Abstract
Idiopathic pulmonary fibrosis (IPF) is a fatal disease of the lower respiratory tract with restricted therapeutic options. Repetitive injury of the bronchoalveolar epithelium leads to activation of pulmonary fibroblasts, differentiation into myofibroblasts and excessive extracellular matrix (ECM) deposition resulting in aberrant wound repair. However, detailed molecular and cellular mechanisms underlying initiation and progression of fibrotic changes are still elusive. Here, we report the generation of a representative fibroblast reporter cell line (10-4ABFP) to study pathophysiological mechanisms of IPF in high throughput or high resolution in vitro live cell assays. To this end, we immortalized primary fibroblasts isolated from the distal lung of Sprague-Dawley rats. Molecular and transcriptomic characterization identified clone 10-4A as a matrix fibroblast subpopulation. Mechanical or chemical stimulation induced a reversible fibrotic state comparable to effects observed in primary isolated fibroblasts. Finally, we generated a reporter cell line (10-4ABFP) to express nuclear blue fluorescent protein (BFP) under the promotor of the myofibroblast marker alpha smooth muscle actin (Acta2) using CRISPR/Cas9 technology. We evaluated the suitability of 10-4ABFP as reporter tool in plate reader assays. In summary, the 10-4ABFP cell line provides a novel tool to study fibrotic processes in vitro to gain new insights into the cellular and molecular processes involved in fibrosis formation and propagation.
Keywords: idiopathic pulmonary fibrosis, lung, myofibroblast, TGF-β, extracellular matrix, alpha smooth muscle actin
Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive, irreversible and usually fatal lung disease with poor prognosis. IPF is characterized by subpleural fibrosis, subepithelial fibroblast foci, and microscopic honeycombing (Raghu et al., 2011; Wuyts et al., 2013; Lederer and Martinez, 2018; Sgalla et al., 2018). Various risk factors, including air pollution and smoking, have been associated with the development of IPF (Selman and Pardo, 2001; Kage and Borok, 2012; Liang et al., 2016; Kasper and Barth, 2017; Richeldi et al., 2017). In recent years, it has become clear that IPF is also strongly associated with genetic aberrations (Armanios et al., 2007; Seibold et al., 2011; Ryu et al., 2014; Stuart et al., 2015; Evans et al., 2016). Hence, an approach in understanding IPF pathogenesis is to consider it as a three-stage process: predisposition, initiation, and progression.
The conceptual model for the pathogenesis of IPF postulates that recurrent micro-injuries to the bronchoalveolar epithelium, superimposed on accelerated epithelial aging, result in aberrant wound repair. The reduced renewal capacity of bronchoalveolar stem cells, including alveolar type II cells, leads to reduced alveolar-epithelial cell proliferation, and secretion of profibrotic mediators (Selman and Pardo, 2001; Plantier et al., 2011; Kage and Borok, 2012; Ryu et al., 2014; Chambers and Mercer, 2015; Liang et al., 2016; Xu et al., 2016; Kasper and Barth, 2017; Richeldi et al., 2017; Lederer and Martinez, 2018). The main and most studied profibrotic cytokine is transforming growth factor beta 1 (TGF-β1). Several other cytokines play a major role in immune and inflammation responses for fibrosis formation, including interleukins (IL) like IL-13 (Zhu et al., 1999; Kolodsick et al., 2004; O’Reilly, 2013), IL-33 (Yanaba et al., 2011; Luzina et al., 2012, 2013) and IL-4 (Huaux et al., 2003; Saito et al., 2003), tumor necrosis factor alpha (TNFα) (Sime et al., 1998; Oikonomou et al., 2006; Epstein Shochet et al., 2017) as well as thymic stromal lymphopoietin (TSLP) (Datta et al., 2013; Lee et al., 2017). Profibrotic cytokines promote fibroblast activation and proliferation (Sime et al., 1997; Hinz, 2009; Luzina et al., 2015).
Fibroblast activation results in altered and increased ECM production, deposition, and accumulation (Liu et al., 2010; Bagnato and Harari, 2015). This causes remodeling processes of the pulmonary interstitium, forming scar tissue and modifying its mechanical properties (Booth et al., 2012; Parker et al., 2014). Scarring is accompanied by a strong increase in the tissue stiffness and an overall thickening of the alveolar septae (Horowitz and Thannickal, 2006; Hinz, 2009; Richeldi et al., 2017). During this progression phase, the matrix stiffness can increase from ∼ 0.5 to 15 kPa in healthy lung tissue to up to 100 kPa in fibrotic tissue depending on the measured lung compartment (Liu et al., 2010; Booth et al., 2012). The pathologically stiff matrix further propagates remodeling independent of epithelial cell dysfunction. In a feed-forward loop, increased matrix stiffness promotes additional differentiation of fibroblasts to myofibroblasts and matrix deposition (Hinz, 2009; Enomoto et al., 2013; Burgess et al., 2016; Tschumperlin et al., 2018). Overall, these processes result in the destruction of the overall alveolar architecture, leading to a strong impairment of lung function and eventually resulting in the death of the patient (Wuyts et al., 2013).
Studies regarding the onset and progress of IPF are mainly conducted in animal models by the application of fibrosis inducing agents like bleomycin (Gabazza et al., 2004; Xiao et al., 2006; Löfdahl et al., 2018). However, animal models of IPF do not fully recapitulate human pathophysiology (Lederer and Martinez, 2018). These models only partially mimic the events hallmarking IPF but it’s challenging to gain a deep insight into cellular and molecular processes. Hence, studies investigating molecular alterations of affected cell populations in response to pro-fibrotic stimuli are essential to get a more in-depth understanding of key processes responsible for development and progression of IPF. In this context, in vitro studies mimicking the in vivo situation hold great promise to elucidate molecular mechanisms underlying IPF initiation and progression. Sophisticated “lung on a chip” approaches recapitulating the alveolar microenvironment were developed and optimized by different groups (Huh, 2015; Stucki et al., 2018; Felder et al., 2019). These enable co-culture of differentiated alveolar epithelial and mesenchymal cells at air-liquid conditions whilst mimicking breathing motion and blood flow. However, the impact of these in vitro models depends on the use of cells representative of the in vivo situation.
In order to promote in vitro models for studying fibrotic processes, we generated an immortalized pulmonary fibroblast reporter cell line (10-4ABFP) using CRISPR/Cas9 gene-editing. 10-4ABFP cells express nuclear blue fluorescent protein (BFP) under the promotor of the myofibroblast marker alpha smooth muscle actin (Acta2). To this end, we isolated primary cells from the distal lung of Sprague-Dawley rats and immortalized them using a recently described technology (Kuehn et al., 2016). We characterized several clones and validated selected clones for suitability in fibrosis studies, directly comparing responsiveness to either mechanical or chemical stimuli to responses observed in primary isolated fibroblasts. We identified clone 10-4A as a matrix fibroblast subpopulation that can be (reversibly) induced to a fibrotic state comparable to primary isolated fibroblasts. The 10-4A clone was then used for generation of a reporter cell line (10-4ABFP) expressing nuclear BFP under the promotor of the myofibroblast marker alpha smooth muscle actin (Acta2) using CRISPR/Cas9 technology. Finally, we evaluated the use of 10-4ABFP cells as screening tool in plate reader assays. In summary, the 10-4ABFP cell line provides a novel tool to study fibrotic processes in an in vitro co-culture system at high resolution and/or high throughput and thereby enables new insights into the cellular and molecular processes involved in fibrosis formation and propagation.
Materials and Methods
Chemicals and Antibodies
Human TGF-β1 was obtained from Proteintech (cat. # HZ-1011, Manchester, United Kingdom), rat IL-13 (cat. # 1945-RL-025) and rat TNF-α (cat. # 510 RT) from R&D Systems (Minneapolis, MN, United States), rat IL-33 (cat. # ab200250) from Abcam (Cambridge, United Kingdom) and rat IL-1β (cat. # 80023-RNAE) from Sino Biological (Vienna, Austria). All other chemicals were obtained from Sigma-Aldrich GmbH (Steinheim, Germany) if not stated otherwise. The following primary and secondary antibodies were used for immunofluorescence staining: αSMA (1:200, cat. # ab5694; Abcam; RRID:AB_2223021), vimentin (1:500, cat. # ab73159; Abcam; RRID:AB_1271458), EpCAM (1:200, cat. # ab71916; Abcam, RRID:AB_1603782), ABCa3 (1:500, cat. # ab24751; Abcam, RRID:AB_448287), Aqp5 (1:200, cat. # ab92320; Abcam, RRID:AB_2049171), caveolin 1 (1:200, cat. # ab2910, Abcam, RRID:AB_303405), CD45 (1:500, cat. # 12-0461-80, Thermo Fisher Scientific, Bonn, Germany, RRID:AB_2572560).
Alexa Fluor® 488 goat anti-chicken (1:300, cat. # A11039; Thermo Fisher Scientific, RRID:AB_142924); Alexa Fluor® 568 goat anti-rabbit (1:300, cat. # A11011; Thermo Fisher Scientific, RRID:AB_143157) Alexa Fluor® 488 goat anti-mouse (1:300, cat. # A11029; Thermo Fisher Scientific, RRID:AB_138404).
Cell Isolation and Cultivation
All lung cells were isolated from 12 to 14-week-old male Sprague-Dawley rats.
Primary alveolar type II (ATII) cells were isolated according to a modified protocol described by Jansing et al. (2018) In short, rats were anesthetized with ketamine (10%) and xylazil (2%) and injected with heparin (400 IU/kg). Lungs were perfused, removed, washed with BSS-A supplemented with EGTA, BSS-A w/o EGTA and BSS-B solution. The tissue was incubated with 0.5 mg/ml elastase (Elastin Products Co., Owensville, MO, United States) for 20 min. Then, 2 mg/ml DNase were added and the tissue was minced with sharp scissors into bits of about 1 mm3. The enzymatic reaction was stopped by adding FCS (GIBCO® life technologies, Carlsbad, CA, United States) (37°C, 2 min). The digested tissue was filtered through gauze and nylon meshes (mesh sizes: 100, 40, and 10 μm) and the cell filtrate was centrifuged for 8 min at 130 rcf. For further cell separation, density gradient centrifugation was applied by mixing the cells in OptiPrepTM Density Gradient medium (1.077 g/mL) diluted in BSS-B. The cells were centrifuged for 20 min at 200 rcf. The layer containing ATII cells was collected and supplemented with BSS-B to a total volume of 40 ml. Cells were centrifuged at 130 rcf for 8 min, resuspended in MucilAirTM cell culture medium and 1 × 106 cells/cm2 were seeded apically on 0.4 μm transparent Transwell® filter inserts (Sarstedt, Nümbrecht, Germany). Purity of the ATII cells (>90%) was determined by staining with 0.4 μM Lyso Tracker Red DND 99 for 10 min (Thermo Fisher Scientific, Waltham, MA, United States) and more specifically with an ABCa3 staining. The amount of LTR positive cells was determined using a Countess II FL Automated Cell Counter (Thermo Fisher Scientific, Waltham, MA, United States).
Primary lung fibroblasts and distal lung cells were isolated according to the method of Dobbs et al. (1986) with minor modifications as previously described (Miklavc et al., 2010).
For primary isolated fibroblasts further modifications were applied:
Anesthesia, lung perfusion and removal followed the protocol described for isolation of AT2 cells. The tissue was then incubated with 0.5 mg/ml elastase (Elastin Products Co., Owensville, MO, United States) and 0.05 mg/ml trypsin at 37°C for 30 min. 2 mg/ml DNase were added, the enzymatic reaction was stopped by FCS and the digested tissue was filtered through gauze and nylon meshes (mesh sizes: 100, 70, and 40 μm). For purification of primary fibroblasts, cell suspensions were depleted of leukocytes using anti-CD45 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) before fibroblasts were isolated using anti- CD90.1 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to manufacturer’s instructions. Cells were seeded on polydimethylsiloxane (PDMS) gels or plastic substrate in MucilAirTM culture medium containing 25.6 μg/ml Gentamicin ± 5 ng/mL TGF-β1 at a density of 1 to 5 × 105 cells/cm2. Cells were cultured at 37°C, 5% CO2 and 95% humidity for up to 14 days. Culture media were changed every 2 days, with TGF-β1 being added freshly to the medium in corresponding experiments.
Generation of Immortalized Cell Lines
Immortalization was performed by InSCREENeX (Braunschweig, Germany) as previously described (Lipps et al., 2018). In short: Immortalization genes were incorporated with third generation self-inactivating lentiviral vectors. Gene expression is controlled by an internal SV 40 promoter. Integration of the transgenes was verified by PCR and subsequent gel electrophoresis.
Overall, 15 immortalized cell clones were generated, all displaying characteristics of different cell populations of the distal lung. The incorporated genes used for the immortalization process for the cell line 10-4A are TAg, ID2, ID3, Rex, Nanog, and E7.
10-4A cells were maintained in the chemically defined, standardized, cell culture medium MucilAirTM (Epithelix, Genève, Switzerland) containing 25.6 μg/ml Gentamicin (Thermo Fisher Scientific) at 37°C, 5% CO2 and 95% humidity. Cells were detached upon reaching 80% confluence using TrypLE (Thermo Fisher Scientific), centrifuged, resuspended in cell culture medium ± 5 ng/mL TGF-β1 and seeded on PDMS gels or plastic dishes at a density of 0.5 × 103 to 40 × 103 cells/cm2. Culture media were changed every 2 days, with TGF-β1 being added freshly to the medium in corresponding experiments. Cells from passage 11 to 25 were used for all experiments.
PDMS Gel Preparation and Coating of Cell Culture Dishes
PDMS gels were prepared with the Sylgard 527 Silicon Dielectric Gel Kit (Dow Europe GmbH, Wiesbaden, Germany) as previously described (Palchesko et al., 2012) with minor modifications. In brief, component A and B were thoroughly mixed in a 1:1 ratio and added to 24 Well Culture Plates (Sarstedt, Nümbrecht, Germany) or ibiTreat μSlide 8 well (ibidi GmbH, Gräfelfing, Germany), respectively. Culture containers were kept under vacuum for 2 h to remove potential air inclusions and then incubated at room temperature for 48 h for polymerization. Fully polymerized PDMS gels were sterilized in a UV Crosslinker (GE Healthcare Europe GmbH, Freiburg im Breisgau, Germany) for 30 min and then coated with a 0.01% w/v polydopamine solution [50 mM Tris–HCl, pH = 8.5, 0.01% (w/v) Dopamine Hydrochloride] for 1 h and a 38 μg/ml rat tail collagen I solution (Advanced BioMatrix Inc., San Diego, CA, United States), diluted in Dulbecco PBS (Biochrom, Berlin, Germany; pH 7.4) over night at 37°C, respectively.
Identical coating conditions were used for culture plastic dishes w/o PDMS to ensure comparability.
RNA Isolation, cDNA Synthesis and qPCR
Total RNA was isolated using the my-Budget RNA Mini Kit (Bio-Budget Technologies GmbH, Krefeld, Germany) with an additional DNA removal step using the RNase free DNase Set (QIAGEN GmbH, Hilden, Germany). cDNA synthesis was performed using the SuperScript® VILOTM cDNA Synthesis Kit (Thermo Fisher Scientific) and cDNA was diluted in a 1:3 ratio with DEPC treated H2O (Carl Roth, Karlsruhe, Germany) prior qPCR.
Amplification was performed on a StepOnePlus qPCR cycle (Applied Biosystems, Foster City, CA, United States) using EvaGreen QPCR Mix II (Bio-Budget Technologies). The following QuantiTect® Primer assays (QIAGEN GmbH, Hilden Germany) were used: Rn_Plin2_2_SG (QT01624329), Rn_Acta2_1_SG (QT01615901), Rn_Col1a1_ 1_SG (QT01081059), Rn_Sftpc_1_SG (QT00179368), Rn_ ABCa3_1_SG (QT01587936), Rn_Vim_1_SG (QT00178724), Rn_HOPX_1_SG (QT00182693), Rn_Cav1_1_SG (QT00 398181), and Rn_Hmbs_1_SG (QT00179123). The relative quantification of mRNA expression was performed according to the method of Pfaffl (2001).
Illumina Library Preparation and Sequencing
The Sequencing library preparation has been done using 200 ng of total RNA input with the TruSeq RNA Sample Prep Kit v2-Set B (RS-122–2002, Illumina Inc., San Diego, CA, United States) producing a 275 bp fragment including adapters in average size. In the final step before sequencing, eight individual libraries were normalized and pooled together using the adapter indices supplied by the manufacturer. Pooled libraries have then been clustered on the cBot Instrument from Illumina using the TruSeq SR Cluster Kit v3 – cBot – HS (GD-401–3001, Illumina Inc., San Diego, CA, United States) sequencing was then performed as 50 bp, single reads and 7 bases index read on an Illumina HiSeq2000 instrument using the TruSeq SBS Kit HS- v3 (50-cycle) (FC-401–3002, Illumina Inc., San Diego, CA, United States).
mRNA-Seq Bioinformatics Pipeline
RNA-Seq reads were aligned to the rat genome using the STAR Aligner v2.5.2a (Dobin et al., 2013) with the Ensembl 84 reference genome1. Sequenced read quality was checked with FastQC v0.11.22 and alignment quality metrics were calculated using the RNASeQC v1.18 (Deluca et al., 2012). Following read alignment, duplication rates of the RNA-Seq samples were computed with bamUtil v1.0.11 to mark duplicate reads and the dupRadar v1.4 Bioconductor R package for assessment (Sayols et al., 2016). The gene expression profiles were quantified using Cufflinks software version 2.2.1 (Trapnell et al., 2013) to get the Reads Per Kilobase of transcript per Million mapped reads (RPKM) as well as read counts from the feature counts software package (Liao et al., 2014). The matrix of read counts and the design file were imported to R, normalization factors calculated using trimmed mean of M-values (TMM) and subsequently voom normalized, before subjected to downstream descriptive statistics analysis.
Western Blot
Cells were washed twice with PBS, collected with RIPA Buffer (Sigma-Aldrich) and sonicated (Sonifier 250, Branson Ultrasonics Corporation, Danbury, CT, United States) prior loading on gels. The protein concentration was determined by Pierce BCA Protein Assay (Thermo Fisher Scientific). Protein Loading Buffer and NuPAGE reducing agent (Thermo Fisher Scientific) were added in a 1:5 and 1:10 ratio, respectively. Samples were incubated at 70°C for 10 min, separated by SDS-PAGE and blotted on a nitrocellulose membrane. Immunodetection of αSMA and HSP90 was performed using Anti-alpha smooth muscle actin (1:200, cat # ab5694; Abcam, RRID:AB_2223021) and HSP90 α/β antibodies (F-8) (1:500, cat. # sc-13119; Santa Cruz Biotechnology, Dallas, TX, United States, RRID:AB_675659) in combination with fluorescent labeled secondary antibodies [1:20,000; IRDye® 800CW Donkey anti-Rabbit (cat. # 926-32213, RRID:AB_621848), IRDye® 680RD Donkey anti-Mouse (cat. # 926-68072, RRID:AB_10953628)] diluted in Intercept® Blocking Buffer (cat. # 927-60001) (all from LI-COR Biosciences, Lincoln, NE, United States). Primary antibodies were incubated over night at 4°C, secondary antibodies for 1 h at RT, respectively. Membranes were analyzed with the Odyssey Fc Imaging System (LI-COR Biosciences).
Immunofluorescence
For immunofluorescence staining, cells were washed with DPBS (Biochrom, Berlin, Germany; pH 7.4) and fixed in a 4% paraformaldehyde solution (dissolved in DPBS) for 10 min followed by a 1 min incubation in ice-cold 99.8% MeOH. Cells were permeabilized in a 0.2% w/v saponin solution (dissolved in DPBS) containing 10% FBS (Thermo Fisher Scientific) and 50 mM HEPES. Subsequently, cells were stained for 1 h with primary antibodies, washed twice with DPBS and stained with secondary antibodies diluted in saponin solution for 1 h. Images were taken on an iMIC digital microscope (FEI Munich GmbH, Gräfelfing, Germany) with an Olympus UApo/340 40x/1.35 Oil Iris, Infinity/0.17 lens (Olympus Europa SE & Co. KG, Hamburg, Germany) and the corresponding software (Live Aquisition v2.6.0.14).
Generation of the Acta2-BFP Reporter Cell Line
CRISPR/Cas9 dependent gene editing of 10-4A cells was performed according to the method described in Ran et al. (2013).
A 20 bp single guide RNA (sgRNA) (AAACAGGAGT ATGACGAAGC), binding at the end of the coding region of the Acta2 gene was designed by using the R&D Benchling software3. The sgRNA was cloned into SpCas9(BB)-2A-GFP (Addgene plasmid ID: 48138). A donor vector was designed to allow for in-frame fusion of a T2A-BFP-NLS (BFP from Evrogen, Heidelberg, Germany) cassette at the 3′ end of the Acta2 gene. Primers used were listed in Table 1. DNA sequences flanking the sgRNA cutting site at the 5′and 3′ end were amplified from genomic DNA isolated from 10-4A cells (isolated with DNeasy Blood ans Tissue Kit, QIAGEN GmbH), and subcloned into the targeting pGEX-6P-1 Vector (GE Healthcare, 28-9546-48) using the In-Fusion kit (Clontech, Mountain View, CA, United States). 10-4A cells were co-transfected with the sgRNA/SpCas9(BB)-2A-GFP plasmid and the pGEX-6P-1 donor plasmid in a ratio of 2:1 using Lipofectamine LTX (Ratio: LTX Reagent: PLUSTM Reagent, 1:1) (Thermo Fisher Scientific). The efficiency of sgRNA/Cas9-mediated integration of the BFP was evaluated after addition of 5 ng/ml TGF-β1 via fluorescence microscopy. 72 h after transfection, isolation of clonal cells was achieved by fluorescent activated cell sorting (FACS). Cells were selected for GFP and BFP expression using the BD FACSAriaTM III (Becton Dickinson GmbH) with the corresponding BD FACSDivaTM v6.1.3 software. Single cells were seeded in 96-well plates containing cell culture medium.
TABLE 1.
Primers used for cloning of T2A-BFP-NLS and homology arms into donor vector pGEX-6P-1.
| Primer Name | Sequence 5′ → 3′ |
| pGEX-6P-1_fwd | tgtggaattgtgagcggataac |
| pGEX-6P-1_rev | cattatacgcgatgattaattg |
| T2A-BFP-NLS_fwd | tcggctcgtataatggagggcagaggaag |
| tctgct | |
| T2A-BFP-NLS_rev | gctcacaattccacattatacctttctcttcttt |
| tttgga | |
| Acta2_right_homologous_right_arm_fwd | tcggctcgtataatgccctctgtgttgggcag |
| Acta2_right_homologous_right_arm_rev | acttcctctgccctcccacatctgctgga |
| aggtaga | |
| Acta2_left_homologous_right_arm_fwd | caggaaacagtattcgtcacgcccccaccct |
| Acta2_left_homologous_right_arm_rev | aacttccagatccgatgtaaacacatgtata |
| attgtttttacttatccggtcac | |
| pGEX-6P-1_T2A-BFP-NLS_right_arm_rev | cattatacgcgatgattaattgtcaacag |
| pGEX-6P-1_T2A-BFP-NLS_right_arm_fw | gagggcagaggaagtctgctaac |
| pGEX-6P-1_T2A-BFP-NLS_left_arm_fw | tcggatctggaagttctgttccagg |
| pGEX-6P-1_T2A-BFP-NLS_left_arm_rev | gaatactgtttcctgtgtgaaattgttatccg |
Functional Testing of BFP Integration
The DNA of the cell clones was extracted as described above. The region of interest was amplified by PCR and the respective products (WT: 1000 bp, with BFP insert: 1880 bp) were verified by Sanger Sequencing (Eurofins).
Additionally, in order to verify the functionality of the Acta2 coupled BFP reporter system, cells were seeded in plastic or 5 kPa PDMS coated ibiTreat μSlide 8 well at a density of 25000/cm2 in MucilAir ± 5 ng/ml TGF-β1. Pictures were taken with an iMIC Digital Microscope (FEI Munich GmbH). All images were obtained using an Olympus Objective UApo/340 40x/1.35 Oil ∞/0.17 and a 405 nm excitation filter for BFP.
Microplate Reader Assay
Cells were seeded in PDMS coated 96-well plates (Sarstedt, Nümbrecht, Germany). Respective growth factors were added 24 h post-seeding and the BFP signal was measured at indicated time points. For fluorescence measurements, cells were trypsinized, transferred to a collagen (Advanced BioMatrix Inc.) coated black 96-well plate (Sarstedt) and let adhere for 3 h. Cells were stained with Calcein AM (Thermo Fisher Scientific) for 30 min in bath solution (in mM: 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 5 glucose, and 10 HEPES; pH 7.4), washed twice with PBS w/o Ca2+/Mg2+ and maintained in 200 μl bath solution during analysis of BFP signal with the plate reader (Tecan, Salzburg, Austria). Excitation wavelength were 385 nm and 485 nm and BFP and Calcein emissions were collected at 445 nm and 535 nm, respectively. BFP and Calcein signals were background subtracted and the BFP signal was normalized to the Calcein signal to adjust for cell number.
Statistical Analysis
GraphPad Prims7 software (GraphPad, La Jolla, CA, United States) was used for statistical analysis, curve fitting and data representation. Respective tests are given within the figure legend. Data are represented as means ± SEM unless stated otherwise. Statistical significance was determined using the non-parametric Mann–Whitney-U test for comparison of two independent samples at the same time point. The number of experiments (N) indicates individual animals for primary fibroblasts and cells from varying passages for immortalized fibroblasts. Data was considered significant if the p value was < 0.05 and is indicated with an asterisk. Statistical significance is indicated in the graphs as follows: p-values < 0.05: ∗, p-values < 0.01: ∗∗, p-values < 0.001: ∗∗∗.
Results
10-4A Cells Resemble Matrix Fibroblasts but Not Myo-/Lipofibroblasts
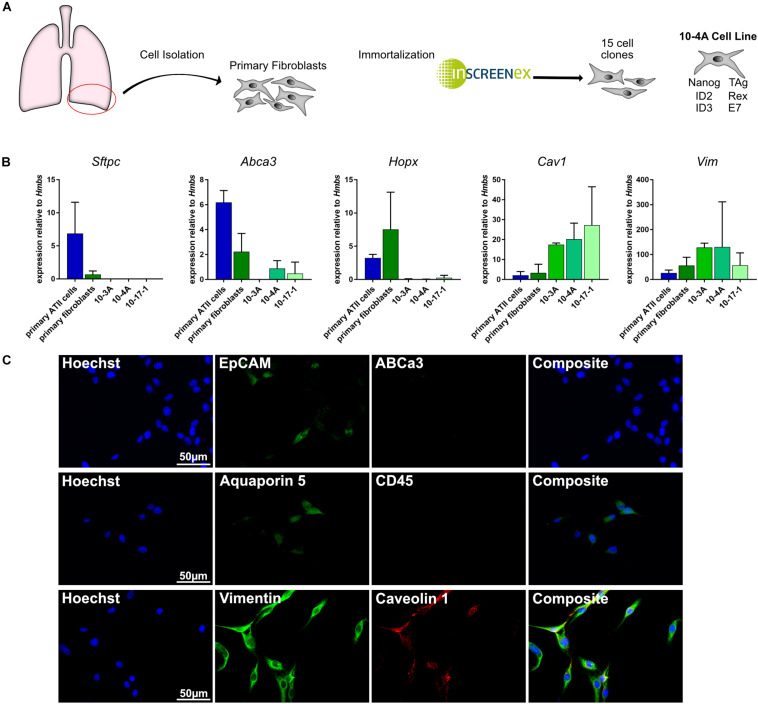
Isolation and immortalization of primary cells from the distal lung yielded 15 individual cell clones (Figure 1A). Subsequently, cell clones were analyzed for phenotypic expression patterns resembling primary epithelial and mesenchymal cells. Expression of marker genes for alveolar type II (ATII) [Abca3, Sftpc (Beers et al., 2017)], alveolar type I (ATI) [Hopx, Aqp5, Cav1 (McElroy and Kasper, 2004; Beers et al., 2017)], pan-epithelial [Epcam (Hasegawa et al., 2017)], leukocyte [CD45 (Barletta et al., 2012)], and mesenchymal [Vim (Cheng et al., 2016)] cells was analyzed on the gene and protein level. Both, semi-quantitative RT-PCR and immunofluorescence data identified the presence of mesenchymal and absence of epithelial and leukocyte cell markers in several clones (Figures 1B,C). Taking into consideration that Cav1 is also expressed in lung fibroblasts (Xiao et al., 2006) the data indicate a fibroblast phenotype of all investigated cell clones.
FIGURE 1.
Gene and protein expression pattern of the immortalized cell lines. (A) Schematic representation of the immortalization process. Right: Genes incorporated in 10-4A cells for immortalization (B) Semi-quantitative RT-PCR of ATII cell (Sftpc, Abca3), ATI cell (Hopx, Cav1), and mesenchymal cell (Vim) marker genes. N = 5 (C) Immunofluorescent staining of 10-4A cells for expression of epithelial cell (EpCAM), alveolar type II cell (ABCa3), alveolar type I cell (aquaporin 5, caveolin 1), leukocyte (CD45), and mesenchymal cell (vimentin) marker expression. 10-4A cells predominantly express mesenchymal marker vimentin and caveolin-1, that is also expressed in lung fibroblasts. Scale bar = 50 μm.
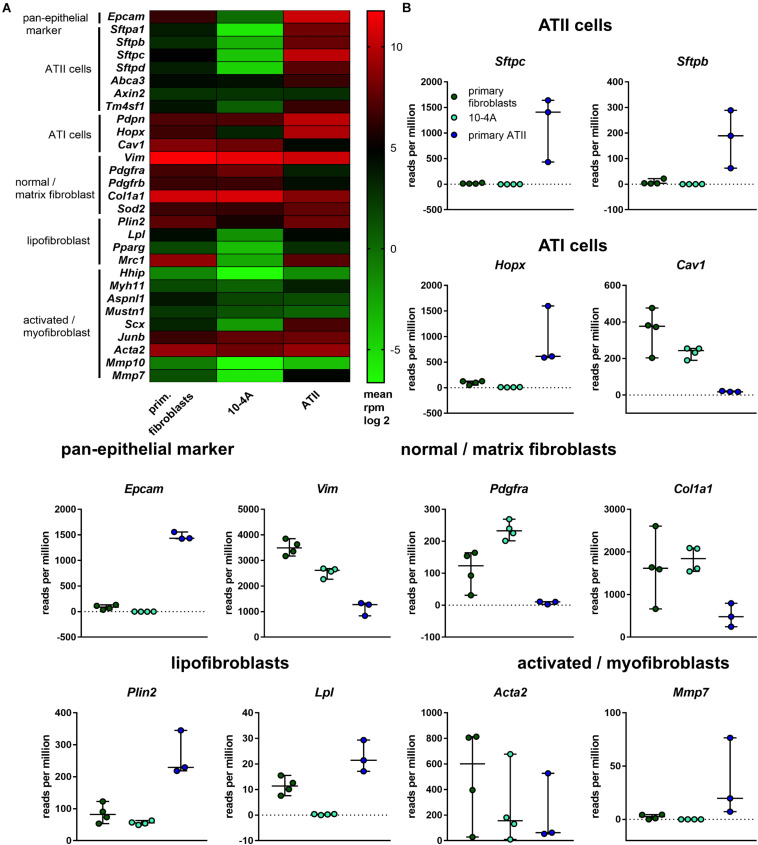
Based on gene expression analysis and protein localization, clone 10-4A was selected for further analysis. In-depth characterization was performed by transcriptomic analysis. Gene expression in 10-4A cells was also compared to expression in healthy primary distal lung fibroblasts and primary ATII cells. The 10-4A cell line exhibits high expression of matrix fibroblast marker genes, in particular Col1a1 and Vim, and low expression of myofibroblast marker genes. Lipofibroblast marker gene expression was low in 10-4A cells when compared to primary fibroblasts (Figure 2). Expression of specific pan- and alveolar epithelial marker genes was very low. Together these data suggest that 10-4A cells exhibit a matrix fibroblast phenotype (Zepp et al., 2017; Xie et al., 2018).
FIGURE 2.
Transcriptomic profile of 10-4A compared to freshly isolated rat fibroblasts and ATII cells. (A) Heat map of selected pan-epithelial, ATII, ATI and different fibroblast subtype marker genes. Values are given as log2-fold change of the mean reads per million base pairs. 10-4A and primary fibroblasts, N = 4, primary ATII cells, N = 3. (B) Detailed presentation of rpm values obtained for selected marker genes.
Interestingly, 10-4A exhibited high expression of Pdgfrα, which is associated with the capability of myofibroblast differentiation (Li et al., 2018).
Increased Substrate Stiffness Exhibits of Myofibroblast Characteristics in 10-4A Cells
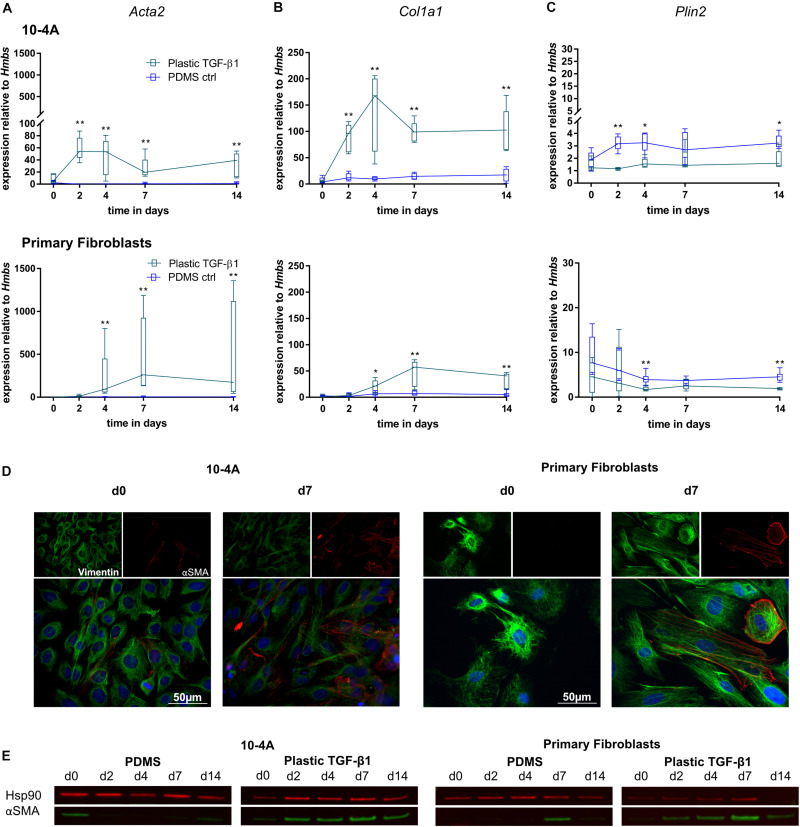
The high expression of Pdgfrα suggested that these cells may constitute a model to study activation/differentiation of fibroblast cells observed in pulmonary fibrosis. To test whether 10-4A cells resemble a suitable surrogate cell model for in vitro fibrosis studies, we first investigated the responsiveness of 10-4A cells to mechanical stimuli (Hinz, 2009). We analyzed expression of myofibroblast marker genes in response to changes in substrate stiffness. Results in 10-4A cells were compared to effects on freshly isolated primary fibroblasts.
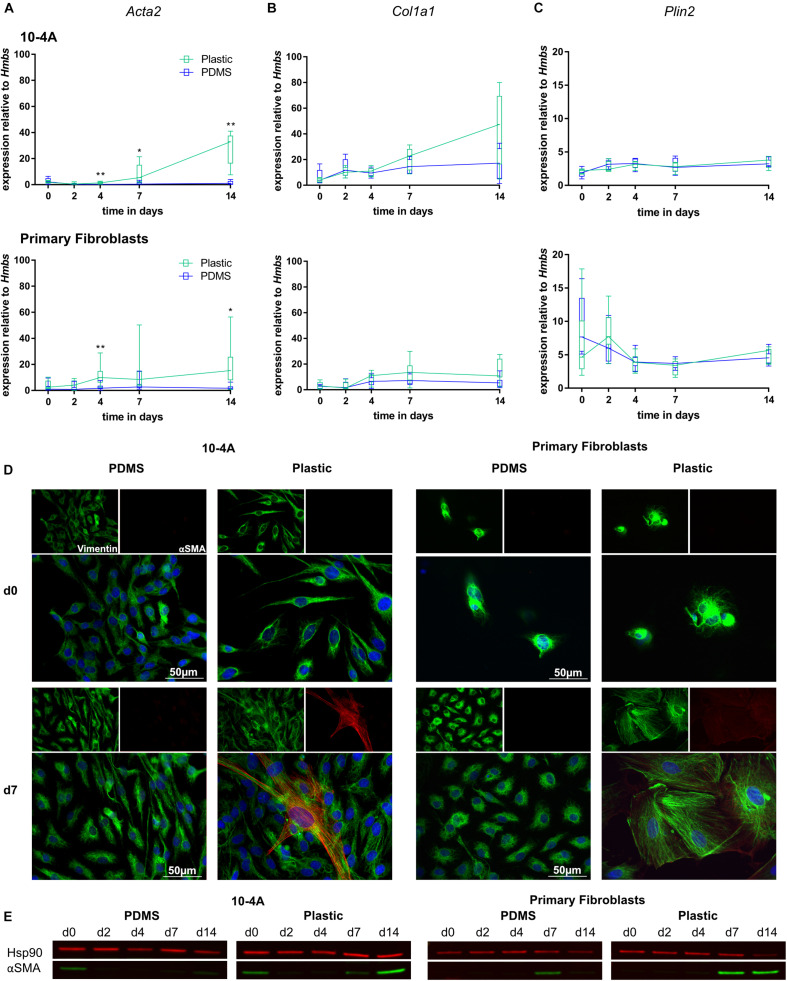
Expression of myofibroblast marker Acta2 and ECM component Col1a1 were unchanged over a 14 days period in 10-4A and primary fibroblasts when maintained on soft PDMS gels with physiological stiffness (Young’s Modulus of 5 kPa) (Palchesko et al., 2012). In line, with maintenance of a quiescent phenotype, Plin2, a marker for lipofibroblasts, did not significantly change (Figures 3A–C). In some cases, a faint alpha smooth muscle actin (αSMA) signal (the product of Acta2) was detected in Western Blots at day 0 in 10-4A.
FIGURE 3.
Mechanical stimulation of 10-4A cells and primary fibroblasts. Semi-quantitative RT-PCR analysis of Acta2 (A), Col1a1 (B), and Plin2 (C) expression in 10-4A cells (top graph) and primary fibroblasts (bottom graph) maintained on soft PDMS or stiff plastic substrate, respectively. Data are expressed as fold expression of the housekeeping gene hydroxymethylbilane synthase (Hmbs). Primary fibroblast data were obtained from seven different animals, 10-4A data from five different passages. Statistical significance for the respective time points was tested with the non-parametric Mann–Whitney-U-Test. Statistical significance is indicated as follows: p-values < 0.05: *, p-values < 0.01: **. Box plots show data as median values, the boxes represent percentiles, the whiskers indicate the minimum/maximum. (D) Immunofluorescence staining of 10-4A cells (left) or primary fibroblasts either seeded on PDMS or plastic directly after adherence (d0) or 7 days post-seeding. Cells were stained for the mesenchymal marker vimentin (green) and the pro-fibrotic protein αSMA (red). Scale bar = 50 μm. (E) Western Blot for αSMA in 10-4A cells and primary fibroblasts cultured over 14 days on soft (PDMS) and stiff (Plastic) matrices, respectively. HSP90 was used as loading control.
In contrast, culture on rigid plastic substrate resulted in significantly increased Acta2 expression in 10-4A (day 4, p = 0.008; day 7, p = 0.03; day 14, p = 0.008) and primary fibroblasts (day 4, p = 0.007; day 7, p = 0.053; day 14, p = 0.03) (Figure 3A). Consistently, αSMA stress fiber formation was detected from day 7 onward, in immunofluorescence experiments and was confirmed by Western Blot (Figures 3D,E). This likely originates from αSMA expressed during cell culture in plastic culture flasks that had not been degraded by the time samples were collected (approx. 6 h after seeding on PDMs substrate). Expression of Col1a1 and Plin2 were not affected by stiff substrate within the 14 days culture period, suggesting that the time course might not be sufficiently long enough for full differentiation and activation of fibroblasts. A weak αSMA signal was detected in Western blots for day 0 samples from 10-4A cells.
TGF-β1 Induces a Transient Myofibroblast Phenotype in 10-4A Cells
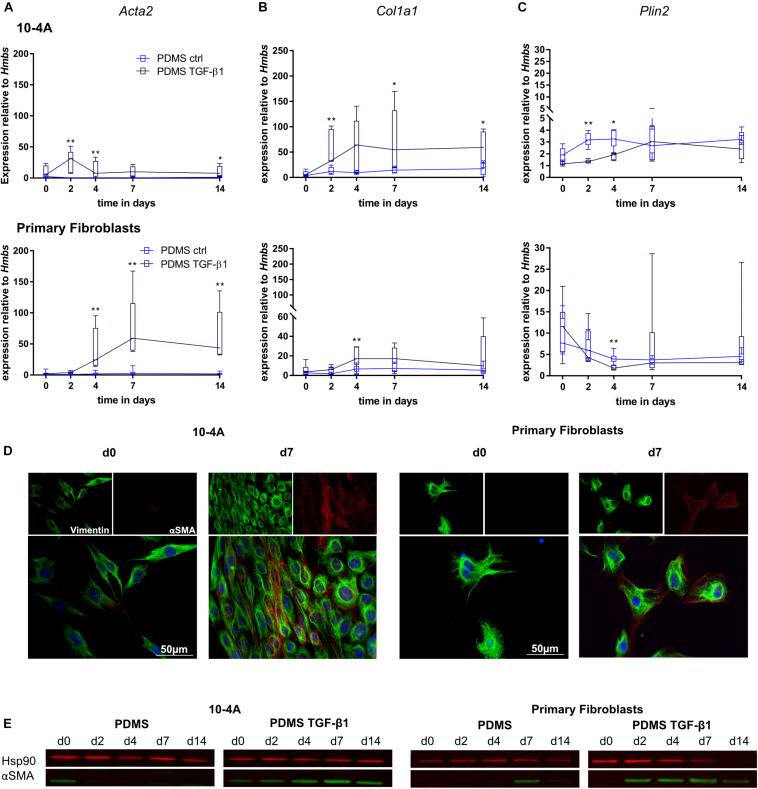
To further characterize the response of 10-4A cells to fibrotic stimuli, we stimulated the cells with the potent profibrotic cytokine TGF-β1.
Transforming growth factor beta 1 (5 ng/ml) treatment resulted in significantly increased Acta2 gene expression after 2 days in 10-4A cells (day 2, p = 0.008; day 4, p = 0.008; day 7, p = 0.057; day 14, p = 0.03) and after 4 days in primary fibroblasts (day 4, p = 0.003; day 7, p = 0.003; day 14, p = 0.003) (Figure 4A). In line, αSMA stress fiber formation was more prominent from day 2 post-seeding onward in 10-4A cells as well as primary fibroblasts (Figures 4D,E). Interestingly, the TGF-β1-induced increase in Acta2 expression followed a transient course in the 10-4A cells, peaking at day 2 after TGF-β1 addition and returning to baseline at day 4–5. A similar trend was noticeable in primary fibroblast post day 7. In contrast to mechanical stimulation, TGF-β1 administration also resulted in significantly increased Col1a1 (day 2, p = 0.008, day 7, p = 0.03, day 14, p = 0.03) and a reduced Plin2 (day 2, p = 0.008, day 4, p = 0.02) gene expression in 10-4A cells (Figures 4B,C) and primary fibroblasts (Col1a1 day 4, p = 0.003, Plin2, day 4, p = 0.003). Together these data suggest that TGF-β1 administration triggers changes observed in fibrosis in 10-4A cells and primary fibroblasts, respectively. However, the effect was not sustained over a 14-day time course, despite constant exposure to TGF-β1.
FIGURE 4.
Chemical stimulation of 10-4A cells and primary fibroblasts with TGF-β1 on a physiological substrate. Semi-quantitative RT-PCR of Acta2 (A), Col1a1 (B), and Plin2 (C) expression in 10-4A cells (top graph) and primary fibroblasts (bottom graph) cultured in the presence or absence of 5 ng/ml TGF-β1 on a soft PDMS substrate, respectively. Data are expressed as fold expression of housekeeping gene Hmbs. Primary fibroblast data were obtained from 5 different animals, 10-4A data from five independent passages. The respective time points were tested with the non-parametric Mann–Whitney-U-Test. Statistical significance is indicated as follows: p-values < 0.05: *, p-values < 0.01: **. Box plots show data as median values, the boxes represent percentiles, the whiskers indicate the minimum/maximum. (D) Immunofluorescence staining of 10-4A cells (left) or primary fibroblasts seeded on PDMS with administration of 5 ng/ml TGF-β1 directly after adherence (d0) or 7 days post-seeding. Cells were stained for the mesenchymal marker vimentin (green) and the pro-fibrotic protein αSMA (red). Scale bar = 50 μm. (E) Western Blot for αSMA in 10-4A cells and primary fibroblasts cultured over 14 days on PDMS substrate in the presence or absence of TGF-β1, respectively. HSP90 was used as loading control.
Combination of Stiff Matrix and TGF-β1 Stimulation Leads to Strong and Persistent Expression of Myofibroblast Markers in 10-4A Cells
In pulmonary fibrosis mechanical and chemical cues act simultaneously. Therefore, we investigated the combination of mechanical and chemical stimulation seeding cells on plastic substrate and stimulated them with TGF-β1.
The combination of mechanical and chemical stimulation resulted in significantly increased Acta2 gene expression in 10-4A cells from day 2 onward (day 2, p = 0.008, day 4, p = 0.008, day 7, p = 0.008, day 14, p = 0.008) and 4 days onward in primary fibroblasts (day 4, p = 0.003, day 7, p = 0.003, day 14, p = 0.003) (Figure 5A). This was also observed on the protein level (Figures 5D,E). Similar changes were observed for Col1a1 (Figure 5B) (10-4A: day 2, p = 0.008, day 4, p = 0.008, day 7, p = 0.008, day 14, p = 0.008; primary fibroblasts: day 4, p = 0.05, day 7, p = 0.003, day 14, p = 0.003). In line, Plin2 expression (Figure 5C) was decreased in both cell types after 4 and 2 days, respectively (10-4A: day 2, p = 0.008, day 4, p = 0.02, day 14, p = 0.03; primary fibroblasts: day 4, p = 0.005, day 14, p = 0.003).
FIGURE 5.
Combination of chemical and mechanical stimulation of 10-4A cells and primary fibroblasts with TGF-β1 on a physiological substrate. Semi-quantitative RT-PCR of Acta2 (A), Col1a1 (B), and Plin2 (C) expression in 10-4A cells (top graph) and primary fibroblasts (bottom graph) cultured on either soft substrate (PDMS) or in the presence of 5 ng/ml TGF-β1 stiff substrate (Plastic), respectively. Data are expressed as fold expression of housekeeping gene Hmbs. Primary fibroblast data were obtained from five different animals, 10-4A data from five independent cell culture experiments. The respective time points were tested with the non-parametric Mann–Whitney-U-Test. Statistical significance is indicated as follows: p-values < 0.05: *, p-values < 0.01: **. Box plots show data as median values, the boxes represent percentiles, the whiskers indicate the minimum/maximum. (D) Immunofluorescence staining of 10-4A cells (left) or primary fibroblasts seeded on PDMS with administration of 5 ng/ml TGF-β1 directly after adherence (d0) or 7 days post-seeding. Cells were stained for the mesenchymal marker Vimentin (green) and the pro-fibrotic protein αSMA (red). Scale bar = 50 μm. (E) Western Blot for αSMA in 10-4A cells and primary fibroblasts cultured over 14 days on PDMS substrate or on Plastic in the presence of TGF-β1, respectively. HSP90 was used as loading control.
Overall, the combination of mechanical and chemical stimulation induced a robust and persistent expression of myofibroblast markers in 10-4A cells and primary fibroblasts. Interestingly the transient effect observed by TGF-β1 treatment alone was counteracted by increased substrate stiffness.
Generation of a 10-4A Reporter Cell Line to Monitor Fibroblast to Myofibroblast Differentiation in Live Cell in vitro Assays
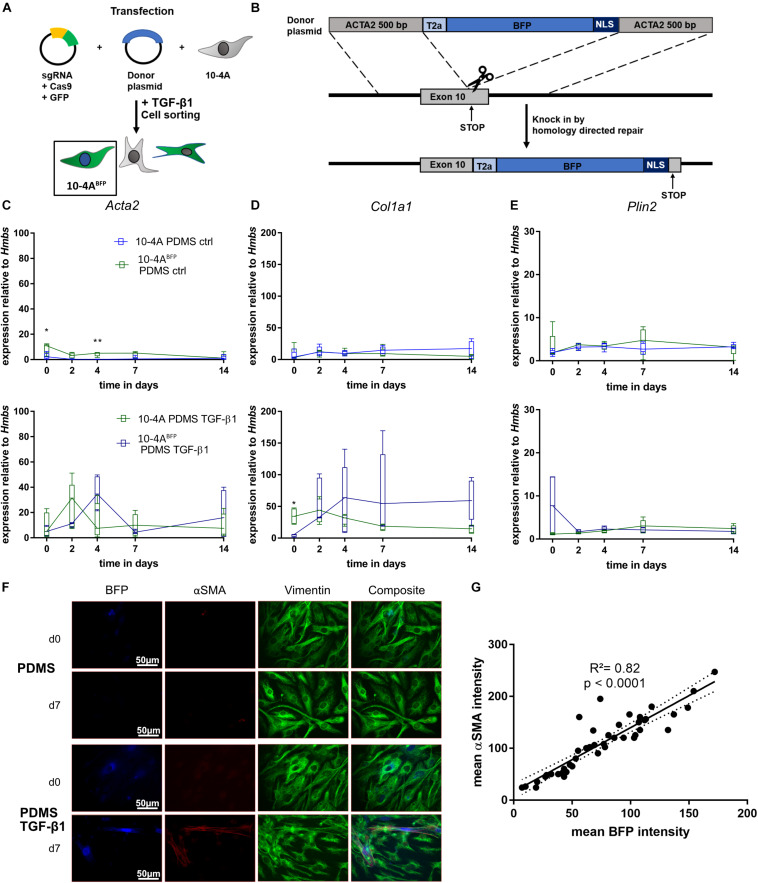
In order to track fibroblast to myofibroblast differentiation within living cells, we designed a reporter system targeting the Acta2 gene locus in 10-4A cells by generating a BFP-reporter of Acta2 expression. We inserted a T2A-BFP-NLS sequence at the end of the Acta2 CDS in 10-4A cells (10-4ABFP). The correct integration of the BFP sequence was verified via PCR and Sanger sequencing. Induction of myofibroblast differentiation (TGF-β1) resulted in a nuclear BFP signal in 10-4ABFP cells (Figures 6A,B).
FIGURE 6.
Generation and verification of an Acta2 coupled BFP reporter cell line. (A,B) Schematic representation of 10-4A cell transfection. Cells were transfected with a plasmid containing Cas9 and the sgRNA binding at the desired gene (exon 10) and a donor plasmid. The donor plasmid contained a BFP flanked by a self-cleaving T2A cassette, a nuclear localization sequence and two 500 bp homologous arms of the sgRNA cutting site. After cell transfection, Acta2 gene expression was induced by addition of 5 ng/ml TGF- β1 and cells were FACS sorted for BFP. Semi-quantitative RT-PCR analysis of Acta2 (C), Col1a1 (D), and Plin2 (E) expression in 10-4A and 10-4ABFP cells with and without administration of 5 ng/ml TGF-β1 on a soft PDMS substrate, respectively. Values are means from five individual culture experiments. The respective time points were tested with the non-parametric Mann–Whitney-U-Test. Statistical significance is indicated as follows: p-values < 0.05: *, p-values < 0.01: **. Box plots show data as median values, the boxes represent percentiles, the whiskers indicate the minimum/maximum. (F) Immunofluorescence staining of 10-4ABFP cells seeded on PDMS with and without administration of 5 ng/ml TGF-β1 directly after adherence (d0) or 7 days post-seeding. Cells were monitored for BFP and stained for the mesenchymal marker vimentin (green) and the pro-fibrotic protein αSMA (red). Scale bar = 50 μm. (G) Correlation of the BFP and αSMA signal intensity within individual cells 7 days post seeding. N = 48 cells. A linear regression line and the corresponding indicator R and p-value show the linear dependency of BFP and αSMA signal.
To exclude changes in Acta2 gene expression arising from genetic modifications, we compared the 10-4ABFP cell line to wildtype 10-4A cells during quiescence (on PDMS, soft matrix) and after TGF-β1 stimulation, respectively. 10-4ABFP cells showed similar responses to PDMS and TGF-β1 stimulation as 10-4A cells when analyzing Acta2, Col1a1, and Plin2 gene expression (Figures 6C–E), indicating no adverse effects of T2A-BFP-NLS cassette integration.
Next, we tested on the protein level, whether the BFP signal correlated with αSMA expression. TGF-β1 treatment resulted in double-positive cells, whereas cells cultured on soft substrate without TGF-β1 were negative for BFP and αSMA (Figure 6F). Correlating the αSMA and BFP signal within individual cells, confirmed a linear correlation between BFP intensity and αSMA expression (R2 = 0.82, p < 0.0001) (Figure 6G).
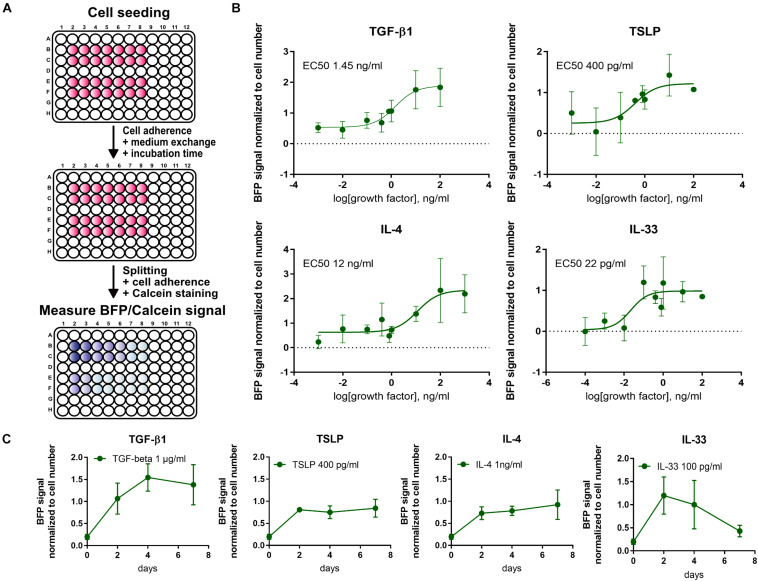
For a high throughput evaluation of fibrotic signals, a fluorescence-based 96-well assay was established. First, a linear correlation between BFP signal and cell count was verified under different culture conditions.
For a more thorough characterization and verification of the 10-4ABFP cells, cytokines elevated during pulmonary fibrosis (TGF-β1, IL-33, IL-4, and TSLP) were screened to establish dose- and time-response curves (Figure 7). EC50 values following a 2-day incubation post-seeding were 1.45 ng/ml for TGF-β1, 400 pg/ml for TSLP, 12 ng/ml for IL-4, and 22 pg/ml for IL-33. Time course analysis of BFP signal expression revealed that the BFP signal was transiently increasing following TGF-β1 and IL-33, respectively, similar to what was observed for wildtype 10-4A cells (Figure 4). In contrast, TSLP and IL-4 show an increase in BFP expression until day 2 and afterward a stable BFP expression up to day 7.
FIGURE 7.
High-throughput screening using the 10-4ABFP cell line. (A) Schematic of plate reader assay to study the fibrotic response of 10-4ABFP to cytokine treatment in live cell assays. For normalization to the cell number, cells were stained with Calcein AM. (B) Dose-response curves of BFP expression in 10-4ABFP cells in response to 2 days exposure with increasing doses of TGF-β1, TSLP, IL-4, and IL-33. (C) Time-response curve of 10-4ABFP cells to continuous exposure to TGF-β1, TSLP, IL-4, and IL-33.
In summary, the 10-4ABFP cell line resembles a valuable tool for high throughput analysis of factors driving fibroblast to myofibroblast differentiation in pulmonary fibrosis, enabling screening with limited sample preparation and processing.
Discussion
Frequently, when studying molecular and cellular mechanisms in pulmonary fibrosis, primary fibroblasts are used for in vitro experiments (Pierce et al., 2007; Huang et al., 2012; Pardo and Selman, 2016). However, use of primary cells is often limited by availability, restricted propagation, ethical hurdles, the cost and difficulty of repetitive cell isolations and often a heterogeneity of isolated cells within or between isolations from different donors (Kaur and Dufour, 2012). Thus, well-characterized, representative cell lines provide a useful alternative for high-throughput, live-cell assays and can be used for in vitro disease modeling. Genetic modifications are easily introduced to study specific signaling pathways. In addition, they offer the opportunity for long-term studies replicating diseases progression in vitro. The aim of our work was to generate a representative cell line to be used for in vitro fibrosis research. To this end, we immortalized primary rat lung cells, screened them for fibroblast characteristics and tested the induction of fibrosis after mechanical as well as chemical stimulation.
Within recent years, various cell and, in particular, fibroblast subpopulations were reported for the distal lung. Diverse roles in development, lung homeostasis, aging and injury repair were attributed to distinct populations, raising the complexity for modeling pulmonary fibrosis (Zepp et al., 2017; Xie et al., 2018). Interestingly, some myo-and lipofibroblast marker genes like Mmp7, Mmp10 or Plin2 (Adams et al., 2019; Reyfman et al., 2019; Strunz et al., 2019) were also expressed in ATII cells, emphasizing the importance to look in detail at a combination of distinct marker genes in order to assign cells to a cellular group. The presence of Epcam in the RNA sequencing data from primary fibroblasts likely indicates small impurities of epithelial cells, whereas the presence of Vim indicates minor impurities in the epithelial cell in the primary cell isolates. The presence of small fractions of other cell types after the isolation of primary lung cells is also described by other groups (Dobbs et al., 1986; Driscoll et al., 2012; Lee et al., 2018).
Regarding the fibroblast subpopulations stated by Xie et al. (2018), our 10-4A cell line neither exhibits a classical myofibroblast nor a lipofibroblast phenotype, but rather a matrix fibroblast phenotype with unexpected high expression of Pdgfrα. On a molecular level, the 10-4A cells could also be clearly differentiated from alveolar epithelial cells. The ATI-related genes Cav1 as well as Pdpn, often used to discriminate ATI and ATII cells, are both also highly expressed in lung fibroblasts (Xiao et al., 2006; Quintanilla et al., 2019). Considering lineage-tracing and CRISPR/Cas9 knock-in experiments, Pdgfrα positive cells have been shown to give rise to either myofibroblasts or lipofibroblasts during development and to be able to differentiate into myofibroblasts in the adult lung (Ntokou et al., 2015; Zepp et al., 2017; Li et al., 2018). This directed differentiation of fibroblasts into myofibroblasts, as described for Pdgfrα positive cells, is one of the major requirements when studying pulmonary fibrosis. Thus, the gene and protein expression pattern of the 10-4A cell line indicates their suitability to study the mechanisms involved in the initiation and progression of pulmonary fibrosis.
Congruent with previous findings, we were also able to verify that myofibroblast differentiation can be induced mechanically (Wipff et al., 2007; Huang et al., 2012; Asano et al., 2017) and/or chemically (Chambers et al., 2003; Thannickal et al., 2003; Goffin et al., 2006; Blaauboer et al., 2011; Shi et al., 2013; El Agha et al., 2017) in 10-4A cells. Moreover, our data confirmed that fibroblasts grown on a substrate with physiological stiffness (Huang et al., 2012; Asano et al., 2017) and serum free medium (Baranyi et al., 2019) stay in a quiescent state over a time-course of 14 days. Since it is well established that fetal bovine serum exerts a heterogeneous influence on cellular behavior in cell culture due to batch-dependent variations and variable growth factor levels (Krämer et al., 2005; Mannello and Tonti, 2009; Baranyi et al., 2019), we decided to use a chemically defined, standardized, serum-free culture medium. Thereby, we were able to exclude potential variations in cellular responses due to heterogeneity of our culture conditions.
With regards to mechanical activation of the 10-4A cell line, the increase in gene and protein expression of αSMA was comparable to what has been described for primary fibroblasts as well as cell lines (Wipff et al., 2007; Huang et al., 2012; Asano et al., 2017). In line, a recent report from Tschumperlin et al., has shown that Col1a1 mRNA levels were stable above a substrate stiffness of 0.1 kPa (Liu et al., 2010).
Transforming growth factor beta 1 in combination with a soft substrate resulted in an increase of Acta2, Col1a1 and decrease of Plin2 gene expression as well as an increase of αSMA on the protein level. These observations are in accordance to already published data in several studies of human (Chambers et al., 2003; Thannickal et al., 2003; Blaauboer et al., 2011; El Agha et al., 2017) and rodent lung fibroblasts (Goffin et al., 2006; Shi et al., 2013; El Agha et al., 2017). Interestingly, continuous stimulation with TGF-β1 induced an early but transient increase in profibrotic marker gene expression. Acta2 expression was downregulated after 4 days in 10-4A cells continuously treated with TGF-β1. This effect was also observed in primary fibroblasts from two out of five animals after 7 days. In accordance, Col1a1 and Plin2 expression also adopt to control conditions for the primary fibroblasts. This indicates, that TGF-β1 stimulation alone, is not sufficient to maintain a myofibroblast phenotype and that changes in substrate stiffness are essential to induce a robust, persistent fibrotic response. The dependence of TGF-β1 on mechanical activation was already described by Shi et al. (2013). However, due to the lack of long-term studies examining the TGF-β1 effect beyond >4 days, comparable data are not available. Likewise, it has been reported, that TGF-β1 requires mechanical induction to be able to regulate myofibroblast differentiation (Hinz, 2009). This is again in accordance with our data, that the combination of chemical and mechanical stimulation is necessary to fully resemble the aspects of myofibroblast differentiation. This is also more likely to reflect the in vivo situation where fibrosis might be triggered by chemical stimuli but shifts to a self-perpetuating state with increasing stiffening of the lung tissue (spreading from fibrotic foci). We limited the observation time to 14 days as cellular overgrowth under fibrotic conditions, due to increased proliferation rates, resulted in inconsistent findings at observation periods beyond 14 days.
In order to accelerate and facilitate the read-out of fibroblast differentiation in a live cell, high-throughput setting, we generated an Acta2-BFP coupled reporter system using CRISPR/Cas9 technology. This reporter system is intended for use in live-cell, long-term screening applications. CRISPR/Cas9 can lead to off-target mutations. Therefore, we have verified correct insertion of BFP-NLS by Sanger sequencing and that 10-4ABFP and 10-4A cells exhibit a similar behavior under physiological cell culture conditions and after fibrotic stimulation. Minor differences in expression of Acta2 and Col1a1 at day 0 might indicate a slightly delayed RNA turnover in 10-4ABFP during adjustment to non-fibrotic culture conditions after transfer from culture flasks (Kitsera et al., 2007). On the protein level, αSMA signal and the respective BFP signal correlated very well, confirming appropriate reporter characteristics. Similarly, the regulation of BFP expression under the αSMA promoter could be confirmed by the targeted induction of the BFP signal after TGF-β1 administration.
Finally, in order to confirm the proper functionality and the suitability to use 10-4ABFP cells in high throughput assays, we performed plate-reader based experiments to analyze the dose- and time-response of Acta2 expression (i.e., myofibroblast induction) in response to exposure to distinct profibrotic cytokines. The effective concentrations were in agreement with previous reports for induction of fibrosis (Saito et al., 2003; Yanaba et al., 2011; Datta et al., 2013; Lee et al., 2017; Jones et al., 2019).
One potential limitation of the presented reporter cell line is its origin from rat rather than human. Several human pulmonary fibroblast cell lines are already available. However, in contrast to the cell line presented here, none of the currently available human cell lines has been characterized in detail with regards to the representation of a specific mesenchymal cell sub-population (Adams et al., 2019; Reyfman et al., 2019; Valenzi et al., 2019; Habermann et al., 2020; Liu et al., 2020; Travaglini et al., 2020). This might not only affect cellular responses to pro-fibrotic mechanical and chemical cues, but also affect epithelial-mesenchymal crosstalk and fibrotic remodeling when used in complex co-culture in vitro models (e.g., lung-on-a-chip). It is also accepted that in vivo animal models do not fully recapitulate all features of IPF pathogenesis (Moore et al., 2013). The reasons are not fully understood. The human lung contains structural and cellular differences to the rodent lung, however, the alveolus is one of the most conserved regions between rodents and human lungs and whether the mesenchymal and epithelial cells believed to be central to development of IPF are different is still a matter of debate. Even lipofibroblasts, a cell that’s presence has been found in rodents but has long been controversial in the human lung (Tahedl et al., 2014), has recently been identified in human lung (Liu et al., 2020; Travaglini et al., 2020). Therefore, we believe that the cell line reported here provides a valuable tool for fibrosis research, offering the opportunity to investigate molecular and cellular responses of a representative mesenchymal cell to mechanical and/or chemical stimuli with high resolution and in high throughput formats.
In summary, our data clearly demonstrate that the 10-4A cell line can be used as a valuable, novel tool for studying the onset and progression of fibrotic changes observed in pulmonary fibrosis. Additionally, the deviated 10-4ABFP reporter cell line is a useful tool for high-throughput live cell, in vitro assays to directly monitor fibrotic changes over time.
Data Availability Statement
The RNA sequencing is deposited in the Gene Expression Omnibus (accession: GSE155743).
Ethics Statement
The animal study was reviewed and approved by the Regierungspräsidium Tübingen.
Author Contributions
JN, AS, and MF conceived and designed the study, interpreted results of the experiments, and edited and revised the manuscript. JN, AS, and VW performed the experiments. JN, KQ, and AS analyzed the data. JN and AS prepared the figures. JN, AS, KQ, and MF drafted the manuscript. All authors approved final version of manuscript.
Conflict of Interest
KQ was employed by the company Boehringer Ingelheim Pharma GmbH & Co. KG, Biberach, Germany. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Achim Riecker and Tatiana Felder for help with primary cell isolation, Susan Tavakoli and Kay Gottschalk for advice with PDMS gel preparation and handling, and Prof. Dr. C. Buske and the Core Facility Cytometry Ulm University for performance of cell sorting. We also want to thank Coralie Viollet, Head of the Genomics Lab at Boehringer Ingelheim, Biberach, for the Illumina library preparation.
Funding. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer(n): 278012962 (GRK “Pulmosens”), 251293561 (SFB1149), and 175083951 (to MF) and a grant from the Ministry of Science, Research and the Arts of Baden-Württemberg (Az: 32-7533.-6-10/15/5) (to MF).
References
- Adams T., Schupp J., Poli S., Ayaub E., Neumark N., Ahangari F., et al. (2019). Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in idiopathic pulmonary fibrosis. bioRxiv [preprint]. 10.1101/759902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M. Y., Chen J. J. L., Cogan J. D., Alder J. K., Ingersoll R. G., Markin C., et al. (2007). Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 356 1317–1326. 10.1056/NEJMoa066157 [DOI] [PubMed] [Google Scholar]
- Asano S., Ito S., Takahashi K., Furuya K., Kondo M., Sokabe M., et al. (2017). Matrix stiffness regulates migration of human lung fibroblasts. Physiol. Rep. 5 1–11. 10.14814/phy2.13281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnato G., Harari S. (2015). Cellular interactions in the pathogenesis of interstitial lung diseases. Eur. Respir. Rev. 24 102–114. 10.1183/09059180.00003214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranyi U., Winter B., Gugerell A., Hegedus B., Brostjan C., Laufer G., et al. (2019). Primary human fibroblasts in culture switch to a myofibroblast-like phenotype independently of TGF Beta. Cells 8:721. 10.3390/cells8070721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta K. E., Cagnina R. E., Wallace K. L., Ramos S. I., Mehrad B., Linden J. (2012). Leukocyte compartments in the mouse lung: distinguishing between marginated, interstitial, and alveolar cells in response to injury. J. Immunol. Methods 375 100–110. 10.1016/j.jim.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers M. F., Moodley Y., Stem R., Hall E. J. S. (2017). When is an alveolar Type 2 cell an alveolar Type 2 Cell? A conundrum for lung stem cell biology and regenerative medicine. Am. J. Respir. Cell Mol. Biol. 57 1–31. 10.1165/rcmb.2016-0426PS [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaauboer M. E., Smit T. H., Hanemaaijer R., Stoop R., Everts V. (2011). Cyclic mechanical stretch reduces myofibroblast differentiation of primary lung fibroblasts. Biochem. Biophys. Res. Commun. 404 23–27. 10.1016/j.bbrc.2010.11.033 [DOI] [PubMed] [Google Scholar]
- Booth A. J., Hadley R., Cornett A. M., Dreffs A. A., Matthes S. A., Tsui J. L., et al. (2012). Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am. J. Respir. Crit. Care Med. 186 866–876. 10.1164/rccm.201204-0754OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess J. K., Mauad T., Tjin G., Karlsson J. C., Westergren-Thorsson G. (2016). The extracellular matrix – the under-recognized element in lung disease? J. Pathol. 240 397–409. 10.1002/path.4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R. C., Leoni P., Kaminski N., Laurent G. J., Heller R. A. (2003). Global expression profiling of fibroblast responses to transforming growth factor-β1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching. Am. J. Pathol. 162 533–546. 10.1016/S0002-9440(10)63847-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R. C., Mercer P. F. (2015). Mechanisms of alveolar epithelial injury, repair, and fibrosis. Ann. Am. Thoracic Soc. 12 S16–S20. 10.1513/AnnalsATS.201410-448MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Shen Y., Mohanasundaram P., Lindström M., Ivaska J., Ny T., et al. (2016). Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-β-Slug signaling. Proc. Natl. Acad. Sci. U.S.A. 113 E4320–E4327. 10.1073/pnas.1519197113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Alexander R., Sulikowski M. G., Nicholson A. G., Maher T. M., Scotton C. J., et al. (2013). Evidence for a functional thymic stromal lymphopoietin signaling axis in fibrotic lung disease. J. Immunol. 191 4867–4879. 10.4049/jimmunol.1300588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca D. S., Levin J. Z., Sivachenko A., Fennell T., Nazaire M. D., Williams C., et al. (2012). RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28 1530–1532. 10.1093/bioinformatics/bts196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs L. G., Gonzalez R., Williams M. C. (1986). An improved method for isolating type II cells in high yield and purity. Am. Rev. Respir. Dis. 134 141–145. 10.1164/arrd.1986.134.1.141 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll B., Kikuchi A., Lau A. N., Lee J., Reddy R., Jesudason E., et al. (2012). Isolation and characterization of distal lung progenitor cells. Methods Mol. Biol. 879 109–122. 10.1007/978-1-61779-815-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E., Moiseenko A., Kheirollahi V., De Langhe S., Crnkovic S., Kwapiszewska G., et al. (2017). Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 20 261.e3–273.e3. 10.1016/j.stem.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N., Suda T., Kono M., Kaida Y., Hashimoto D., Fujisawa T., et al. (2013). Amount of elastic fibers predicts prognosis of idiopathic pulmonary fibrosis. Respir. Med. 107 1608–1616. 10.1016/j.rmed.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Epstein Shochet G., Brook E., Israeli-Shani L., Edelstein E., Shitrit D. (2017). Fibroblast paracrine TNF-α signaling elevates integrin A5 expression in idiopathic pulmonary fibrosis (IPF). Respir. Res. 18:122. 10.1186/s12931-017-0606-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. M., Fingerlin T. E., Schwarz M. I., Lynch D., Kurche J., Warg L., et al. (2016). Idiopathic pulmonary fibrosis: a genetic disease that involves mucociliary dysfunction of the peripheral airways. Physiol. Rev. 96 1567–1591. 10.1152/physrev.00004.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder M., Trueeb B., Stucki A. O., Borcard S., Stucki J. D., Schnyder B., et al. (2019). Impaired wound healing of alveolar lung epithelial cells in a breathing lung-on-a-chip. Front. Bioeng. Biotechnol. 7:3. 10.3389/fbioe.2019.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabazza E. C., Kasper M., Ohta K., Keane M., D’Alessandro-Gabazza C., Fujimoto H., et al. (2004). Decreased expression of aquaporin-5 in bleomycin-induced lung fibrosis in the mouse. Pathol. Int. 54 774–780. 10.1111/j.1440-1827.2004.01754.x [DOI] [PubMed] [Google Scholar]
- Goffin J. M., Pittet P., Csucs G., Lussi J. W., Meister J. J., Hinz B. (2006). Focal adhesion size controls tension-dependent recruitment of α-smooth muscle actin to stress fibers. J. Cell Biol. 172 259–268. 10.1083/jcb.200506179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A. C., Gutierrez A. J., Bui L. T., Yahn S. L., Winters N. I., Carla L., et al. (2020). Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6 10.1101/753806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Sato A., Tanimura K., Uemasu K., Hamakawa Y., Fuseya Y., et al. (2017). Fraction of MHCII and EpCAM expression characterizes distal lung epithelial cells for alveolar type 2 cell isolation. Respir. Res. 18 1–13. 10.1186/s12931-017-0635-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B. (2009). Tissue stiffness, latent TGF-β1 activation, and mechanical signal transduction: implications for the pathogenesis and treatment of fibrosis. Curr. Rheumatol. Rep. 11 120–126. 10.1007/s11926-009-0017-1 [DOI] [PubMed] [Google Scholar]
- Horowitz J. C., Thannickal V. J. (2006). Epithelial-mesenchymal interactions in pulmonary fibrosis. Semin. Respir. Crit. Care Med. 27 600–612. 10.1055/s-2006-957332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Yang N., Fiore V. F., Barker T. H., Sun Y., Morris S. W., et al. (2012). Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am. J. Respir. Cell Mol. Biol. 47 340–348. 10.1165/rcmb.2012-0050OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaux F., Liu T., McGarry B., Ullenbruch M., Phan S. H. (2003). Dual Roles of IL-4 in lung injury and fibrosis. J. Immunol. 170 2083–2092. 10.4049/jimmunol.170.4.2083 [DOI] [PubMed] [Google Scholar]
- Huh D. (2015). A human breathing lung-on-a-chip. Ann. Am. Thoracic Soc. 12 42–44. 10.1513/AnnalsATS.201410-442MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansing N. L., McClendon J., Kage H., Sunohara M., Alvarez J. R., Borok Z., et al. (2018). Isolation of rat and mouse alveolar type II epithelial cells. Methods Mol. Biol. 1809 69–82. 10.1007/978-1-4939-8570-8_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. L., Haak A. J., Caporarello N., Choi K. M., Ye Z., Yan H., et al. (2019). TGFβ-induced fibroblast activation requires persistent and targeted HDAC-mediated gene repression. J. Cell Sci. 132:jcs233486. 10.1242/jcs.233486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage H., Borok Z. (2012). EMT and interstitial lung disease: a mysterious relationship. Curr. Opin. Pulm. Med. 18 517–523. 10.1097/MCP.0b013e3283566721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M., Barth K. (2017). Potential contribution of alveolar epithelial type I cells to pulmonary fibrosis. Biosci. Rep. 37:BSR20171301. 10.1042/bsr20171301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G., Dufour J. (2012). Cell lines: valuable tools or useless artifacts. Spermatogenesis 2 1–5. 10.4161/spmg.19885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsera N., Khobta A., Epe B. (2007). Destabilized green fluorescent protein detects rapid removal of transcription blocks after genotoxic exposure. BioTechniques 43 222–227. 10.2144/000112479 [DOI] [PubMed] [Google Scholar]
- Kolodsick J. E., Toews G. B., Jakubzick C., Hogaboam C., Moore T. A., McKenzie A., et al. (2004). Protection from fluorescein isothiocyanate-induced Fibrosis in IL-13-Deficient, but Not IL-4-Deficient, mice results from impaired collagen synthesis by fibroblasts. J. Immunol. 172 4068–4076. 10.4049/jimmunol.172.7.4068 [DOI] [PubMed] [Google Scholar]
- Krämer D. K., Bouzakri K., Holmqvist O., Al-Khalili L., Krook A. (2005). Effect of serum replacement with Plysate on cell growth and metabolismin primary cultures of human skeletal muscle. Cytotechnology 48 89–95. 10.1007/s10616-005-4074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn A., Kletting S., De Souza Carvalho-Wodarz C., Repnik U., Griffiths G., Fischer U., et al. (2016). Human alveolar epithelial cells expressing tight junctions to model the air-blood barrier. Altex 33 251–260. 10.14573/altex.1511131 [DOI] [PubMed] [Google Scholar]
- Lederer D. J., Martinez F. J. (2018). Idiopathic pulmonary fibrosis. N. Engl. J. Med. 378 1811–1823. 10.1056/NEJMra1705751 [DOI] [PubMed] [Google Scholar]
- Lee D. F., Salguero F. J., Grainger D., Francis R. J., MacLellan-Gibson K., Chambers M. A. (2018). Isolation and characterisation of alveolar type II pneumocytes from adult bovine lung. Sci. Rep. 8:11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. U., Chang H. S., Lee H. J., Jung C. A., Bae D. J., Song H. J., et al. (2017). Upregulation of interleukin-33 and thymic stromal lymphopoietin levels in the lungs of idiopathic pulmonary fibrosis. BMC Pulm. Med. 17:39. 10.1186/s12890-017-0380-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Bernau K., Sandbo N., Gu J., Preissl S., Sun X. (2018). Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response. eLife 7 1–20. 10.7554/eLife.36865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Zhang Y., Xie T., Liu N., Chen H., Geng Y., et al. (2016). Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Na. Med. 22 1285–1293. 10.1038/nm.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lipps C., Klein F., Wahlicht T., Seiffert V., Butueva M., Zauers J., et al. (2018). Expansion of functional personalized cells with specific transgene combinations. Nat. Commun. 9:994. 10.1038/s41467-018-03408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Mih J. D., Shea B. S., Kho A. T., Sharif A. S., Tager A. M., et al. (2010). Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J. Cell Biol. 190 693–706. 10.1083/jcb.201004082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Rowan S. C., Liang J., Yao C., Huang G., Deng N., et al. (2020). Definition and signatures of lung fibroblast populations in development and fibrosis in mice and men. bioRxiv [preprint]. 10.1101/2020.07.15.203141 [DOI] [Google Scholar]
- Löfdahl A., Rydell-Törmänen K., Larsson-Callerfelt A. K., Wenglén C., Westergren-Thorsson G. (2018). Pulmonary fibrosis in vivo displays increased p21 expression reduced by 5-HT2B receptor antagonists in vitro - A potential pathway affecting proliferation. Sci. Rep. 8:1927. 10.1038/s41598-018-20430-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzina I. G., Kopach P., Lockatell V., Kang P. H., Nagarsekar A., Burke A. P., et al. (2013). Interleukin-33 potentiates Bleomycin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 49 999–1008. 10.1165/rcmb.2013-0093OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzina I. G., Pickering E. M., Kopach P., Kang P. H., Lockatell V., Todd N. W., et al. (2012). Full-Length IL-33 promotes inflammation but not Th2 response in vivo in an ST2-Independent fashion. J. Immunol. 189 403–410. 10.4049/jimmunol.1200259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzina I. G., Todd N. W., Sundararajan S., Atamas S. P. (2015). The cytokines of pulmonary fibrosis: much learned, much more to learn. Cytokine 74 88–100. 10.1016/j.cyto.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Mannello F., Tonti G. A. (2009). Concise review: no breakthroughs for human mesenchymal and embryonic stem cell culture: conditioned medium, feeder layer, or feeder-free; medium with fetal calf serum, human serum, or enriched plasma; serum-free, serum replacement nonconditioned medium, o. Stem Cells 25 1603–1609. 10.1634/stemcells.2007-0127 [DOI] [PubMed] [Google Scholar]
- McElroy M. C., Kasper M. (2004). The use of alveolar epithelial type I cell-selective markers to investigate lung injury and repair. Eur. Respir. J. 24 664–673. 10.1183/09031936.04.00096003 [DOI] [PubMed] [Google Scholar]
- Miklavc P., Frick M., Wittekindt O. H., Haller T., Dietl P. (2010). Fusion-Activated Ca2+ entry: an “active zone” of elevated Ca2+ during the postfusion stage of lamellar body exocytosis in rat Type II Pneumocytes. PLoS One 5:e10982. 10.1371/journal.pone.0010982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. B., Lawson W. E., Oury T. D., Sisson T. H., Raghavendran K., Hogaboam C. M. (2013). Animal models of fibrotic lung disease. Ame. J. Respir. Cell Mol. Biol. 49 167–179. 10.1165/rcmb.2013-0094TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntokou A., Klein F., Dontireddy D., Becker S., Bellusci S., Richardson W. D., et al. (2015). Characterization of the platelet-derived growth factor receptor-α-positive cell lineage during murine late lung development. Am. J, Physiol. Lung Cell. Mol. Physiol. 309 L942–L958. 10.1152/ajplung.00272.2014 [DOI] [PubMed] [Google Scholar]
- Oikonomou N., Harokopos V., Zalevsky J., Valavanis C., Kotaniduo A., Szymkowski D. E., et al. (2006). Soluble TNF mediates the transition from pulmonary inflammation to fibrosis. PLoS One 1:e108. 10.1371/journal.pone.0000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly S. (2013). Role of interleukin-13 in fibrosis, particularly systemic sclerosis. BioFactors 39 593–596. 10.1002/biof.1117 [DOI] [PubMed] [Google Scholar]
- Palchesko R. N., Zhang L., Sun Y., Feinberg A. W. (2012). Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS One 7:e0051499. 10.1371/journal.pone.0051499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo A., Selman M. (2016). Lung fibroblasts, aging, and idiopathic pulmonary fibrosis. Ann. Am. Thoracic Soc. 13 S417–S421. 10.1513/AnnalsATS.201605-341AW [DOI] [PubMed] [Google Scholar]
- Parker M. W., Rossi D., Peterson M., Smith K., Sikstrom̈ K., White E. S., et al. (2014). Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J. Clin. Invest. 124 1622–1635. 10.1172/JCI71386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce E. M., Carpenter K., Jakubzick C., Kunkel S. L., Evanoff H., Flaherty K. R., et al. (2007). Idiopathic pulmonary fibrosis fibroblasts migrate and proliferate to CC chemokine ligand 21. Eur. Respir. J. 29 1082–1093. 10.1183/09031936.00122806 [DOI] [PubMed] [Google Scholar]
- Plantier L., Crestani B., Wert S. E., Dehoux M., Zweytick B., Guenther A., et al. (2011). Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax 66 651–657. 10.1136/thx.2010.151555 [DOI] [PubMed] [Google Scholar]
- Quintanilla M., Montero L. M., Renart J., Villar E. M. (2019). Podoplanin in inflammation and cancer. Int. J. Mol. Sci. 20:707. 10.3390/ijms20030707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G., Collard H. R., Egan J. J., Martinez F. J., Behr J., Brown K. K., et al. (2011). An Official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 183 788–824. 10.1164/rccm.2009-040GL [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyfman P. A., Walter J. M., Joshi N., Anekalla K. R., McQuattie-Pimentel A. C., Chiu S., et al. (2019). Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199 1517–1536. 10.1164/rccm.201712-2410OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richeldi L., Collard H. R., Jones M. G. (2017). Idiopathic pulmonary fibrosis. Lancet 389 1941–1952. 10.1016/S0140-6736(17)30866-8 [DOI] [PubMed] [Google Scholar]
- Ryu J. H., Moua T., Daniels C. E., Hartman T. E., Yi E. S., Utz J. P., et al. (2014). Idiopathic pulmonary fibrosis: evolving concepts. Mayo Clin. Proc. 89 1130–1142. 10.1016/j.mayocp.2014.03.016 [DOI] [PubMed] [Google Scholar]
- Saito A., Okazaki H., Sugawara I., Yamamoto K., Takizawa H. (2003). Potential action of IL-4 and IL-13 as fibrogenic factors on lung fibroblasts in vitro. Int. Arch. Allergy Immunol. 132 168–176. 10.1159/000073718 [DOI] [PubMed] [Google Scholar]
- Sayols S., Scherzinger D., Klein H. (2016). dupRadar: a bioconductor package for the assessment of PCR artifacts in RNA-Seq data. BMC Bioinformatics 17:428. 10.1186/s12859-016-1276-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold M. A., Wise A. L., Speer M. C., Steele M. P., Brown K. K., Loyd J. E., et al. (2011). A common MUC5B promoter polymorphism and pulmonary fibrosis. N. Engl. J. Med. 364 1503–1512. 10.1056/NEJMoa1013660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M., Pardo A. (2001). Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir. Res. 3 1–8. 10.1186/rr175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgalla G., Iovene B., Calvello M., Ori M., Varone F., Richeldi L. (2018). Idiopathic pulmonary fibrosis: pathogenesis and management. Respir. Res. 19:32. 10.1186/s12931-018-0730-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Dong Y., Duan Y., Jiang X., Chen C., Deng L. (2013). Substrate stiffness influences TGF-β1-induced differentiation of bronchial fibroblasts into myofibroblasts in airway remodeling. Mol. Med. Rep. 7 419–424. 10.3892/mmr.2012.1213 [DOI] [PubMed] [Google Scholar]
- Sime P. J., Marr R. A., Gauldie D., Xing Z., Hewlett B. R., Graham F. L., et al. (1998). Transfer of tumor necrosis factor-α to rat lung induces severe pulmonary inflammation and patchy interstitial fibrogenesis with induction of transforming growth factor-β1 and myofibroblasts. Am. J. Pathol. 153 825–832. 10.1016/S0002-9440(10)65624-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sime P. J., Xing Z., Graham F. L., Csaky K. G., Gauldie J. (1997). Adenovector-mediated gene transfer of active transforming growth factor- β1 induces prolonged severe fibrosis in rat lung. J. Clin. Invest. 100 768–776. 10.1172/JCI119590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz M., Simon L. M., Ansari M., Mattner L. F., Angelidis I., Mayr C. H., et al. (2019). Longitudinal single cell transcriptomics reveals Krt8+ alveolar epithelial progenitors in lung regeneration. bioRxiv [preprint]. 10.1101/705244 [DOI] [Google Scholar]
- Stuart B. D., Choi J., Zaidi S., Xing C., Holohan B., Chen R., et al. (2015). Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet. 47 512–517. 10.1038/ng.3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki J. D., Hobi N., Galimov A., Stucki A. O., Schneider-Daum N., Lehr C. M., et al. (2018). Medium throughput breathing human primary cell alveolus-on-chip model. Sci. Rep. 8:14359. 10.1038/s41598-018-32523-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahedl D., Wirkes A., Tschanz S. A., Ochs M., Mühlfeld C. (2014). How common is the lipid body-containing interstitial cell in the mammalian lung? Am. J. Physiol. Lung Cell. Mol. Physiol. 307 L386–L394. 10.1152/ajplung.00131.2014 [DOI] [PubMed] [Google Scholar]
- Thannickal V. J., Lee D. Y., White E. S., Cui Z., Larios J. M., Chacon R., et al. (2003). Myofibroblast differentiation by transforming growth factor-β1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. J. Biol. Chem. 278 12384–12389. 10.1074/jbc.M208544200 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini K. J., Nabhan A. N., Penland L., Sinha R., Gillich A., Sit R. V., et al. (2020). A molecular cell atlas of the human lung from single cell RNA sequencing. bioRxiv [Preprint]. 10.1101/742320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumperlin D. J., Ligresti G., Hilscher M. B., Shah V. H. (2018). Mechanosensing and fibrosis. J. Clin. Invest. 128 74–84. 10.1172/JCI93561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzi E., Bulik M., Tabib T., Morse C., Sembrat J., Trejo Bittar H., et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann. Rheumat. Dis. 78 1379–1387. 10.1136/annrheumdis-2018-214865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipff P. J., Rifkin D. B., Meister J. J., Hinz B. (2007). Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J. Cell Biol. 179 1311–1323. 10.1083/jcb.200704042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts W. A., Agostini C., Antoniou K. M., Bouros D., Chambers R. C., Cottin V., et al. (2013). The pathogenesis of pulmonary fibrosis: a moving target. Eur. Respir. J. 41 1207–1218. 10.1183/09031936.00073012 [DOI] [PubMed] [Google Scholar]
- Xiao M. W., Zhang Y., Hong P. K., Zhou Z., Feghali-Bostwick C. A., Liu F., et al. (2006). Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J. Exp. Med. 203 2895–2906. 10.1084/jem.20061536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Wang Y., Deng N., Huang G., Taghavifar F., Geng Y., et al. (2018). Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22 3625–3640. 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Mizuno T., Sridharan A., Du Y., Guo M., Tang J., et al. (2016). Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1:e90558. 10.1172/jci.insight.90558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaba K., Yoshizaki A., Asano Y., Kadono T., Sato S. (2011). Serum IL-33 levels are raised in patients with systemic sclerosis: association with extent of skin sclerosis and severity of pulmonary fibrosis. Clin. Rheumatol. 30 825–830. 10.1007/s10067-011-1686-5 [DOI] [PubMed] [Google Scholar]
- Zepp J., Zacharias W., Frank D. (2017). Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170 1134–1148. 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Homer R. J., Wang Z., Chen Q., Geba G. P., Wang J., et al. (1999). Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 103 779–788. 10.1172/JCI5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA sequencing is deposited in the Gene Expression Omnibus (accession: GSE155743).