Abstract
Background
Intra-tumour heterogeneity in lymphoid malignancies encompasses selection of genetic events and epigenetic regulation of transcriptional programs. Clonal-related neoplastic cell populations are unsteadily subjected to immune editing and metabolic adaptations within different tissue microenvironments. How tissue-specific mesenchymal cells impact on the diversification of aggressive lymphoma clones is still unknown.
Methods
Combining in situ quantitative immunophenotypical analyses and RNA sequencing we investigated the intra-tumour heterogeneity and the specific mesenchymal modifications that are associated with A20 diffuse large B-cell lymphoma (DLBCL) cells seeding of different tissue microenvironments. Furthermore, we characterized features of lymphoma-associated stromatogenesis in human DLBCL samples using Digital Spatial Profiling, and established their relationship with prognostically relevant variables, such as MYC.
Findings
We found that the tissue microenvironment casts a relevant influence over A20 transcriptional landscape also impacting on Myc and DNA damage response programs. Extending the investigation to mice deficient for the matricellular protein SPARC, a stromal prognostic factor in human DLBCL, we demonstrated a different immune imprint on A20 cells according to stromal Sparc proficiency. Through Digital Spatial Profiling of 87 immune and stromal genes on human nodal DLBCL regions characterized by different mesenchymal composition, we demonstrate intra-lesional heterogeneity arising from diversified mesenchymal contextures and impacting on the stromal and immune milieu.
Interpretation
Our study provides experimental evidence that stromal microenvironment generates topological determinants of intra-tumour heterogeneity in DLBCL involving key transcriptional pathways such as Myc expression, damage response programs and immune checkpoints.
Funding
This study has been supported by the Italian Foundation for Cancer Research (AIRC) (grants 15999 and 22145 to C. Tripodo) and by the University of Palermo.
Keywords: Diffuse large B-cell lymphoma, Intra-tumour heterogeneity, Digital spatial profiling, Microenvironment, SPARC
Research in context.
Evidence before this study
The heterogeneity of Diffuse Large B-cell lymphomas (DLBCL) casts its influence over the disease prognostication and therapeutic management of this entity. Increasing amount of evidence underlines the relevance of classification efforts based on the integration of transcriptional and genetic profiles of DLBCL, which reveal a conspicuous diversity in lymphomagenic trajectories and lymphoma-associated microenvironment composition. In this setting, the contribution to the intra-tumor diversity of these aggressive lymphomas of heterogeneous tumor-stromal interfaces within different infiltrated foci is still unknown.
Added value of this study
Through the adoption of a syngeneic aggressive B-cell lymphoma model our study provides experimental evidence that the stromal microenvironment of the hosting tissue can affect gene expression within a clonal lymphoma, generating an additional level of intra-tumour heterogeneity. RNA-seq revealed differential expression of several transcriptional programs, including those involving MYC and DNA damage responses.
In the human DLBCL setting, the study highlights, for the first time, the conspicuous, yet underestimated, intra-lesional heterogeneity of the DLBCL-associated stromatogenesis which confers spatial segregation to MYC expression, a key element in the current diagnostic workout of DLBCL. Digital Spatial Profiling allowed an in depth characterization of the stromal and immune diversity matching MYC with stromal heterogeneity.
The study also sheds light on the influence of the matricellular protein SPARC, a consolidated prognostic stromal feature of human DLBCL, in the stromal-driven intra-tumor heterogeneity. In mice lacking Sparc, the expression of genes from A20 lymphoma cells in distinct tissue compartments was affected, along with the immune contexture of lymphoma infiltrates, hinting to the known role of SPARC in bridging mesenchymal architecture and immune cell activation.
Implication of all the available evidence
This study adds relevant new pieces of information to the DLBCL field, drawing attention to the multifaceted implications of diversified mesenchymal adaptations associated with aggressive lymphoma dissemination. Our finding of MYC modulation at different tissue stromal interfaces opens a new prospect on the complex interpretation of MYC positivity on bioptic samples, which has implications in DLBCL diagnostic/prognostic assessment.
Alt-text: Unlabelled box
Introduction
In the era of liquid biopsy-based analyses of the systemic tumour landscape of patients, the exploitation of events on a subclonal scale has enabled a new level of management of intra-tumour heterogeneity in lymphomas [1]. Although the systemic overview of clonal evolution offers new strategies for prognostication and new windows of opportunity on targeted treatments [2], the nature of local dynamics contributing to such diversification are still poorly defined and the role of tissue stromal and immune microenvironments in influencing clonal diversity at phenotypic, transcriptional, or genetic levels deserves investigation.
Diffuse Large B-cell Lymphoma (DLBCL) represents a highly heterogeneous diagnostic category including aggressive malignant proliferations of mature B cells with variable imprints of germinal centre (GC)-derivation or post-GC differentiation - referred to as cell of origin (COO) - and dramatic variability in mutational signatures [3]. The most recent efforts in the interpretation and classification of DLBCL heterogeneity have relied on the integration of genetic and transcriptional signatures and led to the identification of discrete clusters characterized by different oncogenic trajectories, biologies, and clinical behaviour [4,5]. In the attempt to deconvolute the DLBCL complexity, the nature of associated stromal microenvironment has been investigated, revealing several layers of complexity in the stromal microenvironment, some of which associated with the COO, others providing further diversification [6,7]. From the seminal studies by the Staudt's group, a prognostic significance of the DLBCL-associated stromal microenvironment clearly emerged, which identified among the major determinants the matricellular protein SPARC, by the authors associated with tumour-infiltrating macrophages [6,8]. Indeed, SPARC consistently emerged as the fronting gene of prognostically relevant microenvironment-related signatures in DLBCL [6,7,9], being low SPARC levels associated with poor prognosis in immune-chemotherapy-treated DLBCL patients. Our group has previously demonstrated that Sparc is a major stromal factor supporting bone marrow (BM) B lymphopoiesis and secondary lymphoid organ (SLO) function influencing the mesenchymal architecture of the GC [10,11]. Moreover, we demonstrated that defective Sparc expression in SLO licensed the activation of myeloid elements towards class-I Interferon-driven responses eventually unleashing malignant lymphoproliferation in the setting of persistent immune stimulation [10].
In the present study we investigated whether tissue-specific microenvironmental cues may influence the phenotypic and transcriptional heterogeneity of an established full-blown aggressive clonal DLBCL model based on the A20 cell line, gaming for seeding to different organs [12]. We focused on two levels of microenvironment complexity: one related to the multiple tissue localizations of the lymphomatous cells, namely bone marrow (BM), liver (LI), and spleen (SPL); the other related to Sparc proficiency/deficiency in the stroma. We demonstrated that the A20 transcriptome was substantially modulated by the seeding tissues, which in turn gave raise to diversified stromal modifications. In A20 cells, Myc expression and its related transcriptional programs - along with DNA damage pathways - were affected by the tissue environment. In tissue stroma Sparc-deficiency was associated with the overexpression of Complement-related innate immune/inflammatory programs and of oncogenic/metabolic pathways thus exerting a higher immune pressure on lymphomatous cells. Moreover, through in situ quantitative immunolocalization analyses and Digital Spatial Profiling of 87 immune and stromal mRNA targets, we characterized human DLBCL intra-lesional diversity matching MYC and immune features spatial heterogeneity.
Methods
Mice and cell lines
BALB/cAnNCrl mice were purchased from Charles River Laboratories (Calco, Italy). Sparc−/- mice (on a BALB/c background) were generated and maintained in the Molecular Immunology Unit of the National Cancer Institute, Milan Italy [13]. All the experiments involving animals described in this study were approved by the Ministry of Health (authorization number 1027/2016-PR). BALB/c-derived A20 B lymphoma cell line was obtained from the American Type Culture Collection (Rockville, MD) and maintained in RPMI 1640 supplemented with 10% FCS (GIBCO). For in vivo experiments A20 cell lines were injected i.v. (5*105 cells). To perform multiple analyses and easily follow A20 lymphomatous cells, A20-GFP+ cells were adopted in multiple transplantation experiments (see Supplemental Methods). For independent validation of in situ results relative to A20 tissue infiltrates, we adopted the DLBCL cell lines OPL239 and OPL241. These models were obtained from a 6-month-old Spp1−/−/Faslpr/lpr mouse. Cell lines were characterized by flow cytometry and RT-PCR for DLBCL markers and showed the following phenotype: B220+ Cd19+ IgMneg Bcl6low/neg Bcl2+ Irf4+ Myc+. For in vivo experiments, OPL239 and OPL241 cells were injected i.v. (5*105 cells) and mice were sacrificed as they showed the first signs of distress. Spleen, lymph nodes, bone marrow and liver were analysed by flow cytometry and histology to assess the presence of lymphomatous cells.
Human DLBCL tissue samples
Formalin-fixed and paraffin embedded (FFPE) tissue samples of human DLBCL cases diagnosed between 2017 and 2019 at the San Raffaele Hospital Haematopathology Unit, Milan, Italy and at the Sant'Andrea “La Sapienza” University Hospital, Rome, Italy, were selected for the quantitative in situ immunophenotypical and multiresolution analyses. The cases were selected according to a variable expression of MYC - ranging from 10% to 60% and characterized by zonal/focal distribution. The clinical-pathological characteristics of the 12 cases are summarized in Supplemental Table S1. DLBCL nodal lesions from eight additional cases (4 GCB, 4 ABC; 3 MYC/BCL2 Double Expressors, DE, 2 MYC/BCL2 Double-hit, DH) diagnosed between 2018 and 2019 were selected from the same archives for Digital Spatial Profiling. Samples were collected according to the Helsinki Declaration and the study was approved by the University of Palermo Institutional Review Board (approval number 09/2018).
In situ histomorphological and quantitative immunophenotypical analyses
Tissues dissected from mice were washed in PBS and collected for fixation in 10% neutral buffered formalin overnight, washed in water and paraffin embedded. Bone samples were decalcified using an EDTA-based decalcifying solution (MicroDec EDTA-based, Diapath) for 8 hours, then washed out with water for 1 hour and subsequently processed and embedded in paraffin.
Four-micrometres-thick sections from mouse FFPE tissues were stained for H&E to define tissue infiltration and tumour morphology. Immunohistochemical and immunofluorescence stainings were performed as previously reported and detailed in the Supplemental Methods section [10]. For multiple-marker immunostainings, sections were subjected to sequential rounds of single-marker immunostaining and the binding of the primary antibodies was revealed by the use of specific secondary antibodies conjugated with different fluorophores or enzymes (See Supplemental Methods section). Slides were analysed under a Zeiss Axioscope A1 microscope equipped with four fluorescence channels wide field IF. Microphotographs were collected using a Zeiss Axiocam 503 Colour digital camera with the Zen 2.0 Software (Zeiss). Slide digitalization was performed using an Aperio CS2 digital scanner (Leica Biosystems) with the ImageScope software.
Quantitative analyses of immunohistochemical stainings were performed by calculating the average percentage of positive signals in five non-overlapping fields at high-power magnification (x400) using the Nuclear Hub or Positive Pixel Count v9 Leica Software Image Analysis.
Quantitative analyses of immunofluorescence stainings were performed in five non-overlapping fields at high-power magnification (x400), by isolation of the fluorescent marker component and measurement of its density. Multi-resolution analysis of double-marker immunostained sections has been performed through an ad-hoc developed software tool (See Supplemental Methods) [14].
Digital Spatial Profiling
Digital Spatial Profiling was performed to investigate the immune and stromal composition of spatially resolved DLBCL microenvironments characterized by different mesenchymal composition, namely SMA- or NGFR-rich foci. Multiplexed wide field immunofluorescence (IF) analysis was combined to ROIs definition and segmentation, and in situ mRNA analysis to screen four-micrometer thick tissue sections from formalin-fixed and paraffin-embedded human nodal DLBCL biopsies using a GeoMx Digital Spatial Profiler (DSP) (NanoString, Seattle WA) [15]. The following antibodies were adopted for 4-plex IF tissue imaging: mouse anti-human CD20 (L26 Novocastra, Leica Biosystems), mouse anti-human CD271 (NGFR, MRQ-21 Cell Marque), mouse anti-human SMA (ASM-1 Novocastra, Leica Biosystems). Syto83 was used as nuclear counterstain. Four independent ROIs were profiled for each DLBCL sample. For the determination of the 87-plex customized TAP Human Immuno Oncology panel, mRNA binding DNA probes conjugated with UV photocleavable indexing oligos were hybridized to the tissue as previously reported [15]. The UV photocleavable probes were released from each ROI according to custom masks for UV illumination and digitally counted using the NanoString nCounter Analysis System. Heatmaps showing the unsupervised hierarchical clustering of the ROIs according to the expression of selected genes were generated with the Heatmap function of the R package ComplexHeatmap (http://www.bioconductor.org/packages/devel/bioc/html/ComplexHeatmap.html) on the mean-centered normalized log2-expression values. Genes differentially expressed between DLBCL NGFR-rich and SMA-rich stromal compartments were identified by applying an empirical Bayes test using the Limma R package. Clustering of ROIs according to SPARC expression was generated with the K-means clustering algorithm.
Flow cytometry
To evaluate A20 lymphoma take in WT and Sparc−/− mice in BM, LI, and SPL, and to recover A20 cells from the BM such to perform re-transplantation experiments, cell suspensions were stained with antibodies to CD45, CD19 and B220. A20 cells can be discriminated from the normal B-cell counterpart according to their higher expression of CD45 and B220 combined with a higher side scatter (SSC-A) (Supplemental Fig. S1). To characterize T cell infiltrates in WT and Sparc−/− mice in re-transplantation experiments, BM cell suspensions were stained with antibodies to CD4, CD8, Foxp3, Ki-67, TIM3, ICOS, CD25, PD1, OX40, H-2Kd, I-A/I-E. The antibodies used are detailed in the Supplemental Methods section. Surface staining was performed in phosphate-buffered saline (PBS) supplemented with 2% fetal bovine serum (FBS) for 30 min on ice. Foxp3 intracellular staining was performed according to the manufacturer's instructions (eBioscience). Flow cytometry data were acquired on an LSRFortessa (Becton Dickinson) and analysed with FlowJo software (version 8.8.6, Tree Star Inc.).
Mesenchymal cell purification and in vitro co-culture experiments
Murine BM mesenchymal stromal cell (MSC) cultures were obtained from the trabecular fraction of femurs and tibias of WT and Sparc−/- mice. Briefly, the cellular fraction of the femurs and tibias was washed out and the compact bone was incubated with collagenase I (1 mg/ml) for 1 h at 37°C. After enzyme digestion, the bone suspension was passed through a 70μm filter mesh to remove debris. Cells were seeded in complete medium (MesenCult Basal Medium) at a density of 25*106 cells/ml. Floating cells were removed every 3-4 days. Adherent cells were phenotypically characterized using the following antibodies: CD31, CD45, CD34, cKit, Ter119, CD44, Sca-1, CD29. The antibodies adopted are detailed in the Supplemental Methods Section. BM- and SPL-derived MSCs were defined according to their negativity for hematopoietic and lineage markers (CD45, CD34, CD31, cKit, Ter119) and positivity for the MSC-markers CD44, CD29 and Sca-1. Perycytic mesenchymal cells (An2+, the mouse homolog of NG2) were enriched from BM cell suspensions using anti-An2/NG2 microbeads from Miltenyi. In vitro experiments involving murine BM-derived An2+ pericytes were performed using cells between the 2nd and the 5th passages. For co-culture experiments 105 BM-MSCs, SPL-MSCs or An2+ pericytes were seeded over-night into a 24-well plate and then added with 105 A20 cells. At basal, 48h, and 96h time points, A20 cells were recovered and processed for qPCR analysis. To analyse Myc expression, qPCR primers from TaqMan (Myc: Mm00487804_m1; Hif1α: Mm00468869_m1) were adopted.
RNA sequencing and data analysis
Whole-transcriptome stranded sequencing libraries were generated using the TruSeq Stranded Total RNA with Ribo-Zero in order to remove ribosomal RNA. Sequencing was carried out on the Illumina HiSeq 3000 system requiring 80M reads/sample with paired-end mode. Reads were aligned to the ENSEMBL mouse genome assembly BALB_cJ_v1 using STAR.
Differentially expressed genes were called using DESeq2 R package starting from the count tables generated by HTSeq.
PCA plot was generated with the function plotPCA of the DESeq2 package, using default parameters on the log2 transformed intensity values. Euclidean distance between samples was calculated with the dist function of DESeq2 using default parameters. Gene fusion events were detected in RNA-seq data using the tool FusionCatcher (https://github.com/ndaniel/fusioncatcher). Additional details regarding the RNA sequencing data analysis are reported in the Supplemental Methods section.
RNA sequencing data deposition
The RNA sequencing data sets generated during the current study are available in the GEO (Gene Expression Omnibus) repository under the accession number GSE157920.
Gene set enrichment analysis, hierarchical clustering, and CIBERSORT deconvolution on human DLBCL cases
GSEA [16] was performed on the GSE98588 dataset [4] using the continuous expression level of NGFR gene to label the cases (ranked by Pearson metric method). We then tested roughly 300 gene sets extracted from widely used annotated databases (Kegg, Reactome, MSigDB) including several cellular processes and MYC-related pathways.
Raw data from Chapuy et al [4]. were collected and used to generate expression profiles by RMAExpress (Robust Multi-Array Average). Multiple probes were collapsed to unique genes by selecting the ones with the maximal average expression for each gene. Expression values were log2 transformed. Heatmap and clustering analysis were performed using R statistical software. For hierarchical clustering analysis the Euclidian distances across samples were calculated and used to applied complete cluster aggregation method within pheatmap R package.
For CIBERSORT [9] analyses, we created a customized signature matrix as previously reported [7]. Briefly, we used GEP from 24 different purified cell types collected from published datasets, to generate a microenvironment matrix specific for DLBCL [7]. The customized signature featuring 990 genes characteristic of tumour samples of both ABC and GCB COO categories, immune cells and stromal compartment, was applied to the 137 DLBCLs from Chapuy et al [4]. The 137 cases were divided into two groups according to the median value of SPARC transcript expression from quantile normalized and log2 transformed gene expression measurements.
Statistical analysis
Unless differently specified, data are presented as the mean ± standard error of the mean. Two classes comparisons were performed according to either two-tailed Student's T test or contingency two-sided Fisher's exact test for discrete variables. For multiple classes comparisons, one-way analysis of variance (ANOVA) was performed. A p<0.05 was considered significant and indicated with *; p<0.01 was indicated as ** and p<0.001 as ***.
Results
A20 DLBCL infiltrates display different morphology and phenotype depending on the tissue microenvironment
The tissue microenvironment-induced heterogeneity of A20 lymphomas was initially investigated by histomorphological and immunophenotypical characterization of tissue infiltrates in different sites upon i.v. injection of 5 × 105 cells into BALB/c mice. Following 5 weeks of injection, mice were sacrificed and the A20 cells recovered from the BM, LI, and SPL tissues through flow cytometry sorting (Supplemental Fig. S1) and processed for total RNA-sequencing as described in the Supplemental Methods section. BM, LI, and SPL from 5 other mice were collected for histopathology (Fig. 1A). To specifically assess the relevance of stromal SPARC-induced modifications in A20 transcriptional programs, A20 cells were also injected into Sparc−/− BALB/c hosts (Fig. 1A).
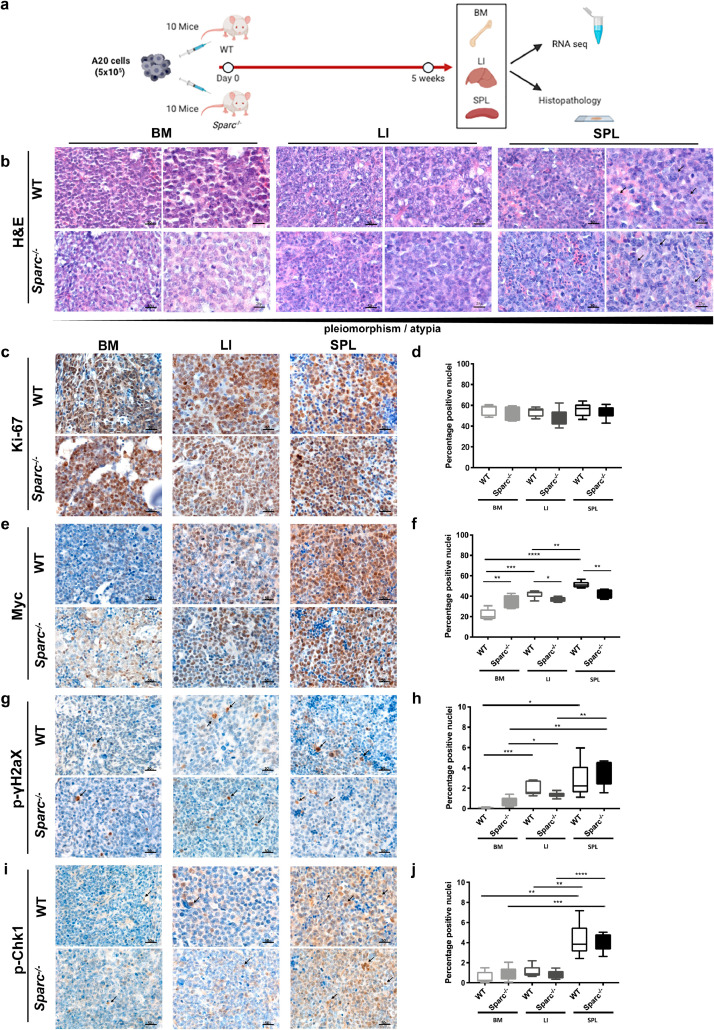
Fig. 1.
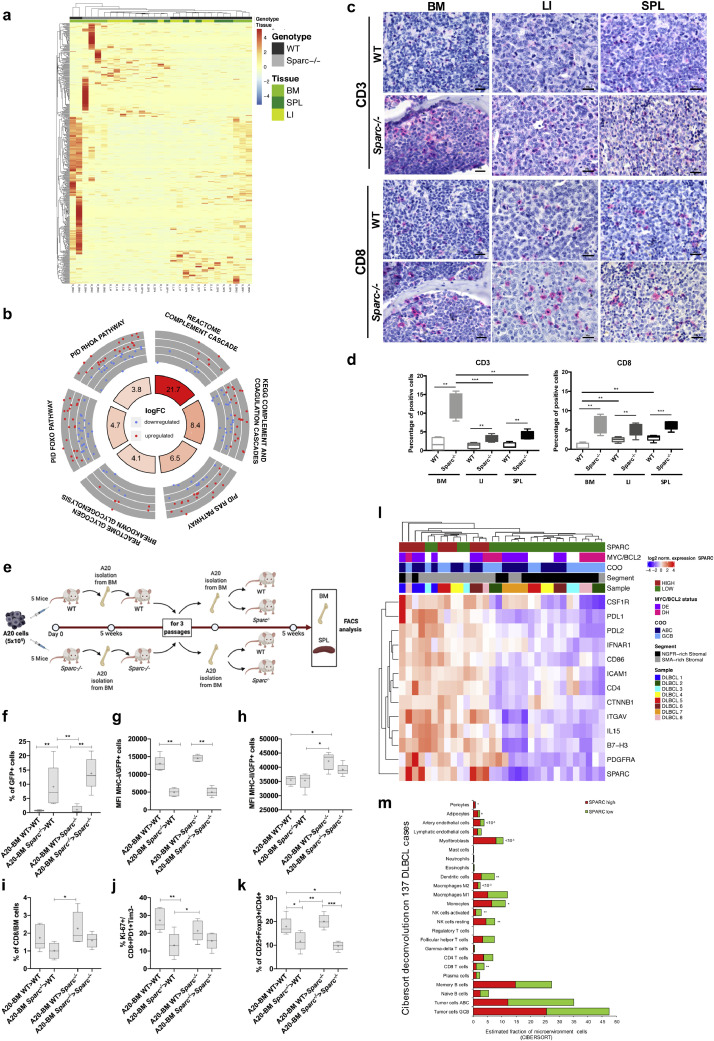
A20 DLBCL tumours display morphological and phenotypical heterogeneity in different tissue sites. (a) Graphical scheme of the in vivo A20 injection and purification experiment aimed at investigating the host tissue microenvironment effect on A20 morphology, phenotype and transcriptome. (b) Representative microphotographs of H&E-stained sections from A20 DLBCL infiltrates showing the increasing degree of pleiomorphism and cytological atypia (arrowheads) of A20 cells from BM to LI and SPL tissues in WT and Sparc−/− hosts. Original magnification x400 and x630. c-d, IHC for Ki-67 (c) and quantitative analysis (d) showing comparable proliferative fractions in BM, LI, and SPL tissue infiltrates. (e-f) IHC for Myc (e) and quantitative analysis (f) highlighting significant differences in Myc nuclear expression among BM, LI, and SPL A20 infiltrates. G-J IHC for p-gH2aX and p-Chk1 DNA damage response markers (g, i) and quantitative analysis (h, j) showing an increase fraction of immunoreactive nuclei in A20 SPL infiltrates as compared with LI and, even more significantly, BM. Original magnifications x400.
A20 cells formed diffuse infiltrates within the BM parenchyma, which mostly consisted of monomorphic medium-sized cells with hyperchromatic nuclei in WT and Sparc−/− hosts (Fig. 1B). In the LI, A20 cells infiltrated the hepatocyte muralia, forming multiple nodular foci mostly composed by atypical large cells with a high degree of pleiomorphism and scattered anaplastic figures (Fig. 1B). The SPL architecture was almost completely effaced by the proliferation of A20 atypical cells infiltrating the red pulp and also extending to white pulp remnants, showing the highest degree of morphological atypia and frequency of mitotic Fig.s with no relevant difference between the WT and Sparc−/− genotypes (Fig. 1B, arrowheads). The observed degree of morphological variation among BM LI and SPL in the A20 tumours was reminiscent of reports on human DLBCL with discordant morphology between lymphoid tissue and BM foci, with BM DLBCL infiltrates showing a lower degree of morphological atypia and smaller cytology [17]. Moreover, it suggested that underlying differences in biological features of the malignant clone could be induced by tissue site-intrinsic adaptations.
To challenge this hypothesis, we assessed the proliferation of A20 cells in the different tissue sites, along with the expression of the key transcription factor Myc, which is known to drive tumour aggressiveness in B-cell lymphomas [18]. Quantitative IHC analysis of the proliferative index of A20 DLBCL infiltrates assessed as the percentage of Ki-67+ cells revealed no significant differences among BM SPL and LI infiltrates in both the WT and Sparc−/− settings (Fig. 1C-D). Notably, A20 DLBCL infiltrates showed consistent variation in the extent of Myc expression, which was significantly higher in the SPL infiltrates than in LI and BM, with a higher trend in WT as compared with Sparc−/− hosts, excepted for the BM where Sparc−/− samples showed a higher expression than WT (Fig. 1E-F). This finding indicates that despite a comparable proliferative attitude, A20 lymphomatous cells populating different tissue microenvironments could be induced to modulate key transcriptional regulators, such as Myc.
Myc expression facilitates the acquisition of a mutated phenotype in malignant B cells, by promoting DNA replicative stress and DNA damage induction. We therefore investigated the expression of DNA damage and replication stress markers phospho (p)−γH2ax and p-Chk1 by in situ quantitative IHC analysis. Consistently with Myc expression, p-γH2ax and p-Chk1 were significantly more expressed within SPL infiltrates as compared with LI and BM and showed a trend towards decrease in the Sparc−/− genotype excepted for Sparc−/− BM infiltrates, which showed slightly higher expression of the two markers (Fig. 1 G-J). Collectively these data point to the selection of different DNA damage-prone phenotypes within different tissue contextures.
To validate the findings of A20 morphological and phenotypical variations upon infiltration of different tissue microenvironments, at the same time taking into account the issue of different immune pressures potentially influencing adaptation to secondary lymphoid (SPL) versus primary hematopoietic (BM) and non-lymphoid (LI) tissues, we extended histopathological and quantitative IHC analyses on BM LI and SPL infiltrates of two DLBCL-like cell lines, OPL239 and OPL241, derived from Spp1−/−/Faslpr/lpr mice, injected into immunodeficient nude hosts (Supplemental Fig. S2). We observed a comparable degree of morphological variation between BM LI and SPL infiltrates, with SPL lymphomas showing the highest degree of pleiomorphism and atypia. Consistently, Myc and DDR markers were induced in the SPL infiltrates as compared with BM and LI, while Ki-67 was comparably high in the three environments (Supplemental Fig. S2).
Diversified mesenchymal adaptations characterize human DLBCL nodal lesions and A20 tissue infiltrates
On the basis of the observed morphological and phenotypic heterogeneity in the A20 lymphomas among different tissue sites, we investigated whether host tissue-related features could contribute to the diversification of the three microenvironments and/or of the two Sparc genotypes.
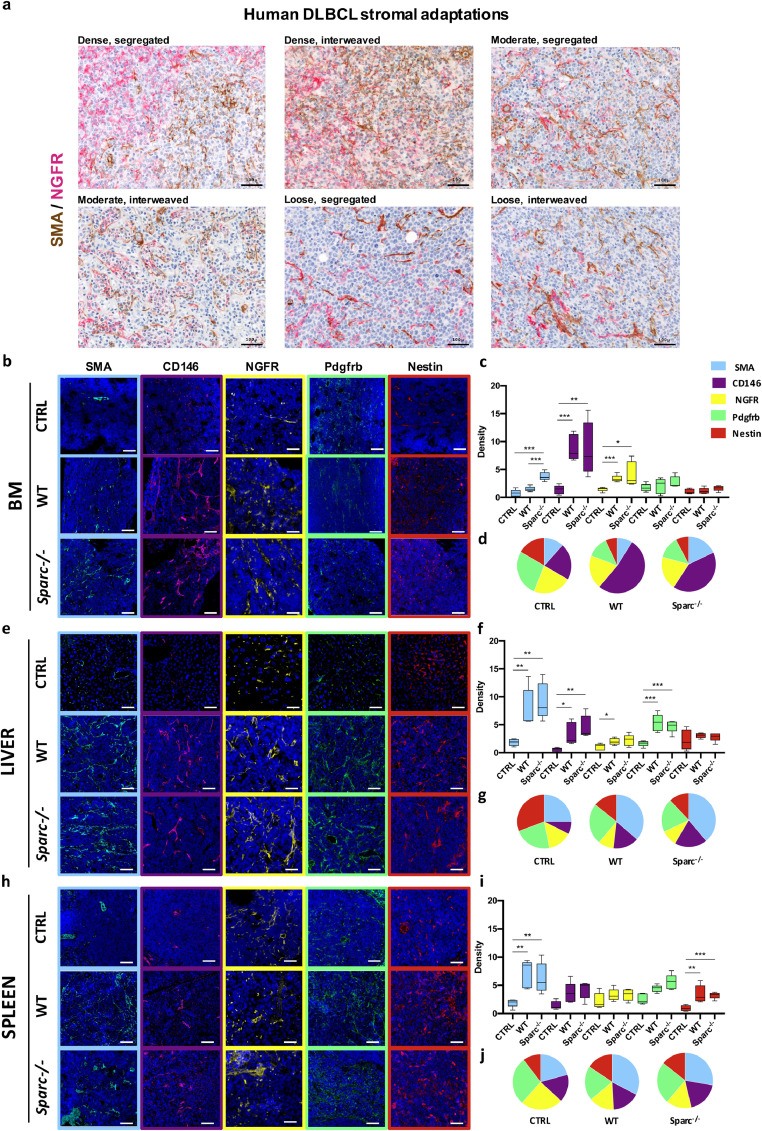
In a small set of nodal DLBCL cases (n=12), comprising cases of GC and non-GC phenotype according to Hans algorithm [19] and selected according to the heterogeneous, focal/zonal, pattern of MYC expression (Supplemental Table S1), we highlighted the presence of different types of mesenchymal cell meshwork within distinct areas of the same nodal lesions (Supplemental Fig. S3A). In these cases, SMA+ myofibroblastic/reticular cells, NGFR+ (CD271) and CD146+ pericytic/mesenchymal stromal cells, and PDGFRβ+ perivascular stromal cells showed disconnected patterns (Supplemental Fig. S3A). Moreover, mesenchymal markers including NGFR, SMA, PDGFRβ, and VCAM-1 that were either spatially associated or mutually exclusive in the GC or peri-follicular areas of non-neoplastic lymph nodes, were aberrantly splitted or focally associated in DLBCL infiltrates (Supplemental Fig. S3B). Of note, within DLBCL nodal infiltrates, prominent heterogeneity in the density and spatial segregation of mesenchymal meshwork formed by different cell types was observed. According to NGFR and SMA double-marker immunostainings, DLBCL lesions showed dense, moderate or low NGFR and SMA mesenchymal meshwork either spatially segregated or interweaved (Fig. 2A).
Fig. 2.
Mesenchymal meshwork heterogeneity in human DLBCL nodal lesions and in A20 infiltrates at different tissue sites. (a) Double-marker IHC for SMA (brown signal) and NGFR (purple signal) in the nodal infiltrates of six representative DLBCL cases (Supplemental Table S1) reflecting the variable degree (dense, moderate, low) of stromal reaction and spatial segregation (segregated, interweaved) of the two mesenchymal meshwork, supporting the engendering of diversified tumour-stroma interfaces. Original magnifications x200. (b-j) IF for Sma (cyan signal), Cd146 (violet signal), Ngfr (yellow signal), Pdgfrb (green signal), and Nestin (red signal) mesenchymal markers (b, e, h), quantitative analysis (histograms, c, f, i) and average relative fractions (pie charts, d, g, j) of uninvolved (CTRL) or infiltrated BM (b-d), LI (e-g), and SPL (h-j) from WT and Sparc−/− hosts, detailing that different combinations of mesenchymal markers are variably induced across the three tissue sites. Original magnifications x200.
Prompted by the evidence of an active and variably intense mesenchymal response to human DLBCL infiltration contributing to intra-tumour heterogeneity, we tested the hypothesis that host tissue adaptation to A20 lymphoma seeding could imply a diversified remodelling of mesenchymal cell meshworks. We thus analysed the density of Sma, Cd146, Ngfr, Pdgfrb and Nestin mesenchymal markers in the BM, LI and SPL A20 infiltrates by quantitative immunofluorescence. In the BM, a prominent induction in Cd146+ and Ngfr+ elements characterized A20 lymphomas in both the WT and Sparc−/− hosts, while Sma+ stromal cells were significantly induced only in Sparc−/− infiltrates (Fig. 2B-D). In LI, A20 tumour microenvironment was marked by a significant induction of Sma+, CD146+, and Pdgfrb+ mesenchymal elements in both genotypes (Fig. 2E-G), while the stromal reaction of SPL foci was characterized by a significant expansion of Sma+ and Nestin+ cells (Fig. 2H-J). Of note, mesenchymal modifications occurring in the BM, LI, and SPL of nude mice infiltrated by OPL239 and OPL241 DLBCL-like lymphomas proved to be consistent with those observed in A20 infiltrates (Supplemental Fig. S4), suggesting that the nature of stromatogenesis is predominantly determined by tissue stromal characteristics. These results suggest that different qualities of stromal adaptation to DLBCL infiltration provide a microenvironment frame for diversification of the malignant clone phenotype.
A20 lymphoma cells gene expression profile is influenced by the tissue microenvironment
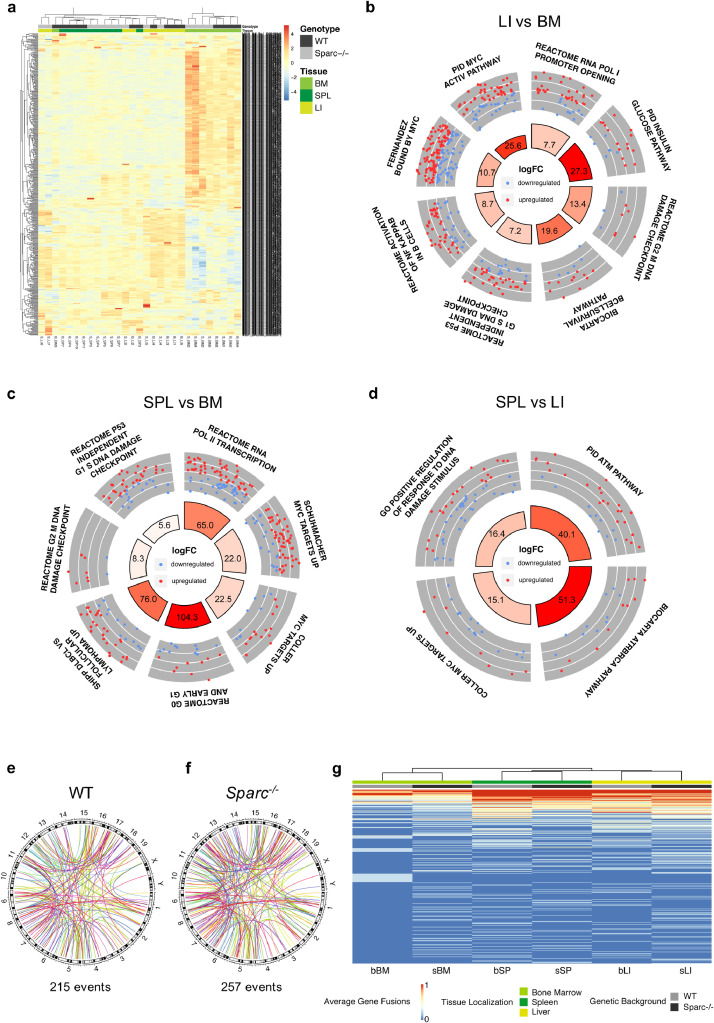
We then analysed by RNA-seq, the transcriptome of A20 cells purified from the three environments of WT and Sparc−/− mice (Supplemental Table S2). Principal component analysis of the samples showed a predominance of tissue site over Sparc genotype in determining the clustering of the A20 lymphoma samples (Supplemental Fig. S5A-B). Indeed, unsupervised clustering analysis of A20 DLBCL transcriptome identified clusters on the basis of the tissue environment (Fig. 3A).
Fig. 3.
Diversified tissue microenvironment imprints on A20 transcriptional activity. (a) Unsupervised hierarchical clustering of the samples according to the expression of the 500 most variable genes among the samples, showing the tissue imprint on clustering. (b-d) Circular layout visualization of the modulation of selected pathways in LI vs. BM (b), SPL vs. BM (c) and SPL vs. LI (d). The height of the bars of the inner ring represents the significance of the term calculated as -log(p-val). The number within each bar refers to the logFC of the gene set. The outer ring displays scatterplots of the logFC for the genes of the pathway. (e-f) Circular layout visualization of the genomic location of the predicted gene fusions (see Supplemental Methods) in WT (e) and Sparc-/- (f) mice. The 19 autosomal and the two sexual mouse chromosomes are shown with the relative chromosome bands. Each colored line represents a fusion event. The number of fusion events is reported under each plot. (g) Unsupervised hierarchical clustering of the gene fusion fraction calculated among all the samples of each genetic background and tissue localization showing a tissue-based imprint on clustering of fusion transcripts (see Supplemental Table S4).
To gain insight into the transcriptional programs modulated among different tissue environments irrespectively of the Sparc genotype, we compared the transcriptome profiles of the A20 cells isolated from different tissues by applying Quantitative Set Analysis of Gene Expression (QuSAGE) [20]. The comparison between LI and BM A20 lymphoma cell transcriptional profiles revealed differences in the expression of programs related with metabolism and transcription (Supplemental Table S3, Supplemental Fig. S5C), and a significant enrichment in pathways related with B-cell survival and activation of Nf-KB in B cells, as well as in pathways related with DNA damage checkpoints (Fig. 3B) in A20 cells purified from LI. Moreover, LI versus BM A20 transcriptome showed enrichment in transcriptional targets of Myc, including genes identified by ChIP as high-affinity targets of Myc (Fig. 3B), pointing to a higher activation of Myc in LI as compared with BM A20 cells. Similarly, the comparison between SPL and BM A20 transcriptome revealed a significant positive enrichment in pathways involved in transcription, DNA damage checkpoint and Myc transcriptional targets (Fig. 3C) in A20 cells purified from the SPL (Supplemental Table S3, Supplemental Fig. S5D). Of note, SPL A20 transcriptome also proved to be enriched in genes reported to be upregulated in DLBCL versus follicular lymphomas [21] (Fig. 3C), which mainly comprised genes involved in glycolytic metabolism, supporting a higher transcriptional activation of programs related with a more aggressive biology. In the comparison between SPL and LI A20 transcriptome, differences in the transcriptional activation of DNA damage-associated programs were observed, with ATM- and ATR-related pathways being more active in SPL A20 infiltrates (Fig. 3D). SPL A20 profiles were also positively enriched in Myc transcriptional targets suggesting that the SPL environment better matched a mutator phenotype (Fig. 3D). At contrast, LI A20 transcriptome showed a positive enrichment in B-cell survival programs, which also characterized the comparison with the BM (Supplemental Table S3, Supplemental Fig. S5E).
These results indicate that the BM LI and SPL microenvironment that we found to be characterized by different stromal adaptations to a same DLBCL infiltration variably promoted or restrained the expression of transcriptional profiles relevant to DLBCL cell biology.
As a control on the effect of transcripts potentially derived from recipient host cells in the analyses, we considered fusion transcript events as a proxy of A20 DLBCL cell specific gene expression. We thus investigated whether a microenvironment-driven clustering of expression data also occurred when only A20 lymphoma-cell specific aberrant gene fusion transcripts were considered. To this aim, fusion events were detected in RNA-seq data (see Supplemental Methods section; Supplemental Table S4), and the distribution and frequency of the fusion events among different sites in the WT and Sparc−/− genotypes were analysed (Fig. 3E-F). Also in this setting, a tissue microenvironment-driven clustering emerged, sporting fusion events that were more or under-expressed in A20 transcriptome from one tissue site and/or host Sparc genotype, and other fusion events that were expressed at similar levels in all the microenvironments (Fig. 3G).
Myc is modulated in DLBCL cells at mesenchymal interfaces
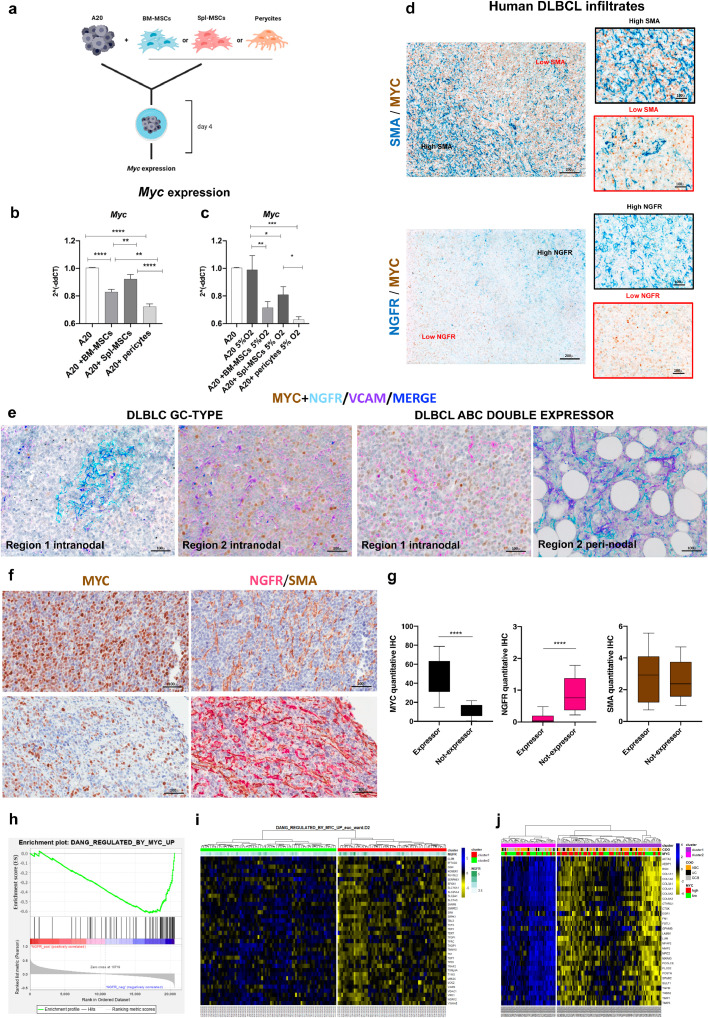
Given the observed heterogeneity in A20 lymphoma transcriptional programs in microenvironments characterized by different stromal adaptations, we investigated the relationship between A20 metabolic transcriptional programs, Myc expression, and the mesenchymal milieu. We focused on the A20 transcriptional modulation in the BM and SPL tissue environments that were characterized by divergent transcriptional hallmark gene sets related with Myc transcriptional activity, oxidative phosphorylation and mTorc1 signaling, DNA damage and replication checkpoints (programs upmodulated in the SPL A20 transcriptomes), hypoxia and Kras signaling pathways (positively enriched in the BM) (Supplemental Fig. S6). The two environments implied different metabolic transcriptional imprints on A20 cells, the SPL licensing glycolytic and ox-phos programs, the BM driving hypoxia-related metabolic rewiring involving Kras signaling. To validate the actual translation of transcriptional modulations at the protein level, we selected six targets among the top 50 up- or down-modulated transcripts in the BM vs SPL comparison (Supplemental Table S5) to be validated by IHC in the A20 and OPL lymphoma infiltrates. The targets included Eif6, a key regulator of cell metabolism and Myc signaling enforcer, which has been demonstrated to stimulate glycolitic programs, acting as a limiting factor in Myc-driven lymphomagenesis [22], the modulator of Rho family gtpases Tiam2, intervening in B-cell proliferation and DLBCL survival [23], and the DNA ligase I (Lig1) involved in DNA replication, recombination and repair. Along with these three targets upmodulated in the transcriptome of A20 cells isolated from SPL, three targets up-modulated in BM A20 transcriptomes were selected, which included the salt-inducible kinase Sik2, which has been reported to control glucose uptake via Glut4 regulation and response to nutrient deprivation [24], the Pi3k regulatory subunit Pik3r2 cooperating with Pi3kβ nuclear activity and promoting tumor progression [25], and the ATP-dependent RNA helicase Ddx39, playing a role in transcription and translation control and associated with Wnt/β−catenin activation [26]. On IHC analysis, the in situ expression of the differential targets in A20 infiltrates proved to be consistent with the RNA-seq data, and also showed consistency in OPL239/241 line infiltrates (Supplemental Fig. S7). On these bases, to functionally test whether DLBCL cells could be influenced by different mesenchymal interactors in their metabolic asset prototypically represented by Myc expression levels, A20 cells were cultured in the presence of total BM- and SPL-derived MSCs (CD45-CD34-Cd31-Ter119-cKit-Cd29+Sca-1+Cd44+) or BM-derived An2+ pericytes (Fig. 4A). In line with the in situ results, BM-MSCs and pericytes significantly down-modulated Myc expression in A20 cells in comparison to splenic MSCs (Fig. 4B). Since GSEA of A20 cells sorted from the BM and SPL showed positive enrichment of hypoxia programs in the BM (Supplemental Fig. S6), we considered that through Hif1α, hypoxia could act as a major direct regulator of Myc activity and expression [27]. Considering the constitutive hypoxic condition of the BM microenvironment [28] to determine the impact of hypoxia on Myc expression in the presence or absence of BM-MSCs and An2+MSCs or SPL-MSCs, we repeated the co-culture experiments under hypoxic conditions (5% of O2). A20 cell co-cultured with all types of MSCs under hypoxic conditions showed a significant down-modulation of Myc (Fig. 4C). However, only in the case of SPL-MSCs we found the correspondent Hif1α upregulation (Supplemental Fig. S8). On the contrary in BM-MSCs or pericytic An2+MSCs co-cultures, Hif1α was not up-modulated by the low oxygen conditions (Supplemental Fig. S8). This suggests that BM mesenchymal cells can restrain Myc expression, at the same time controlling the hypoxic response. This finding hinted to a complex influence of stromal neighbours over the expression of Myc in such high-grade lymphomatous cells.
Fig. 4.
Different mesenchymal contextures correlate with Myc modulation and spatial heterogeneity. (a) Graphical scheme of the in vitro co-culturing experiment with A20 cells and either WT or Sparc−/− total BM-derived MSCs or peri-vascular An2+MSCs (see Methods section for MSC purification and phenotyping). (b) Myc gene expression by Real-time PCR on total RNA isolated from A20 cells at basal conditions and following 4 days co-culture with BM-MSCs, Spl-MSCs or pericytes, showing the significant downmodulation of Myc in the presence of BM-derived MSCs and pericytes but not in co-culture with Spl-derived MSCs. (c) The same co-culture experiment was repeated under hypoxic conditions (5%O2), where Myc downmodulation resulted more marked and detectable also in the presence of Spl-MSCs. (d) Double-marker IHC for SMA (blue signal, top panels) and MYC (brown signal) or NGFR (blue signal bottom panel) and MYC (brown signal) on human DLBCL lymph node infiltrates (Supplemental Table S1) indicating that no MYC expression gradient is evident between SMA-high and SMA-low areas, while an inverse distribution of MYC and NGFR density emerges. Original magnifications x50 and x400 (insets). (e) Representative multiplexed chromogenic and fluorescence immunostainings for MYC (brown signal), NGFR (cyan signal), and VCAM-1 (violet signal) on human DLBCL samples (Supplemental Table S1) showing the different expression of MYC among areas with higher or lower NGFR expression. VCAM-1 highlights the presence of a stromal cell network also in areas devoid of NGFR. Original magnification x200. (f) Representative double-marker IHC (left panels) for SMA (brown signal) and NGFR (purple signal) in infiltrated areas from six MYC-expressor (average expression >40%) or non-expressor (<40%) DLBCL cases. In the same areas MYC expression is shown (right panels). (g) Quantitative IHC analysis of SMA, NGFR and MYC in five medium-power (x200) microscopic fields of the six cases dichotomized according to MYC average expressions, showing that while SMA did not significantly vary in the two conditions, NGFR+ stroma was significantly more represented in non-expressor infiltrates. (h) GSEA output showing the positive enrichment of MYC-upregulated target genes, in human DLBCL cases characterized by lower (above median) NGFR expression, using GSE98588 dataset (NES, normalized enrichment score; FDR, false discovery rate). (i) Heatmap depicting hierarchical clustering analysis on 36 MYC-upregulated target genes extracted from GSE98588 dataset. Signal intensities ranked from highest (blue) to lowest (yellow) are indicated as row z-score. On the top, information including clusters and NGFR expression (greenscale) as continuous variable. (j) Heatmap related to the expression of the 30 genes characteristic of stromal compartment in DLBCL from GSE98588 dataset. Signal intensities ranked from highest (blue) to lowest (yellow) are indicated as row z-score. On the top, information including clusters (derived from hierarchical clustering analysis), cell of origin (COO) and MYC expression are depicted. The analysis shows that stromal gene-based clustering of the 137 cases into two groups determines differential enrichment of MYC-high and -low cases.
Prompted by the observed Myc modulation in in vitro co-culture experiments, we investigated whether the intra-tumour mesenchymal heterogeneity that we highlighted in the small cohort of human nodal DLBCLs could relate with in situ MYC expression. To this aim, we applied an ad-hoc-developed software tool for multiresolution spatial analysis (see Supplemental Methods section) to immunostained sections for the myofibroblastic/reticular cell marker SMA and for the MSC/pericytic marker NGFR in combination with MYC, and evaluated the degree of spatial association between the two mesenchymal phenotypes and MYC expression in lymphomatous infiltrates. Interestingly, while SMA+ stroma distribution did not show a significant degree of association with MYC, NGFR+ mesenchymal foci were negatively associated with MYC expression, suggesting that a topographic compartmentalization relative to mesenchymal cell distribution and/or phenotype acquisition could occur (Fig. 4D, Supplemental Fig. S9). Indeed, in MYC-expressing DLBCLs, areas characterized by an NGFR-rich meshwork showed a significantly lower MYC expression as compared with areas lacking NGFR but still showing a dense stromal meshwork as highlighted by VCAM-1 (Fig. 4E). Remarkably, in cases in which the nodal DLBCL proliferation extended to the peri-nodal adipose tissue highly rich in NGFR+ perivascular mesenchymal elements, a neat downmodulation of MYC expression was observed, which provided an insight into the dynamical switch of MYC expression under specific stromal microenvironment conditions (Fig. 4E). We further extended the combined analysis of MYC and SMA/NGFR stroma association by evaluating the same areas of 6 DLBCL cases classified as MYC expressors (average expression >40% MYC by quantitative IHC software analysis) or non-expressors (average expression <40% MYC), and observed that the NGFR meshwork was more represented in the non-expressor condition (Fig. 4F-G).
Consistently with the observed negative association between NGFR+ stroma and MYC expression, GSEA for MYC transcriptional targets on 137 DLBCL cases [4] categorized according to NGFR expression levels into NGFR-high and -low showed a significant enrichment of upregulated MYC targets in NGFR-low cases (NES=-1.88, FDR=0.01, Fig. 4H). Hierarchical clustering of the 137 cases [4] according to the same gene set showed that the two clusters delineated by different MYC target expression showed different NGFR expression levels (Fig. 4I). Moreover, zooming out on whole transcriptome, we performed hierarchical clustering analysis of 30 stromal genes previously described as specific of the mesenchymal compartment in DLBCL [7] and observed a significantly different distribution of cases stratified by MYC median expression between the two clusters characterized by high or low levels of the 30-gene mesenchymal signature (Fisher's exact test, p=0.01, Fig. 4J), which pointed to clear-cut differences in the mesenchymal milieu of cases with divergent MYC expression levels.
Altogether these data point to topographic influences over relevant DLBCL phenotypes such as MYC expression, related with mesenchymal determinants, the mechanisms of which deserve to be thoroughly dissected.
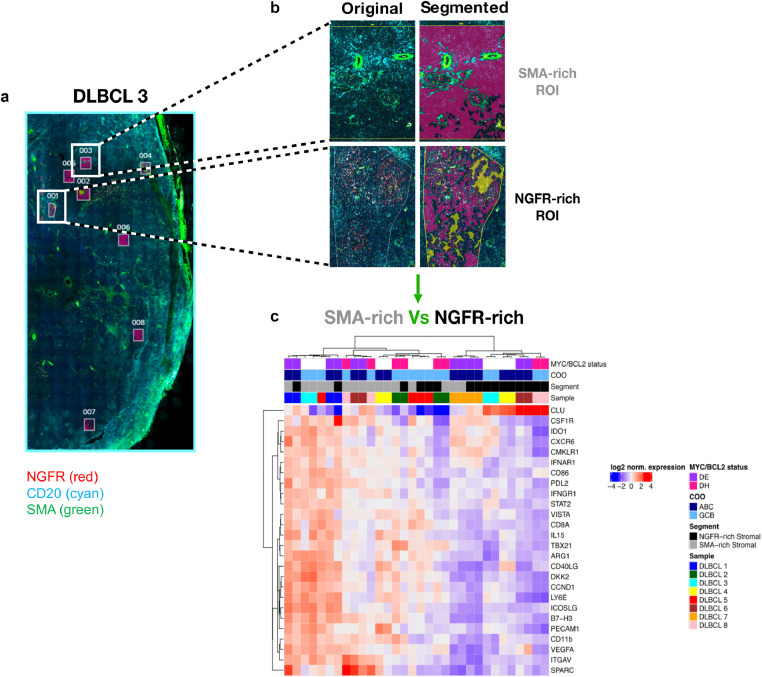
Digital Spatial Profiling of SMA- and NGFR-rich mesenchymal foci highlights immune/stromal intra-lesional heterogeneity of human DLBCL
To experimentally investigate the actual intra-lesional variations in the stromal and immune microenvironment composition of DLBCL infiltrates according to the presence of different mesenchymal meshworks, we selected eight nodal DLBCL cases, which included 4 ABC-DLBCL (three of which MYC/BCL2 double-expressors) and 4 GCB-DLBCL (two of which MYC/BCL2 double-hit). Based on multispectral immunofluorescence for CD20, NGFR, SMA and DNA, we selected four different Regions of Interest (ROIs) for each DLBCL lesion based on the prevalent expression of either NGFR+ or SMA+ stromal meshworks. The selected ROIs were segmented for profiling of the stromal (i.e. CD20-) subcompartments according to the expression of a customized version of the Human Immuno-Oncology RNA Panel including 87 immune and stromal genes using a Nanostring GeoMx [15] (Fig. 5A-B). A list of the 87 genes and of their main biological pathways according to Gene Ontology Annotation (https://www.ebi.ac.uk/GOA/) is reported in Supplemental Table S6. According to DSP, we found 26 genes differentially expressed between SMA-rich and NGFR-rich stromal ROIs (Fig. 5C, Supplemental Table S7). Interestingly, the 26 differentially expressed genes included genes related with negative control of T-cell activation by immune checkpoints such as PDL2, B7-H3, and VISTA, myeloid-derived suppressor and M2 hallmarks IDO-1, ARG1, CSF1R, VEGF-A, and mesenchymal and ECM markers SPARC, DKK2, ITGAV, CLU, PECAM1. From this analysis, SMA-rich stromal foci emerged as strikingly enriched in immunoregulatory and vascular stroma-associated transcripts, as compared with NGFR-rich foci characterized by higher expression of the FDC-related marker CLU. Notably, from the unsupervised hierarchical clustering analysis according to the 26 differentially expressed genes ROIs predominantly clustered according to the SMA or NGFR status rather than sample ID, COO or DE/DH status, which indicated that intra-lesional heterogeneity in immune and stromal programs prevailed over inter-case variations.
Fig. 5.
Digital Spatial Profiling for immune and stromal genes in SMA- and NGFR-rich foci of eight human DLBCL cases.
Identification of differentially expressed genes by digital spatial profiling on nodal DLBCL SMA-rich and NGFR-rich stromal ROIs. (a) Representative multiplexed immunofluorescence for CD20 (cyan signal), NGFR/CD271 (red signal), SMA (green signal) and Syto83 (blue signal) on a human DLBCL case (out of the eight profiled, see Methods Section) showing the selection of ROIs corresponding to SMA-rich and NGFR-rich stromal (CD20-) foci, for segmentation and gene expression analysis. (b) Unsupervised hierarchical clustering of SMA-rich and NGFR-rich segmented ROIs from eight human nodal DLBCL samples with different cell of origin (COO) and MYC/BCL2 status, according to a custom immune and stromal mRNA panel (see Methods Section), showing the distribution of 26 differentially expressed genes. SMA-rich stromal ROIs emerge as positively enriched in immunoregulatory and vascular stroma-associated transcripts, as compared with NGFR-rich ROIs.
Stromal Sparc proficiency tunes the immune pressure on A20 lymphomas
Following the analyses of transcriptional and phenotypic heterogeneity related with tissue microenvironment-intrinsic determinants, we finally investigated whether Sparc proficiency could also play a role in modulating lymphoma dynamics. The transcriptional differences between A20 cells from WT and Sparc−/− hosts were mostly contributed by genes differentially expressed between the WT and Sparc−/− BM samples (Fig. 6A), consistently with the prominent expression of Sparc in the BM stroma [29]. We performed QuSAGE analysis by comparing A20 transcriptomes from WT and Sparc−/− infiltrates including all tissue sites (Supplemental Table S3, Supplemental Fig. S10A). In the Sparc−/− host, a significantly higher activation of the Ras and Rhoa GTPase pathways was detected in association with increased activity of Foxo and Glycogenolysis pathways (Fig. 6B), suggesting a different type of metabolic adaptation to environment sensing. Moreover, A20 transcriptome in the Sparc-deficient environment harboured a positive enrichment in genes involved in Complement activation (Fig. 6B), which is in line with our previous demonstration of defective stromal Sparc enhancing the expression of cell-intrinsic innate immune/inflammatory programs also involved in tumour-stroma interactions [30,31]. Consistently with an environment more prone to immune activation, A20 infiltrates in Sparc−/− hosts showed a higher density of infiltrating T cells assessed by CD3 and CD8 quantitative IHC, as compared with the WT counterpart (Fig. 6C-D). The difference was particularly prominent in the BM microenvironment, suggesting a more profound alteration of the BM immune contexture in the absence of Sparc. To evaluate whether the differences observed in the T cell infiltration of Sparc−/− and WT BM environment could result in a different immune pressure on lymphomatous cells, A20 (GFP+) cells were injected intravenously into WT and Sparc−/− hosts and then recovered from the BM for serial i.v. transplants into either WT or Sparc−/− syngeneic hosts according to the donor genotype (Fig. 6E). After 4 passages, A20 cells conditioned by either WT or Sparc−/− microenvironment were compared for growth into both WT and Sparc−/− recipients, through FACS analysis of A20 cells in the BM and SPL (Fig. 6E). Moreover, the expression of MHC class-I and class-II on A20 cell surface and the frequency and activation of CD8+ infiltrating T cells, were analysed. We observed an increased take of A20 cells conditioned within the Sparc-deficient BM environment (A20-BMSparc−/−) regardless their final transplant into WT or Sparc−/− recipients (Fig. 6F). This was associated with a significant down-modulation of MHC-I, but not of MHC-II, on the cell surface of A20-BMSparc−/− compared to A20-BMWT (Fig. 6G-H). Although the frequency of total CD8+ T cells only barely differed among BM samples infiltrated by A20-BMSparc−/− and A20-BMWT (Fig. 6I), the frequency of activated proliferating CD8+PD-1+Tim-3-Ki-67+ T cells was lower in the presence of A20-BMSparc−/− cells (Fig. 6J), indicating a lower degree of effector T cell activation in samples with lower MHC-I levels. Notably, in the same samples, a lower frequency of CD4+CD25+Foxp3+ regulatory T cells was detected (Fig. 6K), further supporting the selection of a less immunogenic profile in the Sparc−/− BM microenvironment. The extremely low fractions of GFP+ A20 cells detected in the SPL of the same hosts (Supplemental Fig. S10B) indicated that BM microenvironment priming of A20 cells mainly fostered BM infiltration, as also indicated by the progressive gain in BM homing by BM-sorted A20-GFP cells along passages (Supplemental Fig. S10C), in line with the centrality of tissue environment-specific determinants in the stromal/immune adaptation to lymphoma growth.
Fig. 6.
Stromal Sparc status conditions immune pressure and escape. (a) Heat map showing the unsupervised hierarchical clustering of the samples according to the top 500 deregulated genes between A20 lymphoma cells injected in WT and Sparc−/− mice. Genes were ranked according to absolute value of their log2FC between cells isolated from WT and Sparc−/− tissues. The heatmap highlights the prominent contribution of BM samples to WT vs. Sparc−/− differential gene expression. (b) Circular layout visualization of the differences in the activities of genes belonging to gene sets involved in Ras, Rhoa, Foxo signalling pathways, glycogenolysis, and Complement cascade, between A20 cells isolated from WT and Sparc−/− mice as assessed by QuSAGE. The height of the bars of the inner ring represents the significance of the term calculated as -log(p-val). The number within each bar refers to the logFC of the gene set. The outer ring displays scatterplots of the logFC for the genes of the pathway. (c) IHC for CD3 (upper panels) and CD8 (lower panels) in BM LI and SPL A20 DLBCL infiltrates of WT and Sparc−/− mice, highlighting significant differences in the density of infiltrating T cells in the two stromal settings. Original magnification x400. (d) quantitative analysis of CD3 and CD8 IHC in BM, LI, and SPL A20 DLBCL infiltrates in WT and Sparc−/− mice. (e) Graphical scheme of the in vivo experiment of A20 injection, BM purification and re-transplantation into WT and Sparc−/− hosts, aimed at investigating the effects of immune priming by WT and Sparc−/− BM microenvironment on A20 lymphomas. (f-k) Flow cytometry analyses of the infiltrating A20 (GFP+) cell fraction (f); MHC class-I (g) and class–II (h) mean fluorescence intensity (MFI) on A20 (GFP+) cells; total (i) and activated (j) proliferating CD8+ T cell effectors and regulatory T cells (k), in the BM of WT and Sparc−/− final recipients of A20 cells conditioned by three passages into WT and Sparc−/− BM. (l) Unsupervised hierarchical clustering of SMA-rich and NGFR-rich segmented stromal ROIs from eight human nodal DLBCL samples as shown in Fig. 5A (see and Methods Section), categorized as SPARC-high and -low, showing the distribution of 13 differentially expressed genes. SPARC-high ROIs are largely overlapping with SMA-rich ROIs and show positive enrichment in transcripts involved in regulation of T-cell activation, M2 polarization, and vascular stromatogenesis. (m) CIBERSORT deconvolution analysis output of the relative frequencies of immune and stromal cell populations according to the levels of SPARC expression in the GSE98588 dataset of human DLBCLs, showing that SPARC-low cases are significantly enriched in CD8, activated NK, and Dendritic cell populations, as compared with SPARC-high cases.
The observed stronger immune pressure exerted on A20 lymphomatous infiltrates when in a Sparc-defective stromal setting prompted us to extend the analysis of SPARC intra- and inter-tumor heterogeneity to human DLBCL setting. We took advantage of the DSP experiment on the stromal SMA- and NGFR-rich ROIs of nodal DLBCL lesions, which showed that SPARC was listed among differentially expressed genes. We performed unsupervised hierarchical clustering analysis of the stromal ROIs according to immune and stromal genes that were significantly associated with SPARC expression (Fig. 6L, Supplemental Table S8). The analysis identified two main clusters characterized by SPARC-high and –low expression, respectively. The clusters differed for the expression of the immune checkpoints PDL1, PDL2 and B7-H3, for the M2 receptor CSF1R, and for the expression of the stromal markers ITGAV, ICAM1 and PDGFRA, all upregulated in SPARC-high ROIs. Moreover, CTNNB1 positively correlated with SPARC, in line with the consolidated role of SPARC in enforcing beta-catenin signaling in stromal cells [32]. This result indicated that intra-lesional foci characterized by different SPARC expression levels showed different immunological and stromal milieus. We finally investigated whether human DLBCL cases characterized by different SPARC expression levels could display signs of a different T cell infiltration. We applied the CIBERSORT deconvolution algorithm (see Methods section) to the transcriptome of the 137 DLBCL dataset adopted for MYC GSEA analysis [4], using a customized gene signature matrix generated as previously reported [7] and detailed in the Methods section. The CIBERSORT output revealed that CD8 T cells, activated NK cells and Dendritic cells frequencies were significantly higher in low-SPARC, as compared with high-SPARC DLBCL cases (Fig. 6M, Supplemental Table S9). Altogether, these evidences further underline the relationship between the control of immune activation and SPARC expression in the DLBCL microenvironment, which outlines the engendering of immune editing dynamics that may be influenced by SPARC at both the intra- and inter-tumor heterogeneity level.
Discussion
The recent focus on the integration among mutational landscape and gene expression profiles identified DLBCL subsets independent from the cell of origin, with distinct mutational trajectories and druggable pathways, including clusters identified by recurrent EZH2 and other chromatin modifier gene mutations or by genetic lesions in JAK/STAT and NF-kB pathways [4,5]. Yet, these achievements did not shed light on the possible transcriptional and/or phenotypic heterogeneity that is coded at the topographic level.
We described here, in a syngeneic model of DLBCL, that the expression of biologically relevant features, such as Myc and its target genes, and the modulation of DNA damage-associated programs are consistently influenced by the tissue microenvironment. This is in line with the recently reported evidence of diversified subclonal complexity within lymphoid and non-lymphoid tissues in spontaneous Myc-driven lymphomas, as a result of immune editing [33]. The differences in the stromatogenesis observed in the BM, LI, and SPL A20 DLBCL tumours, which were validated in two independent cell-line-based DLBCL-like models, point to a still neglected contribution of resident mesenchymal populations to the intra-tumour diversification of these high-grade malignancies. In solid cancers, specifically in pancreatic adenocarcinoma, cancer-associated fibroblasts have been recently demonstrated to modulate the acquisition of proliferative and/or epithelial-to-mesenchymal transition phenotypes in malignant cells, shaping intra-tumour heterogeneity within the glandular component [34].
Along with the tissue-site-related modulations of A20 transcriptome and phenotypic features, we herein also identified heterogeneous mesenchymal foci within the same human DLBCL nodal lesions, which related differently to MYC expression in lymphomatous cells. Within secondary lymphoid organs, resident mesenchymal cells represent a highly diversified constellation of non-hematopoietic elements that finely tune the functional differentiation and activity of the immune cells taking part into the on-going immune responses [35]. Among these populations, FDCs and perivascular stromal cells display high phenotypical homology with bone marrow-derived mesenchymal stromal cells expressing NGFR and CD146 MSC markers [30,36]. We found that DLBCL infiltrates characterized by the presence of NGFR+ mesenchymal foci displayed MYC downregulated expression within the same areas, as compared with areas characterized by a predominance of other mesenchymal phenotypes such as SMA+ and/or VCAM-1+ reticular elements. The finding that the MYC heterogeneous expression may associate with diversified stromal interfaces claims for a consideration regarding the actual accuracy of the determination of MYC expression in the diagnostic algorithm of DLBCL [37] and allows speculating on the possibility of different paces in the disease progression and/or treatment response influenced by topographic stroma determinants. Our Digital Spatial Profiling experiment probing the immune and stromal profile of DLBCL ROIs characterized by different SMA- or NGFR-rich meshworks clearly indicated that a conspicuous extent of variation exists at the intra-tumor heterogeneity level, which has been so far unrecognized in studies focusing on DLBCL microenvironment. Indeed, our findings may have interesting clinical/therapeutic implications as they advance the hypothesis that the outcome of immunomodulatory treatments, immune checkpoint inhibitors or anti-angiogenic strategies may determine and be affected by intra-lesional selection events.
In our experiments we also adopted Sparc−/− hosts in the attempt to gain insight into the biological bases of the reported stromal Sparc prognostic influence in DLBCL. Indeed, within a signature of 30 stromal genes correlated with prognosis in haematological malignancies [9], SPARC showed the strongest association with a favourable prognosis in the DLBCL setting (Supplemental Table S10). The positive prognostic influence cast by SPARC has been correlated with its myeloid cell-intrinsic expression [38] and related with the ABC setting, in which oncogenic activation of inflammatory pathways has been described [39]. Consistently with SPARC influencing the tumour inflammatory microenvironment through a myeloid cell-related function, we have previously demonstrated that the abundance of Sparc in the stromal milieu of cancer dictates the pro-inflammatory or suppressive phenotype of infiltrating myeloid cells [39]. In the present A20 model, we observed that lymphomatous cells residing in Sparc-defective environment showed transcriptional activation of pathways related with complement cascade triggering and activation. This finding is consistent with the preferential induction of complement recognition factor C1q and C5a effector expression in the BM microenvironment of mice with genetically driven myeloproliferation in the absence of stromal Sparc [40]. Among other pathways induced in A20 transcriptome in the Sparc−/− setting, were those associated with Ras, Rhoa and Foxo activity, which have been implicated in the pathogenesis of ABC-DLBCL [41,42]. Modulation of Ras, Rhoa, and Foxo1 signalling is strongly implicated in BCR activity [43,44], which has been reported to crucially influence the fitness of Myc-driven high grade B-cell clones [45]. The observed difference in the transcriptional activity of these key pathways could indicate different assets of environment sensing and response by A20 cells embedded in WT and Sparc−/− stroma. Further corroborating the hypothesis of a tumour/stroma interface altered by Sparc deficiency, A20 infiltrates in the Sparc−/− setting showed a denser degree of T-cell infiltration. These results found correlative evidence in the spatial profiling of 87 immune and stromal genes in human DLBCL foci characterized by different mesenchymal composition and classified according to SPARC-high or –low expression. The expression of SPARC, which proved to be significantly higher in foci with a SMA-rich mesenchymal meshwork, as compared with NGFR-rich foci, was correlated with the expression of the major immune checkpoints PDL1 and PDL2 and with the M2 receptor CSF1R, extending the concept of a different immune control over T-cell activation to SPARC-related intra-tumor heterogeneity. Moreover, at the inter-tumor variation level, human DLBCL transcriptional deconvolution analyses by CIBERSORT indicated a significant enrichment in CD8 and NK activated effectors in the setting of low SPARC expression. Finally, upon multiple passages in Sparc−/− or WT BM, A20 cells were differently enriched in MHC-I-negative populations in a way that was dependent on the genotype of the priming environment, indicating that a stronger selective pressure towards immune escape was exerted by the Sparc−/− microenvironment. This evidence adds a piece of information to the recent characterization of the genetic landscape of MHC deficiency in DLBCL [46], suggesting that a level of regulation exerted by the stromal microenvironment may intervene determining topographic biases in MHC-negativity selection.
In conclusion, our study provides experimental evidence that stromal microenvironment generates topological determinants of intra-tumour heterogeneity in DLBCL that involve key transcriptional pathways potentially conferring spatial segregation to prognostically relevant features, such as Myc expression and/or DDR programs. We envisage the upcoming emergence of new classification/prognostication efforts based on the representation of spatially resolved mesenchymal-immune ecosystems.
Funding Sources
This Study has been supported by the Italian Foundation for Cancer Research (AIRC) (grants 15999 and 22145 to C. Tripodo) and by the University of Palermo. The funding sources had no role in the study design, data collection, data analysis, interpretation or writing of the report.
Declaration of Competing Interest
The Authors have no competing interest to declare.
Acknowledgements
The Authors acknowledge Dr Stefano Casola (IFOM, the FIRC Institute for Molecular Oncology) for expert advice on experimental design and results interpretation, and Dr Simona De Summa (Giovanni Paolo II Cancer Institute, Bari) for helpful discussion on bioinformatics analyses. The Authors are also grateful to Dr Nadia Castioni for technical support.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.103055.
Contributor Information
Mario Paolo Colombo, Email: mariopaolo.colombo@istitutotumori.mi.it.
Claudio Tripodo, Email: claudio.tripodo@unipa.it.
Appendix. Supplementary materials
References
- 1.Rossi D, Diop F, Spaccarotella E. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017;129(14):1947–1957. doi: 10.1182/blood-2016-05-719641. [DOI] [PubMed] [Google Scholar]
- 2.Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor evolution as a therapeutic target. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0343. [DOI] [PubMed] [Google Scholar]
- 3.Pasqualucci L, Dalla-Favera R. Genetics of diffuse large B-cell lymphoma. Blood. 2018;131(21):2307–2319. doi: 10.1182/blood-2017-11-764332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapuy B, Stewart C, Dunford AJ. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz R, Wright GW, Huang DW. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz G, Wright G, Dave SS. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciavarella S, Vegliante MC, Fabbri M. Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue. Ann Oncol. 2019;30(12):2015. doi: 10.1093/annonc/mdz386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer PN, Fu K, Greiner T. The stromal cell marker SPARC predicts for survival in patients with diffuse large B-cell lymphoma treated with rituximab. Am J Clin Pathol. 2011;135(1):54–61. doi: 10.1309/AJCPJX4BJV9NLQHY. [DOI] [PubMed] [Google Scholar]
- 9.Gentles AJ, Newman AM, Liu CL. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sangaletti S, Tripodo C, Portararo P. Stromal niche communalities underscore the contribution of the matricellular protein SPARC to B-cell development and lymphoid malignancies. Oncoimmunology. 2014;3:e28989. doi: 10.4161/onci.28989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sangaletti S, Tripodo C, Cappetti B. SPARC oppositely regulates inflammation and fibrosis in bleomycin-induced lung damage. Am J Pathol. 2011;179(6):3000–3010. doi: 10.1016/j.ajpath.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varghese B, Widman A, Do J. Generation of CD8+ T cell-mediated immunity against idiotype-negative lymphoma escapees. Blood. 2009;114(20):4477–4485. doi: 10.1182/blood-2009-05-223263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangaletti S, Stoppacciaro A, Guiducci C, Torrisi MR, Colombo MP. Leukocyte, rather than tumor-produced SPARC, determines stroma and collagen type IV deposition in mammary carcinoma. J Exp Med. 2003;198(10):1475–1485. doi: 10.1084/jem.20030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sciortino G, Tegolo D, Valenti C. Automatic detection and measurement of nuchal translucency. Comput Biol Med. 2017;82:12–20. doi: 10.1016/j.compbiomed.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Merritt CR, Ong GT, Church SE. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020;38(5):586–599. doi: 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kremer M, Spitzer M, Mandl-Weber S. Discordant bone marrow involvement in diffuse large B-cell lymphoma: comparative molecular analysis reveals a heterogeneous group of disorders. Lab Invest. 2003;83(1):107–114. doi: 10.1097/01.lab.0000050762.61660.27. [DOI] [PubMed] [Google Scholar]
- 18.Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Hematol Am Soc Hematol Educ Program. 2013;2013:575–583. doi: 10.1182/asheducation-2013.1.575. [DOI] [PubMed] [Google Scholar]
- 19.Hans CP, Weisenburger DD, Greiner TC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 20.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res. 2013;41(18):e170. doi: 10.1093/nar/gkt660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shipp MA, Ross KN, Tamayo P. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8(1):68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 22.Miluzio A, Beugnet A, Grosso S. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell. 2011;19(6):765–775. doi: 10.1016/j.ccr.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Bunting KL, Soong TD, Singh R. Multi-tiered reorganization of the genome during B Cell affinity maturation anchored by a germinal center-specific locus control region. Immunity. 2016;45(3):497–512. doi: 10.1016/j.immuni.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakamoto K, Bultot L, Goransson O. The salt-inducible kinases: emerging metabolic regulators. Trends Endocrinol Metab. 2018;29(12):827–840. doi: 10.1016/j.tem.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Vallejo-Diaz J, Chagoyen M, Olazabal-Moran M, Gonzalez-Garcia A, Carrera AC. The opposing roles of PIK3R1/p85alpha and PIK3R2/p85beta in cancer. Trends Cancer. 2019;5(4):233–244. doi: 10.1016/j.trecan.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhang T, Ma Z, Liu L. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/beta-catenin pathway. Cell Death Dis. 2018;9(6):675. doi: 10.1038/s41419-018-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer JA, Ferraro F, Roussakis E. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508(7495):269–273. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripodo C, Sangaletti S, Guarnotta C. Stromal SPARC contributes to the detrimental fibrotic changes associated with myeloproliferation whereas its deficiency favors myeloid cell expansion. Blood. 2012;120(17):3541–3554. doi: 10.1182/blood-2011-12-398537. [DOI] [PubMed] [Google Scholar]
- 30.Piconese S, Costanza M, Tripodo C. The matricellular protein SPARC supports follicular dendritic cell networking toward Th17 responses. J Autoimmun. 2011;37(4):300–310. doi: 10.1016/j.jaut.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Sangaletti S, Tripodo C, Santangelo A. Mesenchymal transition of high-grade breast carcinomas depends on extracellular matrix control of myeloid suppressor cell activity. Cell Rep. 2016;17(1):233–248. doi: 10.1016/j.celrep.2016.08.075. [DOI] [PubMed] [Google Scholar]
- 32.Nie J, Sage EH. SPARC inhibits adipogenesis by its enhancement of beta-catenin signaling. J Biol Chem. 2009;284(2):1279–1290. doi: 10.1074/jbc.M808285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milo I, Bedora-Faure M, Garcia Z. The immune system profoundly restricts intratumor genetic heterogeneity. Sci Immunol. 2018;3(29) doi: 10.1126/sciimmunol.aat1435. [DOI] [PubMed] [Google Scholar]
- 34.Ligorio M, Sil S, Malagon-Lopez J. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019;178(1):160–175. doi: 10.1016/j.cell.2019.05.012. e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodda LB, Lu E, Bennett ML. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity. 2018;48(5):1014–1028. doi: 10.1016/j.immuni.2018.04.006. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sacchetti B, Funari A, Michienzi S. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 37.Di Napoli A, Remotti D, Agostinelli C. A practical algorithmic approach to mature aggressive B cell lymphoma diagnosis in the double/triple hit era: selecting cases, matching clinical benefit: A position paper from the Italian Group of Haematopathology (G.I.E.) Virchows Arch. 2019;475(4):513–518. doi: 10.1007/s00428-019-02637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry AM, Cardesa-Salzmann TM, Meyer PN. A new biologic prognostic model based on immunohistochemistry predicts survival in patients with diffuse large B-cell lymphoma. Blood. 2012;120(11):2290–2296. doi: 10.1182/blood-2012-05-430389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, Calado DP, Wang Z. An oncogenic role for alternative NF-kappaB signaling in DLBCL revealed upon deregulated BCL6 expression. Cell Rep. 2015;11(5):715–726. doi: 10.1016/j.celrep.2015.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripodo C, Burocchi A, Piccaluga PP. Persistent Immune Stimulation Exacerbates Genetically Driven Myeloproliferative Disorders via Stromal Remodeling. Cancer Res. 2017;77(13):3685–3699. doi: 10.1158/0008-5472.CAN-17-1098. [DOI] [PubMed] [Google Scholar]
- 41.Voena C, Chiarle R. RHO Family GTPases in the Biology of Lymphoma. Cells. 2019;8(7) doi: 10.3390/cells8070646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szydlowski M, Kiliszek P, Sewastianik T. FOXO1 activation is an effector of SYK and AKT inhibition in tonic BCR signal-dependent diffuse large B-cell lymphomas. Blood. 2016;127(6):739–748. doi: 10.1182/blood-2015-06-654111. [DOI] [PubMed] [Google Scholar]
- 43.Saci A, Carpenter CL. RhoA GTPase regulates B cell receptor signaling. Mol Cell. 2005;17(2):205–214. doi: 10.1016/j.molcel.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Srinivasan L, Sasaki Y, Calado DP. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139(3):573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varano G, Raffel S, Sormani M. The B-cell receptor controls fitness of MYC-driven lymphoma cells via GSK3beta inhibition. Nature. 2017;546(7657):302–306. doi: 10.1038/nature22353. [DOI] [PubMed] [Google Scholar]
- 46.Ennishi D, Takata K, Beguelin W. Molecular and Genetic Characterization of MHC Deficiency Identifies EZH2 as Therapeutic Target for Enhancing Immune Recognition. Cancer Discov. 2019;9(4):546–563. doi: 10.1158/2159-8290.CD-18-1090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.