Abstract
BACKGROUND
Cervical cancer (CC) remains a leading cause of gynaecological cancer-related mortality world wide and constitutes the third most common malignancy in women. The RAIDs consortium (http://www.raids-fp7.eu/) conducted a prospective European study [BioRAIDs (NCT02428842)] with the objective to stratify CC patients for innovative treatments. A “metagene” of genomic markers in the PI3K pathway and epigenetic regulators had been previously associated with poor outcome [2].
METHODS
To detect new, more specific, targets for treatment of patients who resist standard chemo-radiation, a high-dimensional Cox model was applied to define dominant molecular variants, copy number variations, and reverse phase protein arrays (RPPA).
FINDINGS
Survival analysis on 89 patients with all omics data available, suggested loss-of-function (LOF) or activating molecular alterations in nine genes to be candidate biomarkers for worse prognosis in patients treated by chemo-radiation while LOF of ATRX, MED13 as well as CASP8 were associated with better prognosis. When protein expression data by RPPA were factored in, the supposedly low molecular weight and nuclear form, of beta-catenin, phosphorylated in Ser552 (pβ-Cat552), ranked highest for good prognosis, while pβ-Cat675 was associated with worse prognosis.
INTERPRETATION
These findings call for molecularly targeted treatments involving p53, Wnt pathway, PI3K pathway, and epigenetic regulator genes. Pβ-Cat552 and pβ-Cat675 may be useful biomarkers to predict outcome to chemo-radiation, which targets the DNA repair axis.
FUNDING
European Union's Seventh Program for research, technological development and demonstration (agreement N°304,810), the Fondation ARC pour la recherche contre le cancer.
Keywords: Cervical cancer, Molecular landscape, Molecular and protein biomarkers for chemo-radiation efficiency, Beta-catenin pβ-cat552 and pβ-cat675
Keywords: Abbreviations:CC, Cervical cancer; HPV, Human papillomavirus; LOF, Loss-of-function; RPPA, Reverse phase protein array; WES, Whole exome sequencing; CNV, copy number variation; PFS, Progression-free survival; SCC, squamous cell carcinoma; LMW, low molecular weight; TCF4, transcription factor 4
INTRODUCTION
Cervical cancer (CC) remains,after breast cancer, the second most common malignancy in women [11]. Although patients with CC exhibit differences in their clinical course, infection by high-risk Human Papilloma Virus (HPV) remains an important initiating event in CC tumourigenesis [12], and one of the most important risk factors for developing CC [13]. The global incidence is approximately 500 000 cases per year and the mortality of this virally initiated disease is in the order of 50% of patients worldwide [11]. Advances in biomarker-driven cancer therapy development are hampered by the complexity of the human genome and the high inter- and intra-patient variability in molecular alterations.
RESEARCH IN CONTEXT.
The molecular landscape of cervical cancer [1,2] similarly to that of squamous cell carcinoma of head and neck [3,4] has been characterized and actionable or targetable genomic alterations have been identified. However, single targeted therapies have very limited activity in unselected patients. Even in selected patients [5], activity is limited, owing to the fact that advanced disease has multiple, heterogeneous targets [6].
Evidence before this study
According to the ESMO glossary, ‘targetable genomic alteration’ encodes an altered protein against which a drug exists or can be synthesized and an ‘actionable genomic alteration’ includes both targetable alterations and genomic alterations that cannot be directly targeted but that leads to dysregulation of a pathway in which there are possible targets.
According to Galot et al. the percentage of patients in a biomarker driven trial that had an ‘actionable genomic alteration’ identified through screening programs ranged from 46% to 63%. However, the number of patients who were finally treated with a matched targeted therapy across three international trials was low: 13%, 16%, and 19% respectively [7]. It was 27% in a most recent trial publication, probably related to the extension of the screening panels. Different reasons explain these low enrolment rates: tumour tissue issues, rapid decline in the patients’ performance status in line with rapidly progressing disease, possibly related to a multiplicity of driver alterations, the non-detection of a targetable alteration or the limitations of access to relevant drugs. With pre-planned access, still only 12% of the patients were finally enrolled in a recently published trial - only 18% of the screened tumours had been found to have a genomic alteration that matched one of the 30 treatment arms [7,8].
The summary on present achievements of biomarker-driven studies is the low number of patients who benefit from this approach. This suggests that for heterogeneous cancers with multiple potential oncogenic drivers, biomarkers assessed only at the DNA level in a panel assay, may 1/ not establish the main tumour drivers and, 2/ not reliably predict drug responses. For that reason, we took-into-account not only the genotype but also the phenotype (e.g. gene expression/proteomic profiles) of cancer cells as well as the multiplicity of potential driver alterations. We initiated BioRAIDs, a supervised longitudinal study with pretreatment cervical cancer sample collection and clinical annotation (NCT02428842) [9]. PIK3CA mutations and/or gene amplifications were the most frequently diagnosed oncogenic alteration, present in 40% of BioRAIDs patients [2]. The most frequently diagnosed suppressor gene alterations were loss-of-function (LOF) mutations in KMT2A-D (Lysine methyl transferase) gene leading to defective histone H3K4 methylation. The cumulative frequency of tumours harboring any suppressor gene alterations in the epigenetic pathway (involving KMT2C, KMT2D, EP300, ARID1A, ARID2, ATRX, CREBBP, KMT2A, KDM5C) was 45% of which 32% also had alterations in PI3KCA.
Added value of this study
While current treatment strategies are still mostly based on tumour location and disease stage and very few on tumour biology [10], we set out to identify upfront the molecular alterations that will need innovative therapies by assessing a European cervical cancer patient population. To do this, we tested the lead molecular alterations based not only on DNA alterations but also on activated protein expression profiles associated with cure or progression following standard (chemo-radiation) therapy. Molecular testing for multiplicity of driver alterations was carried out on a supervised patient population with pre-therapy frozen and fixed tumour biopsies as well as six-monthly liquid biopsies, allowing to follow viral presence and persistent molecular alterations over time (NCT02428842). A boosting implementation of Cox models, compatible with high-dimensional settings, allowed to integrate relevant information on molecular variants (based on whole exome sequencing (WES)), copy number variations as well as reverse phase protein arrays (RPPA), together with clinical data. From our dedicated biomarker screening trial, we identified 16 molecular alterations (based on WES analysis) and 30 activated proteins to be associated with progression-free survival up to 24 months following standard chemo-radiation. Patients with two or more actionable genomic alteration, previously associated with poor outcome, progressed earlier. Most significant beneficial markers (ATRX, MED13) may serve as biomarkers in favor of chemo-radiation, while most recurrent deleterious markers (TP53 and CREBBP) suggest the need for additional innovative therapies.
Implication of all the available evidence
The present findings are to our knowledge the first systematic approach towards the understanding of multiple governing alterations in cervical cancer, treated by the current best standard clinical approach, which has been developed in careful clinical trials over the past decades.
Integration of individual tumour specific constellations based on multiple tumour cell genotypic and phenotypic driver alterations with outcome data leads to a better understanding of relevant mechanisms that govern clinical control (or not) of cervical cancer treated by chemo-radiation.
Pre-treatment awareness of type and constellation of deleterious genetic alterations will assist the design of innovative umbrella type platform trials.
Alt-text: Unlabelled box
BioRAIDs, a supervised longitudinal study with pretreatment sample collection and clinical annotation (NCT02428842) [9] allowed the identification of molecular pathways related to poor outcome. PIK3CA mutations and/or gene amplifications were the most frequently diagnosed oncogenic alteration, present in 40% of BioRAIDs patients. The most frequently diagnosed suppressor gene alterations were loss-of-function (LOF) mutations in KMT2A-D (Lysine methyl transferase) genes leading to defective histone H3K4 methylation. The cumulative frequency of tumours harboring any suppressor gene alterations in the epigenetic pathway (involving KMT2C, KMT2D, EP300, ARID1A, ARID2, ATRX, CREBBP, KMT2A, KDM5C) was 45% of which 32% also had alterations in PI3KCA [2].
The present manuscript attempts to further detail the relevance of specific molecular alterations. The relative impact of individual markers on CC response and outcome following chemo-radiation was assessed using an integrative approach.
MATERIALS and METHODS
Patients
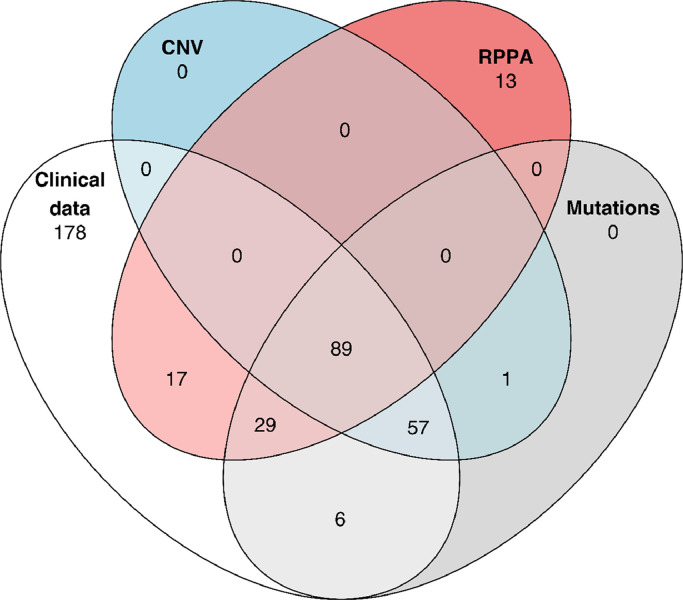
Patients included in this study had been enrolled in the EU-funded prospective CC BioRAIDs study (NCT02428842) run by the RAIDs Network (Rational Molecular Assessment and Innovative Drug Selection, www.raids-fp7.eu). The clinical protocol together with tumour sampling procedures, quality control of samples and treatment was conducted in 18 European centers from seven European countries. Study results have been previously published [2,9,14]. The study has been conducted in accordance with the ethics principles of the Declaration of Helsinki and a signed informed consent for the participation in the study was a prerogative. All patients had pretreatment mandatory frozen tumour, blood and serum sampling as well as mandatory magnetic resonance imaging. Pet scan imaging was optional and is available in half of the population. Genomics analyses on patient derived samples have already been published [2]. Several data-frames were compiled to ask or validate specific questions, based on shared patients and data types. Four subpopulations for whom omics and reverse phase protein array (RPPA) data are available were considered more specifically in this manuscript: 181 pts with mutation data (Supplementary Table 1), 146 with mutation and copy number variation (CNV) data, 135 with RPPA data and 89 patients with full molecular profile (Table 1 and Fig. 1).
Table 1.
Patients’ characteristics of the different BioRAIDs subpopulations.
| Clinical data | Mutation data | Mutation and CNV data | RPPA data | Mutation, CNV and RPPA data | ||
|---|---|---|---|---|---|---|
| Number of patients | 376 | 181 | 146 | 135 | 89 | |
| FIGO 2018 | I/II | 290 (77%) | 133 (73%) | 110 (75%) | 97 (72%) | 66 (74%) |
| III/IV | 86 (23%) | 48 (27%) | 36 (25%) | 38 (28%) | 23 (26%) | |
| PFS Event | 108 (29%) | 60 (33%) | 50 (34%) | 46 (34%) | 32 (36%) | |
| HPV Clade | Clade 7/HPV negative | 104 (28%) | 54 (30%) | 41 (28%) | 42 (31%) | 27 (30%) |
| Clade 9/Others | 220 (58%) | 126 (70%) | 104 (71%) | 93 (69%) | 62 (70%) | |
| NA | 52 (14%) | 1 (<1%) | 1 (<1%) | 0 | 0 | |
| Histology | Squamous | 308 (82%) | 148 (82%) | 118 (81%) | 112 (83%) | 72 (81%) |
| Adenocarcinoma | 43 (11%) | 19 (10%) | 16 (11%) | 15 (11%) | 11 (12%) | |
| Adenosquamous | 15 (4%) | 11 (6%) | 9 (6%) | 7 (5%) | 5 (6%) | |
| Mixed/undifferentiated. | 9 (2%) | 3 (10%) | 3 (2%) | 1 (<1%) | 1 (1%) | |
| NA | 1 (<1%) | 0 | 0 | 0 | 0 | |
| Initial Treatment | Chemoradioation | 242 (64%) | 112 (62%) | 89 (61%) | 89 (66%) | 55 (62%) |
| Surgery | 76 (20%) | 35 (19%) | 30 (21%) | 21 (16%) | 16 (18%) | |
| NACT | 58 (15%) | 34 (19%) | 27 (18%) | 25 (19%) | 18 (20%) | |
CNV= copy number variation; FIGO 2018 integrating lymph nodes status under IIIC; PFS=Progression free survival; HPV type (based on hybridisation test)2: Clade 9 (HPV 16.31.33.35.52.58) & Clade 7 (HPV 18.39.45.59.68); NACT=Neaodjuvant chemotherapy, NA=not available.
Fig. 1.
Venn diagram illustrating the number of patients for the different combinations of omics types and clinical data. (CNV = copy number variation; RPPA = reverse phase protein array).
DNA sequencing & bioinformatics
Paired-end whole exome sequencing (WES) and paired-end targeted gene panel sequencing were performed on a HiSeq2500 platform. The sequencing was performed to reach an average depth of coverage of ~150 × for whole exome sequencing and ~730 × for targeted sequencing. The data were further processed by the Institut Curie bioinformatics pipeline. Somatic alterations (point mutations, insertions/deletions and copy number changes) were identified (Supplementary Table 2) from the aligned sequences of matched-samples using dedicated tools, detailed in Scholl et al. [2]
CoxBoost analysis [15] was used to fit a Cox proportional hazards model by component wise likelihood-based-boosting. This type of analysis is suited for models with high number predictors, typically omics covariates. Analysis was limited to previously curated gene variants of documented clinical significance. The analysis furthermore focused on molecular alterations that were detected in a sizable proportion of patients (>5%), to ensure estimation of the stability and relevance of the procedure.
Reverse phase protein array (RPPA)
For RPPA analyses, the samples were processed as previously described [16] and printed onto nitrocellulose covered slides (Supernova, Grace Biolabs) using a dedicated arrayer (2470 arrayer, Aushon Biosystems). Five serial dilutions, starting at 2000 µg/ml and two technical replicates per dilution were printed for each sample. Arrays were labeled with 194 specific, or without primary antibody (as negative control), as described previously described [16]. All primary antibodies used in RPPA have been previously tested by Western Blotting to assess their specificity for the protein of interest. Raw data were normalized using Normacurve [17], which normalizes for fluorescent background per spot, a total protein stain and potential spatial bias on the slide. Next, each RPPA slide was median centered and scaled (divided by median absolute deviation). We then corrected for remaining sample loading effects individually for each array by correcting the dependency of the data for individual arrays on the median value of each sample, over all 194 arrays, using a linear regression.
STATISTICAL ANALYSIS
The endpoint of interest is progression-free survival (PFS), defined as the minimal time of relapse or death, with administrative censoring at 24 months. All analyses were performed using R version 3.6.1 software.
ROLE of FUNDERS
The funders had no role in study design, data collection, data analysis, interpretation or writing of the report.
RESULTS
Patient characteristics with a complete dataset (n = 89)
Clinical and patho-biological covariates on the complete BioRAIDS population (n = 376) have been previously published [2].
Here we focused first on a patient population (n = 89) with complete information for all data types (mutations, amplifications/deletions and protein expression and phosphorylation patterns). For subsets of the initial population (Table 1), there were no significant changes in treatment allocation or in clinical and patho-biological parameters such as FIGO-2018 stage (I-II vs III-IV), HPV type {negative and viral clade 7 (HPV18,39,45,59,68) as opposed to viral clade 9 (HPV 16,31,33,35,52,58)}. While 62–66% of patients were allocated to chemo-radiation as a first treatment, many patients had external beam radiation with or without platinum or brachytherapy as a follow-on treatment after surgery or neoadjuvant chemotherapy. This led to close to 90% of patients having received radiation therapy as previously published [2].
Prognostic biomarkers for standard treatment in CC: integrating clinical, mutations, CNV, as well as activated protein expression
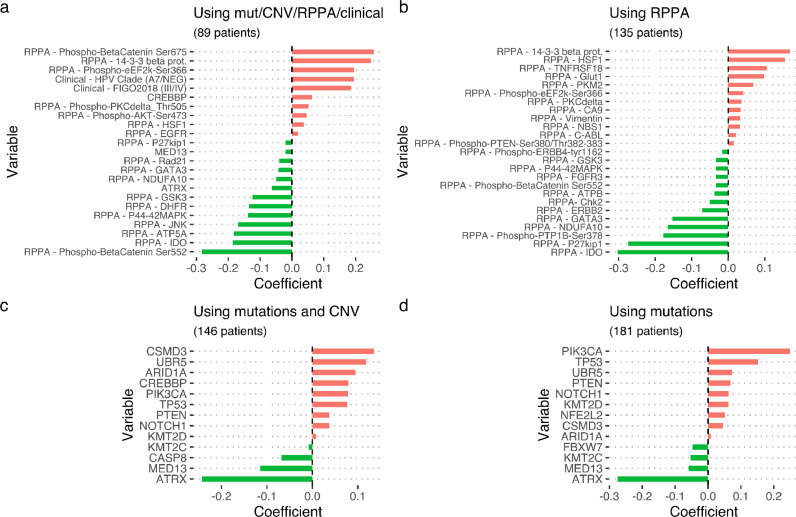
Using this complete cohort of 89 patients, the CoxBoost approach provides non-zero ranked biomarker effect estimates summarized in (Fig. 2a). A positively weighted biomarker in this test corresponds to a higher risk of event occurrence as measured by PFS and therefore represents a deleterious biomarker.
Fig. 2.
Coefficient values in Cox boost of frequent genetic variants associated with worse or better prognosis using all available omics types (mutational, copy number variation and RRPA variables) clinical data (panel a); clinical data with RPPA variables (panel b); gene and copy number variants and mutations (panel c) and mutations only (panel d). (mut = mutations; CNV = copy number variations; RPPA = reverse phase protein array).
When RPPA data were factored into the CoxBoost analysis of mutations and copy number variants in a patient population with information for all omics types, a number of activated phospho-proteins outranked molecular alterations. Intriguingly, beta-catenin, when phosphorylated in Ser552 (pβ-Cat552), ranked highest as a better prognosis indicator, while another post translationally modified form (pβ-Cat675) of the same protein, ranked second highest for worse prognosis. Protein 14–3–3 β, an abundant chaperon protein and supposedly responsible for pβ-Cat552 nuclear exit [18], ranked highest for worse prognosis (Fig. 2a).
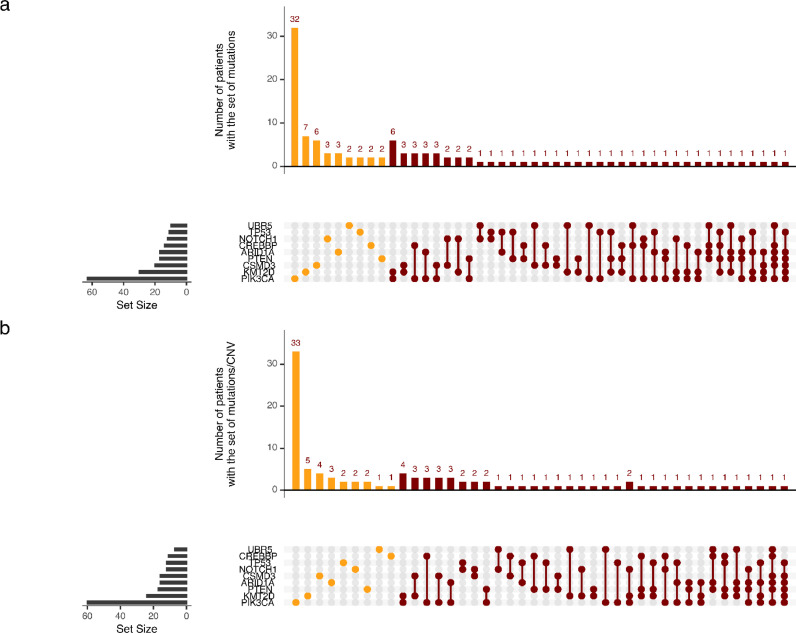
When patients with genetic variants together with copy number variations (n = 146; Fig. 2c) or genetic variants only (n = 181, Fig. 2d) were analyzed separately, other genetic markers with significant alterations for outcome based on WES were detected. Genetic variants consistently associated with better prognosis were alterations in ATRX and MED13, while LOF of TP53 and CREBBP remained the dominant genetic parameters, significantly associated with worse prognosis. Only variants represented in at least 5% of patients were tested. A better prognosis was associated with significant alterations in the following genes: FBXW7, KMT2C (MLL3) while alterations in: CSMD3, UBR5, PIK3CA, NOTCH1, NFE2L2, ARID1A, KMT2D (MLL2), and PTEN were associated with a higher risk for recurrence.
Phosphorylation patterns in β-Cat at ser552 is a good prognostic indicator in an enlarged patient population (n = 135)
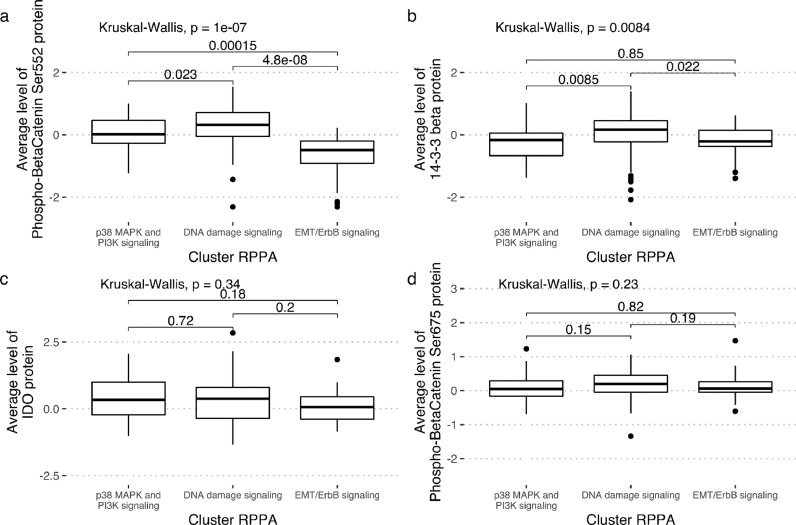
To confirm these protein activation-related findings, we focused on a patient set of 135 patients with data on both clinical outcome and on RPPA expression (Fig. 2b). Furthermore, looking at the previously identified RPPA clusters [2], pβ-Cat552 appeared significantly less expressed in the EMT cluster, as compared to the two other clusters (Fig. 3a), lending credence to its relevance as a good prognostic indicator. Protein 14–3–3 β appeared significantly enriched in the “DNA damage signaling” cluster (Fig. 3b). In addition, no association was found between the levels of IDO protein (Fig. 3c) or the pβ-Cat675 form (Fig. 3d) with any of the three clusters.
Fig. 3.
Expression levels of phosphobeta-catenin-Ser552 (panel a) and 14–3–3 protein (panel b), IDO (panel c) and phosphobeta-catenin-Ser675 (panel d) per RPPA cluster.
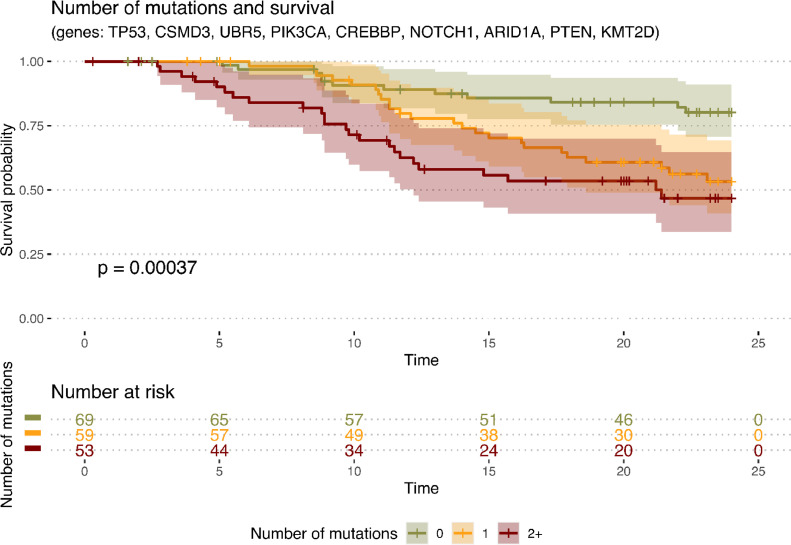
Tumour genetic heterogeneity and timing of recurrence
Patients with co-occurences of several deleterious genetic markers (previously identified as associated with worse prognosis in Fig. 2) were grouped according to the number of mutations they carry. Mutations could be inferred from a population of 181 patients (as shown in Fig. 1 and 2d) and mutations (only) were taken-into-account in this analysis. Kaplan Meier survival curves, based on the number of the deleterious markers identified in a specific tumour, with PFS estimates at 24 months showed that tumours carrying ≥ 2 of relevant genetic appeared to be at a higher risk for earlier recurrence, as compared to those with one or none of the previously documented clinically significant gene variants (Fig. 4, p-value=0.00037). While the notion that tumour heterogeneity at start, carries a higher risk, is already well established, these findings will be retested in larger populations in the future to better define individual risk.
Fig. 4.
Kaplan Meier progression free survival curves as a function of tumour heterogeneity at start (number of mutations per patients from a defined list of genes), limited to the following molecular alterations that were detected in a sizable proportion of patients (>5%). The probability of survival is the probability that the members of each group did not experience death or relapse at each time point.
Several molecular alterations may co-exist and affect outcome in individual patients (Fig. 5a and 5b). When we collated these various genomic alterations associated with treatment resistance on a per patient basis, a large fraction of the population (n = 53) had single alterations. Alterations below the 5% frequency level have not been accounted for. Among the single alterations, PIK3CA was dominant with n = 33 cases. However, 48 (54%) patients had more than one of those genes which ranked high for a poor prognostic signal.
Fig. 5.
Pattern of frequencies of molecular alterations of significance by individual patient. Panel a with mutations only and Panel b integrating mutations and CNV.
DISCUSSION
Recent estimations of benefit of biomarker-driven clinical studies repeatedly document the low number of patients to have benefited from this approach [7]. This suggests that, for heterogeneous cancers with multiple potential oncogenic drivers, biomarkers assessed mostly by alterations in a limited gene panel at the DNA level, may not be able to predict drug responses reliably. Molecular alterations in cancer tissues have profound effects on RNA expression, which in turn lead to modifications in protein expression and activation patterns. Integrating WES and RPPA data and using a CoxBoost analysis for progressive enrichment of biomarkers, we identified 16 molecular alterations and 30 activated proteins that were associated with good or poor outcome up to 24 months, for patients having received standard chemo-radiation. Furthermore, patients diagnosed with two or more of these molecular alterations progressed earlier. The present integration of genotype and phenotype led to a highly interesting finding, namely that two different post-translational modifications of the same protein: beta-catenin, exhibit opposite effect on the PFS.
In agreement with the present integrative results, what is the published evidence that alterations in ATRX, MED13, and possibly CASP8 (LOF) (Fig. 2) might render tumour cells more sensitive to this treatment?
ATRX: This gene was first discovered as the genetic cause of the α-thalassemia, mental retardation, X-linked (ATRX) syndrome [19,20]. It is required for efficient replication of a subset of genomic loci and involved in maintaining telomere structural integrity in embryonic stem cells [21]. ATRX has thus important cellular functions, being involved in meiotic spindle organization, DNA repair, chromatin organization and remodeling as well as nucleosome assembly [21]. ATRX loss was shown to induce genomic instability [22] and ATRX-deficient mouse tumours were shown to be highly aggressive. Nevertheless, a better response to immune check point inhibitors was observed in mice harboring ATRX-deficient tumours [22]. ATRX gene mutations have also been associated with good prognosis by extensive studies in neural tumours like gliomas and neuroblastoma, but, to our knowledge, no studies with respect to ATRX and cervical cancer are presently available. Thus, this finding in cervical carcinoma is of high interest [23].
MED13 in association with MED12, CDK8, and Cyclin C constitutes a four-subunit “kinase” module that exists in variable association with a 26-subunit Mediator core [24]. Genetic and biochemical studies have established the Mediator kinase module to function in developmental and oncogenic signaling through Mediator, and much of its function in signal-dependent gene regulation is thought to derive from its resident CDK8 kinase activity which has been also been linked to Wnt signaling [25,26,27]. Mediator is recruited to promoters by transcriptional activators or nuclear receptors (ER, AR among others) to induce gene expression and serves as a scaffold for the assembly of a preinitiation complex. The mediator function is involved in the regulated transcription of nearly all RNA polymerase II-dependent genes [25,28]. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Beyond CDK8, little is presently known regarding the functional convergence and divergence related to MED12/12 L and MED13/13 [24]. However, genetic ablation of MED12 or MED13 in mice similarly conferred early embryonic lethality, excluding the possibility of functional redundancy among their corresponding paralogs during development [25]. These data are in line with the present results suggesting that cancer cell loss of MED13 function might increase susceptibility to cancer cell damage by chemo-radiation.
Caspase-8 has been originally identified as an essential player of death receptor-induced apoptosis. Emerging evidence suggested that the retention of caspase-8 in glioblastoma may interfere with the sensitivity to radio and chemotherapeutic approaches through multiple pathways, including improvement of the DNA damage repair and the activation of NF-κB and cytokine production. Caspase-8 contributes to the functionality of the DNA damage response. Caspase-8 LOF on the other hand promotes genomic instability and tumour development. Intriguingly, it has been suggested that the inhibition of Caspase-8, although detrimental for apoptosis induction, may enhance the sensitivity of cancer cells to DNA damaging agents, most likely independent of apoptosis, and may therefore represent a valuable therapeutic strategy [29]. Loss of Caspase-8 function was associated with good prognosis in the cancer genome atlas (TCGA) in head and neck squamous cell carcinoma [4]. As a result, tumours with loss of Caspase-8 function may be more likely to benefit from chemoradiation, in accordance with the present results, while they might not be sensitive to immune checkpoint inhibition.
While alterations in ATRX, MED13 and CASP8 were association with treatment response, our results suggested that LOF alterations in TP53 and CREBBP may be top ranking markers that impact cervical cancer cells to resist chemo-radiation.
TP53: In physiological conditions, the encoded p53 protein responds to cellular stresses by inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. In the present patient cohort, the TP53 bi-allelic gene loss was highly detrimental but present at a low frequency, in opposition to other malignancies such as in head/neck squamous cell carcinomas (HNSCC), or high grade serous carcinoma of the ovary, which are primarily driven by complete loss of the gene TP53. In HNSCC, a subgroup of patients with dismal prognosis after chemoradiation was characterized by TP53 mutations. The prognostic impact was stronger for nonsense/frameshift TP53 mutations associated with either expression of a truncated p53 protein or complete loss of p53 expression compared with missense mutations resulting in overexpression of mutated p53 [30]. Similarly, in diffuse intrinsic pontine gliomas (DIPG), inactivation by mutation of TP53 was shown to contribute to radio-resistance [31]. In cervical cancer, the p53 protein is more commonly ubiquitinated and degraded by HPVE6, leading to carcinogenesis. While the HPV virus mediated inactivation of TP53 protein, together with the inactivation of RB (retinoblastoma) protein by HPVE7, are the most frequent driver protein alterations in cervical cancer, only the cervical cancer patients with a complete bi-allelic TP53 gene loss had a dismal prognosis. Furthermore, HPV positivity and TP53 loss are mutually exclusive patterns in cervical cancer patients (Kamal et al., submitted).
Novel cancer therapeutics that supposedly reactivate a variant tumour p53 protein include small molecule drug candidates (such as APR-246), presently in clinical development for myelodysplastic syndromes, acute myeloid leukemia, as well as solid tumours (NCT03268382) [32]. A phase Ib/II clinical trial in TP53 mutated high-risk hematological cancers is ongoing, combining APR-246 and azacytidine (NCT03931291). Such a combination might be worth considering for chemoradiation resistant cervical cancer patients with loss of function in both p53 and epigenetic regulating genes.
CREBBP was first isolated as a nuclear protein that binds to cAMP-response element binding protein (CREB), and together with EP300 was shown to encode lysine acetyl-transferases (KAT) with critical roles in embryonic development, growth control, and homeostasis by coupling chromatin remodeling to transcription factor recognition [33].
CREBBP alterations have been documented in many cancers, and specifically in 5% of SCC of the uterine cervix [30], which is comparable to the 8% LOF by mutation or non-frame-shift deletion, reported here. CREBBP loss in lung cancer was shown to reduce histone acetylation and transcription of cellular adhesion genes, while driving tumorigenesis [34]. These effects could be partially restored by HDAC inhibition, which exhibited enhanced effectiveness in CREBBP-deleted tumours, suggesting a role for treatment by a HDAC inhibitor such as Vorinostat, while CREBBP/EP300 bromo‑domain inhibitors are also under investigation [35].
In a previous publication, we had tested the relative role of PIK3CA alone or PIK3CA and a “metagene” of epigenetic regulators. While PIK3CA alone did not carry a bad prognosis, it did so in association with alterations in epigenetic regulators [2]. Fig. 5 details the patients with single or multiple molecular alterations (above the 5% frequency level) that were present in the population. These results may assist to define the patient population for future molecular targeting.
Two different post-translational modifications on beta-catenin appeared to be high-profile parameters related to outcome, with opposite prognosis, depending on the protein isoform as distinguished by its specific phosphorylation pattern. The supposedly active nuclear form of beta catenin, which according to Goretsky et al. [18], is phosphorylated in Ser 552 (pβ-Cat552) appeared associated with good outcome, while the predominance of a phosphorylated epitope in Ser 675 (pβ-Cat675), which is located just outside the armadillo repeat at the beginning of the C terminal domain, was associated with poor outcome in our dataset. Truncations in this C terminal domain was shown to lead to loss of function [36]. Crystallographic studies have suggested that binding of E-cadherin or APC occurs in the armadillo structure, whereas helix C (containing Ser 675) appears to interact with Chibby and ICAT (alias CTNNBIP1), which is a physiological inhibitor of Wnt signals that prevents the binding of TCF to beta catenin [37]. Of interest is the finding that high protein 14–3–3 expression which cooperates with Chibby to regulate subcellular distribution and signaling activity [38] in the CoxBoost analysis came up close together and on the same side with the high levels in the Helix C (pβ-Cat675) chain. We confirmed our results on a larger patient population (n = 135) for whom RPPA and clinical, but not genetic data, were presently available. Furthermore, the (pβ-Cat552) isoform associated with response to chemoradiation was inversely correlated with the RPPA cluster of proteins associated with EMT. Our findings, based on quality controlled fresh frozen clinical samples, were puzzling in view of the experimentally derived data by Goretsky et al. [18] who suggested that, during Wnt-activated signaling, beta-catenin undergoes partial site-directed cleavage prior to a nuclear localization of a low molecular weight form (LMW) which is supposedly the pβ-Cat552 isoform. Furthermore their findings suggested that the LMW pβ-Cat552 isoform supposedly binds transcription factor 4 (TCF4) and drives transcription in chromatin-bound fractions. Overexpression in vitro of a double truncated form of beta-catenin, (reducing it to the LMW form), enhanced transcriptional activation, cell proliferation and growth of tumour xenografts, while a substitution of Ser 552 to Alanine abrogated all these effects.
Wnt-activated signaling is commonly associated with cancer progression. However, if pβ-Cat552 is indeed associated with cell proliferation, as suggested by Goretsky et al., cells overexpressing pβ-Cat552 are likely to be more sensitive to chemo-radiation and thus be associated with a better prognosis in this population treated by chemo-radiation. Moreover, according to our biostatistics analysis, pβ-Cat552 was lower in the EMT cluster, and EMT is often a hallmark of stem cells that proliferate less and are resistant to chemo-radiation. While cancer cells are thought to proliferate and to invade, it is tempting to speculate that the pβ-Cat552 phosphorylation form might specifically orientate towards proliferation and not invasion, while the predominance of the pβ-Cat675 form might be more permissive to invasion. The bioinformatics analysis does not associate this latter form with any of the RPPA clusters, but it is not anti-correlated with EMT. In cancer cells, many additional alterations are at play and inactivation of UBR5 (an E3 ligase), normally directing the beta-catenin protein to the proteasome pathway, may be of relevance. Alternatively, similarly to the accumulation of a non-functional defective p53 protein in the case of LOF TP53 mutations, pβ-Cat552 might accumulate when it cannot function and/or resist degradation, or degradation enzymes may be inactive. While the subcellular location of the activated protein forms of beta-catenin as well as their functionality and their kinetics remain to be assessed in the context of cancer treatment, different drugs relevant to target aberrantly active beta-catenin signaling have been suggested, such as Bortezomib [18]. Wnt pathway targeting has also been recently reviewed in a context of head and neck squamous cell carcinoma [39]. Furthermore, evidence from a human retinal epithelial cell model showed silencing of endogenous TF (tissue factor) to significantly suppress the Wnt/β-catenin signal transduction cascades, suggesting that the regulation of TF on VEGF expression may be mediated by activation of the Wnt/β-catenin signaling pathway [40]. Tisotumab Vedotin, directly targets tissue factor and was shown safe and effective in SCCC [41]. Targeted therapy towards TF and Wnt signaling and its downstream proteins is promising for interventions of pathological processes involving TF-regulated angiogenesis and inflammation.
IDO (indole amine 2,3 oxygenase) is thought to play a role in a variety of pathophysiological processes such as antimicrobial and anti-tumour defense, neuropathology, immunoregulation and antioxidant activity. This enzyme catalyzes the first step of the catabolism of the essential amino acid tryptophan along the kynurenine pathway [42]. In addition, through its expression in dendritic cells, monocytes, and macrophages, this enzyme modulates T-cell behavior by its peri‑cellular catabolization of the essential amino acid tryptophan. IDO is thought to act either as a suppressor of anti-tumour immunity or involved in anti-tumour defense while high IDO in the present analysis was an important predictor of good prognosis in the context of response to chemo-radiation therapy [43,44]. Furthermore, mutations in the gene UBR5 (Ubiquitin Protein Ligase E3 Component N-Recognin 5) was associated with poor prognosis. Since IDO is an important enzyme in the tryptophan metabolism and the gene UBR5 being related to tryptophan metabolism, these two factors might contribute to radiation-induced immune check point activation which could potentially affect the response to radiation [45].
In conclusion, this is to our knowledge the first omics derived evidence for the relevance of two different post translationally modified forms of beta-catenin as potential biomarkers for chemo-radiation. Our data is consistent with the idea shared by others [46,38] that beta-catenin expression/activation together with p53 [30] and epigenetic alterations (in particular CREBBP and possibly UBR5, CSMD3, KMT2D) may be at the heart of cervical cancer control/progression. The relevance of Wnt pathway activity and CTNNB1 mutations and activations in cancer is not new, but elucidation of the intimate control mechanisms, leading to the nuclear translocation of an active form, is recent and our findings call for additional mechanistic studies, on post-translational modifications of beta catenin together with cell invasive behavior.
Our study comes with some limitations, notably the need for validation in larger cohorts with clinical, molecular alterations, conjointly with protein expression/activation. The reproducibility of the set of biomarkers identified by the boosting approach will be under further scrutiny. Large aggregated studies are mandatory to confirm relevant constellations of molecular alterations and decipher multiple functional drivers that can be targeted. Focus on individual patient-specific molecular information will lead to innovative combined treatments to be explored in umbrella type platform trials.
FUNDING SOURCES: This project has received funding from the European Union's Seventh Program for research, technological development and demonstration under grant agreement No 304,810 and the Fondation ARC pour la recherche contre le cancer. The funders had no role in study design, data collection, data analysis, interpretation or writing of the report.
AUTHORS contribution ITEMS
Conception and design: Suzy M Scholl and Maud Kamal
Development of methodology: Jonas Beal and Aurélien Latouche
Acquisition of data: Leanne de Koning, Marina Popovic, Anne de la Rochefordière, Fabrice Lecuru, Virginie Fourchotte, Charlotte Ngo, Anne Floquet, Els Berns, Gemma Kenter, Heiko von der Leyen, Vincent Puard
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Jonas Beal, Elodie Girard, Attila Kereszt, Balazs Balint, Pierre Gestraud, Aurelien Latouche, Suzy Scholl, Maud Kamal
Writing, review, and/or revision of the manuscript: Suzy M Scholl, Jonas Beal, Maud Kamal, Aurélien Latouche
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Heiko von der Leyen, Charlotte Lecerf
Study supervision: Sergio Roman, Aurelien Latouche, Roman Rouzier
DECLARATION OF INTERESTS
All authors report no conflict of interest.
ACKNOWLEDGEMENTS
We thank the RAIDs consortium (http://www.raids-fp7.eu/consortiumRAIDs consortium http://www.raids-fp7.eu/consortium): Pierre Fumoleau, Linda Larbi Chérif, Christophe Le Tourneau, Ivan Bièche, Emmanuelle Jeannot, Diana Bello Roufai, Aljosa Mandic, Nina Samet, Choumouss Kamoun, Windy Rondof, Sebastien Armanet, Alexandra Rohel, Souhir Neffati, Marie-Emmanuelle Legrier, Sinette Ngoumou Mabiala, Sylvain Dureau, Coralie Errera, Marius Craina, Madalin Margan, Sanne Samuels, Henry Zijlmans, Peter Hillemanns, Sorin Dema, Alis Dema, Goran Malenkovic, Branislav Djuran, Frédéric Guyon, Pierre Emmanuel Colombo, Michel Fabbro, Christine Kerr, Eleonor Rivin del Campo, Charles Coutant, Frédéric Marchal, Nathalie Mesgouez-Nebout, Jean Guillaume Feron, Philippe Morice, Eric Deutsch, Pauline Wimberger, Jean-Marc Classe, Mathieu Minsat, Istvan Nagy, Nicolas de Saint-Jorre, Alexia Savignoni, Patricia Tresca, Noreen Gleeson, Philippe Hupe, Emmanuel Barillot, Fanny Coffin, Bastiaan Nuijen, Alexandre Boissonnas, Marc Billaud, Laurence Lafanechere, Kirsten Ruigrok-Ritstier, Andrea Slocker, Michele Mondini, Maud Bossard, Sjoerd Rodenhuis, Rene Medema, Anika Havemeier, Thomas Fink, Amelie Michon, Christine Kubiak, Corine Beaufort, Judit Cseklye, Dora Latinovics, Peter Bihari, Isabel Brito, Bérengère Ouine, Elaine Del Nery, Jos Beijnen, Dominique Koensgen, Daniela Bruennert, Slavica Knezevic, Milos Lucic, and Natalja ter Haar for their precious help in the conduct of the RAIDs project.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103049.
Appendix. Supplementary materials
References
- 1.Cancer Genome Atlas Research Network. Albert Einstein College of Medicine. Analytical Biological Services. Barretos Cancer Hospital. Baylor College of Medicine. Beckman Research Institute of City of Hope Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi: 10.1038/nature21386. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholl S., Popovic M., de la Rochefordiere A., Girard E., Dureau S., Mandic A. Clinical and genetic landscape of treatment naive cervical cancer: alterations in PIK3CA and in epigenetic modulators associated with sub-optimal outcome. EBioMedicine. 2019 May;43:253–260. doi: 10.1016/j.ebiom.2019.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017 Dec 14;171(7) doi: 10.1016/j.cell.2017.10.044. 1611-1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015 Jan 29;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Tourneau C., Delord J.-.P., Gonçalves A., Gavoille C., Dubot C., Isambert N. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015 Oct;16(13):1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 6.McGranahan N., Swanton C. Clonal Heterogeneity and Tumor Evolution: past, Present, and the Future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. 09. [DOI] [PubMed] [Google Scholar]
- 7.Galot R., Le Tourneau C., Guigay J., Licitra L., Tinhofer I., Kong A. Personalized biomarker-based treatment strategy for patients with squamous cell carcinoma of the head and neck: EORTC position and approach. Ann Oncol. 2018;29(12):2313–2327. doi: 10.1093/annonc/mdy452. 01. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty K.T., Gray R., Chen A., Li S., Patton D., Hamilton S.R., et al. THE MOLECULAR ANALYSIS FOR THERAPY CHOICE (NCI-MATCH) TRIAL: LESSONS for GENOMIC TRIAL DESIGN. J Natl Cancer Inst [Internet]. [cited 2020 Apr 23]; Available from:https://academic.oup.com/jnci/advance-article/doi/10.1093/jnci/djz245/5699915 [DOI] [PMC free article] [PubMed]
- 9.Ngo C., Samuels S., Bagrintseva K., Slocker A., Hupé P., Kenter G. From prospective biobanking to precision medicine: bIO-RAIDs – an EU study protocol in cervical cancer. BMC Cancer [Internet] 2015 Nov 4:15. doi: 10.1186/s12885-015-1801-0. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4632364/ [cited 2020 Apr 23]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarkar R., Patil D., Crook T., Datta V., Bhalerao S., Dhande S. Encyclopedic tumor analysis for guiding treatment of advanced, broadly refractory cancers: results from the RESILIENT trial. Oncotarget. 2019 Sep 24;10(54):5605–5621. doi: 10.18632/oncotarget.27188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015 Mar 1;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 12.Schiffman M.H., Bauer H.M., Hoover R.N., Glass A.G., Cadell D.M., Rush B.B. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993 Jun 16;85(12):958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- 13.Wentzensen N., Vinokurova S., von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004 Jun 1;64(11):3878–3884. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 14.Samuels S., Balint B., von der Leyen H., Hupé P., de Koning L., Kamoun C. Precision medicine in cancer: challenges and recommendations from an EU-funded cervical cancer biobanking study. Br J Cancer. 2016 Dec 6;115(12):1575–1583. doi: 10.1038/bjc.2016.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder H., Schumacher M. Allowing for mandatory covariates in boosting estimation of sparse high-dimensional survival models. BMC Bioinformatics. 2008 Jan 10;9(1):14. doi: 10.1186/1471-2105-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meseure D., Vacher S., Lallemand F., Alsibai K.D., Hatem R., Chemlali W. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br J Cancer. 2016 14;114(12):1395–1404. doi: 10.1038/bjc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troncale S., Barbet A., Coulibaly L., Henry E., He B., Barillot E. NormaCurve: a SuperCurve-based method that simultaneously quantifies and normalizes reverse phase protein array data. PLoS ONE. 2012;7(6):e38686. doi: 10.1371/journal.pone.0038686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goretsky T., Bradford E.M., Ye Q., Lamping O.F., Vanagunas T., Moyer M.P. Beta-catenin cleavage enhances transcriptional activation. Sci Rep. 2018 Jan 12;8(1):1–15. doi: 10.1038/s41598-017-18421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons R.J., Picketts D.J., Villard L., Higgs D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995 Mar 24;80(6):837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 20.Wada T., Kubota T., Fukushima Y., Saitoh S. Molecular genetic study of japanese patients with X-linked alpha-thalassemia/mental retardation syndrome (ATR-X) Am J Med Genet. 2000 Sep 18;94(3):242–248. [PubMed] [Google Scholar]
- 21.Watson L.A., Goldberg H., Bérubé N.G. Emerging roles of ATRX in cancer. Epigenomics. 2015;7(8):1365–1378. doi: 10.2217/epi.15.82. [DOI] [PubMed] [Google Scholar]
- 22.Koschmann C., Calinescu A.-.A., Nunez F.J., Mackay A., Fazal-Salom J., Thomas D. ATRX loss promotes tumor growth and impairs nonhomologous end joining DNA repair in glioma. Sci Transl Med. 2016 Mar 2;8(328) doi: 10.1126/scitranslmed.aac8228. 328ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fazal Salom J., Bjerke L., Carvalho D., Boult J., Mackay A., Pemberton H. PDTM-33. ATRX LOSS CONFERS ENHANCED SENSITIVITY TO COMBINED PARP INHIBITION AND RADIOTHERAPY IN PAEDIATRIC GLIOBLASTOMA MODELS. Neuro Oncol. 2018 Nov 5;20(suppl_6):vi210–vi211. [Google Scholar]
- 24.Bourbon H.-.M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008 Jul;36(12):3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark A.D., Oldenbroek M., Boyer T.G. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol. 2015;50(5):393–426. doi: 10.3109/10409238.2015.1064854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature. 2008 Sep 25;455(7212):547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Li X., Kong X., Luo Q., Zhang J., Fang L. MiRNA-26b inhibits cellular proliferation by targeting CDK8 in breast cancer. Int J Clin Exp Med. 2014;7(3):558–565. [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai K.-.L., Sato S., Tomomori-Sato C., Conaway R.C., Conaway J.W., Asturias F.J. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol. 2013 May;20(5):611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boege Y., Malehmir M., Healy M.E., Bettermann K., Lorentzen A., Vucur M. A Dual Role of Caspase-8 in Triggering and Sensing Proliferation-Associated DNA Damage, a Key Determinant of Liver Cancer Development. Cancer Cell. 2017 11;32(3) doi: 10.1016/j.ccell.2017.08.010. 342-359.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinhofer I., Stenzinger A., Eder T., Konschak R., Niehr F., Endris V. Targeted next-generation sequencing identifies molecular subgroups in squamous cell carcinoma of the head and neck with distinct outcome after concurrent chemoradiation. Ann Oncol. 2016;27(12):2262–2268. doi: 10.1093/annonc/mdw426. [DOI] [PubMed] [Google Scholar]
- 31.Werbrouck C., Evangelista C.C.S., Lobón-Iglesias M.-.J., Barret E., Teuff G.L., Merlevede J. TP53 pathway alterations drive radioresistance in Diffuse Intrinsic Pontine Gliomas (DIPG) Clin Cancer Res [Internet] 2019 Jan 1 doi: 10.1158/1078-0432.CCR-19-0126. https://clincancerres.aacrjournals.org/content/early/2019/08/31/1078-0432.CCR-19-0126 [cited 2020 Jun 22]. Available from: [DOI] [PubMed] [Google Scholar]
- 32.Gourley C., Green J., Gabra H., Vergote I., Basu B., Brenton J.D. PISARRO: a EUTROC phase Ib study of APR-246 in combination with carboplatin (C) and pegylated liposomal doxorubicin (PLD) in platinum sensitive relapsed high grade serous ovarian cancer (HGSOC) JCO. 2016 May 20;34(15_suppl) 5571–5571. [Google Scholar]
- 33.Attar N., Kurdistani S.K. Exploitation of EP300 and CREBBP Lysine Acetyltransferases by Cancer. Cold Spring Harb Perspect Med. 2017 Mar 1;7(3) doi: 10.1101/cshperspect.a026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia D., Augert A., Kim W, Eastwood E., Wu N., Ibrahim A.H. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov. 2018;8(11):1422–1437. doi: 10.1158/2159-8290.CD-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou L., Xiang Q., Xue X., Zhang C., Li C., Wang C. Y08197 is a novel and selective CBP/EP300 bromodomain inhibitor for the treatment of prostate cancer. Acta Pharmacol. Sin. 2019 Nov;40(11):1436–1447. doi: 10.1038/s41401-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox R.T., Pai L.M., Kirkpatrick C., Stein J., Peifer M. Roles of the C terminus of Armadillo in Wingless signaling in Drosophila. Genetics. 1999 Sep;153(1):319–332. doi: 10.1093/genetics/153.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing Y., Takemaru K.-.I., Liu J., Berndt J.D., Zheng J.J., Moon R.T. Crystal Structure of a Full-Length β-Catenin. Structure. 2008 Mar 11;16(3):478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F.-.Q., Mofunanya A., Harris K., Takemaru K.-.I. Chibby cooperates with 14-3-3 to regulate β-catenin subcellular distribution and signaling activity. J Cell Biol. 2008 Jun 30;181(7):1141–1154. doi: 10.1083/jcb.200709091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paluszczak J. The Significance of the Dysregulation of Canonical Wnt Signaling in Head and Neck Squamous Cell Carcinomas. Cells. 2020 Mar 15;9(3) doi: 10.3390/cells9030723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Sang A., Zhu M., Zhang G., Guan H., Ji M. Tissue factor induces VEGF expression via activation of the Wnt/β-catenin signaling pathway in ARPE-19 cells. Mol Vis. 2016;22:886–897. [PMC free article] [PubMed] [Google Scholar]
- 41.Hong D.S., Concin N., Vergote I., de Bono J.S., Slomovitz B.M., Drew Y. Tisotumab Vedotin in Previously Treated Recurrent or Metastatic Cervical Cancer. Clin Cancer Res. 2020 Mar 15;26(6):1220–1228. doi: 10.1158/1078-0432.CCR-19-2962. [DOI] [PubMed] [Google Scholar]
- 42.Metz R., Duhadaway J.B., Kamasani U., Laury-Kleintop L., Muller A.J., Prendergast G.C. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound 1-methyl-tryptophan. Cancer Res. 2007 Aug 1;67(15):7082–7087. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- 43.Uyttenhove C., Pilotte L., Théate I., Stroobant V., Colau D., Parmentier N. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003 Oct;9(10):1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 44.van Baren N., Van den Eynde B.J. Tryptophan-degrading enzymes in tumoral immune resistance. Front Immunol. 2015;6:34. doi: 10.3389/fimmu.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kesarwani P., Prabhu A., Kant S., Kumar P., Graham S.F., Buelow K. Tryptophan metabolism contributes to radiation-induced immune checkpoint reactivation in glioblastoma. Clin Cancer Res. 2018 Aug 1;24(15):3632–3643. doi: 10.1158/1078-0432.CCR-18-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S., Jeong S. Mutation Hotspots in the β-Catenin Gene: lessons from the Human Cancer Genome Databases. 2019 Jan 7;42(1):8–16. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.