Abstract
Muscle-invasive bladder cancer (MIBC) is characterized by high recurrence and rapid progression. Progression is linked to changes in glycan structures and altered levels of glycosyltransferases. The relationship of mRNA expression by glycosyltransferase genes B4GALT1, EXT1, MGAT5B, and POFUT1 to the probability of surviving MIBC after radical cystectomy has not yet been investigated.
mRNA expression was analyzed using qRT-PCR in formalin-fixed and paraffin-embedded tumor samples (n = 105; 74% male patients and 26% female patients; median age = 72 years), correlated with histopathological variables, and evaluated by means of multivariable Cox regression analysis regarding to overall survival (OS), cancer-specific survival (CSS), and disease-free survival (DFS).
Multivariable Cox regression analysis identified POFUT1 mRNA expression as superior prognostic marker, compared with currently used histological tumor stage methods, for CSS by MIBC patients following radical cystectomy. Thus, the patients with low POFUT1 mRNA were at a 4.9-fold greater risk for cancer-specific death according to the multivariable analysis (p = 0.0001). Low mRNA levels predicted poor survival according to the Kaplan-Meier analysis ((POFUT1:OS p = 0.0014; CSS p = 0.0007; DFS p = 0.0088); (EXT1:OS p = 0.0150; CSS p = 0.0130; DFS p = 0.0286); (B4GALT1:CSS p = 0.0134; DFS p = 0.0493)). A subgroup analysis of patients without lymph node metastasis (pN−; n = 73) indicated that low expression of POFUT1 predicted reduced OS (p = 0.0073), CSS (p = 0.0058,) and DSS (p = 0.0079).
Low levels of POFUT1 mRNA are an independent prognostic indicator for OS and CSS in MIBC patients following radical cystectomy. This finding demonstrates the importance of altered glycosylation for the progress of MIBC.
Abbreviations: B4GALT1, β-1,4-galactosyltransferase 1; VI, blood vessel invasion; CSS, cancer-specific survival; DFS, disease-free survival; EXT1, exostosin-1; FFPE, formalin-fixed and paraffin-embedded; GT(s), glycosyltransferase(s); LI, lymphatic vessel invasion; MGAT5B, mannosyl (α-1,3-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase 5B; MIBC, muscle-invasive bladder cancer; OS, overall survival; POFUT1, protein O-fucosyltransferase 1; RC, radical cystectomy; UCa, urothelial bladder cancer
Keywords: Muscle-invasive bladder cancer, Glycosyltransferases, mRNA expression, Prognostic marker, qRT-PCR, FFPE tumor samples
Highlights
-
•
Low POFUT1 mRNA expression is associated with a higher risk for overall and cancer-specific death in MIBC treated with RC.
-
•
MIBC patients with pN0 histology and, decreased POFUT1 mRNA levels showed poor outcome for OS, CSS and, DFS.
-
•
POFUT1 mRNA is an independent prognostic indicator for OS and CSS in multivariable analysis of MIBC patients following RC.
Introduction
Urothelial bladder carcinoma (UCa) is the fifth most common malignant cancer worldwide [1], being characterized by poor survival rates for patients with muscle-invasive bladder cancer (MIBC) [2,3]. Because it is associated with high rates of recurrence and rapid progression and requires intensive monitoring, MIBC tends to be very expensive to treat [4]. The choice of therapy after surgery is guided by histopathological parameters that are, however, subject to considerable intra- and inter-observational variability [5]. Furthermore, owing to the limited knowledge available regarding molecular and biologic variants of the disease, potential therapy responders have not yet been identified. Previous studies have shown that qRT-PCR investigations of formalin-fixed and paraffin-embedded (FFPE) tissue may be applicable to MIBC biomarker research, in particular for the identification of patients at high risk for progression or invasion [[6], [7], [8], [9]].
The altered glycosylations can serve to distinguish between normal and malignant conditions [10,11]. Glycosylations are highly dynamic post-translational structures on proteins or lipids [12,13], carried out by glycosyltransferases (GTs) [14,15]. Proteins with identical sequences may acquire various glycan structures during glycosylation that impart such distinct properties as stability, folding, localization, and ligand specificity [16], thereby affecting biological processes including protein trafficking, cell-cell and cell-matrix interactions, differentiation, and immune response [[17], [18], [19]].
To date, only a few distinctive GT expressions have been identified in UCa clinical tumor samples. Most studies of these issues have focused on O-glycosylations and the expression of sialylated Tn antigen and/or its corresponding GT, ST6GALNAC1 [[20], [21], [22], [23], [24]]. Another sialyl-related GT, ST6GAL1, appears to be inactivated epigenetically in MIBC and has already been reported in several cancers [25]. In addition, sialylated Lewis X and A antigens may be predictors of poor clinical outcomes [[26], [27], [28]]. Concerning N-glycosylations, the expression of GT MGAT5 is associated with low potential for malignancy [29,30]. Also, the downregulation of the B3GNT2 transcript has been shown to correlate with cancer progression [31]. Other significant predictors of metastasis and poor survival are hyaluronic acid synthases and hyaluronidase-1 expression in bladder tumor samples [32].
The GT POFUT1 (protein O-fucosyltransferase 1) catalyzes the O-linkage of fucose to serine or threonine on target proteins. This reaction, which takes place at condensed epidermal growth factor-like repeats, is required for proper ligand-receptor interactions and signal transduction. Though more than 100 identified proteins contain such repeats and, therefore, are predicted to be modified by POFUT1, only a few targets have been identified so far. Among these, the Notch receptors are the most studied target of POFUT1 and are reported to be rich in O-fucosylated proteins [33].
The data for the glycosylation-based markers in MIBC are incomplete, and further investigation is needed [5].
To our knowledge, the mRNA gene expression of POFUT1 and other GTs, such as B4GALT1 (β-1,4-galactosyltransferase 1), EXT1 (exostosin-1), and MGAT5B (mannosyl (α-1,3-)-glycoprotein β-1,2-N-acetylglucosaminyltransferase 5B), has been little explored in the context of MIBC. However, studies already exist proofing the impact of those GTs on cancer malignancy, based on altered gene expression for other entities as oral, breast, renal and liver cancer [[34], [35], [36], [37]]. The aim of this study, accordingly, was to investigate the expression of POFUT1, B4GALT1, EXT1, and MGAT5B in MIBC patients following RC with regard to the histopathology and survival data.
Materials and methods
Tissue samples
FFPE tumor tissue samples were obtained from 105 patients (74% male and 26% female, with a median age of 72 years; median follow-up was at 22 months) who had undergone RC at the Clinic for Urology and Urosurgery at the University Hospital Mannheim. The histopathological data were collected by the Institute of Pathology at the University Hospital Mannheim and re-evaluated by the Institute of Pathology at the University Hospital Erlangen according to the most recent TNM classification (2017) and the WHO 2016 classification of genitourinary tumors.
The study was approved by the review board of the University Hospital Mannheim, under Numbers 2013-517N-MA and 2016-814R-MA, in accordance with the Declaration of Helsinki. All patients provided written informed consent to participate.
Gene expression experiments
Primers and dual-labelled probes (label: 5′-FAM, 3′-BHQ1) for B4GALT1, EXT1, MGAT5B and, POFUT1 were designed using the Primer-BLAST tool available from NCBI (The National Center for Biotechnology Information; https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and, were validated for qRT-PCR using UCa cell line T24 (Suppl. Table 1). The tended amplicon sequences were confirmed by Sanger sequencing (Sequiserve GmbH, Vaterstetten, Germany). The amplification efficiencies (99%–110%) and intra- and inter-assay variations (0.46%–2.63%) fell within the expected range.
The mRNA was extracted from 10 μm sections of FFPE tissue and processed using a commercially available bead-based extraction method (XTRACT kit; STRATIFYER Molecular Pathology GmbH, Cologne, Germany) according to the manufacturer's protocol. The sections were taken from a paraffin block containing a tumor area of at least 5 × 5 mm and a total tumor content of at least 50%. If the tumor content was less than 50%, samples were incubated in Neoclear (Merck, Darmstadt, Germany) for 4 min; they were then macro-dissected to enrich the tumor RNA. The RNA was eluted with 50 μl elution buffer for analysis.
Reverse transcription was performed using the Superscript III® reverse transcriptase kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).. mRNA expression was measured using qRT-PCR with TaqMan Fast advanced Master Mix (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) on the StepOnePlus PCR system (Applied Biosystems, Darmstadt, Germany). qRT-PCR setting: (1) 20 s at 95 °C, (2) 40 cycles of 3 s at 95 °C and of 30 s at 60 °C. Calmodulin2 (CALM2) was measured as a reference gene [6,7,9]. The means were calculated from the technical duplicates of each gene.
Immunohistochemistry
FFPE tissue was cut at 4 μm. Antigen retrieval was performed using pH 9 and pressure cooker for 20 min. POFUT1 polyclonal antibody (Abcam, #ab74302, Berlin, Germany) was applied at 1:50 dilution. Staining was performed on Autostainer 480-2D (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
The normalized expression data of the GT genes were correlated with the appropriate clinicopathological parameters and analyzed with the Kruskal-Wallis-Test. The relationship between GT gene expression and patients' probability of survival was predicted using the log-rank test and presented as Kaplan-Meier plots for overall survival (OS), cancer-specific survival (CSS), and disease-free survival (DFS). Cut-offs were determined by partition, with a minimum number of ≥48 patients and a minimum group distribution of ≥25%. To identify the independence and predictable impact of each GT gene expression, uni- and stepwise multivariable Cox regressions were applied. All of the statistical tests were calculated with SAS JMP version 13, the plots having been designed using GraphPad prism version 8. All p-values were two-sided; p-values of <0.05 were considered statistically significant.
Results
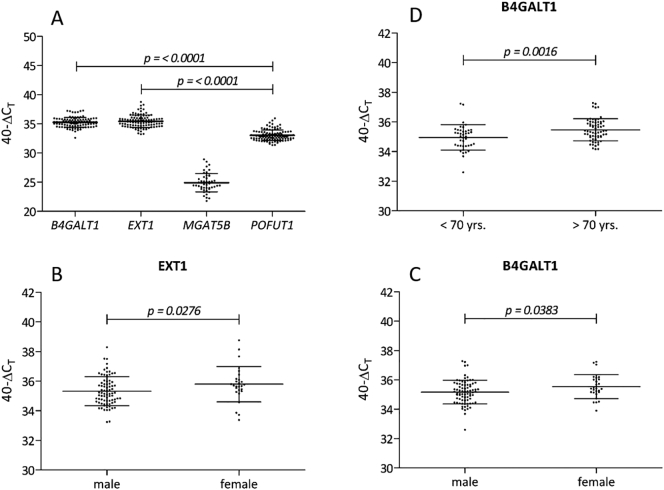
The distribution of the mRNA expression levels of B4GALT1, EXT1, and POFUT1 is shown in Fig. 1A. POFUT1 was expressed at significantly lower levels throughout the population with respect to B4GALT1 and EXT1 (p ≤ 0.0001). With respect to MGAT5B, 25% of the samples were undetectable, 70% exceeded and only 5% were below the LOD. Even assuming that 95% of the patients exhibited the lowest detectable level of gene expression, MGAT5B expression did not correlate significantly with either survival or progression of MIBC (data not shown). Therefore, MGAT5B was excluded from further analysis.
Fig. 1.
Distribution of GT expression in MIBC patients and significant correlations with clinicopathological parameters.
(A) Overall mRNA expression in the entire population of MIBC patients (n = 105) shows that POFUT1 expression was lower than expression of EXT1 and B4GALT1 (p ≤ 0.0001). Since 95% of the MGAT5B signals were below the limit of detection or not detectable, expression of this gene could not be further analyzed. (B; C) EXT1 (p = 0.0276) and B4GALT1 (p = 0.0383) expression were reduced in the male than in the female patients. (D) With regard to the age of the patients, B4GALT1 expression was lower among patients under 70 years of age than those over 70 (p = 0.0016). TaqMan qRT-PCR data were normalized to CALM2 by 40-∆CT and plotted on the Y-axis. Mean and standard deviations are indicated in the scatter plots. Significances were calculated using the Kruskal-Wallis-Test and considered significant if p < 0.05.
The association of gene expression with such clinicopathological parameters as age, gender, pT-stage, pN-stage, lymphatic vessel invasion (LI), blood vessel invasion (VI), and tumor size was assessed with respect to the clinical characteristics listed in Table 1. B4GALT1 and EXT1 mRNA expression correlated significantly with the patients' gender (Fig. 1B, C), measuring lower in men than women (B4GALT1 p = 0.0383; EXT1 p = 0.0276). Moreover, B4GALT1 mRNA expression was lower in patients under the age of 70 years than in those over 70 (p = 0.0016) (Fig. 1D). POFUT1 mRNA expression showed no significant correlation with any of the clinicopathological parameters. Lastly, a positive correlation was found among the mRNA expression profiles of the GTs (ρ = 0.5476–0.6676; p < 0.0001; see Supplementary Fig. 1).
Table 1.
Clinical and pathological characteristics of studied tissue.
| n = 105 |
Median age |
||||
|---|---|---|---|---|---|
| 72 years | |||||
| High grade tumors | |||||
| Gender | M | F | |||
| 74% (78) | 26% (27) | ||||
| pT-stage | T2a/b | T3a/b | T4a/b | ||
| 26% (27) | 51% (54) | 23% (24) | |||
| pN-stage | pN0 | pN1–3 | pNX | ||
| 69% (73) | 23% (24) | 8% (8) | |||
| Lymphovascular invasiona | LI+/VI− | LI−/VI− | VI+/LI− | LI+/VI+ | N/A |
| 26% (27) | 46% (49) | 2% (2) | 23% (24) | 3% (3) | |
| Tumor size | ≥3 cm | <3 cm | |||
| 71% (75) | 29% (30) | ||||
| Histology | Urothelial | Squamous | Neuroendocrine | Mixed | |
| 88% (93) | 6% (6) | 2% (2) | 4% (4) | ||
(n) number of subjects; (m) male; (f) female; (T2a/b) tumor invades (a) inner half or (b) outer half of muscularis propria bladder wall; (T3a/b) tumor invades perivesical tissue, (a) microscopically or (b) macroscopically; (T4a/b) tumor invades (a) prostate, uterus, vagina or (b) pelvic or abdominal wall; (pNX) regional lymph nodes cannot be evaluated histologically; (pN0) no regional lymph node metastasis; (pN1) metastases in a single lymph node, 2 cm or less in greatest dimension; (pN2) Metastases in a single lymph node, more than 2 cm but not more than 5 cm in greatest dimension, or multiple lymph nodes, none more than 5 cm in greatest dimension; (pN3) Metastasis in a lymph node more than 5 cm in greatest dimension; (LI+) tumor invades lymphatic vessel; (LI−) no lymphatic vessel invasion; (VI+) tumor invades blood vessel; (VI−) no blood vessel invasion; (N/A) data not available.
In case of Lymphovascular invasion, LI and VI were considered as separated groups (LI+ = includes all patients with LI+/VI− and LI+/VI+; LI− = LI−/VI− and LI−/VI+; VI+ = VI+/LI− and VI+/LI+; VI− = VI−/LI− and VI−/LI+).
Lower levels of GT gene expression predicted poor survival outcomes for MIBC patients
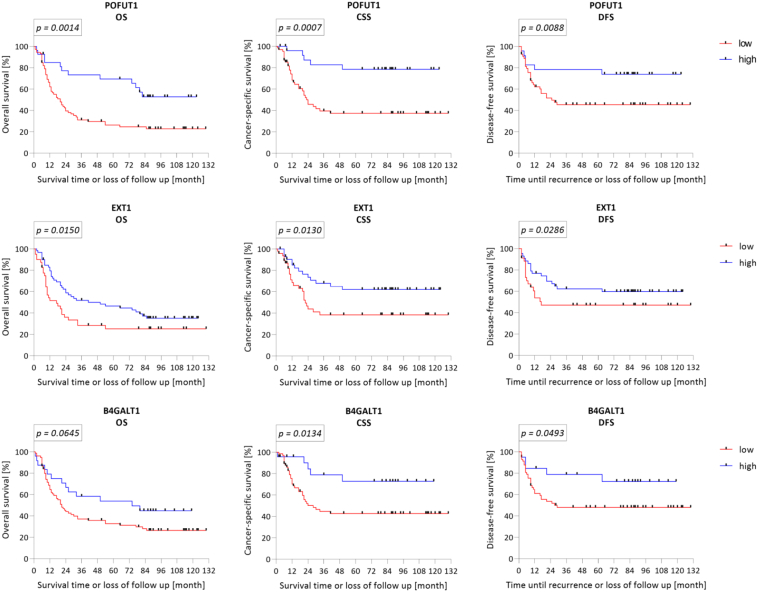
A decreased expression of B4GALT1, EXT1, and POFUT1 correlated significantly with decreased OS, CSS, and DFS among the MIBC patients (Table 2; Fig. 2). The greatest significance was observed in the CSS analysis (POFUT1 p = 0.0007; EXT1 p = 0.0130; B4GALT1 p = 0.0134), with a 5-year survival rate of 41%, 44%, and 45% associated with POFUT1, EXT1, and B4GALT1, respectively. Less significance was observed in OS analysis for POFUT1 (p = 0.0014; 5-year survival rate: 36%) or EXT1 (p = 0.0150; 5-year survival rate: 55%), while B4GALT1 only showed a tendency. Reduced mRNA levels of POFUT1 (p = 0.0088), EXT1 (p = 0.0286), and B4GALT1 (p = 0.0493) correlated with a decreased probability of DFS, specifically, 5-year survival rates of 40%, 17%, and 32%, respectively.
Table 2.
Survival and recurrence data of studied tissue.
| n = 105 | |||||
|---|---|---|---|---|---|
| Overall median follow-up | 22 months | ||||
| Recurrence | Yes | No | N/A | ||
| 46% (48) | 43% (45) | 11% (12) | |||
| Cancer-specific survival | Deceased | Alive | N/A | Deceased median follow-up | Alive median follow-up |
| 38% (40) | 50% (52) | 12% (13) | 12 months | 81 months | |
| Overall survival | Deceased | Alive | N/A | Deceased median follow-up | Alive median follow-up |
| 66% (69) | 33% (35) | 1% (1) | 13 months | 90 months | |
(n) number of subjects; (N/A) data not available.
Fig. 2.
Kaplan-Meier prediction of GT mRNA expression in MIBC patients.
Normalized qRT-PCR data of GT mRNA expression are plotted as Kaplan-Meier curves in relation to the patients' survival probabilities. Cut offs were determined by partition with a minimal group distribution of ≥25%. Overall survival (OS), cancer-specific survival (CSS), and disease-free survival (DFS) were predicted by log-rank test and considered significant if p < 0.05. As the graphs show, in each case, reduced mRNA gene expression predicted poor probability of survival for MIBC patients. POFUT1 and EXT1 were significant for OS (POFUT1 p = 0.0014; EXT1 p = 0.0150), CSS (POFUT1 p = 0.0007; EXT1 p = 0.0130), and DFS (POFUT1 p = 0.0088; EXT1 p = 0.0286). B4GALT1 was significant for CSS (p = 0.0134) and DFS (p = 0.0493).
Uni- and multivariable analyses of survival data indicated that POFUT1 is an independent predictor of MIBC
The dependence of the GT gene expression on clinicopathological parameters, as age, gender, pT-stage, pN-stage, lymphatic vessel invasion (LI), blood vessel invasion (VI), and tumor size, was examined by means of uni- and multivariable Cox proportional hazard analyses (Table 3). In the OS, CSS and DFS investigation, the univariable approach showed significance for the following established prognostic indicators: pT-stage (OS (T3/2) HR: 2.6371; p = 0.0023; OS (T4/2) HR: 3.6504; p = 0.0005; CSS (T3/2) HR: 4.1017; p = 0.0025; CSS (T4/2) HR: 6.0953; p = 0.0007; DFS (T3/2) HR: 2.3974; p = 0.0234; DFS (T4/2) HR: 2.8052; p = 0.0363), pN-stage (OS HR: 2.2057; p = 0.0073; CSS HR: 2.5128; p = 0.0122), LI (OS HR: 2.9219; p ≤ 0.0001; CSS HR: 4.5440; p ≤ 0.0001; DFS HR: 2.9295; p = 0.0009), and VI (OS HR: 2.2386; p = 0.0037; CSS HR: 2.6483; p = 0.0044; DFS HR: 2.3559; p = 0.0209). Patients belonging to the low-mRNA-expression group for POFUT1 showed a 4.4-fold greater risk of a shortened lifespan with regard to CSS (p = 0.0003). By contrast, the risk of a shortened lifespan was 2.6- and 2.9-fold greater for OS (p = 0.0008) and DFS (p = 0.0058), respectively. Furthermore, a decreased EXT1 mRNA expression belonged to the higher-risk group with regard to OS (HR: 1.7791; p = 0.0192), CSS (HR: 2.223; p = 0.0137), and DFS (HR: 1.9499; p = 0.0343). Even in the case B4GALT1, reduced gene expression was associated with the higher-risk patient group, and predicted decreased CSS (HR: 3.0541; p = 0.0079) and DFS (HR: 2.2927; p = 0.0397) but not decreased OS.
Table 3.
Uni- and stepwise multivariable Cox regression of glycosyltransferases.
| Survival | Variables | Univariable |
Multivariable |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| POFUT1 |
EXT1 |
B4GALT1 |
|||||||||
| HR | p-Value | HR | p-Value | HR | p-Value | HR | p-Value | ||||
| OS | GT | POFUT1− | POFUT1+ | 2.6512 | 0.0008 | 3.4104 | 0.0004 | ||||
| EXT1− | EXT1+ | 1.7791 | 0.0192 | 1.5835 | ns | ||||||
| B4GALT1− | B4GALT1+ | 1.7468 | 0.0566 | ns | |||||||
| Age | Continuous range | 2.7024 | 0.0910 | 3.9969 | 0.0173 | ||||||
| Gender | Female | Male | 1.2930 | 0.3506 | |||||||
| pT-stage | T3a/b | T2a/b | 2.6371 | 0.0023 | 2.3534 | 0.0152 | |||||
| T4a/b | T2a/b | 3.6504 | 0.0005 | ||||||||
| T4a/b | T3a/b | 1.3842 | 0.2562 | ||||||||
| pN-stage | pN+ | pN− | 2.2057 | 0.0073 | |||||||
| LI | LI+ | LI− | 2.9219 | <0.0001 | 2.5102 | 0.0009 | 2.7513 | <0.0001 | |||
| VI | VI+ | VI− | 2.2386 | 0.0037 | 1.9521 | 0.0425 | |||||
| TS | ≥3 cm | <3 cm | 1.4033 | 0.2063 | |||||||
| CSS | GT | POFUT1− | POFUT1+ | 4.4066 | 0.0003 | 4.9940 | 0.0001 | ||||
| EXT1− | EXT1+ | 2.2231 | 0.0137 | 2.0873 | 0.0235 | ||||||
| B4GALT1− | B4GALT1+ | 3.0541 | 0.0079 | 3.0721 | 0.0076 | ||||||
| Age | Continuous range | 0.6495 | 0.5516 | ||||||||
| Gender | Female | Male | 1.8607 | 0.0714 | |||||||
| pT-stage | T3a/b | T2a/b | 4.1017 | 0.0025 | 4.1072 | 0.0029 | |||||
| T4a/b | T2a/b | 6.0953 | 0.0007 | 4.2936 | 0.0079 | ||||||
| T4a/b | T3a/b | 1.4861 | 0.2766 | ||||||||
| pN-stage | pN+ | pN− | 2.5128 | 0.0122 | |||||||
| LI | LI+ | LI− | 4.5440 | <0.0001 | 4.4159 | <0.0001 | 4.5255 | <0.0001 | 4.4968 | <0.0001 | |
| VI | VI+ | VI− | 2.6483 | 0.0044 | |||||||
| TS | ≥3 cm | <3 cm | 1.1127 | 0.7508 | |||||||
| DFS | GT | POFUT1− | POFUT1+ | 2.9687 | 0.0058 | 2.4454 | 0.0455 | ||||
| EXT1− | EXT1+ | 1.9499 | 0.0343 | ns | |||||||
| B4GALT1− | B4GALT1+ | 2.2927 | 0.0397 | ns | |||||||
| Age | Continuous range | 0.8699 | 0.8399 | ||||||||
| Gender | Female | Male | 1.5387 | 0.2128 | |||||||
| pT-stage | T3a/b | T2a/b | 2.3974 | 0.0234 | |||||||
| T4a/b | T2a/b | 2.8052 | 0.0363 | ||||||||
| T4a/b | T3a/b | 1.1701 | 0.6918 | ||||||||
| pN-stage | pN+ | pN− | 1.8703 | 0.0841 | |||||||
| LI | LI+ | LI− | 2.9295 | 0.0009 | 2.6977 | 0.0027 | |||||
| VI | VI+ | VI− | 2.3559 | 0.0209 | |||||||
| TS | ≥3 cm | <3 cm | 1.0311 | 0.9274 | |||||||
Overall survival (OS); cancer-specific survival (CSS); disease-free survival (DFS); glycosyltransferase (GT); high gene expression is indicated by “+” and low gene expression by “−”; tumor stage (pT); lymph node metastasis present (pN+); lymph node metastasis absent (pN−); lymphatic vessel invasion present (LI+); lymphatic vessel invasion absent (LI−); blood vessel invasion present (VI+); blood vessel invasion absent (VI−); tumor size (TS); not significant (ns); significant p-values are displayed in bold.
The multivariable analysis (Table 3) identified POFUT1 as a predictor of OS (HR: 3.4104; p = 0.0004) with a strong independence assumption together with age, pT stage, LI, and VI. CSS (HR: 4.9940; p = 0.0001) also showed a strong independence with pT stage and LI, while with regard to DFS (HR: 2.4454; p = 0.0455) POFUT1 was independent together with LI. The multivariable CSS analysis of B4GALT1 and EXT1 showed significant outcomes (EXT1 HR: 2.0873, p = 0.0235; B4GALT1 HR: 3.0721, p = 0.0076) together with LI. Regarding OS and DFS, mRNA expression of B4GALT1 and EXT1 were inconsistent with the multivariable model, and therefore not independent.
In addition, the survival of patient subgroups was analyzed to show the impact of GT gene expression on survival outcomes for patients whose cancers were less aggressive, such as those at a lower pT-stage (T2/3), those not showing lymph node metastasis (pN−), and those suffered by male patients (male). Patients with lower POFUT1 gene expression in all of the subgroups experienced significantly shortened lifespans (OST2/3 p = 0.0056; OSpN− p = 0.0073; OSmale p = 0.0101; CSST2/3 p = 0.0121; CSSpN− p = 0.0058; CSSmale p = 0.0088; DFST2/3 p = 0.0433; DFSpN− p = 0.0079), except for DFSmale subgroup (Table 4). EXT1 reduced gene expression correlated with worse outcomes with respect to OS (OST2/3 p = 0.0256; OSmale p = 0.0380) and CSS (CSST2/3 p = 0.0319; CSSpN− p = 0.0386; CSSmale p = 0.0102) subgroups, however, had no impact with regard to DFS in any subgroup. B4GALT1 mRNA expression showed the strongest correlation within the OSmale (p = 0.0362), CSSmale (p = 0.0097), and DFSmale (p = 0.0112) subgroup, while decreased gene expression was associated with a lower probability of survival. No significant correlations in this regard were observed for the subgroups “T2/3” or “pN−” (Suppl. Table 2).
Table 4.
POFUT1 survival analysis of patient subgroups with less aggressive characteristics.
| Subgroup | Survival | Gene | p-Value | p-Value strength | Group size [n] | Group distribution |
5-year survival rate |
||
|---|---|---|---|---|---|---|---|---|---|
| Low [%] | High [%] | Low [%] | High [%] | ||||||
| OS | POFUT1 | 0.0014 | ** | 104 | 73 | 27 | 36 | 78 | |
| CSS | POFUT1 | 0.0007 | *** | 92 | 71 | 29 | 41 | 73 | |
| DFS | POFUT1 | 0.0088 | ** | 86 | 72 | 28 | 40 | 61 | |
| T2/3 | OS | POFUT1 | 0.0056 | ** | 80 | 30 | 70 | 28 | 73 |
| T2/3 | CSS | POFUT1 | 0.0121 | * | 71 | 34 | 66 | 44 | 76 |
| T2/3 | DFS | POFUT1 | 0.0433 | * | 71 | 31 | 69 | 44 | 76 |
| pN− | OS | POFUT1 | 0.0073 | ** | 72 | 69 | 31 | 40 | 78 |
| pN− | CSS | POFUT1 | 0.0058 | ** | 65 | 66 | 34 | 35 | 59 |
| pN− | DFS | POFUT1 | 0.0079 | ** | 64 | 69 | 31 | 44 | 75 |
| male | OS | POFUT1 | 0.0101 | * | 77 | 71 | 29 | 27 | 67 |
| male | CSS | POFUT1 | 0.0088 | ** | 68 | 74 | 26 | 42 | 84 |
| male | DFS | POFUT1 | 0.0502 | ns | 63 | 75 | 25 | 43 | 83 |
Partition test for cut offs and log rang test were applied. The group distributions and 5-year survival rates are divided into groups representing high and low POFUT1 gene expression. Overall survival (OS); cancer-specific survival (CSS); disease-free survival (DFS); tumor stage T2a/b and T3a/b (T2/3); lymph node metastasis absent (pN−); only male patients within this subgroup (male); not significant (ns); significant p-values are displayed in bold.
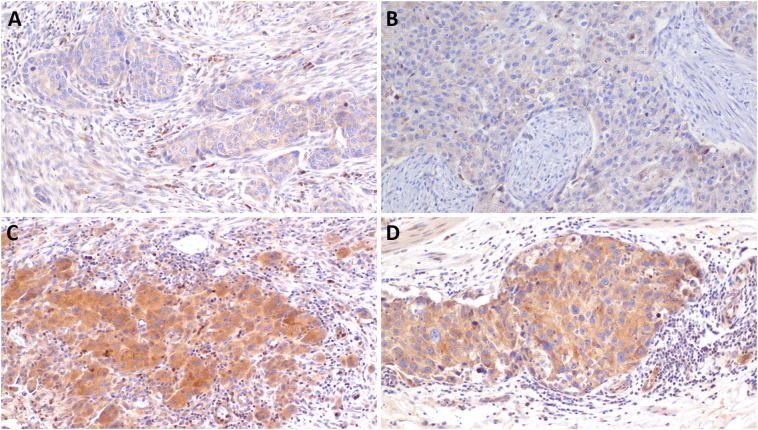
Protein expression of POFUT1 in MIBC
Immunohistochemistry confirmed protein expression of POFUT1 in association with mRNA expression. Representative FFPE tissues, showed high and low POFUT1 mRNA expression are shown in Fig. 3.
Fig. 3.
POFUT1 protein expression of high and low mRNA expressing FFPE tissue.
Examples of different cases with low (A, B) and high (C, D) expression of POFUT1 mRNA that was reflected at the protein level as shown by Immunohistochemistry for POFUT1. Adjacent positive immune cells also stain positive and serve as an internal positive control. Original magnification 200×.
Discussion
The aim of this study was to investigate the prognostic and clinical impact of the GTs B4GALT1, EXT1, and POFUT1 retrospectively in MIBC patients who had been treated with RC. To our knowledge, the expression of mRNA by these genes has not been examined previously in the context of MIBC. In order to evaluate the translational benefit of these genes, mRNA expression was compared with relevant clinicopathological parameters by means of multivariable analyses.
The data indicated poor OS, CSS, and DFS outcomes for patients whose tumor samples showed relatively low levels of POFUT1 gene expression. Multivariable Cox regression analysis also showed POFUT1 expression to be superior as a strong prognostic indicator to the established clinicopathological prognostic parameters (OS (pT-stage; LI; VI); CSS (pT-stage; LI)). These findings were confirmed by the survival analysis of subgroups of patients showing less progressive features (T2/3; pN−; male), with reduced expression of POFUT1 being associated with a shortened life span in MIBC patients following RC. However, the strong prognostic influence of POFUT1 and LI or rather VI was not expected for single marker. Indeed, the lymphovascular invasion showed less effective properties as prognostic biomarker for transurethral resection or biopsy samples originate from high grading T1 UCa, and is associated with understaging, and increased risks of recurrence and progression. However, in MIBC patients treated with RC LVI is linked to aggressive disease and can predict survival [38]. Nevertheless, this increasing effect might also be described by incomplete data of the studied tissue, and could be improved by increasing cases. Nevertheless, POFUT1 protein levels represented an association to the examined mRNA level, exemplified on four cases. Previous reports of a correlation between differential POFUT1 gene expression and cancer progression and elevated levels of POFUT1 mRNA in both tumor tissues and plasma specimens of lung cancer patients suggest that this gene plays an important role in lung carcinogenesis [39] and may be useful in plasma-based early detection of lung cancer [40]. In the case of breast cancer, overexpression of POFUT1 protein in FFPE samples has been associated with poor prognoses [41]. In a study of frozen breast cancer tissue, however, Milde-Langosch et al. correlated elevated expression of POFUT1 mRNA with positive prognoses in terms of DFS and OS, a finding confirmed by our findings on the same molecular level [42]. In the case of colorectal cancer, elevated POFUT1 mRNA and protein levels have been identified as potential drivers of tumor progression [43,44]. By contrast to our results, POFUT1 was overexpressed in other entities, for example, lung, breast, and colorectal tumors. Interestingly, previous data showed that NOTCH1 serves as a tumor suppressor gene in the bladder and that loss of this pathway promotes the emergence of mesenchymal and invasive aspects of tumors [45]. The findings presented here suggest that reduced levels of POFUT1 mRNA indicate a poor prognosis and, therefore, may represent an additional factor contributing to Notch1 inactivation in MIBCs.
mRNA expression analysis showing lower levels of B4GALT1 in men than women and this finding can be supported by survival subgroup analysis in male patients. A similar pattern of mRNA expression was found in a comparison of patients younger than and older than 70 years of age. In addition, impaired CSS and DFS were associated with decreased B4GALT1 mRNA expression. However, the multivariable Cox regression analysis indicated that B4GALT1 mRNA expression did predicted neither OS, CSS, nor DFS. In a previous study, analysis of B4GALT1 protein expression on tissue microarrays was used to predict outcomes for MIBC patients; specifically, high levels of B4GALT1 protein expression were shown to be independent indicators of poor OS. This group also found that B4GALT1 expression correlated with responses to adjuvant chemotherapy in T3/4 or pN+ tumors as well as with the presence of immunological inhibitory receptor ligands [46]. Although the results presented here are not entirely consistent with this finding, the disagreement may be attributable to differences between the two studies with respect to molecular detection levels or the cohorts analyzed, while the proportion of patients treated with adjuvant platinum-based chemotherapy.
Furthermore, MIBC patients with relatively low EXT1 mRNA expression levels had poor prognoses with regard to OS, CSS, and DFS. EXT1, then, did not prove to be a prognostic indicator according to the multivariable Cox analysis. Indeed, the levels of EXT1 mRNA expression were found to be less in men than women, and this finding was confirmed by the subgroup survival analyses, at least for OS and CSS. Other researchers found, in a retrospective study of biomarkers, that a significant decrease in EXT1 expression was associated with metastatic breast cancer, so this gene has already shown promise as a predictor of metastasis [35]. We likewise found decreased EXT1 mRNA expression to be associated with poor survival outcomes. It has also been suggested that EXT1 may suppress tumors based on the correlation between loss of heterozygosity at one or several EXT1 loci with the formation of multiple cartilaginous tumors [47,48]. In light of these findings, then, downregulation of this gene may have tumor-driving properties in MIBC.
Conclusions
In sum, expression of POFUT1 mRNA appears to serve as an independent prognostic indicator of OS and CSS for patients with MIBC after treatment with RC. In the study, the observed decrease in expression of this gene was associated with poor survival prognosis. This result may be due, at least in part, to the inactivity of Notch1 associated with low levels of POFUT1 mRNA and protein expression in MIBC. The present retrospective, single-center study was exploratory in nature, and there is need for further validation of the findings by prospective multicenter studies. In particular, the impact of GTs on the development of resistance to platinum-based chemotherapies and immunotherapies warrants future study.
The following are the supplementary data related to this article.
Spearman correlation of POFUT1, EXT1, and B4GALT1. Normalized GT mRNA expression data are plotted in relation to each other. The correlation coefficient was calculated using Spearman's correlation (ρ). All correlations displayed a positive relationship. There was a fairly strong correlation between POFUT1 and B4GALT1 and a moderate one between POFUT1 and EXT1 as well between EXT1 and B4GALT1. The probability that these results would occur randomly is p ≤ 0.0001.
TaqMan qRT-PCR assay parameters of designed GT primer and probes.
EXT1 and B4GALT1 survival analysis of patients with less aggressive characteristics.
Data accessibility
The datasets generated and analyzed for this study are available from the corresponding author on reasonable request.
Funding sources
This project was funded by the Foundation for Cancer and Scarlet Research Mannheim, the Albert and Anneliese Konanz Foundation Mannheim, and the Medical Faculty Mannheim at the University of Heidelberg.
CRediT authorship contribution statement
Sarah Wahby: project planning, qRT-PCR experiments, data collection, data analysis, and manuscript writing
Jakob Heinkele: sample acquisition, clinical data collection, and follow-up
Alexander Fierek: sample acquisition, clinical data collection, and follow-up
Jonas Jarczyk: sample acquisition, clinical data collection, and follow-up
Cleo- Aron Weis: tissue fixation, embedding, sectioning, staining, and image acquisition
Markus Eckstein: tissue fixation, embedding, sectioning, staining, and image acquisition
Thomas Martini: sample acquisition, clinical data collection, and follow-up
Stefan Porubsky: tissue fixation, embedding, sectioning, staining, and image acquisition
Mathias Hafner: project supervision and manuscript writing
Philipp Erben: project development, project supervision, data analysis, and manuscript writing.
All of the authors contributed to the drafting and reviewing of the manuscript and approved the submitted and final version of it.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Annette Steidler and Lena Hoffmann for technical and intellectual support and Dr. Ralph Markus Wirtz for a critical discussion of the issues raised by our study. In addition, we are thankful to the Foundation for Cancer and Scarlet Research Mannheim and the Albert and Anneliese Konanz Foundation Mannheim for the funding of this project.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016 Jan 1;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg G.D., Trump D.L., Cummings K.B. Metastatic bladder cancer. Natural history, clinical course, and consideration for treatment. Urol. Clin. North Am. 1992 Nov;19(4):735–746. [PubMed] [Google Scholar]
- 3.Liebert M., Seigne J. Characteristics of invasive bladder cancers: histological and molecular markers. Semin. Urol. Oncol. 1996 May;14(2):62–72. [PubMed] [Google Scholar]
- 4.Svatek R.S., Hollenbeck B.K., Holmäng S., Lee R., Kim S.P., Stenzl A. The economics of bladder cancer: costs and considerations of caring for this disease. Eur. Urol. 2014 Aug;66(2):253–262. doi: 10.1016/j.eururo.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Sapre N., Herle P., Anderson P.D., Corcoran N.M., Hovens C.M. Molecular biomarkers for predicting outcomes in urothelial carcinoma of the bladder. Pathology (Phila.) 2014 Jun;46(4):274–282. doi: 10.1097/PAT.0000000000000110. [DOI] [PubMed] [Google Scholar]
- 6.Eckstein M, Wirtz RM, Gross-Weege M, Breyer J, Otto W, Stoehr R, et al. mRNA-expression of KRT5 and KRT20 defines distinct prognostic subgroups of muscle-invasive urothelial bladder cancer correlating with histological variants. Int J Mol Sci. 2018 30;19(11):3396. [DOI] [PMC free article] [PubMed]
- 7.Eckstein M., Wirtz R.M., Pfannstil C., Wach S., Stoehr R., Breyer J. A multicenter round robin test of PD-L1 expression assessment in urothelial bladder cancer by immunohistochemistry and RT-qPCR with emphasis on prognosis prediction after radical cystectomy. Oncotarget. 2018 Mar 13;9(19):15001–15014. doi: 10.18632/oncotarget.24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sikic D., Wirtz R.M., Wach S., Dyrskjøt L., Erben P., Bolenz C. Androgen receptor mRNA expression in urothelial carcinoma of the bladder: a retrospective analysis of two independent cohorts. Transl. Oncol. 2019 Apr;12(4):661–668. doi: 10.1016/j.tranon.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martini T, Heinkele J, Mayr R, Weis C-A, Wezel F, Wahby S, et al. Predictive value of lymphangiogenesis and proliferation markers on mRNA level in urothelial carcinoma of the bladder after radical cystectomy. Urol Oncol. 2018;36(12):530.e19–530.e27. [DOI] [PubMed]
- 10.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. U. S. A. 2002 Aug 6;99(16):10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobata A. A retrospective and prospective view of glycopathology. Glycoconj. J. 1998 Apr 1;15(4):323–331. doi: 10.1023/a:1006961532182. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham S., Gerlach J.Q., Kane M., Joshi L. Glyco-biosensors: recent advances and applications for the detection of free and bound carbohydrates. Analyst. 2010 Sep 20;135(10):2471–2480. doi: 10.1039/c0an00276c. [DOI] [PubMed] [Google Scholar]
- 13.Reinders J., Sickmann A. Modificomics: posttranslational modifications beyond protein phosphorylation and glycosylation. Biomol. Eng. 2007 Jun;24(2):169–177. doi: 10.1016/j.bioeng.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Kuzmanov U., Kosanam H., Diamandis E.P. The sweet and sour of serological glycoprotein tumor biomarker quantification. BMC Med. 2013;11:31. doi: 10.1186/1741-7015-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiro R.G. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002 Apr;12(4):43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 16.Dwek R.A. Glycobiology: toward understanding the function of sugars. Chem. Rev. 1996 Jan 1;96(2):683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 17.Crocker P.R., Feizi T. Carbohydrate recognition systems: functional triads in cell—cell interactions. Curr. Opin. Struct. Biol. 1996 Oct 1;6(5):679–691. doi: 10.1016/s0959-440x(96)80036-4. [DOI] [PubMed] [Google Scholar]
- 18.Feizi T. Carbohydrate-mediated recognition systems in innate immunity. Immunol. Rev. 2000 Feb;173:79–88. doi: 10.1034/j.1600-065x.2000.917310.x. [DOI] [PubMed] [Google Scholar]
- 19.Helenius A., Aebi M. Intracellular functions of N-linked glycans. Science. 2001 Mar 23;291(5512):2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira J.A., Videira P.A., Lima L., Pereira S., Silva M., Carrascal M. Overexpression of tumour-associated carbohydrate antigen sialyl-Tn in advanced bladder tumours. Mol. Oncol. 2013 Jun;7(3):719–731. doi: 10.1016/j.molonc.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severino P.F., Silva M., Carrascal M., Malagolini N., Chiricolo M., Venturi G. Expression of sialyl-Tn sugar antigen in bladder cancer cells affects response to Bacillus Calmette Guérin (BCG) and to oxidative damage. Oncotarget. 2017 Aug 15;8(33):54506–54517. doi: 10.18632/oncotarget.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peixoto A., Fernandes E., Gaiteiro C., Lima L., Azevedo R., Soares J. Hypoxia enhances the malignant nature of bladder cancer cells and concomitantly antagonizes protein O-glycosylation extension. Oncotarget. 2016 Sep 27;7(39):63138–63157. doi: 10.18632/oncotarget.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa C., Pereira S., Lima L., Peixoto A., Fernandes E., Neves D. Abnormal protein glycosylation and activated PI3K/Akt/mTOR pathway: role in bladder cancer prognosis and targeted therapeutics. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0141253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotton S., Azevedo R., Gaiteiro C., Ferreira D., Lima L., Peixoto A. Targeted O-glycoproteomics explored increased sialylation and identified MUC16 as a poor prognosis biomarker in advanced-stage bladder tumours. Mol. Oncol. 2017 Aug;11(8):895–912. doi: 10.1002/1878-0261.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antony P., Rose M., Heidenreich A., Knüchel R., Gaisa N.T., Dahl E. Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer. 2014 Dec 2;14:901. doi: 10.1186/1471-2407-14-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinagawa T, Hoshino H, Taga M, Sakai Y, Imamura Y, Yokoyama O, et al. Clinicopathological implications to micropapillary bladder urothelial carcinoma of the presence of sialyl Lewis X-decorated mucin 1 in stroma-facing membranes. Urol Oncol. 2017;35(10):606.e17–606.e23. [DOI] [PubMed]
- 27.Numahata K., Satoh M., Handa K., Saito S., Ohyama C., Ito A. Sialosyl-Le(x) expression defines invasive and metastatic properties of bladder carcinoma. Cancer. 2002 Feb 1;94(3):673–685. doi: 10.1002/cncr.10268. [DOI] [PubMed] [Google Scholar]
- 28.Saito K., Fujii Y., Kawakami S., Hayashi T., Arisawa C., Koga F. Increased expression of sialyl-Lewis A correlates with poor survival in upper urinary tract urothelial cancer patients. Anticancer Res. 2003 Aug;23(4):3441–3446. [PubMed] [Google Scholar]
- 29.Takahashi T., Hagisawa S., Yoshikawa K., Tezuka F., Kaku M., Ohyama C. Predictive value of N-acetylglucosaminyltransferase-V for superficial bladder cancer recurrence. J. Urol. 2006 Jan;175(1):90–93. doi: 10.1016/S0022-5347(05)00044-3. (discussion 93) [DOI] [PubMed] [Google Scholar]
- 30.Ishimura H., Takahashi T., Nakagawa H., Nishimura S.-I., Arai Y., Horikawa Y. N-acetylglucosaminyltransferase V and beta1-6 branching N-linked oligosaccharides are associated with good prognosis of patients with bladder cancer. Clin. Cancer Res. 2006 Apr 15;12(8):2506–2511. doi: 10.1158/1078-0432.CCR-05-1938. [DOI] [PubMed] [Google Scholar]
- 31.Gromova I., Gromov P., Celis J.E. A novel member of the glycosyltransferase family, β3Gn-T2, highly downregulated in invasive human bladder transitional cell carcinomas. Mol. Carcinog. 2001 Oct 18;32(2):61–72. doi: 10.1002/mc.1065. [DOI] [PubMed] [Google Scholar]
- 32.Morera D.S., Hennig M.S., Talukder A., Lokeshwar S.D., Wang J., Garcia-Roig M. Hyaluronic acid family in bladder cancer: potential prognostic biomarkers and therapeutic targets. Br. J. Cancer. 2017 Nov 7;117(10):1507–1517. doi: 10.1038/bjc.2017.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haltom A.R., Jafar-Nejad H. The multiple roles of epidermal growth factor repeat O-glycans in animal development. Glycobiology. 2015 Oct;25(10):1027–1042. doi: 10.1093/glycob/cwv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokota S., Ogawara K., Kimura R., Shimizu F., Baba T., Minakawa Y. Protein O-fucosyltransferase 1: a potential diagnostic marker and therapeutic target for human oral cancer. Int. J. Oncol. 2013 Dec;43(6):1864–1870. doi: 10.3892/ijo.2013.2110. [DOI] [PubMed] [Google Scholar]
- 35.Taghavi A., Akbari M.E., Hashemi-Bahremani M., Nafissi N., Khalilnezhad A., Poorhosseini S.M. Gene expression profiling of the 8q22-24 position in human breast cancer: TSPYL5, MTDH, ATAD2 and CCNE2 genes are implicated in oncogenesis, while WISP1 and EXT1 genes may predict a risk of metastasis. Oncol. Lett. 2016 Nov;12(5):3845–3855. doi: 10.3892/ol.2016.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie H., Zhu Y., An H., Wang H., Zhu Y., Fu H. Increased B4GALT1 expression associates with adverse outcome in patients with non-metastatic clear cell renal cell carcinoma. Oncotarget. 2016 May 31;7(22):32723–32730. doi: 10.18632/oncotarget.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao H.-J., Chen Y.-J., Zuo D., Xiao M.-M., Li Y., Guo H. Quantitative proteomic analysis for high-throughput screening of differential glycoproteins in hepatocellular carcinoma serum. Cancer Biol. Med. 2015 Sep;12(3):246–254. doi: 10.7497/j.issn.2095-3941.2015.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathieu R., Lucca I., Rouprêt M., Briganti A., Shariat S.F. The prognostic role of lymphovascular invasion in urothelial carcinoma of the bladder. Nat. Rev. Urol. 2016 Aug;13(8):471–479. doi: 10.1038/nrurol.2016.126. [DOI] [PubMed] [Google Scholar]
- 39.Leng Q., Tsou J.-H., Zhan M., Jiang F. Fucosylation genes as circulating biomarkers for lung cancer. J. Cancer Res. Clin. Oncol. 2018 Nov;144(11):2109–2115. doi: 10.1007/s00432-018-2735-0. [DOI] [PubMed] [Google Scholar]
- 40.Leng Q., Lin Y., Zhan M., Jiang F. An integromic signature for lung cancer early detection. Oncotarget. 2018 May 15;9(37):24684–24692. doi: 10.18632/oncotarget.25227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan G, Tian L, Yu Y, Li F, Wang X, Li C, et al. Overexpression of Pofut1 and activated Notch1 may be associated with poor prognosis in breast cancer. Biochem Biophys Res Commun. 2017 09;491(1):104–11. [DOI] [PubMed]
- 42.Milde-Langosch K., Karn T., Schmidt M. zu Eulenburg C, Oliveira-Ferrer L, Wirtz RM, et al. Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res. Treat. 2014 Jun;145(2):295–305. doi: 10.1007/s10549-014-2949-z. [DOI] [PubMed] [Google Scholar]
- 43.Komor M.A., de Wit M., van den Berg J. Martens de Kemp SR, Delis-van Diemen PM, Bolijn AS, et al. Molecular characterization of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression. Int. J. Cancer. 2019 Aug;14:1979–1992. doi: 10.1002/ijc.32627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chabanais J., Labrousse F., Chaunavel A., Germot A., Maftah A. POFUT1 as a promising novel biomarker of colorectal cancer. Cancers. 2018 Oct;30:10(11). doi: 10.3390/cancers10110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rampias T., Vgenopoulou P., Avgeris M., Polyzos A., Stravodimos K., Valavanis C. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat. Med. 2014 Oct;20(10):1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 46.Xie H., Zhu Y., Zhang J., Liu Z., Fu H., Cao Y. B4GALT1 expression predicts prognosis and adjuvant chemotherapy benefits in muscle-invasive bladder cancer patients. BMC Cancer. 2018 May 24;18(1):590. doi: 10.1186/s12885-018-4497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daakour S., Hajingabo L.J., Kerselidou D., Devresse A., Kettmann R., Simonis N. Systematic interactome mapping of acute lymphoblastic leukemia cancer gene products reveals EXT-1 tumor suppressor as a Notch1 and FBWX7 common interactor. BMC Cancer. 2016 May 26;16:335. doi: 10.1186/s12885-016-2374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ropero S., Setien F., Espada J., Fraga M.F., Herranz M., Asp J. Epigenetic loss of the familial tumor-suppressor gene exostosin-1 (EXT1) disrupts heparan sulfate synthesis in cancer cells. Hum. Mol. Genet. 2004 Nov 15;13(22):2753–2765. doi: 10.1093/hmg/ddh298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spearman correlation of POFUT1, EXT1, and B4GALT1. Normalized GT mRNA expression data are plotted in relation to each other. The correlation coefficient was calculated using Spearman's correlation (ρ). All correlations displayed a positive relationship. There was a fairly strong correlation between POFUT1 and B4GALT1 and a moderate one between POFUT1 and EXT1 as well between EXT1 and B4GALT1. The probability that these results would occur randomly is p ≤ 0.0001.
TaqMan qRT-PCR assay parameters of designed GT primer and probes.
EXT1 and B4GALT1 survival analysis of patients with less aggressive characteristics.
Data Availability Statement
The datasets generated and analyzed for this study are available from the corresponding author on reasonable request.