Both In Vitro and In Vivo
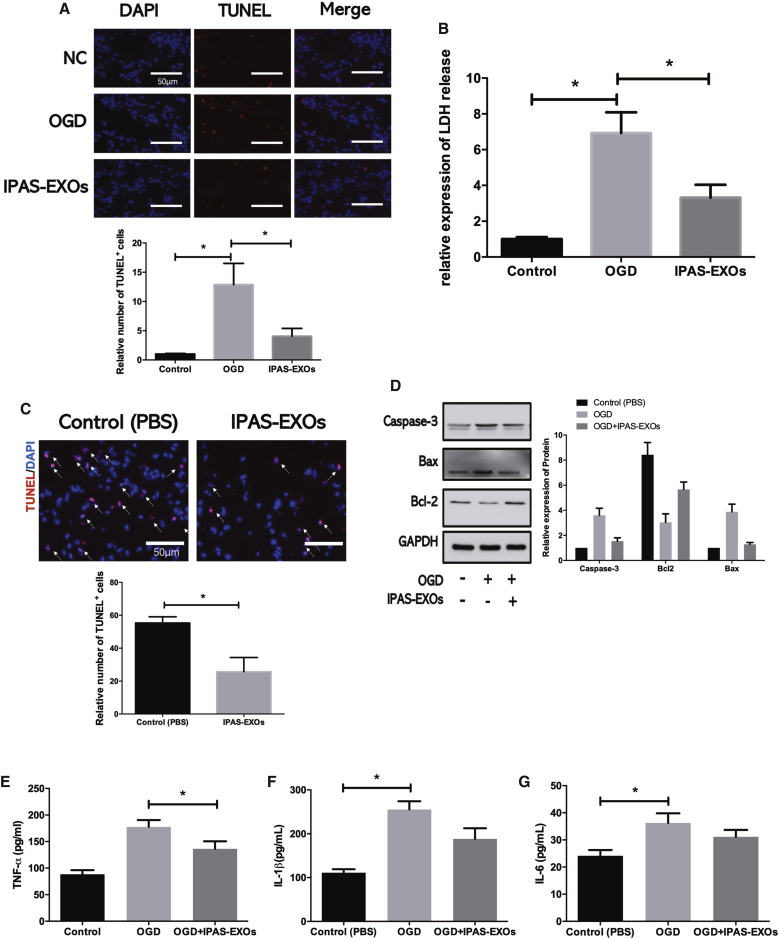
To examine whether IPAS-EXOs were internalized by neurons to provide neuroprotection, TUNEL staining and LDH assays were performed and demonstrated that OGD led to an increase in neuronal apoptosis, which was ameliorated by coculturing neurons with IPAS-EXOs (Figures 2A and 2B). Furthermore, the IPAS-EXO group had fewer apoptotic neurons than did the PBS group (Figure 2C). Western blotting showed that compared with the PBS group, the OGD-induced levels of caspase-3 and Bax, which are apoptosis-related proteins, were significantly reduced in the IPAS-EXO group. Furthermore, IPAS-EXO overexpression significantly promoted Bcl-2 expression, which functions to inhibit apoptosis, in OGD-treated neurons (Figure 2D). In addition, tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β levels were significantly reduced in neurons after treatment with OGD IPAS-EXOs compared with the OGD PBS group, as measured by enzyme-linked immunosorbent assay (ELISA) (Figures 2E–2G). Next, to quantify the protective effect of IPAS-EXOs in neurons, we pretreated neurons with IPAS-EXOs at a range of concentrations (5, 10, 20, and 40 μg/mL) at 6 h before OGD. Cell survival was assessed by Cell Counting Kit-8 (CCK-8) assays. OGD-treated neurons that were pretreated with IPAS-EXOs exhibited significantly increased cellular viability compared with that of the OGD-treated group pretreated with PBS, with 20 μg/mL of IPAS-EXOs having the strongest neuroprotection effect (Figure S1A). We next examined the effects of IPAS-EXOs in ischemic mice established via middle cerebral artery occlusion (MCAO). Dil-labeled exosomes were detected in ischemic mouse brains after 3 days of IPAS-EXO injections (20 μg/mL). Compared with the PBS group, treatment with IPAS-EXOs significantly reduced the infarct volume at 3 days after MCAO (Figure S1B). The modified neurological severity scores (mNSSs) further showed that treatment with IPAS-EXOs greatly attenuated neurobehavioral deficits at 3 days after MCAO compared with PBS treatment (Figure S1C). Collectively, these results demonstrate that IPAS-EXOs inhibited OGD-induced neuronal apoptosis.
Figure 2.
IPAS-EXOs Provide Neuroprotection Both In Vitro and In Vivo
(A) TUNEL staining showing apoptotic rates of neurons treated with IPAS-EXOs after OGD. The bar graph shows the number of TUNEL+ neurons. ∗p < 0.05. (B) LDH assays showing the cellular viability of neurons treated with IPAS-EXOs after OGD. ∗p < 0.05. (C) Staining and statistical analysis of TUNEL+ cells in mice treated with PBS or IPAS-EXOs. ∗p < 0.05. (D) The effect of IPAS-EXOs on the relative expression of apoptosis-related proteins in OGD-treated neurons was analyzed by western blotting. ∗p < 0.05. (E) The effect of IPAS-EXOs on levels of TNF-α in OGD-treated neurons was analyzed by ELISA. ∗p < 0.05. (F) The effect of IPAS-EXOs on levels of IL-6 in OGD-treated neurons was analyzed by ELISA. ∗p < 0.05. (G) The effect of IPAS-EXOs on levels of IL-1β in OGD-treated neurons was analyzed by ELISA. ∗p < 0.05.
IPAS-EXOs Are Internalized by Neurons and Ameliorate Ischemic-Induced Neuronal Damage via Regulating Autophagy Both In Vitro and In Vivo
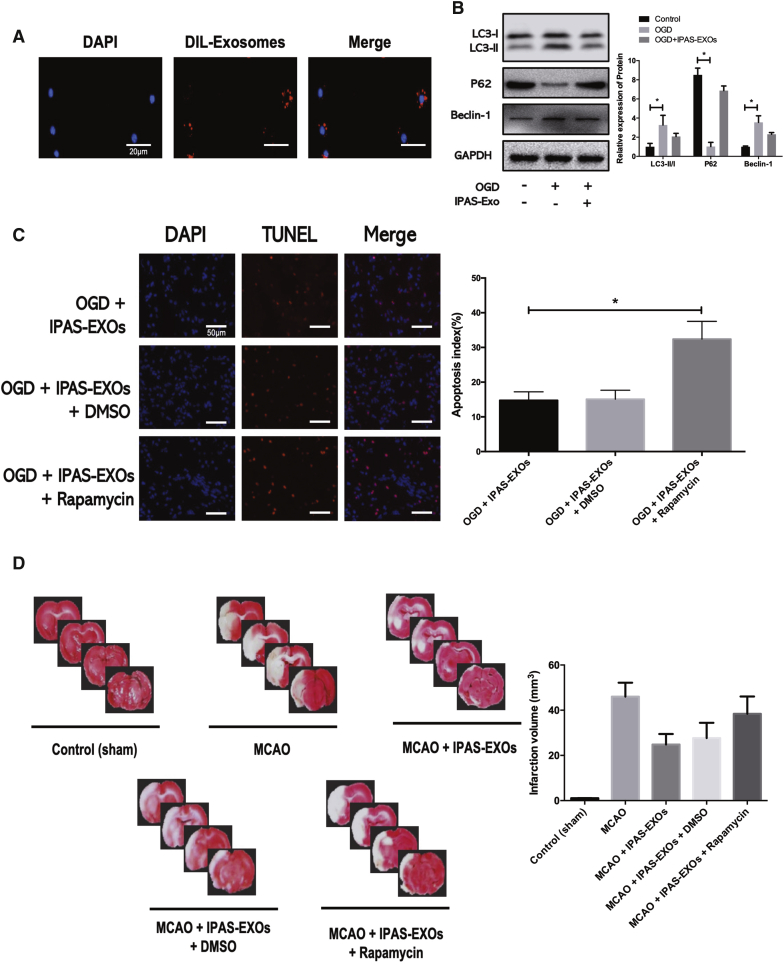
To further investigate the effects of IPAS-EXOs on neuronal apoptosis after OGD culturing in vitro, DiI-stained IPAS-EXOs (red) and neurons were co-incubated for 24 h. Confocal laser-scanning microscopy showed that co-incubated IPAS-EXOs were localized within the cytoplasm of MAP2-positive neurons (green), indicating that IPAS-EXOs were internalized by DAPI-labeled neurons (Figure 3A). Western blotting showed that IPAS-EXOs reduced the ratio of LC3-II/LC3-I to Beclin-1 (two autophagy-related proteins) and increased the expression level of p62 (Figure 3B). These results indicated that IPAS-EXOs inhibited OGD-induced neuronal death by regulating autophagy. To further determine the effects of IPAS-EXOs on neuronal autophagy, we used the known autophagic activator, rapamycin, to induce autophagy. LDH analysis showed that IPAS-EXOs inhibited neuronal apoptosis, whereas rapamycin promoted apoptosis. As shown in Figure 3C, when autophagy was activated via rapamycin, the capacity of IPAS-EXOs to inhibit neuronal apoptosis was significantly impaired. To determine whether IPAS-EXOs could also exert neuroprotection by regulating neuronal autophagy in vivo, IPAS-EXOs were injected into the tail veins of MCAO-induced ischemic mice. Three days after MCAO, coronary brain sections were collected, and the infarct volume was measured via 2,3,5-triphenyltetrazolium chloride (TTC) staining. TTC staining showed that the infarct volume in the MCAO group was significantly larger than that in the control group; in contrast, IPAS-EXO treatment significantly reduced the MCAO-induced infarct volume, whereas this effect was reversed by autophagic activation via rapamycin (Figure 3D). TUNEL staining was also used to detect neuronal apoptosis in brain tissues, and the results confirmed that rapamycin reversed the anti-apoptotic effects of IPAS-EXOs in neurons (Figure S2). Therefore, these results indicate that IPAS-EXOs inhibited ischemia-induced neuronal apoptosis, both in vitro and in vivo, by regulating autophagy.
Figure 3.
IPAS-EXOs Are Internalized by Neurons and Ameliorate Ischemia-Induced Neuronal Damage via Regulating Autophagy Both In Vitro and In Vivo
(A) Confocal imaging showing that Dil-labeled exosomes (red) were internalized by neurons (green) in vitro. (B) The effects of IPAS-EXOs on the levels of autophagy-related genes (Beclin-1, LC3-I, LC3-II, and p62) in OGD-treated neurons were analyzed by western blotting. (C) Activation of autophagy reversed the pro-proliferation effect of IPAS-EXOs on neurons after OGD. ∗p < 0.05. (D) Infarct volumes of mice in each group were measured by TTC staining.
circSHOC2 Is Highly Expressed in IPAS-EXOs and Confers Neuroprotection
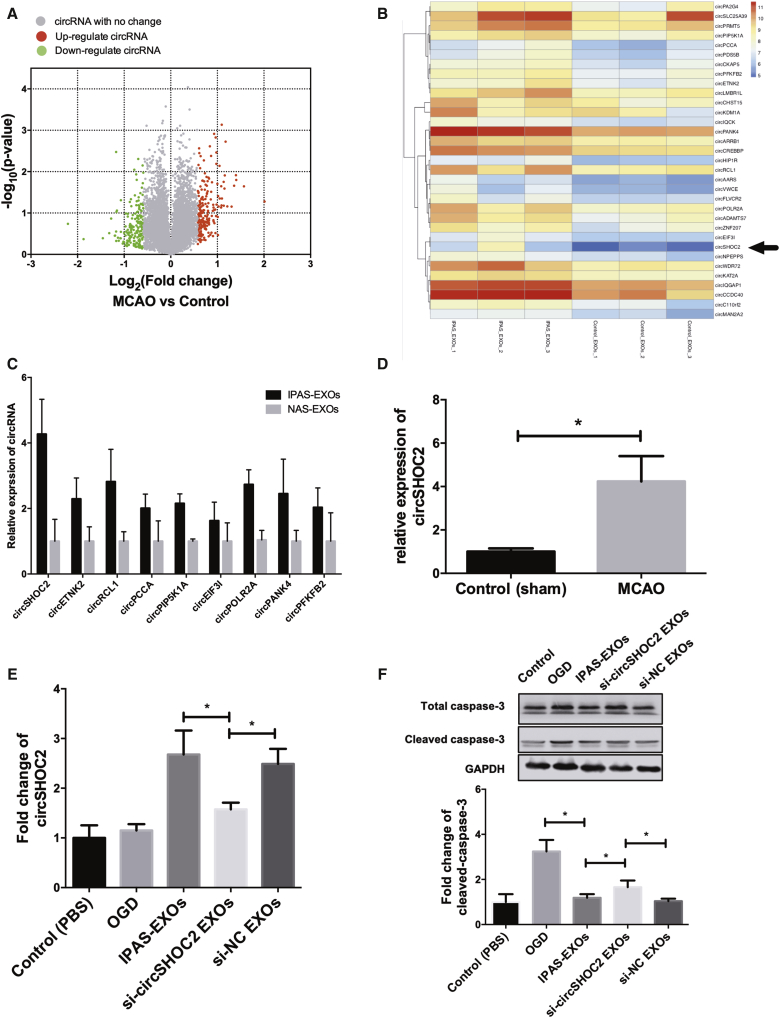
Exosomes are enriched with circRNAs and are able to deliver circRNAs from host cells to target cells to further regulate physiological functions within recipient cells. We identified specific circRNAs that were expressed in IPAS-EXOs (Figures 4A and 4B). The top 10 upregulated circRNAs were selected for validation of circRNA microarray results via quantitative real-time PCR (Figure 4C). The results showed that circSHOC2, circRCL1, circPOLR2A, and circPANK4 levels were upregulated 4.3-, 3.5-, 3.0-, and 2.1-fold, respectively, in the IPAS-EXO group compared with the PBS group. As circSHOC2 was the most prominent circRNA among them, we focused on circSHOC2 for further analysis. We confirmed that the expression level of circSHOC2 was significantly increased in IPAS-EXOs compared with those from normal astrocytes (Figure 4D). Therefore, we hypothesized that IPAS-EXOs exert neuroprotective effects in the ischemic mouse brain via the transfer of circSHOC2 into neurons. To test our hypothesis, we performed a series of validation assays using loss-of-function and gain-of-function experiments. Transfection of Lipofectamine 3000 carrying small-interfering RNAs (siRNAs) that specifically target circSHOC2 were utilized to knock down circSHOC2 expression in astrocytes. Quantitative real-time PCR showed that the expression of circSHOC2 was downregulated not only in IPAS-EXOs but also in neurons treated with si-circSHOC2 (i.e., constructs containing a circSHOC2 siRNA) IPAS-EXOs (Figure 4E). As shown in Figure 4F, western blotting revealed that caspase-3 expression was reduced in the IPAS-EXO-treated ischemic group, whereas treatment with si-circSHOC2 IPAS-EXOs rescued this ischemia-induced caspase-3 expression. An LDH assay also demonstrated that treatment with si-circSHOC2 IPAS-EXOs partly reversed the neuroprotective effects of IPAS-EXOs under ischemic conditions (Figure S3A). Importantly, treatment with either IPAS-EXOs or si-circRNA IPAS-EXOs had no effect on neuronal death under normal conditions (Figure S3B). Furthermore, we overexpressed circSHOC2 using circRNA mimics. As depicted in Figure S3C, the number of TUNEL-positive cells was reduced in the IPAS-EXO-treated group, whereas the number of apoptotic cells was comparatively increased via treatment with si-circSHOC2 IPAS-EXOs. Taken together, these results suggest that IPAS-EXOs reduced OGD-induced neuronal apoptosis via a circSHOC2-dependent mechanism.
Figure 4.
circSHOC2 Is Highly Expressed in IPAS-EXOS and Confers Neuroprotection
(A) Differential expression levels of IPAS-EXOS-derived circRNAs were analyzed by circRNA microarray analysis. (B) A heatmap assessing variations in astrocyte-derived exosomal expression between normal astrocytes and IPASs. (C) Quantitative real-time PCR further validated the top nine upregulated exosomal miRNAs in IPAS-EXO groups. (D) Quantitative real-time PCR analysis was used to determine the expression levels of circSHOC2 in the ischemic cortex of mice from MCAO groups and the control group. ∗p < 0.05. (E) Expression of circRNAs in neurons treated with IPAS-EXOs, circSHOC2-knockdown exosomes (si-circSHOC2-EXOs), or control siRNA-NC EXOs after OGD. ∗p < 0.05. (F) Expression of total caspase-3 and cleaved caspase-3 in neurons treated with M2-EXOs, si-circSHOC2-EXOs, or control siRNA-NC EXOs after OGD. ∗p < 0.05.
circSHOC2 Inhibits OGD-Induced Neuronal Apoptosis via Promoting Autophagy
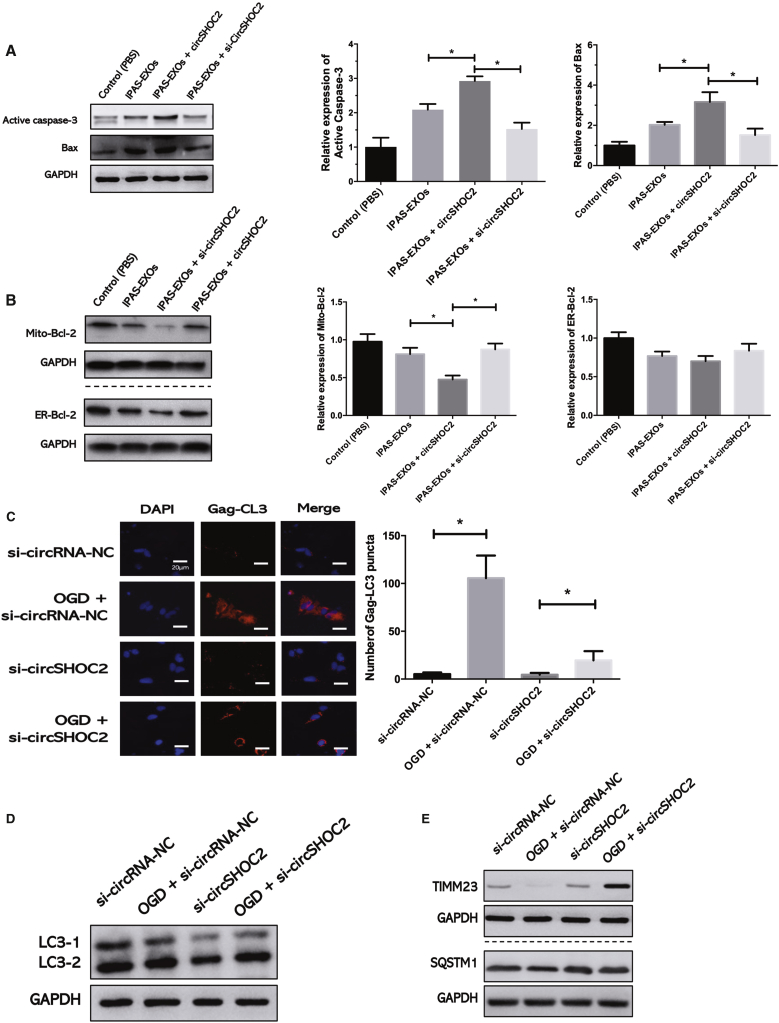
Next, we explored the mechanism underlying circSHOC2-mediated neuroprotection after focal ischemia. As determined by western blot analysis, we confirmed that the apoptotic genes Bax and caspase-3 were significantly increased in the ischemic brain at 24 h after si-circSHOC2 treatment (Figure 5A). Interestingly, si-circSHOC2 treatment only inhibited mitochondria-targeted Bcl-2 expression and not endoplasmic reticulum (ER)-targeted Bcl-2 (Figure 5B). Therefore, we next investigated whether autophagy was an upstream signaling target of these apoptotic genes. Past studies have suggested a link between Toll-like receptor (TLR) signaling and autophagy in neurons and other cell types via activation of downstream signaling pathways. To further test the effects of circSHOC2 on autophagy, we transfected cells with si-circSHOC2 and analyzed autophagy under OGD conditions. The expression levels of the mitochondrial proteins TIMM23 and sequestosome 1 (SQSTM1) were analyzed by western blotting as a readout of autophagy. We observed that si-circSHOC2 treatment significantly blocked OGD-induced Gag-LC3 accumulation (Figure 5C). OGD-activated lipid conjugation of free LC3-I to the phagophore membrane to generate LC3-II was attenuated following transfection with si-circSHOC2 (Figure 5D). Additionally, degradation of the mitochondrial inner membrane protein TIMM23 following OGD treatment was decreased in si-circSHOC2-transfected cellular extracts (Figure 5E). To further test the effects of circSHOC2 on autophagy, we transfected cells with circSHOC2 mimics and analyzed autophagy under OGD conditions. We observed that the OGD-dependent formation of Gag-LC3 puncta was modestly but significantly increased (Figure S4A) and that LC3-I-to-LC3-II conversion was stimulated (Figure S4B). Moreover, TIMM23 degradation following OGD was more prominent after transfection with a circSHOC2 mimic compared with that of the control group (Figure S4C). These results suggest that circSHOC2 is a novel circRNA that regulates autophagy.
Figure 5.
circSHOC2 Inhibits OGD-Induced Neuronal Apoptosis via Promoting Autophagy
(A) The expression levels of Bax and caspase-3 proteins after si-circSHOC2 treatment. ∗p < 0.05. (B) The effects of a si-circSHOC2 on the expression levels of Bcl-2 protein located in the mitochondria (Mito) and endoplasmic reticulum (ER). ∗p < 0.05. (C) Stable Gag-LC3-expressing cells were co-transfected with a si-circSHOC2 or si-circRNA-NC, and autophagy was tested under OGD or normal conditions for 6 h. Quantitative analysis of the amount of Gag-LC3 accumulation per cell was performed. ∗p < 0.05. (D) Western blot analysis of LC3 in the si-circSHOC2 group and si-circRNA-NC group. ∗p < 0.05. (E) Western blot analysis of TIMM23 and SQSTM1 in the si-circSHOC2 group and si-circRNA-NC group. ∗p < 0.05.
circSHOC2 Acts as a Molecular Sponge for miR-7670-3p in Neurons
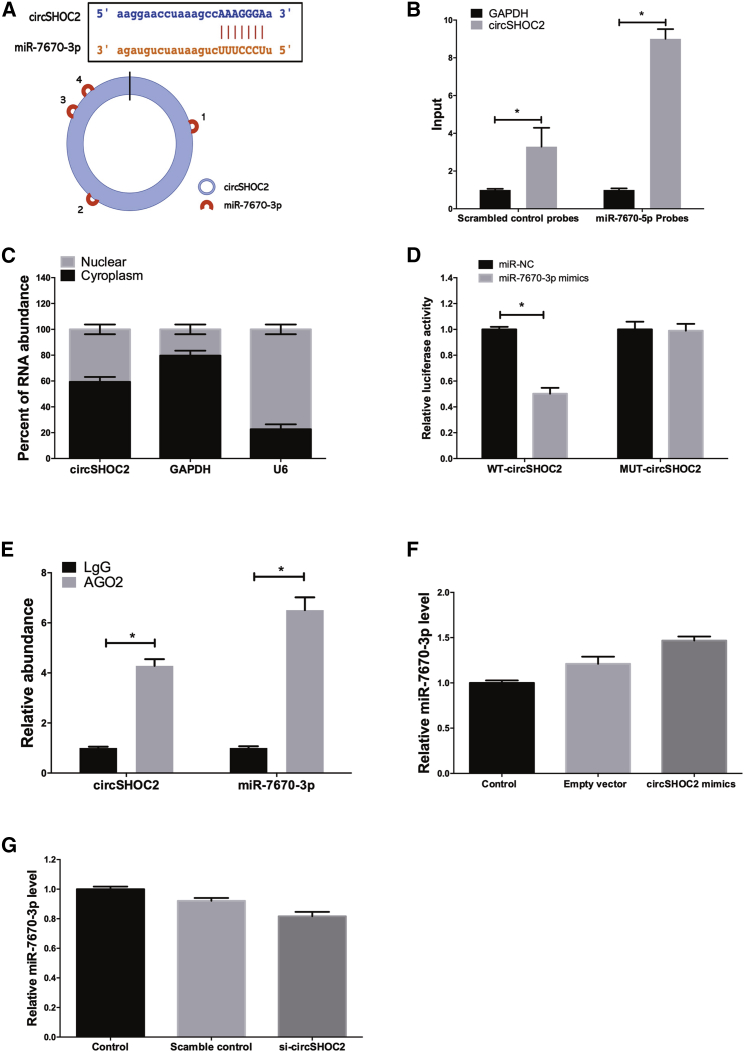
It has been reported that exosmic circRNAs consistently localized in the cytoplasm, and it is established that circRNAs can act as microRNAs (miRNAs) or protein sponges to inhibit their activities.23 We found that circSHOC2 was predominantly localized in the cytoplasm, which indicated that it may function as a miRNA sponge. The bioinformatics software package starBase (http://starbase.sysu.edu.cn) was implemented to identify hypothetical miRNA-binding sites present within the circSHOC2 sequence. We found that circSHOC2 contained four different predicted binding sites for miR-7670-3p, suggesting the possibility of interactions between circSHOC2 and miR-7670-3p (Figure 6A). To test this prediction, we used an inverse pull-down assay to test whether circSHOC2 binds with miR-7670-3p. DNA probes specific to the junction of circSHOC2 pulled down a significantly higher amount of circSHOC2 than did scrambled probes, verifying the efficiency of the probes (Figure 6B). Additionally, we also found co-localization between circSHOC2 and miR-7670-3p in the cytoplasm via quantitative real-time PCR analysis (Figure 6C). A subsequent luciferase reporter assay revealed that the luciferase intensity was reduced after co-transfection of the wild-type (WT) luciferase reporter and miR-7670-3p mimics, whereas the mutated (Mut) luciferase reporter exerted no such effect (Figure 6D). In a further RNA-binding protein immunoprecipitation (RIP) experiment, circSHOC2 and miR-7670-3p simultaneously existed in the production precipitated by anti-argonaute 2 (AGO2) (Figure 6E), which indicated that circSHOC2 directly interacted with miR-7670-3p and could act as a sponge for miR-7670-3p. We further measured the expression levels of miR-7670-3p in neurons with overexpression (Figure 6F) or knockdown of circSHOC2 (Figure 6G), but no significant change was observed in terms of the expression levels of miR-7670-3p. Collectively, these results showed that circSHOC2 may act as a sponge for miR-7670-3p without regulating the expression levels of miR-7670-3p.
Figure 6.
circSHOC2 Acts as a miRNA Sponge for miR-7670-3p in Neurons
(A) circSHOC2 contains four potential binding sites of miR-7670-3p. (B) The levels of total and pulled-down miR-7670-3p and GAPDH by circSHOC2 probes or control probes were analyzed by quantitative real-time PCR. ∗p < 0.05. (C) Quantitative real-time PCR analysis of circSHOC2 in cytoplasmic and nuclear compartments. (D) Dual-luciferase reporter assays showed significant reduction of luciferase activity of the wild-type (WT), while luciferase activity was restored by the mutant sequence. ∗p < 0.05. (E) miR-7670-3p and circSHOC2 simultaneously existed in the production precipitated by anti-AGO2. ∗p < 0.05. (F) Neurons were transfected with a circSHOC2-expression vector. The expression level of miR-7670-3p was analyzed by quantitative real-time PCR. (G) Neurons were transfected with a si-circSHOC2-expression vector. The expression level of miR-7670-3p was analyzed by quantitative real-time PCR.
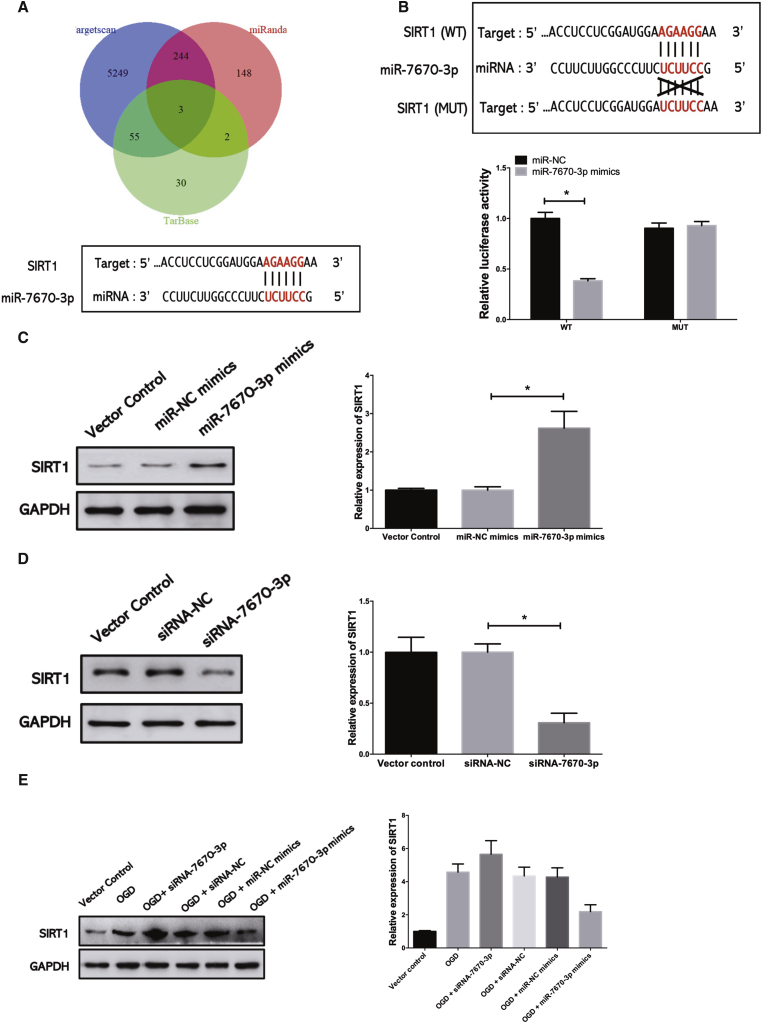
circSHOC2 Reduces OGD-Induced Neuronal Death via the miR-7670a-3p/SIRT1 Axis
To determine whether circSHOC2 sponges miR-7670-3p and further enables the expression of its downstream target, we searched TargetScan for potential target genes of miR-7670-3p, upon which SIRT1 was predicted (Figure 7A). A subsequent luciferase reporter assay revealed decreased luciferase intensity after co-transfection of miR-7670-3p mimics and the WT luciferase reporter, whereas the mutated luciferase reporter exerted no such effect (Figure 7B). Moreover, western blotting was performed to further confirm the results of the luciferase reporter gene assays. We found that miR-7670-3p overexpression remarkably inhibited SIRT1 mRNA and protein expression in mouse neurons (Figure 7C). In contrast, downregulation of miR-320a-5p significantly increased SIRT1 levels (Figure 7D). These data indicated that miR-7670-3p could directly bind to the 3′ UTR of SIRT1 and negatively regulate its expression. Subsequently, we considered whether SIRT1 was involved in the effects of miR-7670-3p in OGD-treated neurons. Western blotting was performed to investigate the effects of OGD on SIRT1 expression in cultured neurons, which revealed that OGD increased SIRT1 mRNA and protein levels. In addition, miR-7670-3p overexpression inhibited the OGD-induced increase in SIRT1 expression, while miR-7670-3p inhibition partially enhanced OGD-induced SIRT1 expression (Figure 7E). Moreover, overexpression of circSHOC2 in neurons caused upregulation of SIRT1; conversely, silencing of circSHOC2 downregulated SIRT1, as revealed by immunoblotting assays (Figure S5A). Furthermore, circSHOC2-induced expression of SIRT1 was attenuated by miR-7670-3p mimics. We next determined the role of the circSHOC2/miR-7670-3p/SIRT1 axis in this process via rescue experiments. Silencing of miR-7670-3p rescued the inhibition of SIRT1 levels caused by circSHOC2 knockdown (Figure S5B). Hence, these data demonstrate that circRNA acted as an anti-autophagic factor via the circSHOC2/miR-7670-3p/SIRT1 regulatory network.
Figure 7.
circSHOC2 Reduces OGD-Induced Neuronal Death via the miR-7670a-3p/SIRT1 Axis
(A) Sequence alignment of predicted mir-7670-3p-binding sites within the SIRT1 sequence. (B) Dual-luciferase reporter assays were conducted to validate the binding effect between miR-7670-3p and SIRT1. ∗p < 0.05. (C) SIRT1 protein levels in neurons transfected with an miR-7670-3p mimic were measured by western blotting. ∗p < 0.05. (D) SIRT1 protein levels in neurons transfected with an siRNA-7670-3p were measured by western blotting. ∗p < 0.05. (E) Neurons were transfected with an siRNA-7670-3p or miR-7670-3p mimic. Neurons were treated with OGD for 6 h. The expression of SIRT1 was analyzed by western blotting.
Discussion
Stroke is a cerebrovascular disease that exhibits a high rate of disability and mortality and is especially prevalent among the elderly population.3,24 Despite various studies elucidating neuroprotective mechanisms of cerebral ischemic tolerance, most of these studies have only investigated neuronal responses after ischemic stroke. Recently, many studies have reported that astrocytes play a key role in the process of ischemic tolerance.25 Emerging data indicate that exosomes released by astrocytes can improve neuronal survival under hypoxic and ischemic conditions. Exosomes can be released from many cell types in the central nervous system.26 Exosomes mediate communication between donor and recipient cells by transferring circRNAs, proteins, and other molecules.27 Recently, exosome-mediated transport has been shown to be involved in several physiological and pathological processes. For example, exosomes released from WT astrocytes promote neuroprotection and plasticity in spinal cord injury.28 In the case of ischemia/hypoxia, exosomes derived from endothelial cells can suppress neuronal death, promote neural plasticity, and improve functional recovery.29 In the current study, we found that exosomes released from IPASs were transmitted to neurons as shuttles carrying circSHOC2, which ameliorated ischemia-induced neuronal apoptosis, indicating that IPAS-EXO-mediated circSHOC2 transport may be sufficient to protect neurons from ischemia-induced damage.
Autophagy is important for the continuous clearance of unnecessary organelles and misfolded proteins in lysosomes, as well as for maintaining differentiation, remodeling, and cellular homeostasis.30 Recently, autophagy has been recognized as a key process in ischemic stroke, in addition to neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease.31,32 It is principally thought that autophagy plays an important role in protecting neurons from inflammation. Recent evidence has also demonstrated an important role of autophagy in ischemic stroke. A previous study showed that metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) downregulation attenuated neuronal death through inhibiting autophagy in cerebral ischemic stroke.33 Therefore, autophagy has been proposed as a neuroprotective mechanism. In the present study, we validated that IPAS-EXOs inhibited OGD-induced neuronal apoptosis via regulating autophagy. We further investigated the mechanisms by which autophagy inhibits neuronal apoptosis. A previous study showed that exosomes mainly exert their functions by transferring exosomal circRNAs or miRNAs to recipient cells.34 In the current study, we found that IPAS-EXOs exerted their protective effects mainly via transfer of circSHOC2 to neurons, rather than by stimulating endogenous circSHOC2 expression within neurons. The effects of circSHOC2 were abolished via an autophagic inhibitor, suggesting that the neuroprotective role of circSHOC2 is mediated, at least in part, through autophagy. Moreover, overexpression of circSHOC2 inhibited Gag-LC3 accumulation, LC3-I-to-LC3-II conversion, and TIMM23 degradation in neurons. To our knowledge, this report is the first to demonstrate that knockdown of circSHOC2 expression inhibits OGD-induced neuronal apoptosis, revealing a novel function of circSHOC2 in the regulation of neuroprotection. Therefore, we think that circSHOC2 treatment may represent an effective strategy to prevent ischemic stroke.
circRNAs often act as miRNA sponges and participate in miRNA-mediated post-transcriptional regulation. miRNAs are non-coding RNAs, as confirmed by luciferase reporter gene tests, and can act as sponges and inhibit the function of circRNAs.35 It has been reported that specific circRNAs destabilize miRNA complexes because of their high complementarities with miRNAs.36 Previous studies have shown that the circRNA HRCR acts as a miR-223 sponge to regulate cardiac hypertrophy and heart failure.37 In the present study, we used the starBase v2.0 target prediction tool to discover potential miRNAs that may bind to circSHOC2. The results of RIP analysis showed that circSHOC2 binds with miR-7670-3p. We verified that circSHOC2 acts as a sponge for miR-7670-3p and promotes SIRT1 expression to reduce neuronal damage. SIRT1 is a valuable candidate gene because it regulates gene expression and cellular metabolism under ischemic stress. In recent years, SIRT1 has been found to exert neuroprotective effects on cerebral ischemic injury. Furthermore, previous studies have shown that SIRT1 regulates autophagy. For example, during cerebral ischemia, Nampt induces autophagy by regulating the TSC2-mTOR-S6K1 signaling pathway in a SIRT1-dependent manner, thereby promoting neuronal survival.23 Other studies have shown that overexpression of SIRT1 can prevent hypokalemia-induced apoptosis in small potassium granule neurons and prevent OGD-induced loss of Bcl-2 expression in neurons.38 To our knowledge, the present study is the first to demonstrate that SIRT1 is a downstream effector of miR-7670-3p, which strongly supports our conclusion that the circSHOC2/miR-7670-3p/SIRT1 axis mitigates ischemia-induced neuronal death.
Conclusions
In summary, the present study demonstrated that IPAS-EXOs may play an important role in providing neuroprotection under ischemic conditions. These findings enhance our understanding of circRNA biology and may help to develop other biomarkers or more effective targets for the treatment of ischemic stroke.
Materials and Methods
Animals and Ethics Statement
Adult male C57BL/6J mice (6–8 weeks old and weighing 16–22 g) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, P.R. China) and were randomly divided into the experimental groups used in this study. All surgical procedures in mice were performed under anesthesia through intraperitoneal injections of sodium pentobarbital (30 mg/kg). All animal experimental designs were approved by the Animal Ethics Committee of Southern Medical University and all experiments conformed to relevant regulatory standards.
Isolation and Culturing of Primary Astrocytes
Primary astrocytes were obtained from the cerebral cortices of C57BL/6 mice at 24 h after birth. Briefly, mouse brain cells were isolated with trypsin-EDTA and were incubated at a density of 6 × 105 cells/cm2 in DMEM. After 7–9 days, astrocytes were purified by shaking at 300 rpm for 4 h at 37°C. Subsequently, the harvested astrocytes were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a humidified incubator at 37°C and 5% CO2.
Primary Neuronal Cultures
Primary neurons were prepared from the cerebral cortices of mouse embryos (16–18 days old). Briefly, trypsin-EDTA-dissociated cortical cells were cultured with DMEM in a humidified incubator at 37°C and 5% CO2 and were then incubated in neural basal medium at 4 h after seeding. Upon reaching 70%–80% confluency, cells were collected and used for further analysis.
Immunostaining and Imaging Analysis
Astrocytes and brain slices were incubated with 0.3% Triton X-100 for 1 h at 4°C and were then incubated with fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse immunoglobulin G (IgG) (Sigma, USA) for 4 h at room temperature. Fluorescent images of the cells were observed under a FV3000 confocal laser scanning microscope (Olympus, Japan).
Isolation of Exosomes
Based on the protocol indicated on the exosome isolation kit (Fab-TACS, IBA Lifesciences, Germany), exosomes were isolated from the supernatant of cultured astrocytes. Before collecting the medium, the IPASs were washed twice with PBS and were then cultured for 48 h. The supernatant was collected and subjected to ultracentrifugation at 2,000 × g for 30 min, 10,000 × g for 30 min, and 100,000 × g for 70 min at 4°C. The exosomes were washed once with PBS and then underwent another ultracentrifugation at 100,000 × g for 70 min, after which they were suspended for further characterization. TEM (Thermo Scientific, USA) was used to identify the characteristics of the isolated exosomes. Nanosizer technology (Malvern Instruments, UK) was then used to evaluate the distribution of diameters of IPAS-EXOs. Western blotting was used to detect the expression levels of exosome-specific biomarkers, such as CD9, CD63, Alix, and TSG101.
Exosomal Labeling
Fluorescent labeling of exosomes was performed according to a previously described protocol.39 Briefly, exosomes were labeled via a DiI fluorescent labeling kit (Sigma-Aldrich, USA). Subsequently, DiI-labeled exosomes were incubated with cultured neurons for 24 h. Then, the cells or brain tissues were fixed and stained with DAPI, and stained images were observed using a confocal microscope. Observation under a fluorescent microscope confirmed that the exosomes were stained by DiI (red).
Oxygen Glucose Deprivation Model
To simulate ischemia-like conditions in vitro, astrocytes were subjected to OGD. Briefly, astrocytes were incubated with deoxygenated DMEM (GIBCO, USA) free of glucose and FBS in a dedicated closed chamber containing an anaerobic gas mixture (95% N2 and 5% CO2) for 6 h. The medium was then removed from the chamber, and the OGD solution in the culture was replaced with ordinary DMEM containing 10% FBS, after which the culture was placed in a CO2 incubator (95% air and 5% CO2). After 24 h, the culture medium collected from the OGD-treated astrocytes was filtered through a 0.22-μm filter (Millipore, USA) to remove debris and dead cells.
CCK-8 Assay
A CCK-8 assay was used to determine neuronal viability. Briefly, 3,000 cells/well were seeded in 96-well plates and incubated overnight in DMEM containing 10% FBS. Neurons were treated with different concentrations of IPAS-EXOs and were then incubated in 10°C CCK-8 solution at 37°C for 3 h. The optical density (OD) value was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, USA). Inhibition of neuronal viability was determined in each group relative to the OD values in the PBS group.
ELISA
At 24 h after IPAS-EXOs were transfected, inflammatory factors, namely TNF-α, IL-6, and IL-1β, in the supernatant were measured via a commercial ELISA kit (R&D Systems, USA). All of the procedures were performed strictly according to the manufacturer’s instructions.
Western Blotting
Western blotting of exosomes was performed as previously described.40 Total protein was extracted from IPAS-EXOs, neurons, or mouse brain tissues using radioimmunoprecipitation assay (RIPA) lysis buffer (Cell Signaling Technology, USA) containing 1 mM PMSF on ice. The concentration of exosomal precipitates was quantified via a bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, USA). SDS-PAGE was used to transfer an equal volume of protein to a polyvinylidene fluoride membrane. The membrane was blocked with 5% skimmed milk and 3% BSA at room temperature for 1 h, and then the membrane was incubated in 5% milk in Tris-buffered saline with Tween 20 (TBST )solution containing the following primary antibodies overnight at 4°C: GFAP (Sigma-Aldrich, USA), CD9 (Abcam, UK), CD63 (Abcam, UK), Alix (Abcam, UK), caspase-3 (Cell Signaling Technology, USA), Bax (Cell Signaling Technology, USA), Bcl-2 (Cell Signaling Technology, USA), Beclin-1 (Santa Cruz Biotechnology, USA), LC3-I (Santa Cruz Biotechnology, USA), LC3-II (Santa Cruz Biotechnology, USA), p62 (Santa Cruz Biotechnology, USA), GAPDH (Santa Cruz Biotechnology, USA), and SQSTM1 (Proteintech, USA). After washing three times with TBST, the membrane was incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-goat secondary antibodies for 1 h at room temperature, and protein bands were detected using enhanced chemiluminescence (ECL) western blot detection reagents (Millipore, USA). The secondary antibodies used for western blotting were anti-rabbit IgG HRP (Cell Signaling Technology, USA) and anti-goat IgG HRP (Abcam, USA). Individual protein bands were quantified by densitometry using ImageJ software.
Immunofluorescence
Immunofluorescence was used to identify astrocytes. Astrocytes were washed with cold PBS, fixed in 4% paraformaldehyde (PFA) for 15 min, blocked in 5% serum at 37°C for 60 min, and then diluted with 1:300 anti-GFAP antibody (Cell Signaling Technology, USA) and S100A10 antibody overnight at 4°C. After being washed three times with PBS, the cells were incubated with a secondary antibody diluted at 1:100 (Cell Signaling Technology, USA) at 37°C for 2 h, and DAPI was used for nuclear staining at 37°C for 2 min. Finally, the samples were washed once with PBS. A confocal microscope (Olympus, Japan) was used to capture immunofluorescence images.
Immunofluorescence was also used to identify whether circRNAs in IPAS-EXOs regulate neuronal autophagy. At 6 h after OGD treatment, neurons that were transfected with specific circRNA siRNA or control mimics of circSHOC2 were cultured on sterile glass. Cells were then transfected with Gag-LC3 plasmid via Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions, and then LC3 spots in neurons were counted by immunofluorescence.
TTC Staining and Measurement of Cerebral Infarction
Mice were decapitated under anesthesia, and their brains were collected and immediately frozen at −20°C for 6 min. Each mouse brain was perfused with 0.9% saline and 4% PFA. After perfusion, the brain was removed, placed in 4% PFA overnight, and then fully dehydrated in 30% sucrose for 2 days. Subsequently, the dehydrated brain tissue was cut into 2-mm slices. Brain slices were incubated in 2% TTC (Sigma-Aldrich, USA) at 37°C for 10 min. After incubation, brain slices were fixed in 4% PFA for 24 h and imaged, and images were then analyzed via ImageJ software. The red area represented the non-infarcted tissue area, whereas the light white area represented the infarcted tissue area. The infarct ratio was calculated by the following formula: corrected percentage of infarct volume = [(contralateral hemisphere volume − ipsilateral non-infarct volume)/contralateral hemisphere volume] × 100%.
Middle Cerebral Artery Occlusion Model
As mentioned previously, a modified mouse model was used to perform permanent focal ischemia to establish a model of MCAO.41 Briefly, mice were anesthetized with 1.5%–2.0% isoflurane and 30%/70% oxygen/nitrous oxide. Subsequently, the common carotid artery, internal carotid artery, and external carotid artery (ECA) were separated. Then, 6-0 nylon filaments coated with silicone rubber were inserted into the ECA and advanced 9–10 mm along the internal carotid artery to carotid artery bifurcation and the origin of the middle cerebral artery. After embolizing for 60 min, the silicone wire was removed. In the sham control group, the internal carotid artery was prepared for the insertion of the filament through surgery, but the filament was not inserted. The neck incision was then sutured, and the mouse was placed into a nursing box at 35°C to restore anesthesia. Immediately after withdrawing the suture, vesicles were injected through the tail vein, and the injection was repeated three times a day for a total of 100 μg of vesicles per day.
Measurements of Neurological Deficits, Behavior, and mNSSs
The severity of the injuries in treated mice were assessed using mNSSs, which represent a composite score of motor, reflex, and balance tests. Neurobehavioral tests were performed before and at 3 days after MCAO by a researcher blinded to the experimental groups. Severity scores ranged from 0 to 14 (with 0 indicating a healthy phenotype, and higher scores indicating more serious injuries). Behavioral tests were conducted before MCAO and at 3, 7, 14, and 21 days after MCAO.
TUNEL Assays
To determine whether treatment with IPAS-EXOs reduced apoptosis of cortical neurons, we performed TUNEL assays. Cells or brain tissues were fixed with 4% PFA. After washing the cells or tissues with PBS, the TUNEL assay was performed using a fluorescein-conjugated probe according to the manufacturer’s protocol. The nuclei were stained with DAPI. The slides were washed and imaged with a fluorescent microscope. Apoptosis was analyzed using an image analyzer to calculate the positive cell rate. In the area around the infarct within brain slices of MCAO model mice, we counted the number of NeuN/TUNEL double-positive cells and NeuN-positive cells in five areas of each brain slice. To assess the proportion of damage, we calculated the ratio of DAPI/TUNEL double-positive cells to DAPI-positive cells.
LDH and Cell Survival Assays
The death of primary neurons was measured by the release of LDH in the culture medium. Briefly, after OGD treatment and transfection, primary neurons were cultured in six-well plates. The neurons were collected, resuspended, and cultured in CO2 in a 96-well plate at 37°C for 30 min. Using an LDH assay kit, the level of LDH release in the supernatant of cultured cells was measured via the absorbance at 490 nm using a microplate reader. The LDH level of the control group was expressed as 100%, and the LDH levels of the other groups were normalized to this value.
RNA Extraction and Quantitative Real-Time PCR
According to the manufacturer’s protocol, total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, USA). The miRNA sequences were obtained from the NCBI database, and the corresponding primers were designed using Primer 5 software. The obtained RNA was reverse transcribed into cDNA using a PrimeScript RT kit (Takara, Japan). Then, cDNA was synthesized by a reverse-assisted first-strand cDNA synthesis kit (Thermo Fisher Scientific, USA). Qualified RNA was reverse transcribed using a reverse transcription kit (Takara, Japan), and amplified via a LightCycler 480 (Roche, Basel, Switzerland) system using SYBR Premix Ex Taq (Takara, Japan). For circRNA, RNase R was used to degrade linear RNA with poly(A), after which it was amplified by divergent primers. Thereafter, cDNA was synthesized from 1 μg of total RNA in a reaction volume of 21 μL using oligo(dT)18 primer and SuperScript reverse transcriptase. PCR was performed using Taq DNA polymerase (Takara, Japan) using 1 μL of first-strand cDNA as a template. The amplification reaction was performed in 30 thermal cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. Qualified RNA was reverse transcribed using a reverse transcription kit (Takara, Japan) and was then amplified on a LightCycler 480 (Roche, Basel, Switzerland) system using SYBR Premix Ex Taq (Takara, Japan). All experiments were repeated three times, and the results were normalized to the expression of β-actin or U6 mRNA. PCR primers are listed in Table S1. The relative mRNA levels were calculated via the 2−ΔΔCT method.
Dual-Luciferase Reporter Assay
According to the manufacturer’s instructions, a dual-luciferase reporter assay was performed in cultured primary neurons. Wide sequences were designed and synthesized based on the predicted binding sites. Additionally, we performed site-directed mutagenesis to further determine the sites of action of circRNAs and the 3′ UTR of the target genes. Sequences were inserted into a luciferase reporter vector. Neurons were harvested and lysed at 48 h post-transfection. After 48 h, the cells were transfected with pmirGLO-circSHOC2-WT (or pmirGLO-circSHOC2-Mut) or pmirGLO-SIRT1-WT (or pmirGLO-SIRT1-Mut), as well as an miR-7670-3p mimic or miR-NC, via Lipofectamine 3000 (Invitrogen, USA). Luciferase activity was assayed using the luciferase reporter assay system (Promega, USA). The results were evaluated through normalization of the firefly luciferase activity with the Renilla luciferase activity. Triplicate wells used for each group.
Cellular Transfections
si-circSHOC2, circSHOC2 mimics, miR-7670-3p siRNA (siRNA-7670-3p), miR-7670-3p mimic vector, or the corresponding negative control (NC) were synthesized by GenePharma (Shanghai, P.R. China). Cells (3 × 105) were seeded into six-well plates and were transfected when they reached 40%–60% confluency. Neurons were then transfected with either the miR-7670-3p-overexpression construct or miR-NC at 50 nM via Lipofectamine 3000 (Invitrogen, USA) and were incubated at 37°C in 5% CO2 in 24-well plates. Three days after transfection, the Gag signals were observed under a fluorescence microscope. No-load siRNA plasmid was used as a control. All steps were performed according to the manufacturers’ instructions.
Analysis of circRNA Expression Profiles
The microarray hybridization and collection of data were performed by KangChen Biotech (Shanghai, P.R. China). The top five upregulated and downregulated circRNAs were identified, and their corresponding hierarchical clustering analysis was performed based on their expression values using Cluster and TreeView software.
RIP
RIP was performed in neurons at 48 h after transfection with an miR-7670-3p-overexpression construct or miR-NC using a Magna RIP kit (Millipore, USA) according to the manufacturer’s protocol. Neurons (3 × 105) were lysed in RNA lysis buffer, and the cell lysate was then coupled to magnetic beads coupled with human anti-AGO2 antibody or control mouse IgG in RIP buffer.
Statistical Analysis
GraphPad 6.0 software was used for statistical analysis. All data are expressed as the mean ± standard deviation (SD). Differences were determined by Student’s t tests for comparisons between two groups and by analyses of variance (ANOVAs) for comparisons among more than two groups. ∗p <0.05 was considered to be statistically significant.
Data Availability
The datasets used or analyzed in this study may be obtained from the authors upon reasonable request.
Author Contributions
W.C. contributed to the figures and tables and to the manuscript. H.W., J.F., and Z.Z. were in charge of experiments and data.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors acknowledge the financial support from the National Nature Science Foundation of China (Grant No. 81671819).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.027.
Supplemental Information
References
- 1.Vilela P., Rowley H.A. Brain ischemia: CT and MRI techniques in acute ischemic stroke. Eur. J. Radiol. 2017;96:162–172. doi: 10.1016/j.ejrad.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein D.L., Zuluaga-Ramirez V., Gajghate S., Reichenbach N.L., Polyak B., Persidsky Y., Rom S. miR-98 reduces endothelial dysfunction by protecting blood-brain barrier (BBB) and improves neurological outcomes in mouse ischemia/reperfusion stroke model. J. Cereb. Blood Flow Metab. 2020;40:1953–1965. doi: 10.1177/0271678X19882264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y.L., Zhang L.X., Liu G.L., Fan Y., Peng Y., Hou W.G. N-Myc downstream-regulated gene 2 (Ndrg2) is involved in ischemia-hypoxia-induced astrocyte apoptosis: a novel target for stroke therapy. Mol. Neurobiol. 2017;54:3286–3299. doi: 10.1007/s12035-016-9814-5. [DOI] [PubMed] [Google Scholar]
- 4.Zhong C.J., Chen M.M., Lu M., Ding J.H., Du R.H., Hu G. Astrocyte-specific deletion of Kir6.1/K-ATP channel aggravates cerebral ischemia/reperfusion injury through endoplasmic reticulum stress in mice. Exp. Neurol. 2019;311:225–233. doi: 10.1016/j.expneurol.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Pekny M., Wilhelmsson U., Tatlisumak T., Pekna M. Astrocyte activation and reactive gliosis—a new target in stroke? Neurosci. Lett. 2019;689:45–55. doi: 10.1016/j.neulet.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Maurya S., Jayandharan G.R. Exosome-associated SUMOylation mutant AAV demonstrates improved ocular gene transfer efficiency in vivo. Virus Res. 2020;283:197966. doi: 10.1016/j.virusres.2020.197966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zha Y., Lin T., Li Y., Zhang X., Wang Z., Li Z., Ye Y., Wang B., Zhang S., Wang J. Exosome-mimetics as an engineered gene-activated matrix induces in-situ vascularized osteogenesis. Biomaterials. 2020;247:119985. doi: 10.1016/j.biomaterials.2020.119985. [DOI] [PubMed] [Google Scholar]
- 8.Suh M., Lee D.S. Brain theranostics and radiotheranostics: exosomes and graphenes in vivo as novel brain theranostics. Nucl. Med. Mol. Imaging. 2018;52:407–419. doi: 10.1007/s13139-018-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang T., Martin P., Fogarty B., Brown A., Schurman K., Phipps R., Yin V.P., Lockman P., Bai S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm. Res. 2015;32:2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P., Pan R., Weaver J., Jia M.J., Yang X., Yang T.H., Liang J., Liu K.J. MicroRNA-30a regulates acute cerebral ischemia-induced blood-brain barrier damage through ZnT4/zinc pathway. J. Cereb. Blood Flow Metab. 2020 doi: 10.1177/0271678X20926787. Published online June 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T., Sun X., Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J. Cell. Biochem. 2020;121:2118–2126. doi: 10.1002/jcb.28239. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H., Zhu L., Bai M., Liu Y., Zhan Y., Deng T., Yang H., Sun W., Wang X., Zhu K. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 13.Nafar F., Williams J.B., Mearow K.M. Astrocytes release HspB1 in response to amyloid-β exposure in vitro. J. Alzheimers Dis. 2016;49:251–263. doi: 10.3233/JAD-150317. [DOI] [PubMed] [Google Scholar]
- 14.Pan Q., Kuang X., Cai S., Wang X., Du D., Wang J., Wang Y., Chen Y., Bihl J., Chen Y. miR-132-3p priming enhances the effects of mesenchymal stromal cell-derived exosomes on ameliorating brain ischemic injury. Stem Cell Res. Ther. 2020;11:260. doi: 10.1186/s13287-020-01761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B., Zhang Y., Du X.F., Li J., Zi H.X., Bu J.W., Yan Y., Han H., Du J.L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27:882–897. doi: 10.1038/cr.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudbergsson J.M., Johnsen K.B. Exosomes and autophagy: rekindling the vesicular waste hypothesis. J. Cell Commun. Signal. 2019;13:443–450. doi: 10.1007/s12079-019-00524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D., Huang S., Zhu J., Hu T., Han Z., Zhang S., Zhao J., Chen F., Lei P. Exosomes from miR-21-5p-increased neurons play a role in neuroprotection by suppressing Rab11a-mediated neuronal autophagy in vitro after traumatic brain injury. Med. Sci. Monit. 2019;25:1871–1885. doi: 10.12659/MSM.915727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X.Q., Liu J.T., Fan L.L., Liu Y., Cheng L., Wang F., Yu H.Q., Gao J., Wei W., Wang H., Sun G.P. Exosomes derived from gefitinib-treated EGFR-mutant lung cancer cells alter cisplatin sensitivity via up-regulating autophagy. Oncotarget. 2016;7:24585–24595. doi: 10.18632/oncotarget.8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang M., Wang H., Jin M., Yang X., Ji H., Jiang Y., Zhang H., Wu F., Wu G., Lai X. Exosomes from miR-30d-5p-ADSCs reverse acute ischemic stroke-induced, autophagy-mediated brain injury by promoting M2 microglial/macrophage polarization. Cell. Physiol. Biochem. 2018;47:864–878. doi: 10.1159/000490078. [DOI] [PubMed] [Google Scholar]
- 20.Yu T., Ding Y., Zhang Y., Liu Y., Li Y., Lei J., Zhou J., Song S., Hu B. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 2019;231:238–245. doi: 10.1016/j.vetmic.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y., Luo W., Li Z., Cao M., Zhu Z., Han C., Dai X., Zhang W., Wang J., Yao H., Chao J. circRNA-012091/PPP1R13B-mediated lung fibrotic response in silicosis via endoplasmic reticulum stress and autophagy. Am. J. Respir. Cell Mol. Biol. 2019;61:380–391. doi: 10.1165/rcmb.2019-0017OC. [DOI] [PubMed] [Google Scholar]
- 22.Kong R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol. Int. 2020;44:1945–1956. doi: 10.1002/cbin.11401. [DOI] [PubMed] [Google Scholar]
- 23.Wang P., Guan Y.F., Du H., Zhai Q.W., Su D.F., Miao C.Y. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. 2012;8:77–87. doi: 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
- 24.Behrouzifar S., Vakili A., Bandegi A.R., Kokhaei P. Neuroprotective nature of adipokine resistin in the early stages of focal cerebral ischemia in a stroke mouse model. Neurochem. Int. 2018;114:99–107. doi: 10.1016/j.neuint.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Sun X., Jung J.H., Arvola O., Santoso M.R., Giffard R.G., Yang P.C., Stary C.M. Stem cell-derived exosomes protect astrocyte cultures from in vitro ischemia and decrease injury as post-stroke intravenous therapy. Front. Cell. Neurosci. 2019;13:394. doi: 10.3389/fncel.2019.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluchino S., Peruzzotti-Jametti L., Frezza C. Astrocyte power fuels neurons during stroke. Swiss Med. Wkly. 2016;146:w14374. doi: 10.4414/smw.2016.14374. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Zhang H., Yang H., Bai M., Ning T., Deng T., Liu R., Fan Q., Zhu K., Li J. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W., Rong Y., Wang J., Zhou Z., Ge X., Ji C., Jiang D., Gong F., Li L., Chen J. Exosome-shuttled miR-216a-5p from hypoxic preconditioned mesenchymal stem cells repair traumatic spinal cord injury by shifting microglial M1/M2 polarization. J. Neuroinflammation. 2020;17:47. doi: 10.1186/s12974-020-1726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao B., Zhou S., Sun C., Cheng D., Zhang Y., Li X., Zhang L., Zhao J., Xu D., Bai Y. Brain endothelial cell-derived exosomes induce neuroplasticity in rats with ischemia/reperfusion injury. ACS Chem. Neurosci. 2020;11:2201–2213. doi: 10.1021/acschemneuro.0c00089. [DOI] [PubMed] [Google Scholar]
- 30.Zhu M., Liu X., Li W., Wang L. Exosomes derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis via activating autophagy. Hum. Exp. Toxicol. 2020;39:1619–1627. doi: 10.1177/0960327120931152. [DOI] [PubMed] [Google Scholar]
- 31.Kizilarslanoğlu M.C., Ülger Z. Role of autophagy in the pathogenesis of Alzheimer disease. Turk. J. Med. Sci. 2015;45:998–1003. doi: 10.3906/sag-1407-75. [DOI] [PubMed] [Google Scholar]
- 32.Esteves A.R., Palma A.M., Gomes R., Santos D., Silva D.F., Cardoso S.M. Acetylation as a major determinant to microtubule-dependent autophagy: relevance to Alzheimer’s and Parkinson disease pathology. Biochim. Biophys. Acta Mol. Basis Dis. 2019;1865:2008–2023. doi: 10.1016/j.bbadis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Li Z., Li J., Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience. 2017;354:1–10. doi: 10.1016/j.neuroscience.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 34.Li T., Gu J., Yang O., Wang J., Wang Y., Kong J. Bone marrow mesenchymal stem cell-derived exosomal miRNA-29c decreases cardiac ischemia/reperfusion injury through inhibition of excessive autophagy via the PTEN/Akt/mTOR signaling pathway. Circ. J. 2020;84:1304–1311. doi: 10.1253/circj.CJ-19-1060. [DOI] [PubMed] [Google Scholar]
- 35.Huang X.Y., Huang Z.L., Huang J., Xu B., Huang X.Y., Xu Y.H., Zhou J., Tang Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S.J., Wang D.D., Zhou S.Y., Zhang Q., Wang J.Y., Zhong S.L., Zhang H.D., Wang X.Y., Xia X., Chen W. Identification of circRNA-miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics. 2020;12:101–125. doi: 10.2217/epi-2019-0058. [DOI] [PubMed] [Google Scholar]
- 37.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 38.Yan W., Fang Z., Yang Q., Dong H., Lu Y., Lei C., Xiong L. SirT1 mediates hyperbaric oxygen preconditioning-induced ischemic tolerance in rat brain. J. Cereb. Blood Flow Metab. 2013;33:396–406. doi: 10.1038/jcbfm.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vashisht M., Rani P., Sunita, Onteru S.K., Singh D. Curcumin primed exosomes reverses LPS-induced pro-inflammatory gene expression in buffalo granulosa cells. J. Cell. Biochem. 2018;119:1488–1500. doi: 10.1002/jcb.26309. [DOI] [PubMed] [Google Scholar]
- 40.Meng L., Liu S., Ding P., Chang S., Sang M. Circular RNA ciRS-7 inhibits autophagy of ESCC cells by functioning as miR-1299 sponge to target EGFR signaling. J. Cell. Biochem. 2020;121:1039–1049. doi: 10.1002/jcb.29339. [DOI] [PubMed] [Google Scholar]
- 41.Deng Y., Chen D., Gao F., Lv H., Zhang G., Sun X., Liu L., Mo D., Ma N., Song L. Exosomes derived from microRNA-138-5p-overexpressing bone marrow-derived mesenchymal stem cells confer neuroprotection to astrocytes following ischemic stroke via inhibition of LCN2. J. Biol. Eng. 2019;13:71. doi: 10.1186/s13036-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used or analyzed in this study may be obtained from the authors upon reasonable request.